Abstract
Medical image acquisition devices are susceptible to producing blurry images due to respiratory and patient movement. Despite having a notable impact on such blind-motion deblurring, medical image deblurring is still underexposed. This study proposes an end-to-end scale-recurrent deep network to learn the deblurring from multi-modal medical images. The proposed network comprises a novel residual dense block with spatial-asymmetric attention to recover salient information while learning medical image deblurring. The performance of the proposed methods has been densely evaluated and compared with the existing deblurring methods. The experimental results demonstrate that the proposed method can remove blur from medical images without illustrating visually disturbing artifacts. Furthermore, it outperforms the deep deblurring methods in qualitative and quantitative evaluation by a noticeable margin. The applicability of the proposed method has also been verified by incorporating it into various medical image analysis tasks such as segmentation and detection. The proposed deblurring method helps accelerate the performance of such medical image analysis tasks by removing blur from blurry medical inputs.
Keywords:
medical image deblurring; dense residual spatial-asymmetric attention; scale-recurrent network; residual learning; deep learning MSC:
68T07
1. Introduction
The perceptual quality of images directly impacts the process of medical image analysis and decision-making maneuvers of medical practitioners [1,2]. Contrarily, acquiring a visually plausible and precise representation of repository organs is a strenuous process. It requires a longer scanning time to capture the image of the complex respiratory system. Slower scanning times combined with respiratory and patient motion impel the medical image acquisition device to capture unclear images with blind-motion blurs [2].
Removing blur from any image is a challenging task. Typically, deblurring refers to a deconvolution operation, where a blurred image comprises a blur kernel with additive sensor noises [3] as follows:
In Equation (1), represents the blurred image, represents the latent sharp image, * represents the convolution operation, represents the blur kernel, and represents sensor noise. The for medical images remains blind in most cases. Arguably, the complex structure and texture of the repository systems with blind blurs make the medical image deblurring far more challenging than for generic images.
In recent years, only a few works in the open literature have attempted to address the challenges of deblurring medical images. Most of these methods leverage classical image processing techniques to reduce blurs by applying sharpening filters [4,5,6,7]. Furthermore, only a few recent methods utilize deep learning for learning deblurring from medical images [8,9]. However, the existing medical image deblurring (MID) methods are domain-oriented and heavily depend on the point spread capacity of the acquisition system. Therefore, the performance of these methods is limited to specific image types (i.e., CT, MRI) and fails to achieve satisfactory performance in diverse data samples.
Oppositely, deep learning-based image deblurring techniques have evolved significantly in the past decade [10]. These methods have illustrated significant improvement in removing deblurs in non-medical images. Notably, the deep deblurring methods outperform traditional counterparts by a considerable margin in diverse data samples [10]. To examine the existing deep deblurring techniques for MID, we trained and tested these methods on medical images collected from multiple image modalities, as shown in Figure 1. It has been found that the existing state-of-the-art (SOTA) deep deblurring and MID methods fail to recover texture and salient information from blurry medical images. Moreover, they are prone to produce visually disturbing artifacts while removing blur from the given image.
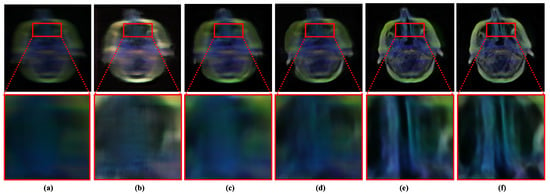
Figure 1.
Performance of existing medical image deblurring methods in removing blind motion blur. The existing deblurring methods immensely failed in removing blur from medical images. (a) Blurry input. (b) Result obtained by TEMImageNet [9]. (c) Result obtained by ZhaoNet [11]. (d) Result obtained by Deep Deblur [12]. (e) Result obtained by SRN Deblur [13]. (f) Proposed Method.
It is worth noting that medical images are typically captured with different image acquisition mechanisms and comprise more salient features than non-medical images. However, removing blur from medical images has a significant impact. It can substantially accelerate the performance of medical image analysis tasks. Despite a wide range of applications and real-world impacts, the performance of existing MID methods is inadequate. They fail to address the large-kernel blind blurs in medical images, as shown in Figure 1. The limitation of existing methods motivated this study to incorporate a robust learning-based MID solution, which can handle the large kernel blurs without explicitly considering image modality.
This study proposes a deep method for learning image acquisition-independent MID from diverse data samples. The proposed method incorporates a scale-recurrent deep network with a novel deep module combining residual dense block [14] and spatial-asymmetric attention [15] (denoted as RD-SAM in later sections). Here, residual-dense learning strives to extract salient features, and the spatial-asymmetric block refines the extracted features with local-global attention. The performance of the proposed deep method has been compared with existing MID methods for different varieties of medical images. The applicability of a deep MID method for accelerating medical image analysis has also been studied by incorporating it into computer-aided diagnosis (CAD) applications, such as segmentation and detection. The main contribution of the proposed study is as follows:
- Propose a scale-recurrent deep network with RD-SAM to accelerate the medical image deblurring. The proposed RD-SAM aims to learn salient features from blurry medical images and refine the features with local-global attention.
- Propose to generalize the MID by learning deblurring from multiple medical image modalities. Additionally, the feasibility of the proposed method has been verified with numerous medical image datasets acquired by different image acquisition techniques.
- Study existing deep learning-based deblurring methods for MID and outperformed them in qualitative and quantitative comparisons.
- Illustrate the feasibility of a robust deblurring method in medical image analysis tasks such as segmentation, detection, etc.
2. Related Works
This section briefly discusses the existing medical image deblurring methods and deep learning-based image deblurring methods.
2.1. Medical Image Deblurring
Most of the existing MID methods have utilized non-learning-based deblurring approaches. Among the existing MID works, Ming et al. [6] introduce a blind deblurring strategy to enhance blurry images without complete knowledge of the underlying point spread function (PSF). Ashish et al. [4] proposed a complex wavelet transform for deblurring ultrasonic and Computed Tomography (CT) images. In a follow-up study [16], they leverage a symmetric Daubechies complex wavelet transform to outperform their counterparts. In another study, K Reddy et al. [17] proposed a modified ADMM for removing blurs from color medical images. Further, Zohair et al. [18] proposed to combine the Laplacian sharpening filter and the iterative Richardson–Lucy algorithm to mitigate blur from CT images. Later, they [19] presented an iterative Landweber algorithm to mitigate artifact-free boundaries and lessen noise amplification for perceiving a faster processing time. Later, Ergun et al. [7] proposed a fast off-resonance frequency deblurring for spiral MR images. Meanwhile, Reza et al. [5] used coherence-enhancing shock filters for sharpening edges of blur images.
Apart from the traditional approaches, a few recent works attempted to utilize machine learning approaches for MID. For instance, Dong et al. [20] proposed to combine the classical Wiener deconvolution framework with deep features for MID. Apart from that, Zhao et al. [11] proposed an end-to-end deep learning-based deblurring method exclusively for optical microscopic imaging systems. In another study, Lin et al. [9] proposed a U-Net-like structure to learn deblurring on atomic-resolution images. It is worth noting that none of the existing methods have been generalized to perform deblurring in diverse data samples collected by different medical image modalities.
2.2. Deep Image Deblurring
Learning-based deblurring methods illustrated significant improvement in recent years. These methods consider image deblurring as an image-to-image translation task. In a recent study, Sun et al. [21] proposed a convolutional neural network (CNN) to predict the field of non-uniform motion blur kernels. Later, Nah et al. [12] proposed a multi-scale CNN to recover the sharp image from blurry inputs. Similarly, YE et al. [22] also utilized a similar multi-scale structure in a coarse-to-refine manner. Zhang et al. [23] also proposed a deep network that fuses multiple CNNs with a spatially variant recurrent neural network (RNN). Apart from that, a few recent studies [24,25] also adopt the concept of generative adversarial networks (GAN) to learn image deblurring in non-medical images.
The deep image deblurring methods have illustrated domination in non-medical image deblurring. However, none of these deep methods have attempted to deblur the medical images. Arguably, medical image deblurring is far more challenging and comprises more real-world applications than non-medical images. Thus, this study aims to introduce a robust deblurring method for MID. Table 1 illustrates a comparison between existing works and the proposed method.

Table 1.
Comparison between existing methods and the proposed method.
3. Proposed Method
The proposed method considers the MID as an image-to-image translation task. This section details the proposed network, its learning strategy, and the process of generating blurry medical images.
3.1. Deep Network
The proposed method aims to translate a blurry medical image () as . Here, the mapping function () learns to deblur an RGB medical image () as . H and W represent the height and width of the input and output images.
Figure 2 illustrates the architecture of the proposed network (mapping function) . The proposed network comprises three sub-networks to learn MID in different image resolutions. The output of individual branches is upscaled and incorporated into the subsequent sub-network as follows:
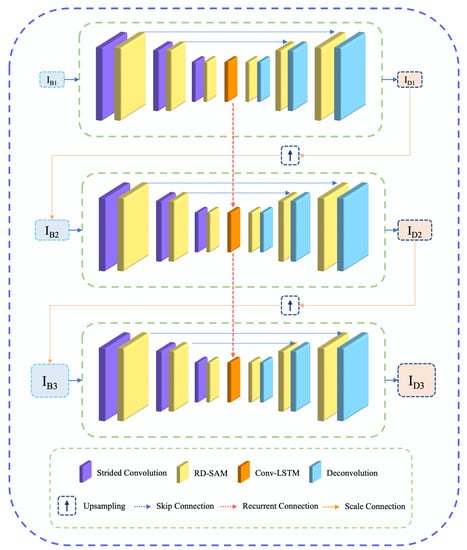
Figure 2.
Overview of the proposed network for learning medical image deblurring. The proposed method comprises a novel RD-SAM block in a scale recurrent network for learning salient features to accelerate deblurring performance.
Here, and present the given blurry input and deblurred output of the scale. presents the the training parameters of proposed network , and h presents the hidden states captured with LSTM [13]. Unlike the previous studies, this study proposes RD-SAM in the encoder and decoder of each sub-network for precise feature learning. Our encoder and decoder have been connected with a skip connection to accelerate residual features.
3.1.1. Residual Dense Spatial-Asymmetric Attention Module (RD-SAM)
Figure 3 illustrates the overview of the proposed RD-SAM. The proposed RD-SAM comprises a residual dense block followed by spatial-asymmetric attention as follows:
In Equation (3), and residual dense block and spatial-asymmetric block pursue local-global attention.
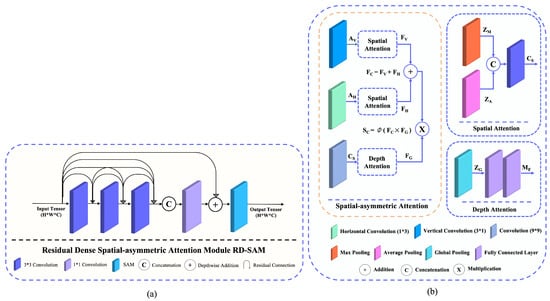
Figure 3.
Overview of proposed RD-SAM. It comprises a residual dense block, followed by a spatial-symmetric attention module. (a) Proposed RD-SAM. (b) Spatial-asymmetric attention module.
It is worth noting, spatial-asymmetric attention is well-known for its capability of refining features. Recently, it has shown significant performance gain in numerous computer vision tasks such as nona-Bayer reconstruction [15] and face anti-spoofing [26]. In this study, we leverage spatial-asymmetric attention to refine extracted feature of the residual dense block as follows:
Here, , , and represent the asymmetric convolution operation, square convolution, and sigmoid activation. Additionally, and present the average pooling and max pooling to generates two 2D feature maps as , . The mapped feature has been concatenated and presented as a 2D map.
Overall, the aggregated bi-directional attention over a given feature of a blurry medical image has been obtained as:
Additionally, a squeeze-extractor descriptor [27] also has been utilized to pursue a global descriptor as follows:
Here, and present consecutive fully connected layers and global pooling operations.
3.1.2. Objective Function
The proposed network parameterized with weights , aims to minimize the training loss by appropriating the given P pairs of training images as follows:
Here, denotes the objective function, which is perceived as:
Here, and present the ground truth image and enhanced medical images.
3.2. Blur Simulation
Training pairs with input and ground truth images are one of the most crucial parts of developing a supervised deep learning-based solution. Regrettably, we could not find any potential dataset with blurry image input and sharp ground truth image pairs for learning MID. To counter the data limitation, we propose synthesizing blurry medical images from existing sharp medical images. The Algorithm 1 depicts the procedure of synthesizing blur-sharp image pairs for learning MID. The blurry input has been generated by applying a blur kernel with a random angle. It is worth noting that the proposed blur simulation method is able to generate the blur-sharp image pairs from any medical images without explicitly knowing its hardware implications.
| Algorithm 1 Simulating motion blurs on medical images |
|
We utilized 18,000 medical images to generate blurry training samples throughout the experiments. Additionally, 1200 (200 for observing convergence and 1000 for testing) medical images have been used to evaluate the deep methods. The following datasets have been utilized to generate blur-sharp image pairs throughout the study: (i) Human Protein atlas [28], (ii) Chest X-ray [29], (iii) CT scan [30], (iv) HRM skin lesion [31], and (iv) Brain MRI [32].
It is worth noting that the blurs in the training pairs generated on-the-fly (during training) utilize Algorithm 1. The proposed blur generation strategy allowed this study to induce an infinite number of blur kernels and the training samples from the available data samples. Such a learning strategy intends to introduce data diversity and avoid overfitting during the training phase [33]. Apart from the training samples, we utilized fixed blur kernels in the validation and testing samples to ensure a fair comparison among all deblurring methods. Figure 4 illustrates the blur-sharp image pairs from our blurry medical image dataset.
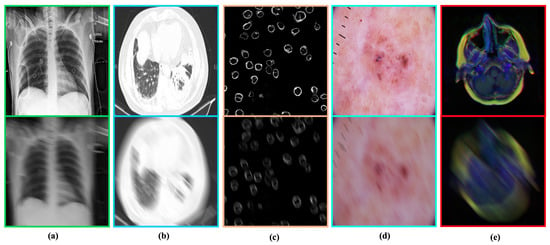
Figure 4.
Blur–sharp image pairs generated by the proposed method. In each pair. Top: Ground-truth image. Bottom: Simulated blurry image. (a) Chest X-ray [29]. (b) COVID-19 CT scan [30]. (c) Human Protein atlas [28]. (d) HRM skin Lesion [31]. (e) MRI images [32].
3.3. Training Details
The proposed method was implemented as an end-to-end convolution network using the PyTorch framework [34]. Thus, it does not require any post-processing to perceive the ultimate outputs. The network was optimized with an Adam optimizer [35] with , , and learning rate = . The network was trained on resized images with dimensions of . However, for testing, we utilized the actual dimensions of the medical images. The batch size for training was fixed at 16 and we trained the network for 150,000 steps. It took around 72 hours to converge on a machine comprising an AMD Ryzen 3200 G central processing unit (CPU) clocked at 3.60 GHz and a random-access memory of 16 GB. A Nvidia Geforce GTX 1060 (6 GB) graphical processing unit (GPU) was utilized for accelerating the training process.
4. Result and Analysis
The proposed method was extensively studied and compared with the potential SOTA deep deblurring method with existing dedicated works for medical image deblurring. Furthermore, we illustrated the impact of deblurring in several medical image analysis tasks.
4.1. Comparison with State-of-the-Art Methods
We evaluated and compared the SOTA deblurring methods. For a fair comparison, we studied the learning-based deep deblurring methods throughout this study. All comparing methods have been trained and tested with our proposed blur-sharp dataset. Please note that our training and testing comprise both RGB and grayscale images. Additionally, they have been trained with their suggested hyperparameters to obtain maximum performance. In our setting, each model took around 2∼4 days for converging with the given data samples. The performance of the deep models has been summarized with three distinct evaluation metrics: (i) PSNR, (ii) SSIM, and (iii) DeltaE. Here, these standards aim to consider specific criteria, while evaluating the generated images. For instance, PSNR considers the signal-to-noise ratio, SSIM evaluates the structural information, and DeltaE quantifies the perceptual quality with color distortion between the generated and target image. The implication of such diverse evaluation metrics allowed us to evaluate the generated images from different imaging perspectives. In addition to the quantitative evaluation, we also performed a visual comparison between the deep deblurring method for visually understanding the performance of these deep methods.
4.1.1. Quantitative Comparison
Table 2 illustrates the objective comparison between deep deblurring methods for MID. We evaluated the performance of each comparing method by utilizing the evaluation metrics. Moreover, we calculated individual scores (i.e., PSNR, SSIM, and deltaE) for all testing images. We compute the mean performance of each comparing method for a specific dataset to observe their performance on that respective modality. Later, we summarized the performance of each comparing method by calculating the mean PSNR, SSIM, and deltaE scores obtained on the individual modality.

Table 2.
Quantitative comparison between deblurring methods in different medical image datasets. The proposed method outperforms existing methods in all evaluating metrics.
In all evaluation criteria, the proposed method outperforms the existing deblurring methods by a notable margin. It also shows that the proposed method is a medical image modality independently and can handle a diverse range of blurry images. As a result, the proposed method demonstrates superior performance across all comparing datasets. On average, our method outperforms its near-performing deep method by 0.86 dB in PSNR, 3% in SSIM, and 0.15 in DeltaE matrics. Overall, the quantitative evaluation confirms that the proposed method can remove blur by recovering structural information, low-noise ratio, and visually plausible sharp medical images compared to the existing methods.
4.1.2. Qualitative Comparison
Figure 5 depicts the qualitative comparison between deep deblurring methods. It can be seen that the proposed method outperforms the existing deblurring methods by recovering salient details. Apart from recovering details, the proposed network can remove maximum blur without illustrating visually disturbing artifacts. The proposed RD-SAM helps us learn salient features with local-global attention, while removing blind blurs from medical images. Overall, the qualitative comparison verifies the feasibility of the proposed method for MID in real-world applications.
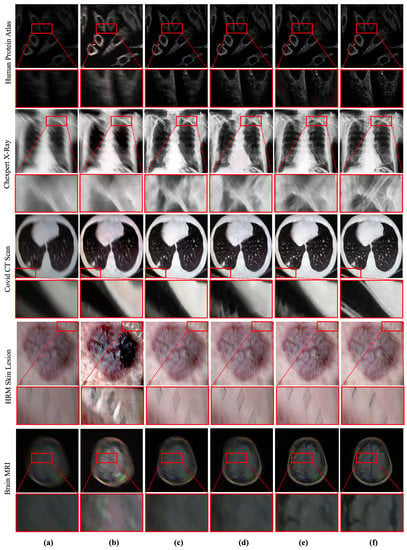
Figure 5.
Qualitative comparison between deep MID methods. The proposed method outperforms the existing methods by recovering salient information from blurry medical images. (a) Blurry input. (b) Result obtained by TEMImageNet [9]. (c) Result obtained by ZhaoNet [11]. (d) Result obtained by Deep Deblur [12]. (e) Result obtained by SRN Deblur [13]. (f) Proposed Method.
4.2. Deblurring in Computer-Aided Diagnosis
The Computer-Aided Diagnosis (CAD) application has gained substantial attention in recent years. In the last decade, several CAD applications, such as segmentation, detection, recognition, etc., illustrated significant impacts in medical image analysis. Nonetheless, the performance of this CAD application can be noticeably affected by the perceptual quality of the medical images. The blind-motion blur can seriously deteriorate the performance of any CAD method. To study the impact of blur in CAD applications, we evaluated the SOTA CAD methods with blurry images in this study. Furthermore, we illustrated that a robust deblurring method such as the proposed method could accelerate the performance of CAD.
4.2.1. Segmentation
Abnormality segmentation is considered among the most prominent CAD applications in medical image analysis. Despite having substantial real-world implications, medical image segmentation suffers from blind-motion blur. As Figure 6 illustrates, the existing segmentation methods’ performance can drastically deteriorate by blind-motion blur. A robust deblurring method can help segmentation methods in improving their performance. Here, we evaluated the SOTA U-net architecture inspecting the impact of blurs in segmentation on MRI images.
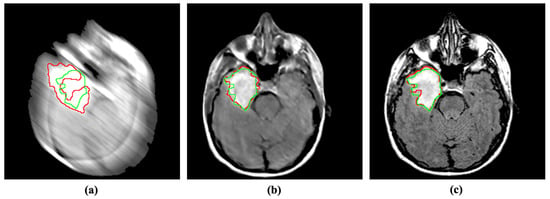
Figure 6.
Incorporating the proposed method for deblurring medical images can improve medical image segmentation performance. The red and green regions represent the area segmented by the deep model and the actual (ground-truth) affected region. (a) Blurry image + U-Net [36]. (b) Deblurred image obtained by proposed method + U-Net [36]. (c) Reference image + U-Net [36].
4.2.2. Detection
Similar to segmentation, abnormality detection is another widely used application in CAD. Such detection methods predict the region of abnormal issues or foreign bodies in a given medical image. Blind motion blurs can deteriorate the performance of such deep networks as well. We evaluated the performance of SOTA Mask R-CNN [37] with blurs in brain tumor detection. Despite being a well establish detection method, Mask R-CNN substantially suffers from blurs in the input image. Inversely, the proposed MID method can help the detection model by enhancing and removing blurs from the given input, as shown in Figure 7.
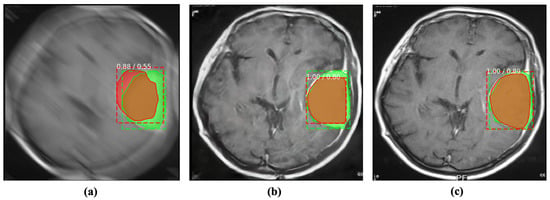
Figure 7.
Incorporating the proposed method for deblurring medical images can improve the performance of abnormality detection. The red and green regions represent the area segmented by the deep model and the actual (ground-truth) affected region. (a) Blurry image + Mask R-CNN [37]. (b) Deblurred obtained by proposed method + Mask R-CNN [37]. (c) Reference image + Mask R-CNN [37].
4.3. Ablation Study
The impact of the proposed components was verified with sophisticated experiments. We remove our scale recurrent subnetworks and the proposed RD-SAM from our network. Later, these components were incorporated into the network architecture to reveal their contribution. We evaluated the performance of our network variant on our testing dataset of 10,000 medical images. Table 3 illustrates the quantitative evaluation of the network variants.

Table 3.
Impact of RD-SAM and scale-recurrent architecture MID. It can be seen that the proposed components drastically improve the performance of MID while handling a diverse range of blind motion blurs.
Apart from the quantitative evaluation, the contribution of the proposed components was also verified with qualitative results. Figure 8 illustrates the visual performance of our proposed network variants. It can be seen that our scale recurrent structure helps us in removing blurs from medical images. Additionally, our RD-SAM enhances the details, while preserving the color information by leveraging local-global attention.
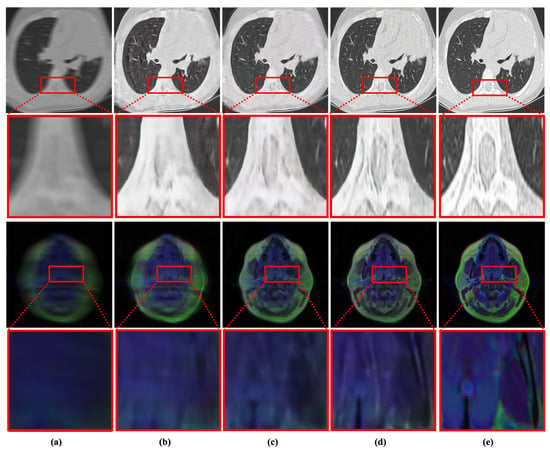
Figure 8.
Qualitative evaluation of the impact of proposed components in MID. (a) Blurry Input. (b) Base network. (c) Base network + scale-recurrent structure. (d) Proposed network with scale-recurrent structure + RD-SAM. (e) Reference image.
4.4. Performance Analysis
Figure 9 illustrates the training progress of the proposed method. The proposed method obtained a stable PSNR performance over training steps. However, the SSIM and deltaE score of the proposed deblurring method deteriorates, while stabilizing the PSNR performance. The MID is considered a challenging-tricky task due to the relationship of blur kernels with sensor noise (). Such complicated behavior of blur kernels can substantially impact structural and perceptual enhancement. This inevitable dilemma drives the MID methods to trade between PSNR and structural enhancement. This study prioritizes the PSNR over other evaluation metrics due to the characteristics of medical images.
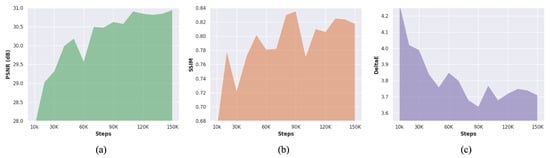
Figure 9.
Learning medical image deblurring over training steps. (a) Steps vs. PSNR. (b) Steps vs. SSIM. (c) Steps vs. DeltaE.
Apart from training performance, the proposed method comprises 24.1 million trainable parameters. The FLOPs and inference time of the proposed method can vary depending on the input dimension. It is worth noting the proposed network is fully convolutional. Thus, it can take any image dimension as an input. However, the proposed network has developed as an end-to-end network for reducing pre or post-processing operations. On static hardware with fixed-dimensioned images, the inference time of the proposed method is expected to remain unchanged. Table 4 depicts the detailed analysis of the proposed deblurring method.

Table 4.
Analysis of the proposed network developed as an end-to-end fully convolutional neural network.
4.5. Discussion
The proposed method reveals several aspects of MID. We illustrated that the scale-recurrent deep structure with RD-SAM block substantially improves the performance in MID. We also demonstrate that MID can be generalized without separating the medical image into different genres. A robust blur generation method with a precise learning strategy can substantially remove blur from medical images. Furthermore, such deblurring methods can improve the performance of CAD applications.
Despite showing improvement over existing methods, one of the limitations of our proposed method is that we trained and tested our network on a synthesized dataset. Regrettably, we could not find any open dataset with blur-sharp images to evaluate our network. However, our data generation methods have been developed based on the suggestion from recent works.
The limitation of the proposed work could drive future MID methods in a new direction. It would be meaningful work to collect a medical image dataset with blur-sharp image pairs. Apart from that, our scale-recurrent network with RD-SAM could be extended for removing blurs from the 3D medical images. However, for such implementation, the input and output layers of the proposed network need to be modified. Additionally, an optimized MID method for edge devices is still due. It could be interesting to evaluate the MID methods in edge devices through a future study.
5. Conclusions
This study proposed a novel learning-based deep method for muti-modal medical image deblurring. The proposed method demonstrates that medical image deblurring can be generalized for multi-modal medical images with a proper learning strategy. This study also introduced a novel scale-recurrent deep network with residual dense block and spatial-asymmetric module. The proposed module aims to learn salient features with local-global attention for recovering detail texture and information while deblurring medical images. The performance of the proposed method has been evaluated with different medical image datasets. The comparison results demonstrated that the proposed method outperforms the existing works in qualitative and quantitative comparisons. The applicability of the proposed method has been evaluated by incorporating it into numerous medical image analysis tasks. The experimental results reveal that the proposed method can substantially improve the performance of such analysis tasks by removing blind-motion blurs from the given image. It has planned to collect real-world blur-sharp medical image pairs and study the performance of the proposed method in 3D images in a future study.
Author Contributions
S.M.A.S., conceptualization, methodology, software, visualization, writing original draft. R.A.N., resources, validation, methodology, writing original draft, project administration. Z.M., data curation, resources, software, editing. J.H., data curation, software, editing. A.A., investigation, funding acquisition, project administration, supervision. S.-W.L., investigation, funding acquisition, supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a national research foundation (NRF) grant funded by the Ministry of Science and ICT (MSIT), South Korea through the Development Research Program (NRF2022R1G1A101022611) and (NRF2021R1I1A2059735).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Code available: https://github.com/sharif-apu/MedDeblur (accessed on 22 November 2022).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sharif, S.; Naqvi, R.A.; Biswas, M. Learning medical image denoising with deep dynamic residual attention network. Mathematics 2020, 8, 2192. [Google Scholar] [CrossRef]
- Sharif, S.; Naqvi, R.A.; Biswas, M.; Loh, W.K. Deep Perceptual Enhancement for Medical Image Analysis. IEEE J. Biomed. Health Inform. 2022, 26, 4826–4836. [Google Scholar] [CrossRef] [PubMed]
- Schuler, C.J.; Hirsch, M.; Harmeling, S.; Schölkopf, B. Learning to deblur. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 38, 1439–1451. [Google Scholar] [CrossRef]
- Khare, A.; Tiwary, U.S. A new method for deblurring and denoising of medical images using complex wavelet transform. In Proceedings of the 2005 IEEE Engineering in Medicine and Biology 27th Annual Conference, Shanghai, China, 17–18 January 2006; pp. 1897–1900. [Google Scholar]
- Amini Gougeh, R.; Yousefi Rezaii, T.; Farzamnia, A. Medical image enhancement and deblurring. In Proceedings of the 11th National Technical Seminar on Unmanned System Technology 2019; Springer: Singapore, 2021; pp. 543–554. [Google Scholar]
- Jiang, M.; Wang, G.; Skinner, M.W.; Rubinstein, J.T.; Vannier, M.W. Blind deblurring of spiral CT images. IEEE Trans. Med. Imaging 2003, 22, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Ahunbay, E.; Pipe, J.G. Rapid method for deblurring spiral MR images. Magn. Reson. Med. Off. J. Int. Soc. Magn. Reson. Med. 2000, 44, 491–494. [Google Scholar] [CrossRef]
- Zhao, H.; Ke, Z.; Chen, N.; Wang, S.; Li, K.; Wang, L.; Gong, X.; Zheng, W.; Song, L.; Liu, Z.; et al. A new deep learning method for image deblurring in optical microscopic systems. J. Biophotonics 2020, 13, e201960147. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Zhang, R.; Wang, C.; Yang, X.Q.; Xin, H.L. TEMImageNet training library and AtomSegNet deep-learning models for high-precision atom segmentation, localization, denoising, and deblurring of atomic-resolution images. Sci. Rep. 2021, 11, 1–15. [Google Scholar] [CrossRef]
- Zhang, K.; Ren, W.; Luo, W.; Lai, W.S.; Stenger, B.; Yang, M.H.; Li, H. Deep image deblurring: A survey. Int. J. Comput. Vis. 2022, 130, 2103–2130. [Google Scholar] [CrossRef]
- Zhang, K.; Zuo, W.; Chen, Y.; Meng, D.; Zhang, L. Beyond a gaussian denoiser: Residual learning of deep cnn for image denoising. IEEE Trans. Image Process. 2017, 26, 3142–3155. [Google Scholar] [CrossRef]
- Nah, S.; Hyun Kim, T.; Mu Lee, K. Deep multi-scale convolutional neural network for dynamic scene deblurring. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 3883–3891. [Google Scholar]
- Tao, X.; Gao, H.; Shen, X.; Wang, J.; Jia, J. Scale-recurrent network for deep image deblurring. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8174–8182. [Google Scholar]
- Zhang, Y.; Tian, Y.; Kong, Y.; Zhong, B.; Fu, Y. Residual dense network for image super-resolution. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 2472–2481. [Google Scholar]
- Sharif, S.; Naqvi, R.A.; Biswas, M. SAGAN: Adversarial Spatial-asymmetric Attention for Noisy Nona-Bayer Reconstruction. arXiv 2021, arXiv:2110.08619. [Google Scholar]
- Khare, A.; Tiwary, U.S.; Jeon, M. Daubechies complex wavelet transform based multilevel shrinkage for deblurring of medical images in presence of noise. Int. J. Wavelets Multiresolution Inf. Process. 2009, 7, 587–604. [Google Scholar] [CrossRef]
- Tech, M.; Scholar, P. A Modified ADMM Approach for Blind Deblurring of Color Medical Images. Int. J. Eng. Res. Technol. (IJERT) 2014, 3, 51–57. [Google Scholar]
- Al-Ameen, Z.; Sulong, G.; Gapar, M.; Johar, M. Reducing the Gaussian blur artifact from CT medical images by employing a combination of sharpening filters and iterative deblurring algorithms. J. Theor. Appl. Inf. Technol. 2012, 46, 31–36. [Google Scholar]
- Al-Ameen, Z.; Sulong, G. Deblurring computed tomography medical images using a novel amended landweber algorithm. Interdiscip. Sci. Comput. Life Sci. 2015, 7, 319–325. [Google Scholar] [CrossRef]
- Dong, J.; Roth, S.; Schiele, B. Deep wiener deconvolution: Wiener meets deep learning for image deblurring. Adv. Neural Inf. Process. Syst. 2020, 33, 1048–1059. [Google Scholar]
- Sun, J.; Cao, W.; Xu, Z.; Ponce, J. Learning a convolutional neural network for non-uniform motion blur removal. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 769–777. [Google Scholar]
- Ye, M.; Lyu, D.; Chen, G. Scale-Iterative Upscaling Network for Image Deblurring. IEEE Access 2020, 8, 18316–18325. [Google Scholar] [CrossRef]
- Zhang, J.; Pan, J.; Ren, J.; Song, Y.; Bao, L.; Lau, R.W.; Yang, M.H. Dynamic scene deblurring using spatially variant recurrent neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 2521–2529. [Google Scholar]
- Kupyn, O.; Budzan, V.; Mykhailych, M.; Mishkin, D.; Matas, J. Deblurgan: Blind motion deblurring using conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8183–8192. [Google Scholar]
- Kupyn, O.; Martyniuk, T.; Wu, J.; Wang, Z. Deblurgan-v2: Deblurring (orders-of-magnitude) faster and better. In Proceedings of the IEEE International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 8878–8887. [Google Scholar]
- Sharif, S.; Kim, S.; Park, S. Learning real-world anti-spoofing leveraging spatial-asymmetric attention. In Proceedings of the Korea Software Conference (KSC2022), Incheon, Republic of Korea, 18–22 September 2022. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based human protein atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 590–597. [Google Scholar]
- Zhao, J.; Zhang, Y.; He, X.; Xie, P. COVID-CT-Dataset: A CT scan dataset about COVID-19. arXiv 2020, arXiv:2003.13865. [Google Scholar]
- Rezvantalab, A.; Safigholi, H.; Karimijeshni, S. Dermatologist level dermoscopy skin cancer classification using different deep learning convolutional neural networks algorithms. arXiv 2018, arXiv:1810.10348. [Google Scholar]
- Buda, M.; Saha, A.; Mazurowski, M.A. Association of genomic subtypes of lower-grade gliomas with shape features automatically extracted by a deep learning algorithm. Comput. Biol. Med. 2019, 109, 218–225. [Google Scholar] [CrossRef]
- Sharif, S.; Mahboob, M. Deep hog: A hybrid model to classify bangla isolated alpha-numerical symbols. Neural Netw. World 2019, 29, 111–133. [Google Scholar] [CrossRef]
- Pytorch. PyTorch Framework Code. 2016. Available online: https://pytorch.org/ (accessed on 24 August 2020).
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).