Abstract
Artificial small RNAs (art-sRNAs), such as artificial microRNAs (amiRNAs) and synthetic trans-acting small interfering RNAs (syn-tasiRNAs), are highly specific 21-nucleotide small RNAs designed to recognize and silence complementary target RNAs. Art-sRNAs are extensively used in gene function studies or for improving crops, particularly to protect plants against viruses. Typically, antiviral art-sRNAs are computationally designed to target one or multiple sites in viral RNAs with high specificity, and art-sRNA constructs are generated and introduced into plants that are subsequently challenged with the target virus(es). Numerous studies have reported the successful application of art-sRNAs to induce resistance against a large number of RNA and DNA viruses in model and crop species. However, the application of art-sRNAs as an antiviral tool has limitations, such as the difficulty to predict the efficacy of a particular art-sRNA or the emergence of virus variants with mutated target sites escaping to art-sRNA-mediated degradation. Here, we review the different classes, features, and uses of art-sRNA-based tools to induce antiviral resistance in plants. We also provide strategies for the rational design of antiviral art-sRNAs and discuss the latest advances in developing art-sRNA-based methodologies for enhanced resistance to plant viruses.
Keywords:
RNA silencing; artificial small RNA; amiRNA; atasiRNA; syn-tasiRNA; antiviral resistance; VSR; plant virus; viroid 1. Introduction
RNA interference (RNAi) is a biological process conserved in most eukaryotes and characterized by the sequence-specific degradation of target RNA by complementary small RNAs (sRNAs) [1]. RNAi pathways are triggered by double-stranded RNA (dsRNA) processed into sRNA duplexes by Dicer ribonucleases [1,2]. One of the strands of the duplex is preferentially loaded into an ARGONAUTE (AGO) protein, and the resulting complex, termed RNA-induced silencing complex (RISC), recognizes and silences complementary target RNA through diverse mechanisms [3,4]. The seminal observation that exogenously applied dsRNA can artificially trigger RNAi of specific genes in Caenorabtitis elegans [1] fueled the development of a plethora of RNAi-based tools in multiple organisms for the study of gene function and for more applied purposes, including medical therapies and diverse biotechnological uses.
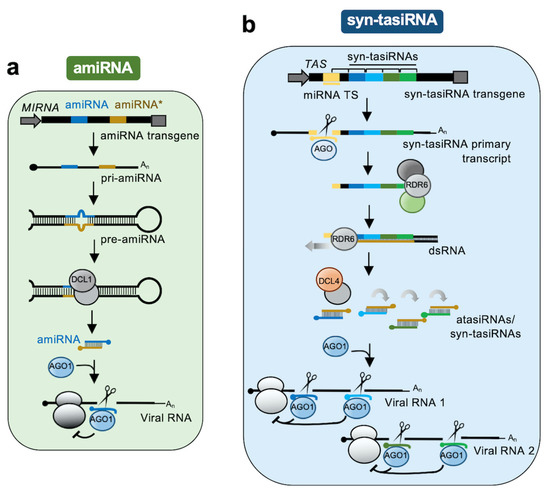
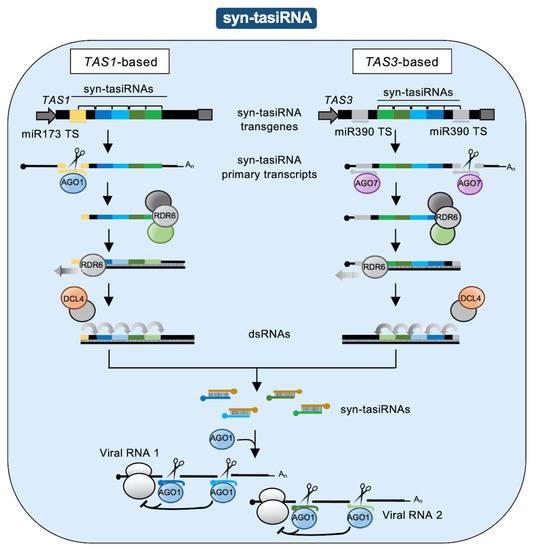
In plants, RNAi tools have been extensively used to confer antiviral resistance. Early RNAi approaches, such as virus-induced gene silencing (VIGS) and hairpin (hp)-based silencing, consisted of the expression of dsRNA or hpRNA precursors, respectively, bearing sequences of the target virus (for a recent review see [5]). Although very popular, these approaches lacked high specificity as the large populations of sRNAs produced from these type of precursors favor the accidental targeting of complementary cellular transcripts [6]. This limitation was overcome with the development of a series of tools based on artificial sRNAs (art-sRNAs) [7], 21-nucleotide sRNAs expressed in planta from endogenous sRNA precursors and computationally designed to silence target RNAs with high specificity [8].
Here, we describe the different classes, features, and uses of art-sRNA-based RNAi (art-sRNAi) tools to target viral RNAs and induce antiviral resistance in plants. We also provide basic rules for art-sRNA design and describe which viral regions are typically targeted. Finally, we discuss the latest art-sRNAi strategies providing enhanced antiviral resistance, such as the high-throughput identification of highly effective antiviral art-sRNAs and their simultaneous co-expression for the multi-targeting of viral RNAs.
5. Application of Art-sRNAs to Control Viral Diseases in Crops
Transgenes producing antiviral art-sRNAs from endogenous sRNA precursors have been introduced in diverse crop species to generate antiviral resistance (Table 1). Regarding amiRNAs, transgenic tomato plants expressing amiRNAs against CMV [28], ToLCNDV [38], or TSWV [14] were resistant, as were barley, maize, rice, and wheat transgenic plants expressing amiRNAs against WDV [40], RBSDV [41], RBSDV/RSV [44], and WSMV [45], respectively. Regarding syn-tasiRNAs, the recent development of highly resistant transgenic tomato plants expressing four different syn-tasiRNAs against TSWV is the only example reported [14]. However, the antiviral resistance of these transgenic crops expressing art-sRNAs has not been examined under field conditions. Indeed, the confirmation that the durability of the antiviral resistance in the field lasts for multiple generations seems necessary before the approval of a transgenic crop for its commercial release. To date, only a few transgenic crops expressing transgenes including short stretches of viral sequences in sense or antisense orientation have been commercially released (for a recent review see [62]). Unfortunately, the current legislations and long paths for the commercialization of a transgenic crop, particularly in those countries where GMO crops are highly regulated, are still barriers to overcome before the release of the first art-sRNA-based crop onto the market.
An alternative to the transgenic expression of antiviral RNAs is their topical delivery into plants. This approach was first used to interfere with Alfalfa mosaic virus (AMV), TEV, or Pepper mild mottle virus (PMMoV) infection by mechanically co-inoculating N. tabacum leaves with naked dsRNAs of virus sequence and their corresponding target virus [63]. Later, the same strategy was also applied successfully to interfere with the infection of several viroids [64]. Since these early works, dsRNAs of viral sequence have been delivered to plants though diverse methods to induce resistance against a large number of plant viruses in multiple model and crop species (reviewed recently in [65,66,67,68]). However, several limitations of these approaches may include the lack of affordable methods for dsRNA production, and the low specificity, potential toxicity, and reduced efficiency of certain dsRNAs. To date, the exogenous application of art-sRNAs to plants has not been reported. In principle, art-sRNA precursors topically delivered into plants will be processed by the endogenous RNAi machinery to produce the antiviral art-sRNAs. Certainly, some of the limitations of the dsRNA approach, such as the lack of efficient production and delivery methods, may also compromise the successful exogenous application of art-sRNA precursors into plants. In this context, several bacterial systems for the efficient production of recombinant RNAs [69,70,71,72] may be used to produce large amounts of art-sRNA precursors in a time- and cost-effective manner. In addition, a possibility to increase the stability and efficient delivery of art-sRNA precursors could be their conjugation to cationic nanoparticles, clay nanosheets, surfactants, or peptide-based RNA delivery systems, as described for other RNAs [66]. For example, sprayed dsRNAs bound to layered double hydroxide (LDH) nanosheets have been successfully used to confer resistance to PMMoV and CMV in N. tabacum [73], in a non-toxic and sustainable manner, and extend the durability of the protection described in previous studies [63]. Thus, the exogenous application of art-sRNA precursors conjugated to new generation nanoparticles may represent a novel, highly efficient, and sustainable strategy to induce antiviral resistance in crops in a GMO-free manner.
6. Concluding Remarks and Future Perspectives
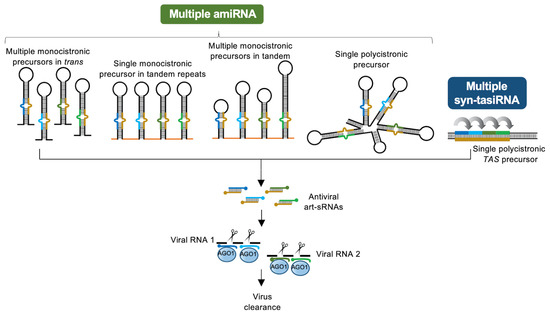
Art-sRNAi tools have been broadly used in plants to confer antiviral resistance against multiple RNA and DNA viruses, and to viroids as well. Currently, the relative simplicity of the webtool-assisted design of highly specific antiviral art-sRNA, combined with the availability of efficient cloning methods, facilitates the design and generation of antiviral art-sRNA constructs for plant delivery. However, one important drawback in the use of art-sRNAi is the difficulty to predict the effectiveness of a particular art-sRNA. Recently described high-throughput systems for rapid in vitro or in vivo screening of the antiviral activity of virus-derived sRNAs or computationally designed art-sRNAs, respectively, seem to have overcome this limitation. Another drawback is the emergence of resistance-breaking virus variants with mutated target sites when using single amiRNAs targeting single sites in viral RNAs. In this case, the artificial multiplexing of amiRNAs in different precursor configurations or the use of syn-tasiRNA precursors, both allowing the co-expression of multiple art-sRNAs, should circumvent this problem. The synchronized targeting of multiple viral RNAs by co-expressed art-sRNAs may minimize the possibility that the virus simultaneously mutates all different target sites to fully escape each art-sRNA, and thus enhance the antiviral resistance.
In the current genome editing era of bacterial CRISPR/Cas-based technologies, we anticipate that art-sRNAi tools will continue to be broadly used to confer antiviral resistance in plants because of their unique features of high simplicity, specificity, and efficacy, as well as for their multiplexing capability and for the availability of high-throughput methodologies for the design, generation, and validation of art-sRNAi constructs. The development of efficient methodologies for the production and topical delivery to plants of art-sRNA precursors, as well as a better knowledge of the basic mechanisms governing art-sRNA biogenesis, mode of action, and viral targeting, are needed to further refine art-sRNAi tools in view of their broader use for enhanced crop protection.
Author Contributions
A.E.C. assisted selecting the bibliography and critically reviewed and edited the manuscript. A.C. selected the bibliography and conceptualized and wrote the article. All authors have read and approved the final manuscript.
Funding
This research was funded by grants RYC-2017-21648 and RTI2018-095118-A-100 from the Ministerio de Ciencia, Innovación y Universidades (MCIU, Spain), Agencia Estatal de Investigación (AEI, Spain), and Fondo Europeo de Desarrollo Regional (FEDER, European Union) to AC.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AB | asymmetric bulge |
| art-sRNA | artificial small RNA |
| art-sRNAi | art-sRNA-based RNA interference |
| AGO | argonaute |
| amiRNA | artificial microRNA |
| atasiRNA | artificial trans-acting small interfering RNA |
| CP | coat protein |
| DCL | Dicer-like |
| dsRNA | double-stranded RNA |
| esiRNA | immunologically effective small interfering RNA |
| hp | hairpin |
| MIRNA | microRNA |
| pri-amiRNA | primary artificial microRNA precursor |
| pri-syn-tasiRNA | primary synthetic trans-acting small interfering RNA precursor |
| RdRp | RNA-dependent RNA polymerase |
| RNAi | RNA interference |
| sRNA | small RNA |
| syn-tasiRNA | synthetic trans-acting small interfering RNA |
| VSR | viral silencing suppressor protein |
References
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Hammond, S.M.; Boettcher, S.; Caudy, A.A.; Kobayashi, R.; Hannon, G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 2001, 293, 1146–1150. [Google Scholar] [CrossRef]
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000, 404, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Gaffar, F.Y.; Koch, A. Catch Me If You Can! RNA Silencing-Based Improvement of Antiviral Plant Immunity. Viruses 2019, 11, 673. [Google Scholar] [CrossRef] [PubMed]
- Senthil-Kumar, M.; Mysore, K.S. Caveat of RNAi in Plants: The Off-Target Effect. In RNAi and Plant Gene Function Analysis: Methods and Protocols; Kodama, H., Komamine, A., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2011; pp. 13–25. ISBN 978-1-61779-123-9. [Google Scholar]
- López-Dolz, L.; Spada, M.; Daròs, J.-A.; Carbonell, A. Fine-Tune Control of Targeted RNAi Efficacy by Plant Artificial Small RNAs. Nucleic Acids Res. 2020, in press. [Google Scholar] [CrossRef]
- Carbonell, A. Artificial small RNA-based strategies for effective and specific gene silencing in plants. In Plant Gene Silencing: Mechanisms and Applications; Dalmay, T., Ed.; CABI Publishing: Wallingford, Oxfordshire, UK, 2017; pp. 110–127. ISBN 978-1-78064-767-8. [Google Scholar]
- Ossowski, S.; Schwab, R.; Weigel, D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. For. Cell Mol. Biol. 2008, 53, 674–690. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.W.; Lin, S.S.; Reyes, J.L.; Chen, K.C.; Wu, H.W.; Yeh, S.D.; Chua, N.H. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 2006, 24, 1420–1428. [Google Scholar] [CrossRef]
- Simon-Mateo, C.; Garcia, J.A. MicroRNA-guided processing impairs Plum pox virus replication, but the virus readily evolves to escape this silencing mechanism. J. Virol. 2006, 80, 2429–2436. [Google Scholar] [CrossRef]
- Lin, S.S.; Wu, H.W.; Elena, S.F.; Chen, K.C.; Niu, Q.W.; Yeh, S.D.; Chen, C.C.; Chua, N.H. Molecular evolution of a viral non-coding sequence under the selective pressure of amiRNA-mediated silencing. PLoS Pathog. 2009, 5, e1000312. [Google Scholar] [CrossRef]
- Lafforgue, G.; Martinez, F.; Sardanyes, J.; de la Iglesia, F.; Niu, Q.W.; Lin, S.S.; Sole, R.V.; Chua, N.H.; Daros, J.A.; Elena, S.F. Tempo and mode of plant RNA virus escape from RNA interference-mediated resistance. J. Virol. 2011, 85, 9686–9695. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Lisón, P.; Daròs, J.-A. Multi-targeting of viral RNAs with synthetic trans-acting small interfering RNAs enhances plant antiviral resistance. Plant J. 2019, 100, 720–737. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.J. Artificial trans-acting small interfering RNA: A tool for plant biology study and crop improvements. Planta 2014, 239, 1139–1146. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cheng, X.; Cai, J.; Zhan, L.; Wu, X.; Liu, Q.; Wu, X. Multiple virus resistance using artificial trans-acting siRNAs. J. Virol. Methods 2016, 228, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Daròs, J.-A. Artificial microRNAs and synthetic trans-acting small interfering RNAs interfere with viroid infection. Mol. Plant Pathol. 2017, 18, 746–753. [Google Scholar] [CrossRef]
- Carbonell, A.; Lopez, C.; Daròs, J.-A. Fast-Forward Identification of Highly Effective Artificial Small RNAs Against Different Tomato spotted wilt virus Isolates. Mol. Plant Microbe Interact. 2019, 32, 142–156. [Google Scholar] [CrossRef]
- Carbonell, A. Secondary Small Interfering RNA-Based Silencing Tools in Plants: An Update. Front. Plant Sci. 2019, 10, 687. [Google Scholar] [CrossRef]
- Felippes, F.F.; Wang, J.W.; Weigel, D. MIGS: miRNA-induced gene silencing. Plant J. Cell Mol. Biol. 2012, 70, 541–547. [Google Scholar] [CrossRef]
- Zhao, M.; San Leon, D.; Mesel, F.; Garcia, J.A.; Simon-Mateo, C. Assorted Processing of Synthetic Trans-Acting siRNAs and Its Activity in Antiviral Resistance. PLoS ONE 2015, 10, e0132281. [Google Scholar] [CrossRef]
- Singh, A.; Taneja, J.; Dasgupta, I.; Mukherjee, S.K. Development of plants resistant to tomato geminiviruses using artificial trans-acting small interfering RNA. Mol. Plant Pathol. 2015, 16, 724–734. [Google Scholar] [CrossRef]
- Singh, A.; Mohorianu, I.; Green, D.; Dalmay, T.; Dasgupta, I.; Mukherjee, S.K. Artificially induced phased siRNAs promote virus resistance in transgenic plants. Virology 2019, 537, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Fahlgren, N.; Hill, S.T.; Carrington, J.C.; Carbonell, A. P-SAMS: A web site for plant artificial microRNA and synthetic trans-acting small interfering RNA design. Bioinformatics 2016, 32, 157–158. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Hao, J.; Li, J.; Baker, B.; Luo, L. Artificial microRNA-mediated resistance to cucumber green mottle mosaic virus in Nicotiana benthamiana. Planta 2019, 250, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Wagaba, H.; Patil, B.L.; Mukasa, S.; Alicai, T.; Fauquet, C.M.; Taylor, N.J. Artificial microRNA-derived resistance to Cassava brown streak disease. J. Virol. Methods 2016, 231, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.G.; Wang, C.H.; Fang, R.X.; Guo, H.S. Artificial MicroRNAs highly accessible to targets confer efficient virus resistance in plants. J. Virol. 2008, 82, 11084–11095. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, H.; Zhang, J.; Zhang, C.; Gong, P.; Ziaf, K.; Xiao, F.; Ye, Z. Expression of artificial microRNAs in tomato confers efficient and stable virus resistance in a cell-autonomous manner. Transgenic Res. 2011, 20, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Ai, T.; Zhang, L.; Gao, Z.; Zhu, C.X.; Guo, X. Highly efficient virus resistance mediated by artificial microRNAs that target the suppressor of PVX and PVY in plants. Plant Biol. 2011, 13, 304–316. [Google Scholar] [CrossRef]
- Mitter, N.; Zhai, Y.; Bai, A.X.; Chua, K.; Eid, S.; Constantin, M.; Mitchell, R.; Pappu, H.R. Evaluation and identification of candidate genes for artificial microRNA-mediated resistance to tomato spotted wilt virus. Virus Res. 2016, 211, 151–158. [Google Scholar] [CrossRef]
- Lafforgue, G.; Martinez, F.; Niu, Q.W.; Chua, N.H.; Daros, J.A.; Elena, S.F. Improving the effectiveness of artificial microRNA (amiR)-mediated resistance against Turnip mosaic virus by combining two amiRs or by targeting highly conserved viral genomic regions. J. Virol. 2013, 87, 8254–8256. [Google Scholar] [CrossRef]
- Kung, Y.J.; Lin, S.S.; Huang, Y.L.; Chen, T.C.; Harish, S.S.; Chua, N.H.; Yeh, S.D. Multiple artificial microRNAs targeting conserved motifs of the replicase gene confer robust transgenic resistance to negative-sense single-stranded RNA plant virus. Mol. Plant Pathol. 2012, 13, 303–317. [Google Scholar] [CrossRef]
- Ali, I.; Amin, I.; Briddon, R.W.; Mansoor, S. Artificial microRNA-mediated resistance against the monopartite begomovirus Cotton leaf curl Burewala virus. Virol. J. 2013, 10, 231. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Ye, J.; Fang, R. Artificial microRNA-mediated virus resistance in plants. J. Virol. 2007, 81, 6690–6699. [Google Scholar] [CrossRef] [PubMed]
- Jelly, N.S.; Schellenbaum, P.; Walter, B.; Maillot, P. Transient expression of artificial microRNAs targeting Grapevine fanleaf virus and evidence for RNA silencing in grapevine somatic embryos. Transgenic Res. 2012, 21, 1319–1327. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.Z.; Han, Q.J.; Jiang, F.; Sun, R.Z.; Fan, Z.H.; Zhu, C.X.; Wen, F.J. Effects of the sequence characteristics of miRNAs on multi-viral resistance mediated by single amiRNAs in transgenic tobacco. Plant Physiol. Biochem. 2014, 77, 90–98. [Google Scholar] [CrossRef]
- Yu, R.; Chen, C.; Cao, W.; Liu, H.; Zhou, S.; Song, Y.; Zhu, C. High-degree and broad-spectrum resistance mediated by a combination of NIb siRNA and miRNA suppresses replication of necrotic and common strains of potato virus Y. Arch. Virol. 2018, 163, 3073–3081. [Google Scholar] [CrossRef]
- Vu, T.V.; Choudhury, N.R.; Mukherjee, S.K. Transgenic tomato plants expressing artificial microRNAs for silencing the pre-coat and coat proteins of a begomovirus, Tomato leaf curl New Delhi virus, show tolerance to virus infection. Virus Res. 2013, 172, 35–45. [Google Scholar] [CrossRef]
- Gago-Zachert, S.; Schuck, J.; Weinholdt, C.; Knoblich, M.; Pantaleo, V.; Grosse, I.; Gursinsky, T.; Behrens, S.-E. Highly efficacious antiviral protection of plants by small interfering RNAs identified in vitro. Nucleic Acids Res. 2019, 47, 9343–9357. [Google Scholar] [CrossRef]
- Kis, A.; Tholt, G.; Ivanics, M.; Varallyay, E.; Jenes, B.; Havelda, Z. Polycistronic artificial miRNA-mediated resistance to Wheat dwarf virus in barley is highly efficient at low temperature. Mol. Plant Pathol. 2016, 17, 427–437. [Google Scholar] [CrossRef]
- Xuan, N.; Zhao, C.; Peng, Z.; Chen, G.; Bian, F.; Lian, M.; Liu, G.; Wang, X.; Bi, Y. Development of transgenic maize with anti-rough dwarf virus artificial miRNA vector and their disease resistance. Chin. J. Biotechnol. 2015, 31, 1375–1386. [Google Scholar]
- Zhang, H.; Feng, H.; Lu, X.; Wang, C.; Yang, W.; Li, F. An asymmetric bulge enhances artificial microRNA-mediated virus resistance. Plant Biotechnol. J. 2019, 18, 608–610. [Google Scholar] [CrossRef]
- Petchthai, U.; Yee, C.S.L.; Wong, S.M. Resistance to CymMV and ORSV in artificial microRNA transgenic Nicotiana benthamiana plants. Sci. Rep. 2018, 8, 9958. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Lin, C.; Du, J.; Song, Y.; Jiang, M.; Liu, H.; Zhou, S.; Wen, F.; Zhu, C. Dimeric artificial microRNAs mediate high resistance to RSV and RBSDV in transgenic rice plants. Plant Cell Tiss Organ. Cult. 2016, 126, 1–13. [Google Scholar] [CrossRef]
- Fahim, M.; Millar, A.A.; Wood, C.C.; Larkin, P.J. Resistance to Wheat streak mosaic virus generated by expression of an artificial polycistronic microRNA in wheat. Plant Biotechnol. J. 2012, 10, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Ravelonandro, M.; Scorza, R.; Briard, P. Innovative RNAi Strategies and Tactics to Tackle Plum Pox Virus (PPV) Genome in Prunus domestica-Plum. Plants 2019, 8, 565. [Google Scholar] [CrossRef]
- Csorba, T.; Kontra, L.; Burgyan, J. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479–480, 85–103. [Google Scholar] [CrossRef]
- Carbonell, A.; Daròs, J.-A. Design, Synthesis, and Functional Analysis of Highly Specific Artificial Small RNAs with Antiviral Activity in Plants. Methods Mol. Biol. 2019, 2028, 231–246. [Google Scholar]
- Carbonell, A.; Takeda, A.; Fahlgren, N.; Johnson, S.C.; Cuperus, J.T.; Carrington, J.C. New generation of artificial MicroRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol. 2014, 165, 15–29. [Google Scholar] [CrossRef]
- Li, J.F.; Zhang, D.; Sheen, J. Epitope-tagged protein-based artificial miRNA screens for optimized gene silencing in plants. Nat. Protoc. 2014, 9, 939–949. [Google Scholar] [CrossRef]
- Li, J.F.; Chung, H.S.; Niu, Y.; Bush, J.; McCormack, M.; Sheen, J. Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 2013, 25, 1507–1522. [Google Scholar] [CrossRef]
- Szittya, G.; Moxon, S.; Pantaleo, V.; Toth, G.; Rusholme Pilcher, R.L.; Moulton, V.; Burgyan, J.; Dalmay, T. Structural and functional analysis of viral siRNAs. PLoS Pathog. 2010, 6, e1000838. [Google Scholar] [CrossRef]
- Pantaleo, V.; Szittya, G.; Burgyan, J. Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J. Virol. 2007, 81, 3797–3806. [Google Scholar] [CrossRef] [PubMed]
- Miozzi, L.; Gambino, G.; Burgyan, J.; Pantaleo, V. Genome-wide identification of viral and host transcripts targeted by viral siRNAs in Vitis vinifera. Mol. Plant Pathol. 2013, 14, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Schuck, J.; Gursinsky, T.; Pantaleo, V.; Burgyan, J.; Behrens, S.E. AGO/RISC-mediated antiviral RNA silencing in a plant in vitro system. Nucleic Acids Res. 2013, 41, 5090–5103. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Carrington, J.C. Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 2015, 27, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Vargason, J.M.; Szittya, G.; Burgyan, J.; Hall, T.M. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 2003, 115, 799–811. [Google Scholar] [CrossRef]
- Cuperus, J.T.; Carbonell, A.; Fahlgren, N.; Garcia-Ruiz, H.; Burke, R.T.; Takeda, A.; Sullivan, C.M.; Gilbert, S.D.; Montgomery, T.A.; Carrington, J.C. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 2010, 17, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Manavella, P.A.; Koenig, D.; Weigel, D. Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. USA 2012, 109, 2461–2466. [Google Scholar] [CrossRef]
- Narjala, A.; Nair, A.; Tirumalai, V.; Hari Sundar, G.V.; Shivaprasad, P.V. A conserved sequence signature is essential for robust plant miRNA biogenesis. Nucleic Acids Res. 2020, 48, 3103–3118. [Google Scholar] [CrossRef]
- Yang, M.; Woolfenden, H.C.; Zhang, Y.; Fang, X.; Liu, Q.; Vigh, M.L.; Cheema, J.; Yang, X.; Norris, M.; Yu, S.; et al. In vivo mRNA structure regulates miRNA cleavage in Arabidopsis | bioRxiv. bioRxiv 2019. [Google Scholar] [CrossRef]
- Khalid, A.; Zhang, Q.; Yasir, M.; Li, F. Small RNA Based Genetic Engineering for Plant Viral Resistance: Application in Crop Protection. Front. Microbiol. 2017, 8, 43. [Google Scholar] [CrossRef]
- Tenllado, F.; Diaz-Ruiz, J.R. Double-stranded RNA-mediated interference with plant virus infection. J. Virol. 2001, 75, 12288–12297. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Martinez de Alba, A.E.; Flores, R.; Gago, S. Double-stranded RNA interferes in a sequence-specific manner with the infection of representative members of the two viroid families. Virology 2008, 371, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Dalakouras, A.; Wassenegger, M.; Dadami, E.; Ganopoulos, I.; Pappas, M.L.; Papadopoulou, K. Genetically Modified Organism-Free RNA Interference: Exogenous Application of RNA Molecules in Plants. Plant Physiol. 2020, 182, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Dubrovina, A.S.; Kiselev, K.V. Exogenous RNAs for Gene Regulation and Plant Resistance. Int. J. Mol. Sci. 2019, 20, 2282. [Google Scholar] [CrossRef]
- Kim, H.; Shimura, H.; Masuta, C. Advancing toward commercial application of RNA silencing-based strategies to protect plants from viral diseases. J. Gen. Plant Pathol. 2019, 85, 321–328. [Google Scholar] [CrossRef]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Xu, Z.P.; Carroll, B.J. Induction of virus resistance by exogenous application of double-stranded RNA. Curr. Opin. Virol. 2017, 26, 49–55. [Google Scholar] [CrossRef]
- Tenllado, F.; Llave, C.; Diaz-Ruiz, J.R. RNA interference as a new biotechnological tool for the control of virus diseases in plants. Virus Res. 2004, 102, 85–96. [Google Scholar] [CrossRef]
- Daros, J.A.; Aragones, V.; Cordero, T. A viroid-derived system to produce large amounts of recombinant RNA in Escherichia coli. Sci. Rep. 2018, 8, 1904. [Google Scholar] [CrossRef]
- Yin, G.; Sun, Z.; Liu, N.; Zhang, L.; Song, Y.; Zhu, C.; Wen, F. Production of double-stranded RNA for interference with TMV infection utilizing a bacterial prokaryotic expression system. Appl. Microbiol. Biotechnol. 2009, 84, 323–333. [Google Scholar] [CrossRef]
- Niehl, A.; Soininen, M.; Poranen, M.M.; Heinlein, M. Synthetic biology approach for plant protection using dsRNA. Plant Biotechnol. J. 2018, 16, 1679–1687. [Google Scholar] [CrossRef]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.J.; Lu, G.Q.; Xu, Z.P. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 2017, 3, 16207. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).