Abstract
Background: Human insulin-like growth factor 1 (hIGF-1) plays a key role in cell proliferation and tissue repair. While plant expression systems offer a cost-effective and scalable alternative for recombinant protein production, the molecular effects of plant-derived hIGF-1 on mammalian cells remain largely unexplored. Methods: In this study, a recombinant fusion protein of hIGF-1 with human Fc (hIGF-1-Fc) was transiently expressed in Nicotiana benthamiana using the geminiviral pBYR2e system and purified by Protein A affinity chromatography. SDS-PAGE and Western blotting confirmed the predicted molecular weight, and LC-MS identified N-glycosylation at the Fc N229 site with plant-type glycans such as GnMXF, GnGnXF, and MMXF. Bioactivity was evaluated using MCF-7 cell proliferation and NIH3T3 wound healing assays. Label-free quantitative proteomics was performed on NIH3T3 fibroblasts to assess molecular changes. Results: hIGF-1 Fc significantly promoted cancer cell migration and fibroblast proliferation. Proteomic profiling revealed an abundance of cytoskeletal proteins such as actin and tubulin and metabolic enzymes related to energy production. Gene ontology and pathway enrichment analyses indicated significant modulation of ribosome biogenesis and carbon metabolism. Conclusions: This study presents the first proteome-level investigation of plant-produced hIGF-1-Fc in mouse fibroblasts and reveals its impact on cytoskeletal organization and metabolic pathways involved in proliferation and wound healing.
1. Introduction
Cellular agriculture represents an innovative approach to producing recombinant proteins or agricultural commodities using cell cultures derived from animals, plants, and microbes [1]. Central to this expression method are growth factors that regulate various cellular processes, including cell growth, tissue repair, and differentiation [2]. They are indispensable in the fields of stem cell research, cancer therapeutics, cell and gene therapy, and in pharmaceutical and cosmetic products. However, their high production costs limit their accessibility, as growth factors account for up to 90% of expenses in the cellular agriculture industry [3].
Among growth factors, human insulin-like growth factors (hIGFs), fibroblast growth factor (FGF), and epidermal growth factor (EGF) are particularly important [4]. hIGF comprises two types—hIGF-1 and hIGF-2 subtypes—and it plays a pivotal role in cell proliferation, survival, and differentiation [5]. This 7.6 kDa protein consists of 70 amino acids with disulfide-linked A and B chains and is primarily secreted by the liver, where it synergizes with growth hormone to mediate anabolic and growth-promoting effects [6]. Recently, hIGF-1 has become an essential supplement in cell culture media formulations, particularly for enhancing growth rates and skeletal muscle development [4]. Additionally, its therapeutic potential in wound healing has been well documented in the literature. For example, hIGF-1 promotes cell migration, proliferation, and extracellular matrix production, all of which are crucial for effective wound repair [7].
Plant molecular pharming has emerged as a promising platform that employs plants as bioreactors to manufacture target proteins in a scalable and economical manner. Plants from the Nicotiana genus are commonly used hosts due to their rapid growth period and ease of genetic manipulation, though other crops like tomato, rice, lettuce, or potato are also viable options. Advantages of this platform include simplified cultivation, a low pathogen contamination risk, rapid mass protein production, scalability, and the ability to produce complex proteins [8]. While plant-based expression of growth factors, e.g., human vascular endothelial growth factor (VEGF) [9], EGF [10], and FGF [11], has been explored, few studies have investigated their functional effects at the molecular level.
Proteomics is a powerful approach to study growth-factor-induced cellular responses. Previous studies have analyzed differential proteome changes and pathway modulation following growth factor treatment [12,13]. For instance, Nagano et al. analyzed the proteomic changes in Swiss 3T3 fibroblasts after long-term exposure to platelet-derived growth factor (PDGF), IGF-1, and EGF [12]. Their findings revealed that PDGF induced the most sustained phospho-signaling and protein synthesis, whereas IGF-1 and EGF contributed to distinct proteomic profiles affecting key regulators of actin cytoskeleton remodeling, translation, and chaperonin activity. Similarly, another research evaluated the acute effects of IGF-1 on murine C2C12 myoblasts, identifying 23 increased and 17 decreased proteins following IGF-1 treatment [13]. These results highlight the impact of IGF-1 on metabolic and structural protein networks in muscle cells. However, to date, no studies have systematically analyzed the proteomic response of fibroblasts to plant-produced hIGF-1.
In this study, we conducted the first proteome-wide investigation of plant-made hIGF-1-Fc in mouse NIH3T3 fibroblasts. The recombinant hIGF-1 was fused with an Fc fragment and expressed in N. benthamiana tobacco plants. The fusion protein was purified via Protein A affinity chromatography, and the purified hIGF-1-Fc was subsequently evaluated for its effects on cell proliferation and wound healing in mammalian cell lines. Additionally, quantitative proteomics was performed to identify differentially expressed proteins in response to hIGF-1-Fc treatment.
2. Materials and Methods
2.1. Host, Expression Vector, and Cell Line
Wildtype N. benthamiana seeds were kindly gifted by Dr. Supaart Sirikantaramas (Faculty of Science, Chulalongkorn University, Bangkok, Thailand). Competent cells, Escherichia coli DH10B (Goldbio, CC-100-5x50) and Agrobacterium tumefaciens GV3101 (Goldbio, CC-105-5x50), were used for subcloning. The pBYR2eK2Md or pBYR2e vector [14,15] was utilized for recombinant protein expression in plants. Antibiotics used for clone selection included kanamycin (Bio Basic, Markham, ON, Canada), rifampicin (Thermo Fischer Scientific, Waltham, MA, USA), and gentamicin (ITW Reagents, Darmstadt, Germany). Restriction enzymes and the T4 ligation kit were purchased from New England Biolabs (Ipswich, MA, USA), and plasmid purification was performed using a DNA-spin™ kit (iNtRON Biotechnology, Seongnam-si, Gyeonggi-do, South Korea). Plasmid DNAs were sent for Sanger sequencing analysis to U2Bio, Thailand. Protein all-blue standards (Bio-Rad Laboratories, Hercules, CA, USA), InstantBlue® Coomassie protein stain (Abcam plc, Cambridge, UK), and goat anti-human IgG-HRP antibody (SouthernBiotech, Birmingham, AL, USA) were used for protein analysis and detection. Mouse embryonic fibroblasts (NIH3T3) and a human breast cancer cell line (MCF-7) were provided by Prof. Jittima Luckanagul (Faculty of Pharmaceutical Sciences, Chulalongkorn University), originally sourced from ATCC (Manassas, VA, USA). All other reagents, DMEM (Biowest, Riverside, MO, USA), FBS (HyClone, Logan, UT, USA), penicillin–streptomycin (Gibco, Waltham, MA, USA), L-glutamine (Gibco), and MTT (Invitrogen, Carlsbad, CA, USA), were procured for cell-based assays.
2.2. Plasmid Construction
The hIGF-1 gene (Uniprot No: P05019, G49-A118) was constructed as a fusion protein with a human IgG Fc fragment at the C-terminus and flanked by XbaI/SacI restriction sites. The hIGF-1-Fc codon sequence was optimized for expression in N. benthamiana and synthesized by Genewiz (Genewiz, Suzhou, China). The optimized gene was then subcloned into the XbaI and SacI sites of the geminiviral vector pBYR2e to generate the pBYR2e/hIGF-1-Fc plasmid (Figure 1). The ligated plasmid was introduced into E. coli DH10B competent cells by heat shock and cultured on LB agar containing 50 mg/L kanamycin overnight at 37 °C. Transformants were screened by PCR, and positive clones were grown in LB broth with 50 mg/L kanamycin. The presence of the hIGF-1-Fc insert was confirmed by DNA sequencing (Supplementary Figure S1).
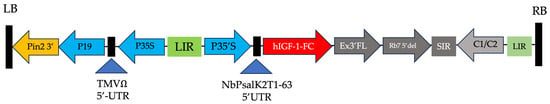
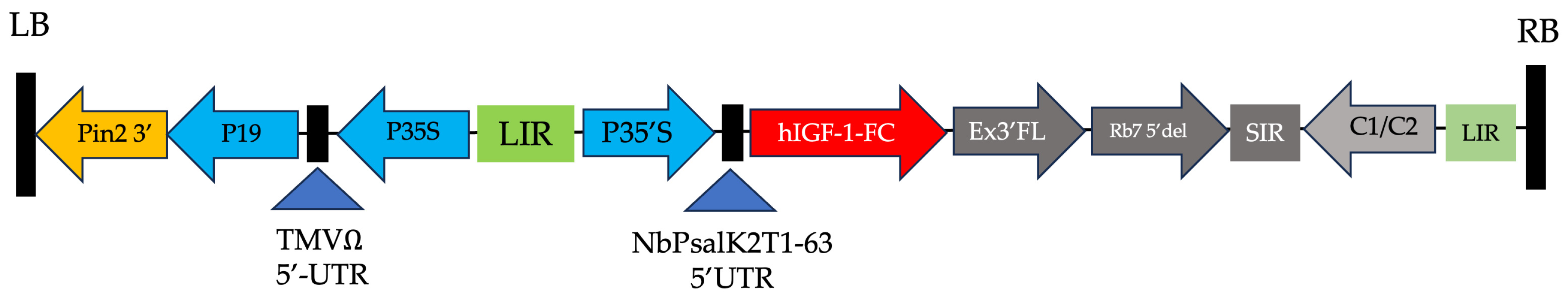
Figure 1.
Schematic of the geminiviral vector containing the hIGF-1-Fc gene. Key elements include the T-DNA left and right borders (LB and RB) for gene insert integration into plant cells, the CaMV 35S promoter (P35S) enhanced by TEV 5′ for translation, and P19 for gene silencing suppression. Additional regions for mRNA stability, replication, expression, and termination include the LIR and SIR of the bean yellow dwarf virus (BeYDV) and BeYDV ORFs C1 and C2. Diagram adapted from previous work on plant-produced growth factors [9,10].
2.3. Transient Expression of hIGF-1 Fusion Protein
Production of recombinant hIGF-1-Fc in tobacco plants was conducted as previously described [11]. The pBYR2e/hIGF-1-Fc vector was transformed into competent A. tumefaciens GV3101 via electroporation. Transformants were selected on LB agar plates containing 50 mg/L kanamycin, 50 mg/L rifampicin, and 50 mg/L gentamicin and incubated for 48 h at 28 °C. Positive colonies were then cultured in LB broth with the same antibiotics overnight at 28 °C. After reaching an OD600 of 0.2, Agrobacterium harboring hIGF-1-Fc gene was infiltrated into four-week-old N. benthamiana plants. Leaves were harvested at 1, 3, 5, and 7 days post-infiltration (dpi) to optimize protein expression levels. Expressed protein abundance was assessed by Western blotting, and band intensity was quantified by ImageJ (version 1.52p, National Institutes of Health, Bethesda, MD, USA) for comparison. The membrane was probed using an HRP-conjugated anti-human IgG Fc antibody to specifically detect the hIGF-1-Fc protein.
2.4. Affinity-Based Purification of Plant-Derived hIGF-1 Fusion Protein
The affinity chromatography purification procedure was adapted from [16]. Approximately 0.25 kg of plant leaves was homogenized in ice-cold PBS buffer pH 7.4 at a 1:2. The crude extract was centrifuged at 15,000 rpm for 45 min at 4 °C, followed by filtration using a 0.45 µm S-Pak membrane filter (Merck, Burlington, MA, USA). The clear supernatant containing the expressed hIGF-1-Fc protein was loaded onto a Protein A gravity-flow chromatography column (Cytiva, Marlborough, MA, USA). The column was washed with PBS buffer pH 7.4, and the bound fusion protein was eluted using 0.1 M glycine buffer pH 2.7. The eluted fraction was subsequently neutralized with 1.5 M Tris-HCl buffer pH 8.8 to reach pH 7.4. The yield of hIGF-1-Fc was measured using a Bradford assay and human IGF-1 ELISA kit (R&D systems, Minneapolis, MN, USA, DY291).
2.5. SDS-PAGE and Western Blotting
Crude extracts and purified hIGF-1-Fc were analyzed by SDS-PAGE and Western blotting, as previously described [17]. Equal amounts of total soluble proteins (6 µg) from leaves harvested at 1, 3, 5, and 7 dpi and 2 µg of pure hIGF-1-Fc were mixed with SDS loading buffer, boiled for 5 min at 95 °C, and separated on 12% SDS-PAGE gels under both non-reducing and reducing conditions. Parallel acrylamide gels were resolved under identical conditions: one was stained with InstantBlue® Coomassie protein stain following the manufacturer’s instructions, while the other was subjected to Western blot analysis. Proteins were transferred onto a nitrocellulose membrane using a wet-transfer system with 1X transfer buffer (25 mM Tris, 192 mM glycine, 15% methanol) operated at 100 V for 90 min at 4 °C. The blots were subsequently blocked with 5% (w/v) skimmed milk in PBS buffer pH 7.4 for 1 h at room temperature and incubated with an HRP-linked goat monoclonal antibody specific to human IgG Fc (SouthernBiotech) at a 1:5000 dilution for 2 h at room temperature with gentle agitation. The target protein bands were detected using a chemiluminescence detection system. Band intensities were quantified using ImageJ (version 1.52p) and reported as raw values without normalization to housekeeping protein.
2.6. LC-MS Glycoprofiling
Protein solution with approximately 15 µg of protein was reduced with 10 mM dithiothreitol (DTT) at 65 °C for 30 min and alkylated with 25 mM iodoacetamide (IAA) at room temperature for 20 min under darkness. Proteins were digested with 0.2 µg of trypsin at 37 °C for 4 h. The reaction was stopped with 1% FA. The solution was then centrifuged at 14,000 rpm for 10 min and transferred to LC-MS vial.
Peptides were analyzed using an Agilent 1290 Infinity II LC system coupled with an Agilent 6545XT Q-TOF mass spectrometer. LC separation was conducted on an AdvanceBio Peptide Mapping column (120 Å, 2.1 × 150 mm, 2.7 µm) at 60 °C. Injection volume was 10 µL. Mobile phase A was 0.1% FA in water, and mobile phase B was 0.1% FA in acetonitrile. LC gradient was set as follows: 0% B for 2 min, 0% to 20% B in 33 min, 20% to 30% B in 20 min, 30% to 50% B in 10 min, 50% to 90% B in 5 min, 90% B for 5 min, 90% to 0% B in 5 min, and 0% B for 5 min, with a constant flow rate of 0.4 mL/min. MS analysis was conducted in positive mode with MS and MS/MS mass range of 100–1700 and 50–1700 m/z, respectively. Acquisition times were 5 and 3 spectra per s for MS and MS/MS, respectively. MS parameters were set as follows: gas temperature at 325 °C, nebulizer at 35 psi, dying gas at 13 L/min, sheath gas temperature at 275 °C, sheath gas flow at 12 L/min, capillary voltage at 4000 V, nozzle voltage at 500 V, and skimmer voltage at 65 V. The top 10 precursor ions per cycle were selected for MS/MS fragmentation. The absolute precursor threshold was 3000 counts. Collision energy (CE) was varied according to the charge state of the peptide. For peptides with charge +1 and +2, the CE was calculated using a formula of (3.1 × ((m/z)/100) + 1), while for peptides with charge ≥+3, the CE was calculated using a formula of (3.6 × ((m/z)/100) − 4.8). Reference mass was monitored at 922.0098 m/z. Data was collected in centroid mode.
2.7. In Vitro Cell Proliferation Assay of Plant-Derived hIGF-1 Fusion Protein
MCF-7 cells were cultured in DMEM supplemented with 10% FBS, 1% penicillin–streptomycin, and 1% L-glutamine. Cells were seeded into a 96-well plate with a density of 5 × 103 cells/well in DMEM and incubated overnight at 37 °C under a humified atmosphere with 5% CO2. An eight-point, ten-fold serial dilution of plant-produced hIGF-1-Fc or Fc, starting from a concentration of 10 µg/mL, was added to the cells. Untreated cells served as the control. After 48 h of treatment, 100 μL of MTT solution (0.50 mg/mL) was added and incubated for 4 h at 37 °C. The solution was carefully removed. Formazan crystals were dissolved by adding 100 µL of DMSO per well. Absorbance was measured at 570 nm using a microplate reader (CLARIOStar, BMG Labtech, Ortenberg, Germany). Cell viability was calculated as a percentage relative to the untreated control cells and represented as dose–response curves. EC50 values were calculated using GraphPad Prism 9.0. The assay was performed following prior MTT protocols [9].
2.8. In Vitro Wound Healing Assay of Plant-Derived hIGF-1 Fusion Protein
The wound healing efficacy of plant-produced hIGF-1-Fc was assessed using an in vitro scratch healing assay based on the method described by [18]. NIH3T3 fibroblasts were seeded at a density of 3 × 104 cells/well. Once a confluent cell monolayer was achieved, a “wound” was created by scratching the cell layer with a sterile 200 µL pipette tip. The wells were washed twice with PBS buffer at pH 7.4 to remove any dislodged cells. Plant-produced hIGF-1-Fc was added to the culture medium at final concentrations of 50 ng/mL and 100 ng/mL, and cells were incubated for 24 h and 48 h. Wound areas were photographed using a phase-contrast microscope (Nikon Corporation, Tokyo, Japan) at 100X magnification. Microscopic images were captured at 0, 24, and 48 h using a Nikon Eclipse Ti-S microscope (Nikon Corporation) equipped with a DS-Ri2 camera and the NIS Element v5.01 software. The ImageJ software version 1.52p was used to measure the wound area and calculate the percentage of wound closure relative to the initial area at 0 h.
2.9. Preparation of Fibroblast Cell Lysates for Proteomics Analysis
NIH3T3 fibroblast cells were seeded into a cell culture dish at a density of 5 × 103 cells/well, then treated with 50 and 100 ng/mL of plant-produced hIGF-1-Fc for 72 h. Cell pellets were collected and extracted with 100 µL of 50 mM ammonium bicarbonate (ABC) buffer with a metal bead in a Mixer Mill 400 (Retsch GmbH, Haan, Germany), following the method and protocol in the shotgun proteomics method and protocol book [19]. The samples were centrifuged at 14,000 rpm for 5 min, and the supernatant was collected. Protein concentration was measured with a Bradford assay and normalized to 2.3 mg/mL protein. The protein solution underwent treatment with 10 mM dithiothreitol (DTT) at 65 °C for 30 min to break disulfide bonds, followed by modification with 25 mM iodoacetamide (IAA) at room temperature for 20 min under darkness. Subsequently, proteins were enzymatically cleaved using 0.5 µg of trypsin at 37 °C for 4 h. To quench the reaction, 10% formic acid (FA) was added, and the mixture was centrifuged at 14,000 rpm for 10 min. The clarified supernatant was transferred into a polypropylene vial and subjected to LC-MS/MS analysis.
2.10. Proteomics LC-MS/MS Acquisition
Peptide samples were analyzed using Agilent 1290 Infinity II LC system coupled with an Agilent 6545XT Q-TOF mass spectrometer. Chromatographic separation was performed on an AdvanceBio Peptide Mapping column (120 Å, 2.1 × 150 mm, 2.7 µm) maintained at 60 °C. For each analysis, 10 µL of peptide solution was injected. The mobile phase consisted of 0.1% formic acid in water (mobile phase A) and 0.1% formic acid in acetonitrile (mobile phase B). The gradient elution protocol began with an isocratic hold at 0% B for 2 min, followed by a linear increase to 20% B in 33 min, then to 30% B over the subsequent 20 min. The gradient was further increased to 50% in 10 min, then to 90% B over 5 min, maintained at 90% B for 5 min, reduced back to 0% B in 5 min, and finally equilibrated at 0% B for 5 min. The flow rate was set at 0.4 mL/min throughout the run. Mass spectrometry was operated in positive mode with a mass range of 100–1700 m/z. Instrument settings included a gas temperature at 325 °C, nebulizer pressure at 35 psi, dying gas flow at 13 L/min, sheath gas temperature at 350 °C with a flow rate of 12 L/min, capillary voltage at 4000 V, nozzle voltage at 500 V, fragmentor voltage at 175 V, and skimmer voltage at 65 V. Data acquisition was performed at a rate of one spectrum per second. During MS/MS acquisition, up to two precursor ions were selected per cycle for fragmentation. Collision energy (CE) was adjusted according to the peptide charge state. For peptides with charge +1 and +2, the CE was calculated using a formula of (3.1 × (m/z)/100) + 1), while peptides with charge +3 or greater, the CE was calculated as (3.6 × (m/z)/100) − 4.8).
2.11. Proteomics Data Analysis and Pathway Analysis
Raw Agilent .d files were converted to .mzML files using the msConvert command in the ProteoWizard software [20] with default settings. Then, the .mzML file was converted to a .mzxML file using the TOPPAS function in the OpenMS software [21]. Data files in .mzxML format were loaded into the MaxQuant software version 2.4 [22]. All default parameters for the Agilent QTOF instrument and LFQ function were applied. Trypsin was selected as a digestive enzyme, and two missed cleavages were allowed. Methionine oxidation and N-terminal acetylation were set as variable modifications, while cysteine carbamidomethylation was a fixed modification. Protein and peptide match was searched with a high confidence level at 0.01 FDR. The mouse protein database was acquired from the UniProt website [23]. Proteins identified by site and reverse peptides were removed. LFQ intensity was used for statistical analysis using MetaboAnalyst version 6.0 [24] (https://www.metaboanalyst.ca/ accessed on 1 July 2025). Data was log 10 transformed and mean-centered.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) [25] and gene ontology (GO) pathway analyses were utilized to identify the possible mechanisms underlying its effects via the online bioinformatics tool [26] (https://www.bioinformatics.com.cn/ accessed on 1 July 2025).
2.12. Statistical Analysis
The day post-infiltration optimization experiment was conducted with three biological replicates. For all subsequent characterization and bioactivity assays, a single large-scale batch of purified hIGF-1-Fc was used to ensure consistency across all tests. Functional assays were performed in triplicate using this protein batch, and data are presented as mean ± standard deviation (SD). An unpaired t-test was conducted to assess differences between individual groups. Additionally, two-way analyses of variance (ANOVA) were performed using the GraphPad Prism 9.0 software (San Diego, CA, USA). A p-value of <0.05 was considered statistically significant.
3. Results
3.1. Effects of Dpi on hIGF-1-Fc Protein Expression
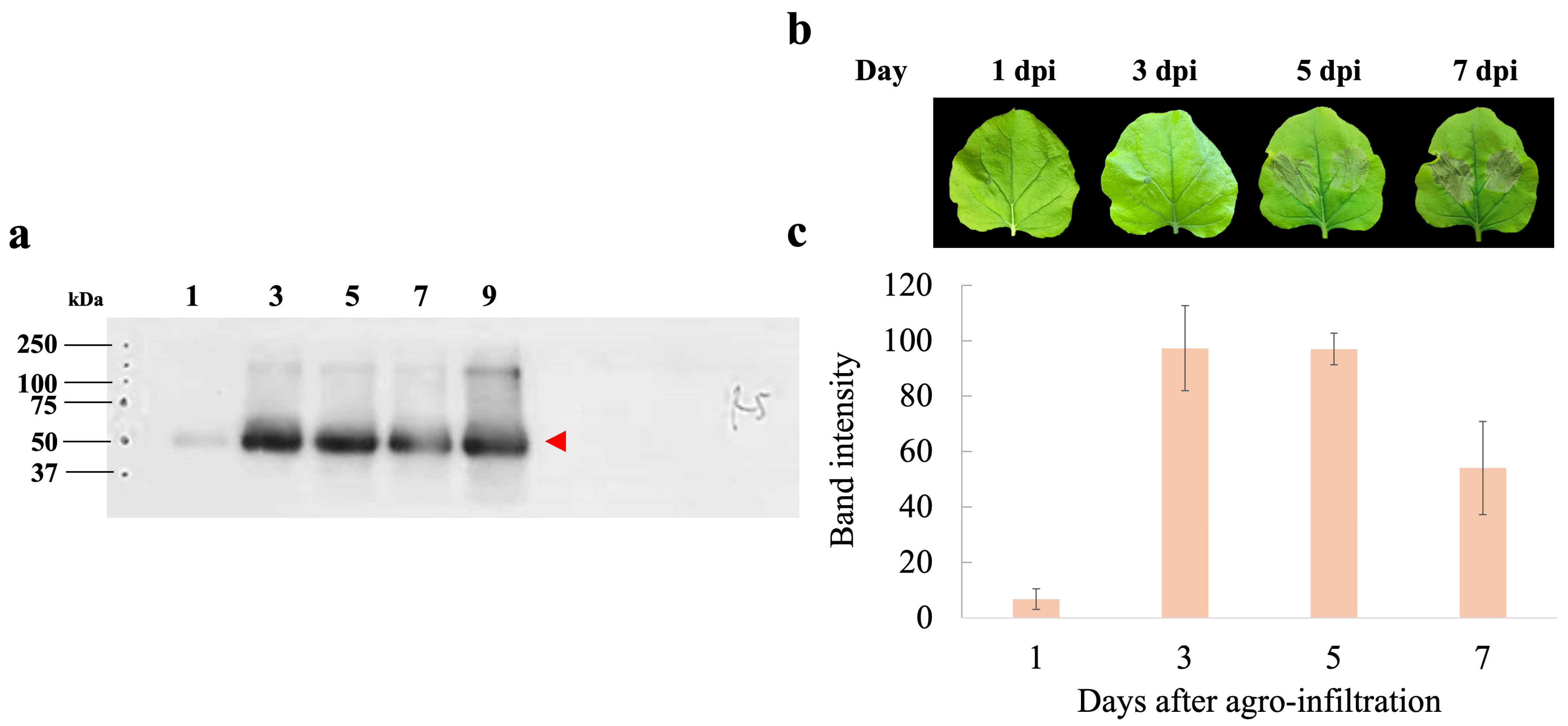
To assess the expression levels of hIGF-1-Fc, infiltrated tobacco leaves were harvested at different dpi (1, 3, 5, and 7 days). The band intensity of the target protein increased over time post-infiltration. Western blot analysis demonstrated that transient expression of hIGF-1-Fc peaked at 3 dpi (Figure 2a), with the corresponding uncropped blots provided in Supplementary Figure S2. Expression levels remained relatively stable up to 5 dpi, followed by a decline at 7 dpi (Supplementary Table S1). The lowest protein expression level was observed at 1 dpi. Morphological assessments of the infiltrated leaf revealed minimal necrosis at 1 and 3 dpi, whereas visible necrotic symptoms appeared at 5 and 7 dpi (Figure 2b). Quantitative analysis of hIGF-1-Fc protein band intensity in crude supernatants showed values of 6.80 at 1 dpi, 97.29 at 3 dpi, 97.00 at 5 dpi, and 54.08 at 7 dpi (Figure 2c). Based on these results, the optimal harvest time for recombinant hIGF-1-Fc production was determined to be 3 dpi.
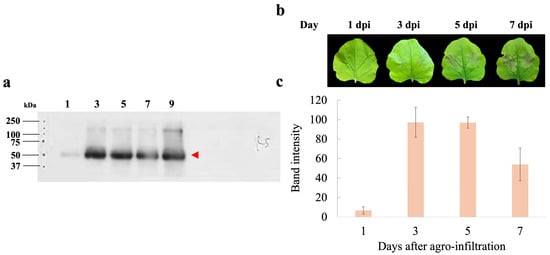
Figure 2.
Days post-infiltration (dpi) optimization of hIGF-1-Fc production in N. benthamiana. (a) Western blotting of crude leaf extracts collected at 1, 3, 5, and 7 days after infiltration. The red arrow indicates the molecular weight of the target protein (approximately 70 kDa). (b) Representative images of leaf phenotypes at different dpi. (c) Bar graph depicting the measured band intensity values from the immunoblots.
3.2. Purification of Plant-Produced hIGF-1-Fc
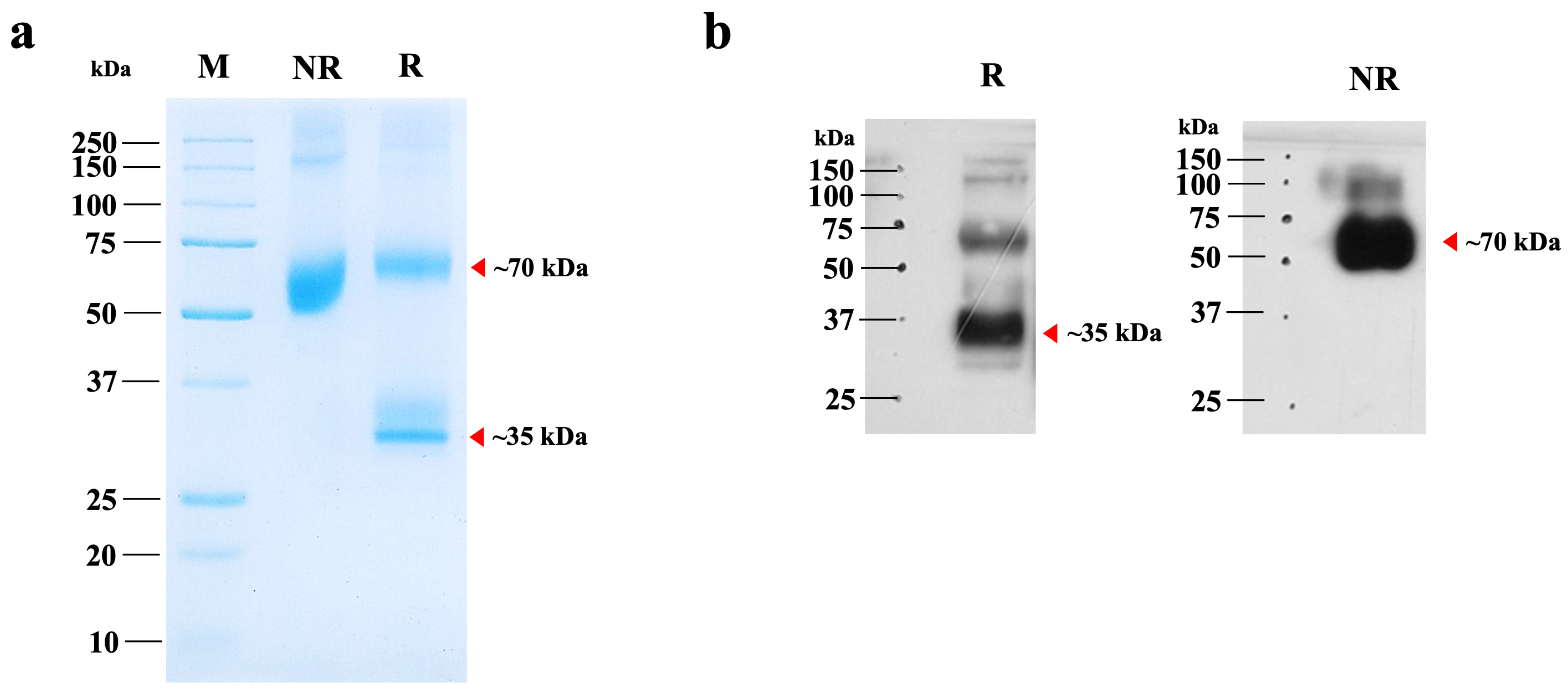
The plant-optimized hIGF-1 gene was fused with an Fc tag to facilitate purification via Protein A affinity chromatography. SDS-PAGE analysis revealed a major band at approximately 70 kDa, corresponding to the dimeric form of hIGF-1-Fc (Figure 3a, lane NR). Under reducing conditions with β-mercaptoethanol, a ~35 kDa protein band representing the monomeric form was detected, along with traces of the multimeric forms (Figure 3a, lane R). Therefore, this finding confirms the successful dimer assembly of the Fc fusion protein in the eukaryotic N. benthamiana system. The purity of plant-produced hIGF-1-Fc was >90% based on visual inspection of stained gels. To further confirm the identity of the target protein, Western blot analysis was conducted. A strong 70 kDa band was detected under non-reducing conditions, while both 35 kDa and minor 70 kDa bands were observed under reducing conditions (Figure 3b and Supplementary Figure S3). These protein band profiles were consistent with those identified in the SDS-PAGE gel. After one-step purification, approximately 0.01 µg of hIGF-1-Fc protein was obtained per g of N. benthamiana leaves.
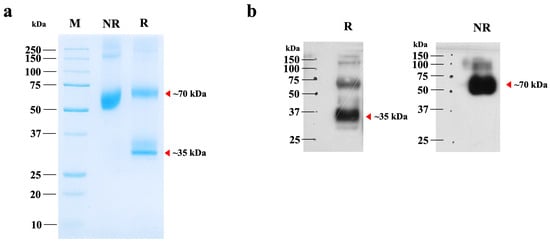
Figure 3.
SDS-PAGE and Western blot analyses of purified hIGF-1-Fc from N. benthamiana. (a) SDS-PAGE analysis of 2 µg of purified hIGF-1-Fc under non-reducing (NR) and reducing (R) conditions. (b) Immunoblot of the same purified sample, probed with an anti-human IgG HRP-conjugated antibody. Lanes NR and R represent non-reducing and reducing conditions, respectively. Red arrows indicate the molecular weights of the hIGF-1-Fc monomer and dimer, as predicted from the canonical sequence.
The apparent molecular weight of hIGF-1-Fc (~70 kDa) on SDS-PAGE gel was slightly higher than the calculated theoretical mass of the unglycosylated dimer (~64 kDa), suggesting the presence of post-translational modifications such as N-glycosylation. To confirm and characterize these modifications, LC-MS-based glycoprofiling was performed.
3.3. Glycoprofiling Analysis of Plant-Produced hIGF-1-Fc
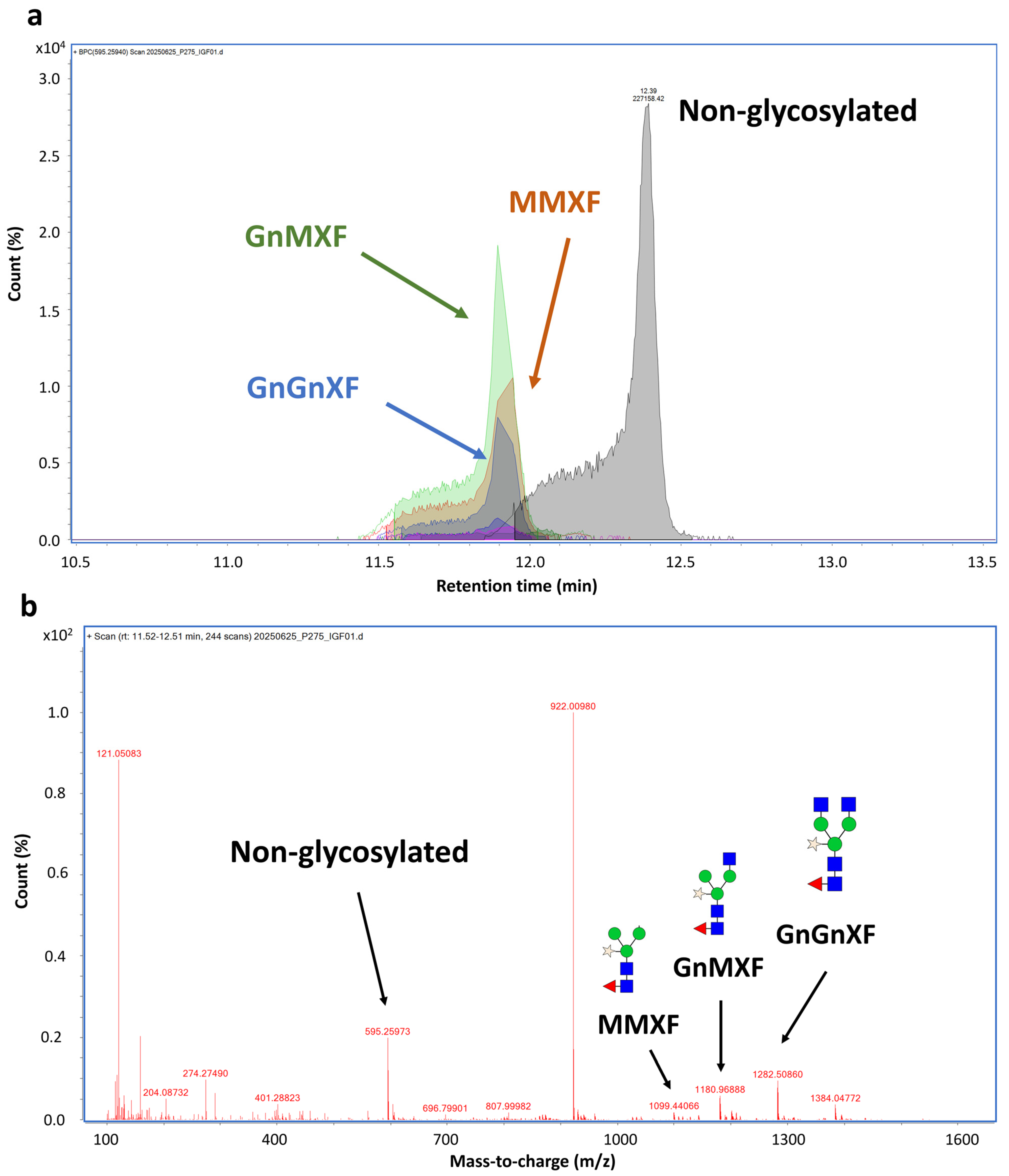
Mass spectrometry analysis confirmed that N-glycosylation was a major post-translational modification contributing to the observed mass shift. From the MS data, multiple glycans were detected at the N229 position, a common glycosylation site in the Fc region. The corresponding peptide, EEQYNSTYR, was found to be attached to different plant-derived glycoforms. The glycosylated species were observed at around 11.5–12.4 min, and its mass difference was calculated for glycan types. Around 63% of the molecule was glycosylated. The glycans detected at this site included GnMXF, GnGnXF, and MMXF (Figure 4 and Supplementary Table S2). Meanwhile, the non-glycosylated peptide was detected at 12.39 min.
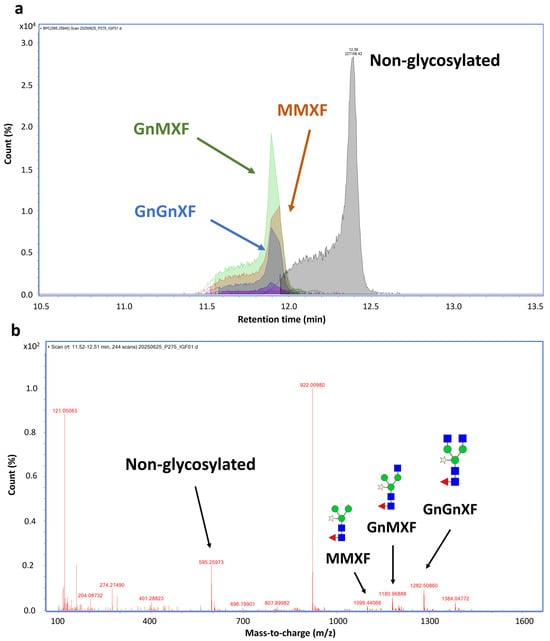
Figure 4.
N-glycosylation analysis at N229 position of hIGF-1-FC. Chromatogram of EEQYNSTYR peptide showing peaks of different plant N-glycan attachments (a). Non-glycosylated peak was eluted at 12.39 min, while glycosylated peaks were eluted faster between 11.9–12.40 min. MS spectrum of glycosylated mass from 1107–1383 m/z. The peaks corresponding to GnGnXF, GnMXF, MMXF, and non-glycosylated forms are shown in blue, green, brown, and gray, respectively (b). Among 63.1% glycosylated peptides, GnMXF, GnGnXF, and MMXF were major forms of attached N-glycans.
3.4. Bioactivity of Plant-Produced hIGF-1-Fc in Mammalian Cell Line
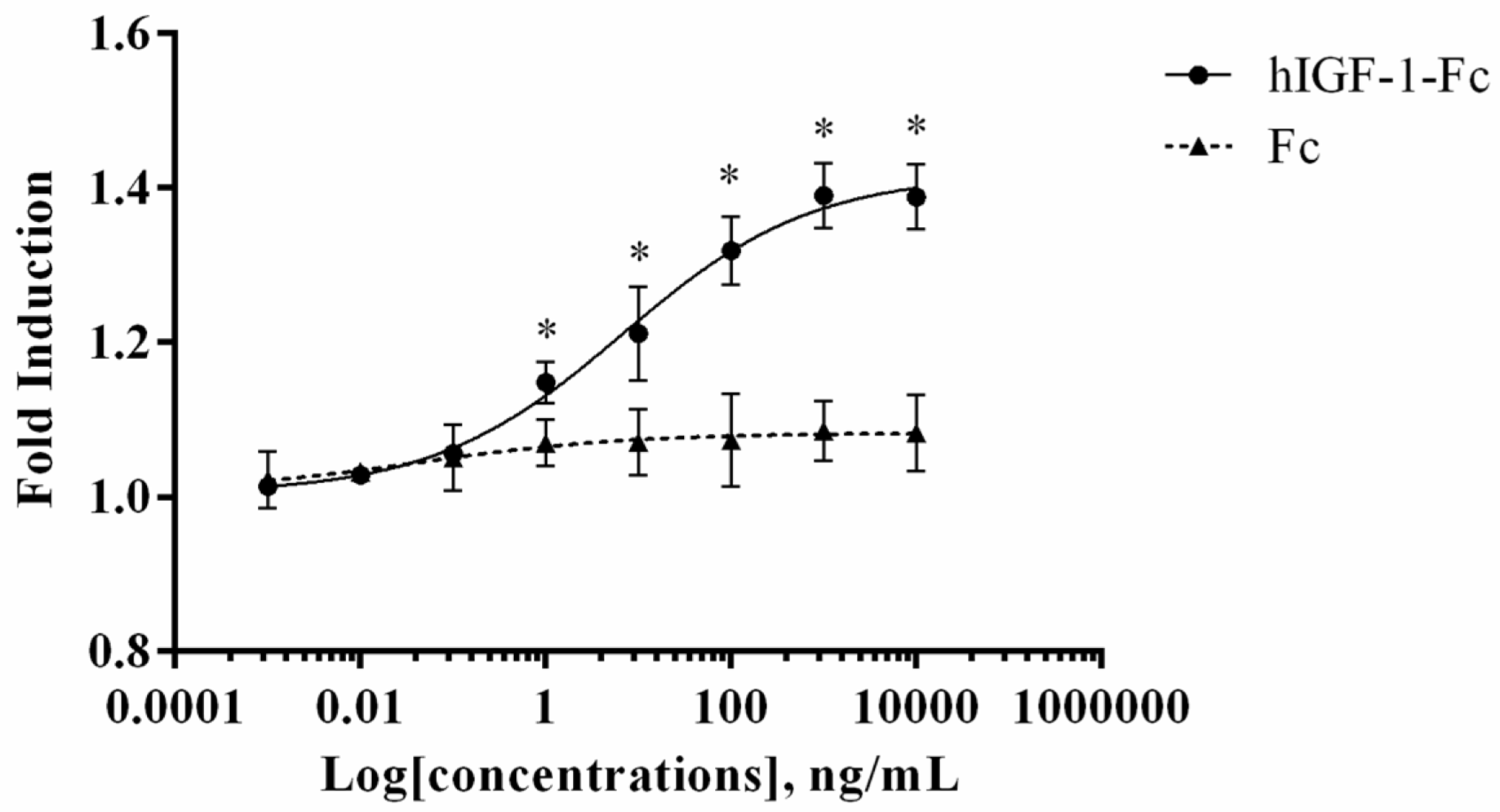
An MTT assay was performed to evaluate the in vitro proliferative activity of plant-produced hIGF-1-Fc in the human breast cancer cell line (MCF-7) and compared to the control Fc fragment. This assay measures the reduction of MTT by metabolically active cells to form insoluble formazan crystals, which are dissolved for absorbance reading to quantify cell proliferation. As shown in Figure 5, treatment with plant-produced hIGF-1-Fc significantly enhanced MCF-7 cell proliferation in a dose-dependent manner (p < 0.05) (Supplementary Tables S3 and S5). A notable increase in fold induction was observed at concentrations ranging from 0.001 ng/mL to 10,000 ng/mL. Dose–response analysis yielded an EC50 value of approximately 6.765 ng/mL for the plant-derived hIGF-1 fusion protein. These results demonstrated the potency of hIGF-1-Fc in stimulating breast cancer cell proliferation.
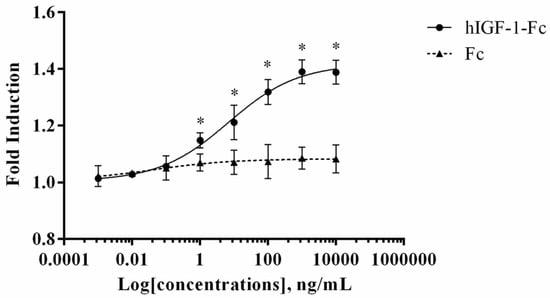
Figure 5.
Dose–response curve of plant-produced hIGF-1-Fc fusion protein in a cell proliferation assay. Increased proliferation of MCF-7 cells was observed with hIGF-1-Fc treatment as compared to the Fc fragment control. Experiments were performed in triplicate, and data are expressed as mean ± SD. * p < 0.05 indicates statistical significance.
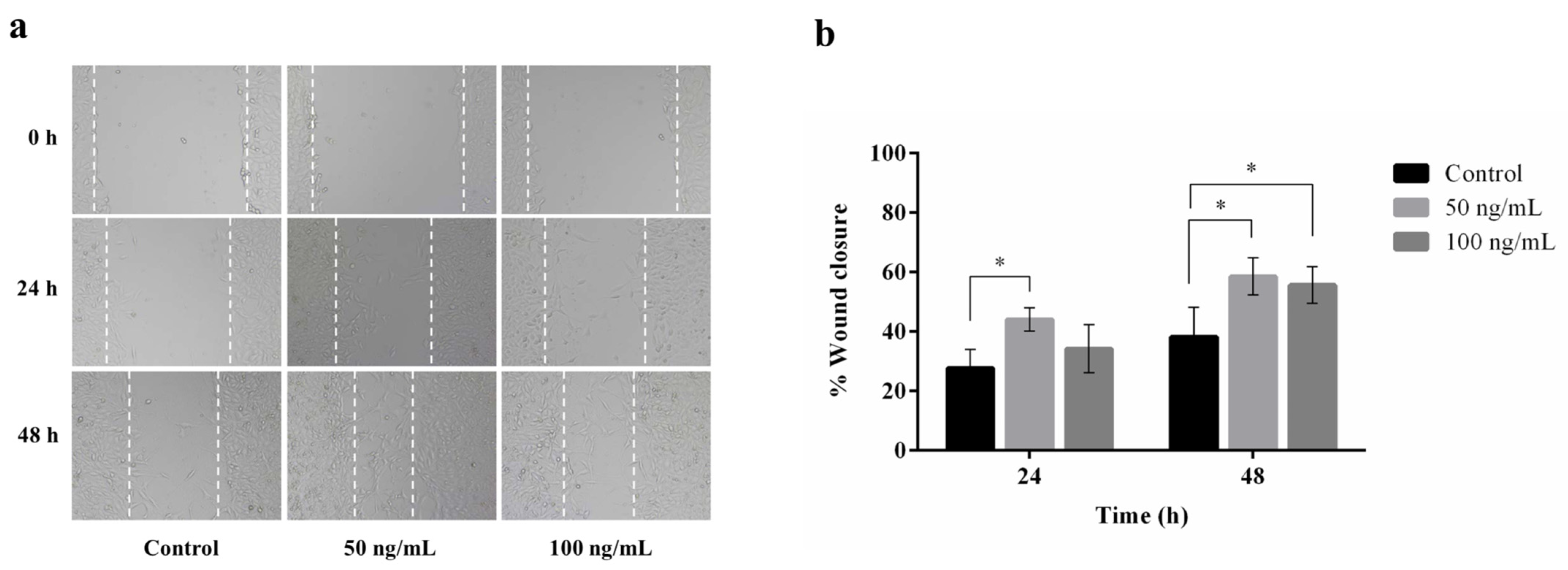
A wound healing assay was carried out to assess the migratory activity of plant-produced hIGF-1-Fc on mouse embryonic fibroblasts (NIH3T3). This assay stimulates in vitro wound healing closure by creating a linear scratch on a confluent monolayer of cells and monitoring cell migration into the scratched area over time. Treatment with plant-produced hIGF-1-Fc at a concentration of 50 ng/mL significantly promoted NIH3T3 cell migration (Figure 6a), with the maximum cell migration distance observed at 48 h (Figure 6b). Statistical analysis showed a significant improvement in wound closure for the hIGF-1-Fc-treated groups (50 and 100 ng/mL) compared to the untreated control group (0 ng/mL) (p < 0.05) (Supplementary Tables S4 and S6). Interestingly, the 50 ng/mL concentration resulted in a higher number of migrating fibroblasts (44.06% wound closure) at 24 h compared to 100 ng/mL (34.25% wound closure). By 48 h, there was no significant difference in cell migration between the 50 ng/mL and 100 ng/mL treatment groups (58.58% vs. 55.63% wound closure).
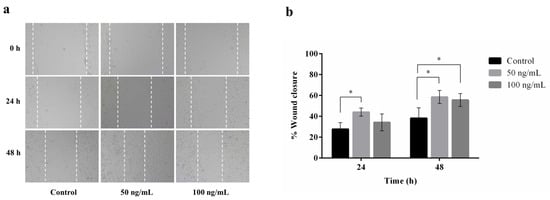
Figure 6.
Effect of plant-produced hIGF-1-Fc on NIH3T3 cell migration. Wound healing assays were conducted with hIGF-1-Fc at concentrations of 50 ng/mL and 100 ng/mL. (a) Representative images of cell migration at 0 h, 24 h, and 48 h. (b) Quantitative analysis of wound closure at different time points. Data are presented as mean ± SD (n = 3). * p < 0.05 indicates statistical significance.
3.5. Proteomics Clustering Analysis
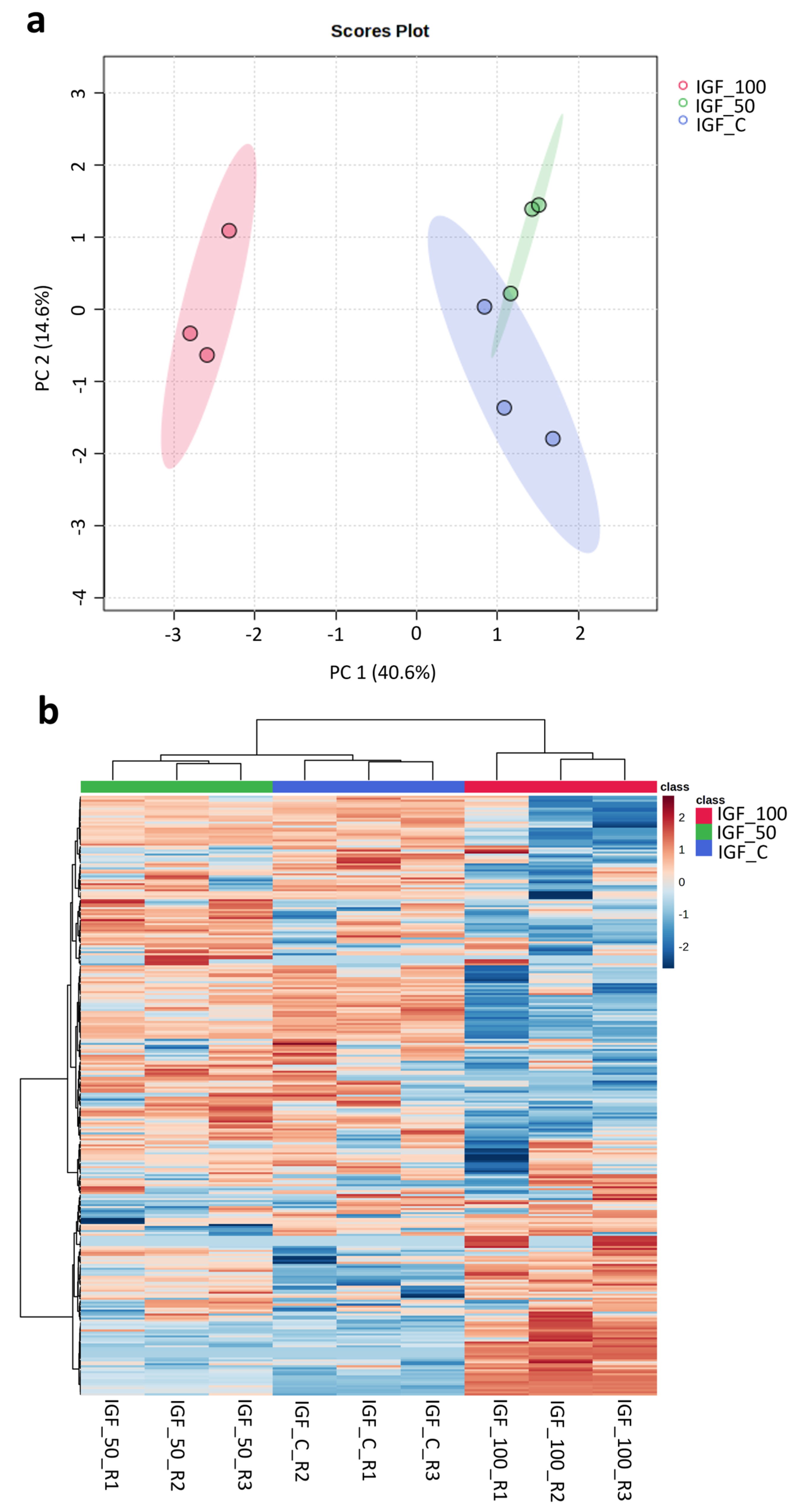
Using mouse NIH3T3 fibroblasts as a model, cells treated with plant-based hIGF-1-Fc at 50 and 100 ng/mL showed differences in cellular protein change from the control. In the principal component analysis (PCA) plot (Figure 7a), hIGF-1-Fc (50 ng/mL)-treated fibroblasts (in green) were slightly separated from the control group (in blue), mainly in the PC2 axis. On the contrary, cells treated with hIGF-1-Fc at 100 ng/mL (in red) exhibited a more pronounced shift from the control along the PC1 direction. The heatmap results (Figure 7b) were well correlated with the PCA data and demonstrated trivial changes between the proteomes of the control and 50 ng/mL hIGF-1-Fc-treated group. However, fibroblasts treated with 100 ng/mL of hIGF-1-Fc displayed obvious proteomic changes, forming a distinct cluster compared to the control group. These results indicate that plant-derived hIGF-1-Fc affected the cellular fibroblast proteome in a dose-dependent manner.
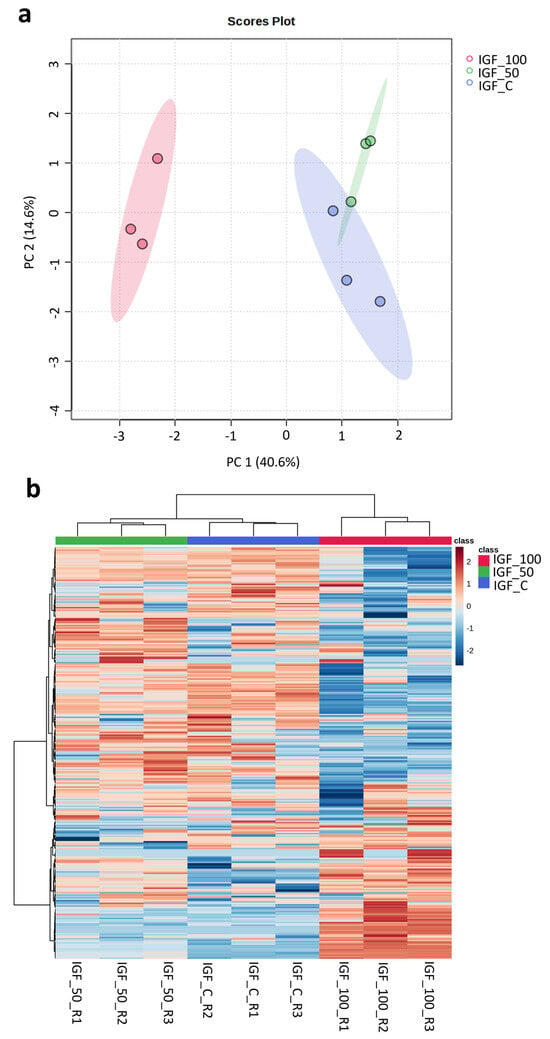
Figure 7.
Proteomics analysis displaying the PCA plot and heatmap of proteomes across control and plant-produced hIGF-1-Fc treatments at concentrations of 50 ng/mL and 100 ng/mL. Colors represent treatment groups: red, IGF-100; green, IGF-50; blue, IGF-C. Three biological replicates were analyzed per treatment. (a) In the PCA plot, control and IGF_50 samples were clustered close together, but three replicates of IGF_100 were clearly separated apart. The colored ellipses around each sample group represent a 95% confidence interval. (b) In the heatmap, control and IGF_50 groups were also clustered together, and IGF_100 group was separated apart. Color gradient indicates protein expression intensity, with red representing high expression and blue representing low expression.
3.6. Proteomics Statistical Analysis
Proteome change was determined using one-way ANOVA across three groups. Table 1 shows the significant proteins identified by ANOVA, followed by Tukey’s post hoc analysis. Among 407 protein groups detected, 91 proteins were significantly different proteins across the three groups, and 315 proteins were not significantly different (Supplementary Table S7). Among the 91 significant proteins, a number of proteins with increased intensity in the plant-produced hIGF-1-Fc (100 ng/mL) group were cytoskeleton proteins, responsible for cell scaffolding and structural support. They were, for example, actin, tubulin, profilin, cofilin, actinin, moesin, and elongation factors. Additionally, the levels of proteins related to the energy production and glycolysis pathway were also increased in the 100 ng/mL hIGF-1-Fc treatment. Those were, for example, phosphoglycerate mutase 1, malate dehydrogenase, hydroxyacyl-coenzyme A dehydrogenase, and isocitrate dehydrogenase. In contrast, the levels of several large ribosomal subunit proteins, such as large ribosomal subunit protein uL2, uL4, uL5, uL22, eL8, eL13, eL18, eL28, and eL31, were detected with a lower level in the plant-based hIGF-1-Fc group. Interestingly, the levels of histone proteins seemed to decrease in the hIGF-1-Fc treatment.

Table 1.
List of significantly different proteins in comparisons between the two treatment groups (hIGF-1 50 and 100 ng/mL) and the untreated control, analyzed by one-way ANOVA with 0.05 FDR. Three biological samples were analyzed. The symbol “–” indicates the absence of a protein class.
3.7. GO Function Enrichment and KEGG Pathway Analysis
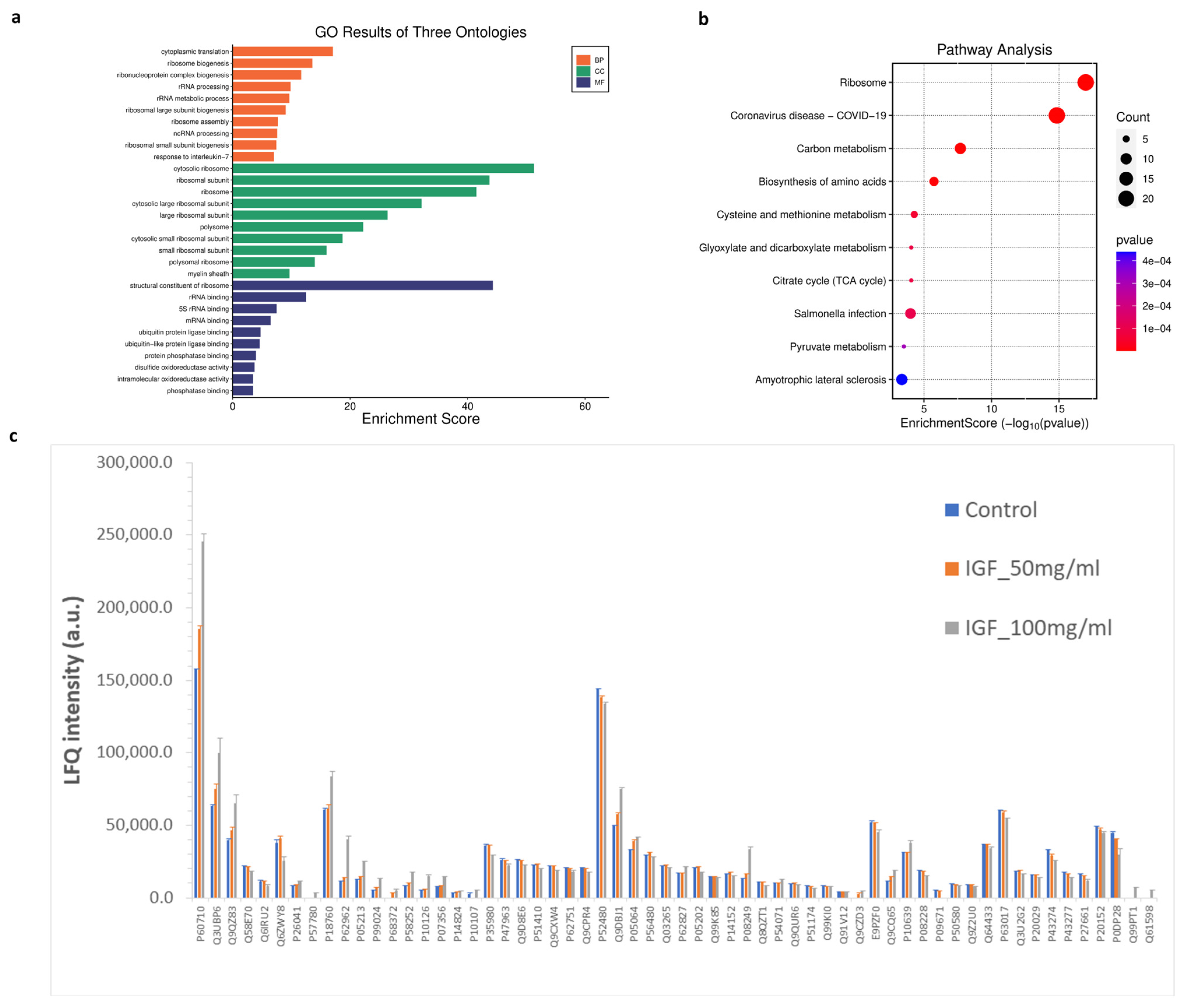
To explore biological pathways correlated to significant proteins, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were conducted. The GO analysis identified enrichment in 1257 biological processes, 238 cellular components, and 160 molecular functions. Additionally, KEGG pathway analysis revealed 150 significantly enriched pathways. The most prominent GO terms and KEGG pathways are illustrated in Figure 8. In biological processes (BP), cytoplasmic translation and ribosome biogenesis were the most affected cellular processes by hIGF-1-Fc treatment (Figure 8a). In the molecular functions (MF) and cell complex (CC) enrichment, proteins associated with ribosome, rRNA binding, mRNA binding, ubiquitin protein ligase, and phosphatase binding were enriched. In KEGG analysis, biological pathways related to ribosome, COVID-19, carbon metabolism, biosynthesis of amino acids, cysteine and methionine metabolism, glyoxylate and dicarboxylate metabolism, citrate cycle (TCA cycle), and pyruvate metabolism were pathways related to the proteins changed upon hIGF-1-Fc treatment (Figure 8b). The bar graph in Figure 8c demonstrates the levels of significant proteins across the three groups in a separate protein class. The results imply that ribosomal proteins, ribosomal biogenesis pathways, and carbon metabolism are highly affected by plant-produced hIGF-1-Fc treatment and could be mechanisms correlated to hIGF-1-promoted cell proliferation and would healing.
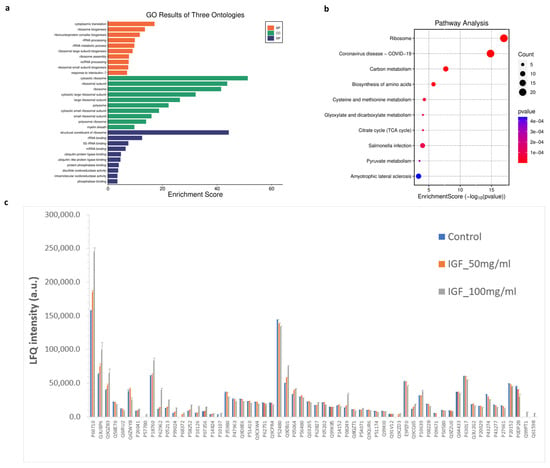
Figure 8.
GO and KEGG enrichment analysis of significantly detected proteins in comparisons between the two treatment groups (plant-produced hIGF-1-Fc: 50 and 100 ng/mL) and the untreated control. (a) Illustrations of GO-enriched biological processes, cellular components, molecular functions, and (b) KEGG enrichment pathway analyses. The color of KEGG terms indicates the p-value, and dot size indicates the number of enriched proteins. (c) Bar graph presenting average LFQ intensity of significant proteins in separated protein class according to gene ontology.
4. Discussion
For several decades, in vitro studies have demonstrated that the hIGF system is a key regulator of cell differentiation, proliferation, and survival [27] hIGF-I, also known as somatomedin C, shares structural similarities with insulin and exhibits significant anabolic effects in humans [28]. Clinically, hIGF-1 has shown therapeutic potential in managing type 1 and 2 diabetes, short stature syndrome, and wound healing [6,29]. Beyond treatment applications, growth factors like hIGF-1 are essential in the cultivated meat industry for promoting mammalian cell growth in bioreactors, ultimately yielding edible meats [2]. In the cosmetics industry, they are incorporated into skincare products to promote skin rejuvenation, collagen production, and improve overall skin health [30,31]. The demand for growth factors across biotechnology, pharmaceutical, and cosmetics sectors continues to grow, yet high production costs limit their accessibility. This study addresses these limitations by using plant-based expression systems as cost-effective bioreactors to produce recombinant hIGF-1 and with the efficacy validated in mammalian cell line models.
Herein, we developed a recombinant hIGF-1 protein conjugated with an Fc fragment at the C-terminus. The fusion protein construct included a flexible (GGGGS)3 Gly-Ser repeat linker between hIGF-1 and Fc [32]. The hIGF-1-Fc was produced in N. benthamiana using a plant geminiviral vector. In particular, we adapted the pBYR2e vector in this study, which has been engineered for high-level protein expression via rolling circle replication [15]. SDS-PAGE under non-reduced conditions showed a distinct band at approximately 70 kDa, matching the predicted mass of the hIGF-1-Fc dimer based on the canonical protein sequence. This result indicates successful expression and correct assembly of the plant-derived Fc fusion protein [16]. After reduction, a monomeric form appeared at around 35 kDa, consistent with disulfide bond cleavage. In addition to the main hIGF-1-Fc bands, we observed multiple bands by Western blot analysis that may indicate the presence of different proteoforms or partially reduced dimers. For example, the ~70 kDa band under reducing conditions may result from incomplete disulfide bond reduction [33,34], though this was not further investigated. Moreover, the detection of faint bands above or below the expected molecular weight may suggest higher-order multimers or degradation. These phenomena are common for Fc fusion proteins and can result from partial proteolysis or heterogeneity in PTMs [35,36]. One potential contributing factor is the absence of a protease inhibitor cocktail during extraction, which may have led to partial degradation and formation of additional proteoforms [37,38]. Although downstream purification improved the purity and removed many non-specific bands (Figure 3), a minor ~35 kDa band remained (Supplementary Figure S3), consistent with monomeric hIGF-1-Fc. Altogether, our findings demonstrate that the plant-based system can effectively produce recombinant proteins in their native multimeric state. However, the presence of putative proteoforms and partially reduced species highlights the need for further optimization. A key limitation of this study is the absence of synthetic controls to confirm antibody specificity. Future studies should employ biochemical characterization of these species and optimize extraction protocols, including the use of protease inhibitors and positive or negative controls to improve product integrity and quality.
The expression level of plant-produced hIGF-1-Fc was optimized by evaluating different dpi for harvest. Similar to earlier studies on transient expression of immunoglobulins and vaccine antigens in tobacco, the optimal harvest period was observed between 3 and 6 dpi [39,40,41,42]. For instance, immune checkpoint antibodies in Nicotiana benthamiana showed a three-fold-higher yield at 4–6 dpi [39,40]. Likewise, expression of the SARS-CoV nucleocapsid protein peaked at 3 dpi with a final yield of 79 µg/g fresh weight [41]. Prior research also explored the impact of signal peptides on expression yields in plants. Panahi et al. demonstrated that including the bacterial Lam B signal sequence slightly reduced the expression level of pro-IGF-1B (30 ng/mg of total protein) compared to constructs without a signal peptide (36–43 ng/mg of total protein) [42]. Furthermore, expression of growth factor FGF without a signal sequence resulted in a five-fold-higher levels than with a signal peptide, suggesting that the effect of signal peptides may vary depending on protein stability across different cellular compartments [11]. Recombinant human IGF-1 has been widely explored in E. coli due to high expression levels, rapid growth, and ease of genetic manipulation. Kim et al. reported the production of IGF-1 fusion protein in E. coli JM109 (at least 30% of the total bacterial protein) in the form of insoluble inclusion bodies. To purify hIGF-1-Fc, we used protein A affinity chromatography and obtained a purity level of >90%. Post-purification yield reached approximately 0.01 µg/g leaf fresh weight, which is lower than the levels reported for pro-IGF-1B in transgenic plants [42]. However, our transient expression system allowed for faster production at 3 dpi compared to their study in stable expression. The findings of this study complement and expand upon those of Musiychuk et al. [43], who demonstrated the production and biological activity of plant-derived hIGF-1 and other erythropoietic growth factors using a TMV-based expression system. While previous work established the feasibility of using plant platforms for cytokine production and confirmed bioactivity through cell proliferation and differentiation assays, the current study provides additional layers of characterization. Specifically, functional activity was assessed in a wound healing model, LC-MS-based glycoprofiling was employed to define Fc glycoform heterogeneity, and label-free quantitative proteomics was performed on NIH3T3 fibroblasts. Furthermore, the Fc-fusion format achieved a simplified one-step purification process. To our knowledge, hIGF-1-Fc fusion has not previously been produced in plants. Although this study successfully expressed hIGF-1-Fc, more efforts are needed to enhance the yield. Strategies could include alternative promoters, optimizing signal sequences, or using different expression vectors [44,45,46].
The MCF-7 breast cancer cell line was employed in this study to evaluate the cell-proliferation-stimulating effects of plant-produced hIGF-1-Fc. Human IGFs are well-known to promote cellular proliferation by driving cell cycle progression and eliciting antiapoptotic effects [47]. Through binding with the hIGF-1 receptor (hIGF-1R), hIGF-1 activates the PI3K and MAPK pathways, both critical for cell division and survival [48,49]. High levels of hIGF-1 have been strongly associated with an increased risk of breast cancer, suggesting its significant role in cancer development and progression [50,51]. In this case, MCF-7 cells provided a relevant in vitro model to test the bioactivity of plant-derived hIGF-1-Fc. After 48 h of treatment, plant-derived hIGF-1-Fc stimulated MCF-7 cell proliferation in a concentration-dependent manner and resulted in an approximate 1.2-fold increase [47]. This confirmed that the hIGF-1 within the fusion construct is biologically active and effectively promotes MCF-7 cell growth. By contrast, cells treated with the plant-derived Fc alone showed no proliferative effect. These data are in line with previous studies demonstrating the mitogenic role of hIGF-1 on MCF-7 cells. The ginsenoside Rg1 promoted MCF-7 cell proliferation by activating the hIGF-1R pathway [52]. Treatment with 1 pM and 1 µM Rg1 led to 1.33- and 1.55-fold increases in cell number, respectively, after 48 h [49]. Likewise, Rajoria et al. (2023) reported that stimulation with 5 nM hIGF-1 significantly increased glycolytic ATP production in MCF-7L cells, suggesting a metabolic shift toward glycolysis to meet the energy demands of proliferation [50]. Together, these studies corroborate the established role of IGF-1 in promoting MCF-7 cell growth and metabolism and further validate the functionality of our plant-produced hIGF-1-Fc.
Moreover, NIH3T3 fibroblast cells were used as a model to mimic natural skin [53,54] and to assess the wound healing potential of plant-produced hIGF-1-Fc. hIGF-1 is a key factor involved in the tissue repair mechanism through the granulation process [55]. The scratch wound assay demonstrated that plant-based hIGF-1-Fc significantly enhanced NIH3T3 cell migration. At 24 h, the wound area was reduced by 44% with the 50 ng/mL hIGF-1-Fc treatment and further progressed to 58% reduction by 48 h. These findings indicate that the recombinant hIGF-1 retains its biological activity to promote healing. Similar studies have shown that other plant-derived growth factors can also induce wound healing [9,10], which is consistent in the current study. On the contrary, non-treated control cells exhibited only 28–38% wound closure. We selected 50 ng/mL and 100 ng/mL IGF-1 concentrations based on the established literature supporting their relevance in fibroblast-based wound healing assays. These doses effectively stimulate key processes such as proliferation and migration without inducing cytotoxic effects [56,57]. The 100 ng/mL concentration is commonly used as a near-maximal dose to capture the full therapeutic potential of IGF-1, while the 50 ng/mL dose provides insight into sub-maximal effects and allows for a basic dose–response comparison [58]. Importantly, these concentrations fall within the physiologically relevant range observed during wound healing and help avoid non-specific or inhibitory effects that may occur at hyper-physiological levels [59].
In this study, proteomics analysis and GO function enrichment and KEGG pathway analysis were also conducted to discern biological mechanisms in correlation with hIGF-1 treatment. With proteomics analysis, cytoskeleton and ribosomal proteins were significantly changed upon 100 ng/mL hIGF-1 treatment. This correlated with the subsequent enrichment analysis, where biological processes of cytoplasmic translation and ribosome biogenesis were enriched. These processes play major roles in protein synthesis, cell proliferation, differentiation, and apoptosis [60]. Additionally, structural constituents of ribosome, rRNA binding, mRNA binding, and ubiquitin protein ligase binding functions were involving molecular functions. These functions play a vital role in regulating various aspects of wound healing through protein modification and degradation [61]. KEGG analysis revealed that the pathways of ribosome, carbon metabolism, biosynthesis of amino acids, citrate cycle (TCA cycle), and pyruvate metabolism were affected by hIGF-1 treatment. These signaling pathways could be key pathways correlated to the promotion of cell proliferation and wound healing processes [62].
Our hIGF-1 fusion protein includes a human Fc fragment containing a conserved N-glycosylation site at position N229. Glycan profiling revealed that approximately 63% of peptides were glycosylated, with GnMXF, GnGnXF, and MMXF as the major plant-type N-glycans attached [63]. Meanwhile, recombinant hIGF-1 produced in E. coli lacks post-translational modifications such as glycosylation, which can influence protein solubility, stability, biological activity, and half-life [29]. Although bacterial expression systems offer speed and high yields, they often require additional refolding steps and lack disulfide bond formation or glycan maturation capabilities [64,65]. In contrast, plant-based systems support disulfide bond formation and N-glycosylation, which allows for production of structurally and functional intact proteins [65]. These host-specific differences may contribute to distinct proteomic responses observed in mammalian cells treated with plant-derived hIGF-1-Fc. While the absence of a commercial hIGF-1 control is a limitation, the inherent functional and structural differences between expression systems justify the biological relevance of our findings.
Relevant proteomics studies using E. coli-derived IGF-1 have demonstrated growth-factor-specific modulation of cellular processes. For example, King et al. [13] reported that IGF-1 stimulation of murine C2C12 myoblasts increased the presence of cytoskeletal proteins such as cofilin and Rho-GDI, along with metabolic enzymes including enolase and Hsp70. Similarly, our proteomic analysis in NIH3T3 fibroblasts treated with plant-derived hIGF-1-Fc revealed increased abundance of actin, tubulin, profilin, cofilin, and elongation factors, as well as enzymes involved in glycolysis and energy production (e.g., phosphoglycerate mutase 1, isocitrate dehydrogenase). These results support a conserved role of IGF-1 in promoting cytoskeletal remodeling and metabolic activity. In the study by Nagano et al. [12], IGF-1 treatment of Swiss 3T3 fibroblasts modulated PI3K/AKT signaling and selectively enhanced levels of ribosomal proteins, translation factors, and actomyosin components. Our data similarly indicate that hIGF-1-Fc treatment reduced the production of large ribosomal subunit proteins (e.g., uL2, uL4, eL8), while pathway enrichment analyses highlighted cytoplasmic translation, ribosome biogenesis, and carbon metabolism as significantly affected processes. Together, these findings demonstrate that IGF-1, regardless of production source, consistently alters cytoskeletal and translational pathways across different cell types. The distinct glycosylation and Fc-fusion features of our plant-derived hIGF-1 may contribute to the specific proteomic signature and various proteoforms, as observed in our study. In addition, further bioinformatics analysis using tools such as KEGG, GO, DAVID, and STRING should be further implemented to supply shotgun proteomics analysis, digging deeply into the biological functions and inclusively discerning the proteome changes according to the hIGF-1-Fc protein treatment. The bioinformatics analysis will also assist the outcome of proteoform formation with hIGF-1-Fc treatment.
5. Conclusions
A transient expression system in N. benthamiana successfully produced a biologically active hIGF-1-Fc fusion protein. This plant-based production platform offers a cost-effective alternative to conventional fermentation methods. The plant-derived hIGF-1-Fc effectively promotes differentiation of breast cancer cells in vitro and enhances wound healing in fibroblasts. Proteomic analysis revealed dose-dependent changes in fibroblast protein expression, significantly affecting cytoskeletal organization, energy metabolism, and ribosomal pathways. At 100 ng/mL, hIGF-1-Fc increased cytoskeletal proteins (actin, tubulin, profilin, cofilin) and metabolic enzymes (phosphoglycerate mutase 1, malate dehydrogenase), indicating enhanced structural remodeling and energy metabolism. Conversely, reduced ribosomal proteins and histones suggest a shift in translational regulation. GO and KEGG pathway analyses further highlighted the enrichment of ribosome biogenesis, carbon metabolism, and amino acid biosynthesis, which may contribute to hIGF-1-induced cell proliferation and wound healing. These findings demonstrate the potential of plant-derived hIGF-1-Fc in modulating key cellular pathways and support its broader application in cell culture and regenerative medicine. Future studies should include direct comparisons with non-plant-derived hIGF-1 to assess efficacy and structural differences, comprehensive proteoform analysis, and mechanistic studies. This research provides a foundation for advancing proteomic insights into plant-based biotherapeutics.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/proteomes13040059/s1, Figure S1: DNA Sequencing chromatograms for hIGF-1-Fc using reverse (78R) and forward (80F) primers; Figure S2: Raw gel bot of day post-infiltration (dpi) optimization of hIGF-1-FC under non-reducing condition for Figure 2a; Figure S3: Raw gel bot for Figure 3b. Lanes II represent reducing condition. Red arrow indicate the predicted molecular weights of the hIGF-1-Fc monomer; Table S1: The band intensity data on hIGF-1-Fc expression; Table S2: Summary of N-glycosylation data; Table S3: Fold induction of hIGF-1-Fc-treated MCF-7 cells; Table S4: The percentage of wound closure on hIGF-1-Fc-treated NIH3T3 cells; Table S5: Statistical comparison of the proliferation of hIGF-1-Fc-treated MCF-7 cells; Table S6: Statistical comparison of the percentage of wound closure of hIGF-1-Fc-treated NIH3T3 cells; Table S7: Raw proteomics data. This manuscript includes Supplementary Material available for reference. Raw proteomics data can be accessed from JPOST repository, identifier ID: JPST003969.
Author Contributions
Conceptualization, W.P. and C.J.I.B.; funding acquisition, W.P.; methodology, K.K., U.N., V.B., I.S., S.Y.N., P.S. and K.R.; formal analysis, U.N., V.B., I.S., P.S. and C.J.I.B.; writing—original draft preparation, C.J.I.B.; writing—review and editing, K.K., S.Y.N. and C.J.I.B. All authors have read and agreed to the published version of the manuscript.
Funding
This project is funded by National Research Council of Thailand (NRCT), Chulalongkorn University (No. N42A670577), and The Second Century Fund (C2F) of Chulalongkorn University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Acknowledgments
We thank Jittima Luckanagul from the Faculty of Pharmaceutical Sciences, Chulalongkorn University, Thailand, for providing the MCF-7 and NIH3T3 cell lines.
Conflicts of Interest
Waranyoo Phoolcharoen is a co-founder and shareholder of Baiya Phytopharm Co., Ltd. San Yoon Nwe, Pipob Suwanchaikasem, Kaewta Rattanapisit, and Christine Joy I. Bulaon are employed by Baiya Phytopharm Co., Ltd. Utapin Ngaokrajang is employed by BGF Plantrix Co., Ltd., a subsidiary of Baiya Phytopharm Co., Ltd. The remaining authors declare no conflicts of interest.
References
- Rischer, H.; Szilvay, G.R.; Oksman-Caldentey, K.-M. Cellular agriculture—Industrial biotechnology for food and materials. Curr. Opin. Biotechnol. 2020, 61, 128–134. [Google Scholar] [CrossRef]
- Ahmad, S.S.; Chun, H.J.; Ahmad, K.; Shaikh, S.; Lim, J.H.; Ali, S.; Han, S.S.; Hur, S.J.; Sohn, J.H.; Lee, E.J.; et al. The roles of growth factors and hormones in the regulation of muscle satellite cells for cultured meat production. J. Anim. Sci. Technol. 2023, 65, 16–31. [Google Scholar] [CrossRef]
- Tripathi, N.K.; Shrivastava, A. Recent Developments in Bioprocessing of Recombinant Proteins: Expression Hosts and Process Development. Front. Bioeng. Biotechnol. 2019, 7, 420. [Google Scholar] [CrossRef]
- Venkatesan, M.; Semper, C.; Skrivergaard, S.; Di Leo, R.; Mesa, N.; Rasmussen, M.K.; Young, J.F.; Therkildsen, M.; Stogios, P.J.; Savchenko, A. Recombinant production of growth factors for application in cell culture. iScience 2022, 25, 105054. [Google Scholar] [CrossRef] [PubMed]
- Bailes, J.; Soloviev, M. Insulin-Like Growth Factor-1 (IGF-1) and Its Monitoring in Medical Diagnostic and in Sports. Biomolecules 2021, 11, 217. [Google Scholar] [CrossRef]
- Laron, Z. Insulin-like growth factor 1 (IGF-1): A growth hormone. Mol. Pathol. 2001, 54, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hu, F.; Li, J.; Chen, L.; Mao, Y.-F.; Li, Q.-B.; Nie, C.-Y.; Lin, C.; Xiao, J. IGF-1 inhibits inflammation and accelerates angiogenesis via Ras/PI3K/IKK/NF-κB signaling pathways to promote wound healing. Eur. J. Pharm. Sci. 2024, 200, 106847. [Google Scholar] [CrossRef] [PubMed]
- Shanmugaraj, B.; Bulaon, C.J.I.; Phoolcharoen, W. Plant Molecular Farming: A Viable Platform for Recombinant Biopharmaceutical Production. Plants 2020, 9, 842. [Google Scholar] [CrossRef]
- Bulaon, C.J.I.; Shanmugaraj, B.; Oo, Y.; Rattanapisit, K.; Chuanasa, T.; Chaotham, C.; Phoolcharoen, W. Rapid transient expression of functional human vascular endothelial growth factor in Nicotiana benthamiana and characterization of its biological activity. Biotechnol. Rep. 2020, 27, e00514. [Google Scholar] [CrossRef]
- Hanittinan, O.; Oo, Y.; Chaotham, C.; Rattanapisit, K.; Shanmugaraj, B.; Phoolcharoen, W. Expression optimization, purification and in vitro characterization of human epidermal growth factor produced in Nicotiana benthamiana. Biotechnol. Rep. 2020, 28, e00524. [Google Scholar] [CrossRef]
- Rattanapisit, K.; Jantimaporn, A.; Kaewpungsup, P.; Shanmugaraj, B.; Pavasant, P.; Namdee, K.; Phoolcharoen, W. Plant-Produced Basic Fibroblast Growth Factor (bFGF) Promotes Cell Proliferation and Collagen Production. Planta Medica Int. Open 2020, 7, e150–e157. [Google Scholar] [CrossRef]
- Nagano, K.; Akpan, A.; Warnasuriya, G.; Corless, S.; Totty, N.; Yang, A.; Stein, R.; Zvelebil, M.; Stensballe, A.; Burlingame, A.; et al. Functional Proteomic Analysis of Long-term Growth Factor Stimulation and Receptor Tyrosine Kinase Coactivation in Swiss 3T3 Fibroblasts. Mol. Cell. Proteom. 2012, 11, 1690–1708. [Google Scholar] [CrossRef][Green Version]
- King, C.C.; Bouic, K.; Friedmann, T. A fractionation method to identify qauntitative changes in protein expression mediated by IGF-1 on the proteome of murine C2C12 myoblasts. Proteome Sci. 2009, 7, 28. [Google Scholar] [CrossRef]
- Chen, Q.; Davis, K.R. The potential of plants as a system for the development and production of human biologics. F1000Research 2016, 5, 912. [Google Scholar] [CrossRef]
- Chen, Q.; He, J.; Phoolcharoen, W.; Mason, H.S. Geminiviral vectors based on bean yellow dwarf virus for production of vaccine antigens and monoclonal antibodies in plants. Hum. Vaccines 2011, 7, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Srisangsung, T.; Phetphoung, T.; Manopwisedjaroen, S.; Rattanapisit, K.; Bulaon, C.J.I.; Thitithanyanont, A.; Limprasutr, V.; Strasser, R.; Phoolcharoen, W. The impact of N-glycans on the immune response of plant-produced SARS-CoV-2 RBD-Fc proteins. Biotechnol. Rep. 2024, 43, e00847. [Google Scholar] [CrossRef] [PubMed]
- Charnsatabut, C.; Suwanchaikasem, P.; Rattanapisit, K.; Iksen, I.; Pongrakhananon, V.; Bulaon, C.J.I.; Phoolcharoen, W. Optimized expression of human interleukin-15 in Nicotiana benthamiana and in vitro assessment of its activity on human keratinocytes. Biotechnol. Rep. 2025, 46, e00889. [Google Scholar] [CrossRef] [PubMed]
- Benington, L.; Mo, J.; Li, M.; Rajan, G.; Locher, C.; Lim, L.Y. In Vitro Assessment of Wound-Healing Efficacy of Stabilized Basic Fibroblast Growth Factor (FGF-2) Solutions. Pharmaceuticals 2024, 17, 247. [Google Scholar] [CrossRef]
- Carrera, M.; Mateos Martín, J. Shotgun Proteomics: Methods and Protocols; Humana: New York, NY, USA, 2020. [Google Scholar]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Röst, H.L.; Sachsenberg, T.; Aiche, S.; Bielow, C.; Weisser, H.; Aicheler, F.; Andreotti, S.; Ehrlich, H.-C.; Gutenbrunner, P.; Kenar, E.; et al. OpenMS: A flexible open-source software platform for mass spectrometry data analysis. Nat. Methods 2016, 13, 741–748. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef]
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Spigelman, A.F.; MacDonald, P.E.; Wishart, D.S.; Li, S.; et al. MetaboAnalyst 6.0: Towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef] [PubMed]
- Salmon, W.D., Jr.; Daughaday, W.H. A hormonally controlled serum factor which stimulates sulfate incorporation by cartilage in vitro. J. Lab. Clin. Med. 1957, 49, 825–836. [Google Scholar] [PubMed]
- Rinderknecht, E.; Humbel, R.E. The amino acid sequence of human insulin-like growth factor I and its structural homology with proinsulin. J. Biol. Chem. 1978, 253, 2769–2776. [Google Scholar] [CrossRef] [PubMed]
- Iranpoor, H.; Omidinia, E.; Vatankhah, V.; Gharanjik, V.; Shahbazi, M. Expression of Recombinant Human Insulin-like Growth Factor Type 1 (rhIGF-1) in Escherichia coli. Avicenna J. Med. Biotechnol. 2015, 7, 101–105. [Google Scholar]
- Miller-Kobisher, B.; Suárez-Vega, D.V.; Velazco de Maldonado, G.J. Epidermal Growth Factor in Aesthetics and Regenerative Medicine: Systematic Review. J. Cutan. Aesthet. Surg. 2021, 14, 137–146. [Google Scholar]
- Quinlan, D.J.; Ghanem, A.M.; Hassan, H. Topical growth factor preparations for facial skin rejuvenation: A systematic review. J. Cosmet. Dermatol. 2023, 22, 2023–2039. [Google Scholar] [CrossRef]
- Lin, J.; Asai, S.; Selicharová, I.; Mitrová, K.; Kaminský, J.; Young, E.; Jiráček, J. Recombinant Insulin-Like Growth Factor 1 Dimers: Receptor Binding Affinities and Activation Abilities. Int. J. Pept. Res. Ther. 2023, 29, 33. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.-Y. A Two-Stage Mechanism for the Reductive Unfolding of Disulfide-containing Proteins. J. Biol. Chem. 1997, 272, 69–75. [Google Scholar] [CrossRef]
- Roszkowski, M.; Mansuy, I.M. High Efficiency RNA Extraction From Sperm Cells Using Guanidinium Thiocyanate Supplemented With Tris(2-Carboxyethyl)Phosphine. Front. Cell Dev. Biol. 2021, 9, 648274. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, F.; Xu, W.; May, K.; Richardson, D.; Liu, H. Disulfide bond assignment of an IgG1 monoclonal antibody by LC-MS with post-column partial reduction. Anal. Biochem. 2013, 436, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Natesan, R.; Dykstra, A.B.; Banerjee, A.; Agrawal, N.J. Heterogeneity in Disulfide Bond Reduction in IgG1 Antibodies Is Governed by Solvent Accessibility of the Cysteines. Antibodies 2023, 12, 83. [Google Scholar] [CrossRef]
- Clemente, M.; Corigliano, M.G.; Pariani, S.A.; Sánchez-López, E.F.; Sander, V.A.; Ramos-Duarte, V.A. Plant Serine Protease Inhibitors: Biotechnology Application in Agriculture and Molecular Farming. Int. J. Mol. Sci. 2019, 20, 1345. [Google Scholar] [CrossRef]
- Ma, J.; Ding, X.; Li, Z.; Wang, S. Co-expression With Replicating Vector Overcoming Competitive Effects Derived by a Companion Protease Inhibitor in Plants. Front. Plant Sci. 2021, 12, 699442. [Google Scholar] [CrossRef]
- Phakham, T.; Bulaon, C.J.I.; Khorattanakulchai, N.; Shanmugaraj, B.; Buranapraditkun, S.; Boonkrai, C.; Sooksai, S.; Hirankarn, N.; Abe, Y.; Strasser, R.; et al. Functional Characterization of Pembrolizumab Produced in Nicotiana benthamiana Using a Rapid Transient Expression System. Front. Plant Sci. 2021, 12, 736299. [Google Scholar] [CrossRef]
- Rattanapisit, K.; Bulaon, C.J.I.; Strasser, R.; Sun, H.; Phoolcharoen, W. In vitro and in vivo studies of plant-produced Atezolizumab as a potential immunotherapeutic antibody. Sci. Rep. 2023, 13, 14146. [Google Scholar] [CrossRef]
- Zheng, N.; Xia, R.; Yang, C.; Yin, B.; Li, Y.; Duan, C.; Liang, L.; Guo, H.; Xie, Q. Boosted expression of the SARS-CoV nucleocapsid protein in tobacco and its immunogenicity in mice. Vaccine 2009, 27, 5001–5007. [Google Scholar] [CrossRef]
- Panahi, M.; Cheng, X.; Alli, Z.; Sardana, R.; Callaghan, M.; Phipps, J.; Altosaar, I. Plant-derived recombinant human insulin-like growth factor precursor prohormone IGF-1B caused differentiation of human neuroblastoma cell lines SH-SY5Y. Mol. Breed. 2003, 12, 21–31. [Google Scholar] [CrossRef]
- Musiychuk, K.; Sivalenka, R.; Jaje, J.; Bi, H.; Flores, R.; Shaw, B.; Jones, R.M.; Golovina, T.; Schnipper, J.; Khandker, L.; et al. Plant-produced human recombinant erythropoietic growth factors support erythroid differentiation in vitro. Stem Cells Dev. 2013, 22, 2326–2340. [Google Scholar] [CrossRef]
- Villao-Uzho, L.; Chávez-Navarrete, T.; Pacheco-Coello, R.; Sánchez-Timm, E.; Santos-Ordóñez, E. Plant Promoters: Their Identification, Characterization, and Role in Gene Regulation. Genes 2023, 14, 1226. [Google Scholar] [CrossRef]
- Rozov, S.M.; Deineko, E.V. Increasing the Efficiency of the Accumulation of Recombinant Proteins in Plant Cells: The Role of Transport Signal Peptides. Plants 2022, 11, 2561. [Google Scholar] [CrossRef]
- Nosaki, S.; Hoshikawa, K.; Ezura, H.; Miura, K. Transient protein expression systems in plants and their applications. Plant Biotechnol. (Tokyo) 2021, 38, 297–304. [Google Scholar] [CrossRef]
- Dupont, J.; Le Roith, D. Insulin-like growth factor 1 and oestradiol promote cell proliferation of MCF-7 breast cancer cells: New insights into their synergistic effects. Mol. Pathol. 2001, 54, 149–154. [Google Scholar] [CrossRef]
- Vanhaesebroeck, B.; Alessi, D.R. The PI3K-PDK1 connection: More than just a road to PKB. Biochem. J. 2000, 346 Pt 3, 561–576. [Google Scholar] [CrossRef] [PubMed]
- Skolnik, E.; Lee, C.; Batzer, A.; Vicentini, L.; Zhou, M.; Daly, R.; Myers, M.; Backer, J.; Ullrich, A.; White, M. The SH2/SH3 domain-containing protein GRB2 interacts with tyrosine-phosphorylated IRS1 and Shc: Implications for insulin control of ras signalling. EMBO J. 1993, 12, 1929–1936. [Google Scholar] [CrossRef] [PubMed]
- Yee, D. The insulin-like growth factor system as a target in breast cancer. Breast Cancer Res. Treat. 1994, 32, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Christopoulos, P.F.; Msaouel, P.; Koutsilieris, M. The role of the insulin-like growth factor-1 system in breast cancer. Mol. Cancer 2015, 14, 43. [Google Scholar] [CrossRef]
- Chen, W.F.; Lau, W.S.; Cheung, P.Y.; Guo, D.A.; Wong, M.S. Activation of Insulin-Like Growth Factor I Receptor-Mediated Pathway by Ginsenoside Rg1. Br. J. Pharmacol. 2006, 147, 542–551. [Google Scholar] [CrossRef]
- Han, H.; Hong, H.; Park, S.M.; Kim, D. Metal–Electrolyte Solution Dual-Mode Electrospinning Process for In Situ Fabrication of Electrospun Bilayer Membrane. Adv. Mater. Interfaces 2020, 7, 2000571. [Google Scholar] [CrossRef]
- Li, X.-J.; Huang, F.-Z.; Wan, Y.; Li, Y.-S.; Zhang, W.K.; Xi, Y.; Tian, G.-H.; Tang, H.-B. Lipopolysaccharide Stimulated the Migration of NIH3T3 Cells Through a Positive Feedback Between β-Catenin and COX-2. Front. Pharmacol. 2018, 9, 1487. [Google Scholar] [CrossRef] [PubMed]
- Achar, R.A.; Silva, T.C.; Achar, E.; Martines, R.B.; Machado, J.L. Use of insulin-like growth factor in the healing of open wounds in diabetic and non-diabetic rats. Acta Cir. Bras. 2014, 29, 125–131. [Google Scholar] [CrossRef]
- Garoufalia, Z.; Papadopetraki, A.; Karatza, E.; Vardakostas, D.; Philippou, A.; Kouraklis, G.; Mantas, D. Insulin-like growth factor-I and wound healing, a potential answer to non-healing wounds: A systematic review of the literature and future perspectives. Biomed. Rep. 2021, 15, 66. [Google Scholar] [CrossRef]
- Yang, L.; Li, X.; Zhang, S.; Song, J.; Zhu, T. Baicalein inhibits proliferation and collagen synthesis of mice fibroblast cell line NIH/3T3 by regulation of miR-9/insulin-like growth factor-1 axis. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3202–3211. [Google Scholar] [CrossRef]
- Huang, Y.-L.; Qiu, R.-F.; Mai, W.-Y.; Kuang, J.; Cai, X.-Y.; Dong, Y.-G.; Hu, Y.-Z.; Song, Y.-B.; Cai, A.-P.; Jiang, Z.-G. Effects of insulin-like growth factor-1 on the properties of mesenchymal stem cells in vitro. J. Zhejiang Univ. Sci. B 2012, 13, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.J.; Lu, M.C.; Chang, H.Y. Sustained Release of Insulin-Like Growth Factor-1 from Bombyx mori L. Silk Fibroin Delivery for Diabetic Wound Therapy. Int. J. Mol. Sci. 2021, 22, 6267. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Liu, Y.; Yu, X.-Y.; Pan, X.; Zhang, Y.; Tu, J.; Song, Y.-H.; Li, Y. Ribosome biogenesis in disease: New players and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 15. [Google Scholar] [CrossRef]
- Zhang, Q.; Gu, R.; Dai, Y.; Chen, J.; Ye, P.; Zhu, H.; He, W.; Nie, X. Molecular mechanisms of ubiquitination in wound healing. Biochem. Pharmacol. 2025, 231, 116670. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, F.; Xu, C.; Zhang, Q.; Ren, H.; Huang, X.; He, C.; Ma, J.; Wang, Z. Metabolic reprogramming in skin wound healing. Burn. Trauma 2024, 12, tkad047. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R. Recent Developments in Deciphering the Biological Role of Plant Complex N-Glycans. Front. Plant Sci. 2022, 13, 897549. [Google Scholar] [CrossRef]
- Ranjbari, J.; Babaeipour, V.; Vahidi, H.; Moghimi, H.; Mofid, M.R.; Namvaran, M.M.; Jafari, S. Enhanced Production of Insulin-like Growth Factor I Protein in Escherichia coli by Optimization of Five Key Factors. Iran. J. Pharm. Res. 2015, 14, 907–917. [Google Scholar] [PubMed]
- Rozov, S.M.; Permyakova, N.V.; Deineko, E.V. Main Strategies of Plant Expression System Glycoengineering for Producing Humanized Recombinant Pharmaceutical Proteins. Biochemistry 2018, 83, 215–232. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).