Abstract
Stress-associated proteins (SAPs) are a class of zinc finger proteins that confer tolerance to a variety of abiotic and biotic stresses in diverse plant species. However, in cucumber (Cucumis sativus L.), very little is known about the roles of SAP gene family members in regulating plant growth, development, and stress responses. In this study, a total of 12 SAP genes (named as CsSAP1-CsSAP12) were identified in the cucumber genome, which were unevenly distributed on six chromosomes. Gene duplication analysis detected one tandem duplication and two segmental duplication events. Phylogenetic analysis of SAP proteins from cucumber and other plants suggested that they could be divided into seven groups (sub-families), and proteins in the same group generally had the same arrangement of AN1 (ZnF-AN1) and A20 (ZnF-A20) domains. Most of the CsSAP genes were intronless and harbored a number of stress- and hormone-responsive cis-elements in their promoter regions. Tissue expression analysis showed that the CsSAP genes had a broad spectrum of expression in different tissues, and some of them displayed remarkable alteration in expression during fruit development. RT-qPCR results indicated that all the selected CsSAP genes displayed transcriptional responses to cold, drought, and salt stresses. These results enable the first comprehensive description of the SAP gene family in cucumber and lay a solid foundation for future research on the biological functions of CsSAP genes.
1. Introduction
Various abiotic stresses, such as salt, light intensity, drought, and extreme temperatures (cold and heat), are important causes of great damage to global crops by affecting their growth and reducing their average yield [1,2]. In order to resist and adapt to various environmental stresses, plants have developed complex molecular mechanisms that regulate the transcriptional levels of a number of stress-related genes to further control the signaling networks involved in stress responses. In recent years, stress-associated proteins (SAPs), a kind of zinc finger proteins (ZFPs), have been widely recognized as key molecular factors participating in the mediation of plant development and stress responses [3,4].
In previous studies, SAP gene family composition has been well characterized in different plant species. A high diversity in gene numbers has been reported, which range from 14 members in Arabidopsis thaliana to 57 members in Brassica napus [5,6,7,8,9,10]. Previous reports have revealed that the classical SAP proteins comprise an N-terminal A20 (ZnF-A20) domain and/or a C-terminal AN1 (ZnF-AN1) domain, which are specific zinc finger domains highly conserved across species [1,8,10,11]. The A20 and AN1 domains were shown to take part in immune response, and can interact with each other for protein–protein interactions and are involved in the transcriptional regulation of stress- and pathogenesis-related genes [12,13,14,15]. Additionally, the A20 and AN1 domains of some SAPs were reported to have ubiquitin ligase activity [16,17,18]. Besides the two zinc finger domains, some SAP proteins were found to possess an extra C-terminal Cys2-His2 domain [3,8,19].
There has been increasing evidence showing that many SAP genes are key regulators of abiotic/biotic stress responses. For example, OsiSAP1 (OsSAP1) is the first SAP gene discovered in rice, and overexpression of OsSAP1 in tobacco and rice increased their resistance to a variety of abiotic stresses by regulating the transcriptional levels of endogenous stress-related genes [12,20,21]. Likewise, overexpression of another rice SAP gene (OsiSAP8) in tobacco and rice also resulted in tolerance to multiple stresses such as salt, drought, and cold [22]. Enhanced stress tolerance was also observed in transgenic plants overexpressing different SAP genes, such as AlSAP from Aeluropus littoralis [23,24], LmSAP from the halophyte Lobularia maritima [25,26], AtSAP9 and AtSAP13 from Arabidopsis thaliana [19,27], TaSAP17-D from Triticum aestivum [28], and PtSAP13 from Populus trichocarpa [11]. In addition, OsSAP1-overexpressing transgenic tobacco plants also displayed an increase in disease resistance against virulent bacterial pathogens with the up-regulation of defense-responsive genes [29]. Recent studies carried out in Solanum lycopersicum L. (tomato) also demonstrated the involvement of SlSAPs in plant response upon different biotic stresses [30,31]. Liu et al. [30] showed that SlSAP4 positively contributes to tomato immunity against B. cinerea by modulating the jasmonate and ethylene signaling pathways, and SlSAP3 positively regulates the immunity against Pseudomonas syringae pv. tomato DC3000 through the salicylic acid (SA) signaling pathway. In addition, SAPs were also found to play vital roles in regulating multiple aspects of biological processes, such as cell elongation [32], cell expansion [33], and glandular trichome development [7].
As one of the most widely distributed vegetable crops in the world, cucumber is sensitive to various environmental stimuli, which may severely threaten the production and quality [34]. Although numerous stress-responsive family genes have been found to play crucial roles in different stress responses of cucumber, such as WRKY [35], bZIP [36], CDPK [37], and NAC [38], the roles of SAP gene family members in response to environmental stresses are still unclear. In this work, we carried out the identification of SAP gene family members in cucumber, and systematically analyzed their chromosomal location, gene duplication events, gene structure, evolutionary relationships, promoter regions, and tissue expression profiles. Additionally, the expression profiles of cucumber SAP genes under various stresses (cold, drought, and salt) were investigated by RT-qPCR. Our findings are expected to shed light on the functional roles of SAP genes in cucumber.
2. Results
2.1. Identification and Chromosomal Location of the SAP Family Genes in Cucumber
In total, 12 SAP genes were identified in cucumber genome, and were named as CsSAP1-CsSAP12 based on their physical locations on the chromosomes (Table 1). The gDNA lengths of CsSAP genes ranged from 414 to 3217 bp and the CDS lengths were from 414 to 870 bp. These CsSAP genes encoded proteins with 137–289 amino acids (aa) in length, with MWs from 14.78 (CsSAP7) to 31.81 (CsSAP6) kDa (Table 1). The predicted pIs and GRAVY values of the CsSAP proteins ranged from 7.95 (CsSAP1) to 9.52 (CsSAP4), and –0.793 (CsSAP2) to –0.332 (CsSAP1), respectively, suggesting that all of them are basic and hydrophilic proteins. In addition, the Plant-mPLoc results showed that all 12 CsSAP proteins were predictably located in the nucleus (Table 1).

Table 1.
Identification and characterization of SAP family genes in cucumber.
The 12 CsSAP genes were distributed on six chromosomes across the cucumber genome. Among them, chromosome 3 harbored the largest number of CsSAP genes (four genes), each of chromosomes 4, 6, and 7 harbored two CsSAP genes, and a single CsSAP gene was located in each of the chromosomes 1 and 5 (Figure 1). In addition, gene duplication analysis showed that CsSAP2 and CsSAP3 were tandemly duplicated genes, while two pairs of CsSAP genes, CsSAP1/CsSAP11 and CsSAP3/CsSAP11, were identified as segmentally duplicated genes (Figure 1).
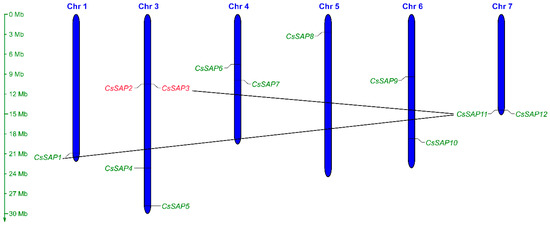
Figure 1.
Distribution of the CsSAP genes among six cucumber chromosomes. The scale on the left represents the length of each chromosome displayed in megabase (Mb). Tandemly duplicated genes are colored with red, and segmentally duplicated genes are linked with black lines.
2.2. Phylogenetic Analysis of SAP Proteins in Cucumber and Different Plant Species
To assess the evolutionary history of the SAP proteins, a phylogenetic tree was created with MEGA 7.0 using the SAP translated protein sequences from cucumber and four other plant species, including A. thaliana [10], S. lycopersicum [1], M. truncatula [3], and M. domestica [8]. As shown in Figure 2, these SAP proteins could be divided into seven groups (Group a–g). There were 21 members in Group a, which was the largest group. These members mainly possessed the AN1 and A20 domains, except for CsSAP7, MtSAP12, and MtSAP13, which only harbored one AN1 domain (Table 1) [3]. In addition, 12, 5, 20, and 12 members fell into groups b, c, d, and e, which also mainly possessed the AN1 and A20 domains. Group f included seven members, which all contained two AN1 domains. There were nine members in group g, which mainly possessed the AN1 and C2H2 domains, and the only exception was AtSAP14, which was lack of the C2H2 domain [10].
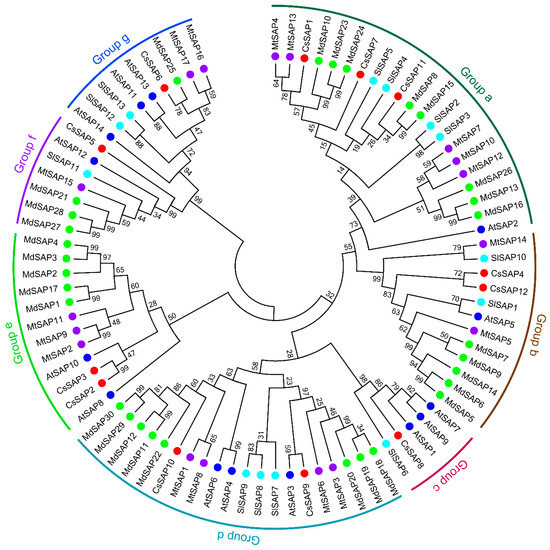
Figure 2.
Phylogenetic analysis of stress-associated proteins (SAP) proteins from cucumber and four other plants. Sequence alignment was performed with full-length SAP amino acid sequences from different plant species using MAFFT with the default parameters, and the neighbor-joining (NJ) phylogenetic tree was created by MEGA 7.0 software using a bootstrap option of 1000 replications. Cs, Cucumis sativus; At, Arabidopsis thaliana; Sl, Solanum lycopersicum; Mt, Medicago truncatula; Md, Malus domestica. The accession numbers of SAPs for phylogenetic analysis are listed in Table S1.
2.3. Conserved Motifs of SAP Proteins in Cucumber
The conserved domains of twelve putative CsSAP proteins were analyzed by Pfam and SMART servers. The results indicated that nine CsSAP proteins possessed both an A20 and an AN1 domain, while other three CsSAP proteins (CsSAP5, CsSAP6, and CsSAP7) contained AN1 domains but no A20 domains (Figure 3A). CsSAP7 contained only a single AN1 domain, while CsSAP5 and CsSAP6 possessed two AN1 domains. Besides two AN1 domains, CsSAP6 harbored two C2H2 domains, which was also observed in some SAP proteins from other plant species, such as SlSAP12 [1], MdSAP25 [8], GhSAP11A and GhSAP11D [9].
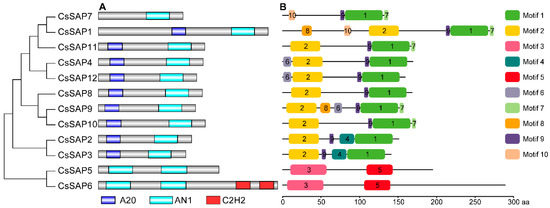
Figure 3.
Conserved motif arrangement in cucumber SAP proteins according to the phylogenetic relationship. (A) Analysis of conserved domains of CsSAP proteins. (B) Conserved motif distributions of the CsSAP proteins annotated with the MEME server. Ten motifs are marked by different colors and their amino acid sequences are shown in Table S2.
The MEME tool was employed to analyze the conserved motifs in the CsSAP proteins, and the motif distributions of these CsSAP proteins were obtained (Figure 3B; Table S2). Motif 2 made up the A20 domain, while motif 1, motif 3, and motif 5 were independently annotated as the AN1 domain (Table S2). Almost all CsSAP proteins had motif 9, except for CsSAP5 and CsSAP6. Motifs 4, 6, 7, 8, and 10 were present in 2, 3, 5, 2, and 2 CsSAP proteins, respectively (Figure 3B). Some evolutionarily well conserved CsSAP proteins harbored unique motifs. For instance, motif 4 was present in CsSAP2 and CsSAP3, while motif 10 was present in CsSAP1 and CsSAP7 (Figure 3B). In addition, besides motif 3 and motif 5, CsSAP5 and CsSAP6 had no additional motifs (Figure 3B).
2.4. Gene Structure of the CsSAP Genes
To investigate the structures of the CsSAP genes, the number and position of introns were predicted by comparing the CDS sequence and respective genomic DNA sequence using the GSDS tool (Figure 4). As a result, a majority of CsSAP genes (9 out of 12) were found to be intronless, two CsSAP genes (CsSAP5 and CsSAP6) harbored one single intron, while CsSAP1 contained three introns (Figure 4).
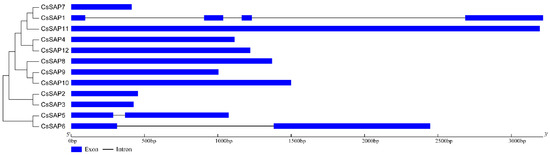
Figure 4.
Compositions of introns and exons of CsSAP genes based on the phylogenetic relationship.
2.5. Promoter Region Analysis of the CsSAP Genes
To study the potential roles of CsSAP genes, a search for cis-elements located at the promoter region of CsSAP genes and responsive to stress and hormones was carried out at the PlantCARE database. A total of seven and nine types of cis-elements associated with stress and hormone responses were identified, respectively (Figure 5). Among the stress-responsive cis-elements, ARE was the most abundant cis-element, with 10 out of the 12 CsSAP genes containing at least one ARE element. Specifically, the promoters of five CsSAP genes contained three or four ARE elements (Figure 5), indicating that they play possible roles in the anaerobic induction process. Additionally, nine kinds of hormone-responsive cis-elements were observed in CsSAP promoters, which were related to different hormones including ABA (ABRE), ethylene (ERE), MeJA (CGTCA-motif), salicylic acid (TCA-element), auxin (AuxRR-core and TGA-element), and gibberellin (P-box, GARE-motif and TATC-box) (Figure 5), suggesting that the CsSAP genes are associated with different hormone responses. In total, 22, 18, and 14 cis-elements were associated with ethylene, ABA, and MeJA responses, respectively, accounting for large proportions of the hormone-responsive cis-elements (Figure 5). Notably, seven TGA-elements, six ABREs, and five EREs were distributed in the promoter regions of CsSAP11, CsSAP8, and CsSAP5, respectively (Figure 5). These findings suggest that the CsSAP genes may play diverse roles in stress and hormone responses.
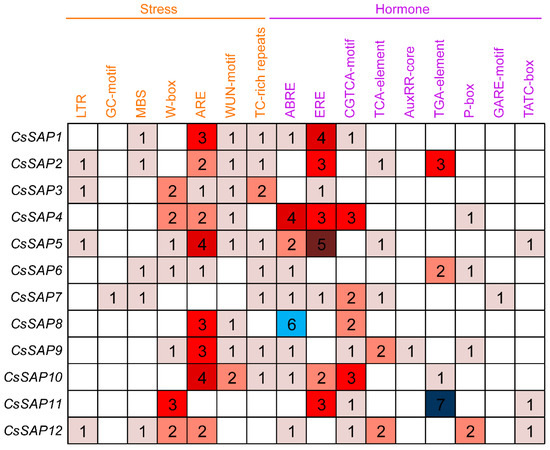
Figure 5.
Types and numbers of stress- and hormone-responsive cis-elements in the promoter regions of CsSAP genes.
2.6. Expression Profiles of CsSAP Genes in Different Tissues and during Fruit Development
The expression profile of CsSAP genes was examined based on the available RNA-seq data in different cucumber tissues and during fruit ripening [39,40]. As shown in Figure 6A, nearly all CsSAP genes were expressed in the tested tissues, except for CsSAP2 and CsSAP3. Among these genes, CsSAP1, CsSAP6, CsSAP7, and CsSAP11 displayed the highest transcriptional levels in unfertilized ovaries; CsSAP4, CsSAP5, CsSAP10, and CsSAP12 exhibited much higher transcript abundance in flowers; while CsSAP8 and CsSAP9 showed the highest mRNA accumulation in roots and basal tendrils, respectively (Figure 6A). To further explore the possible roles of CsSAP genes in fruit development, the transcript levels of CsSAP genes at different fruit development stages were evaluated according to the previously reported RNA-seq data [40]. The transcriptome data showed that several CsSAP genes, such as CsSAP1, CsSAP5, CsSAP7, CsSAP9, CsSAP11, and CsSAP12, displayed remarkable transcriptions at certain time points during fruit development (Figure 6B). The tissue-specific expression patterns suggest that these genes may have different roles depending on the tissues.
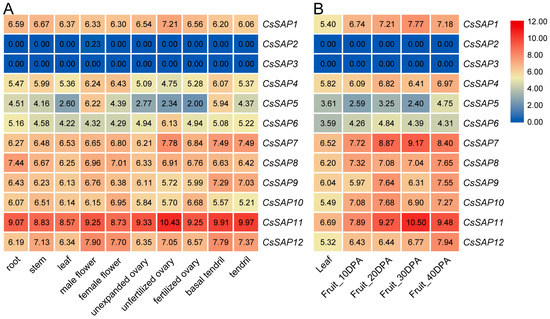
Figure 6.
Expression levels of CsSAP genes in different cucumber tissues (A) and at different stages of fruit ripening (B). The expression levels were calculated as the log2-transformed RPKM+1 values based on data retrieved from available RNA-seq data. DPA, day after pollination.
2.7. Expression Patterns of CsSAP Genes under Abiotic Stress Treatments
To unravel the roles of CsSAP genes in response to environmental stimuli, the expression profiles of six selected CsSAP genes under various abiotic stresses (cold, drought, and salt) were investigated by RT-qPCR. Under cold treatment, the transcript levels of CsSAP5, CsSAP6, CsSAP9, and CsSAP10 were obviously increased at certain time points, particularly CsSAP5, while CsSAP1 and CsSAP7 were significantly inhibited (Figure 7). Under drought stress, all the CsSAP genes were up-regulated at 6 hps and/or 12 hps, followed by a further decrease at 24 hps (Figure 8). For salt stress, the transcription levels of all CsSAP genes were induced at specific time points or during certain periods (Figure 9). Notably, the transcription of CsSAP6 was gradually induced during salt stress, and CsSAP5 and CsSAP9 were highly responsive to salt stress, with the transcript levels being up-regulated and peaking at a later time point (24 hps) (Figure 9). These results indicated that the CsSAP genes might have important effects when cucumber plants are exposed to different environmental stresses.
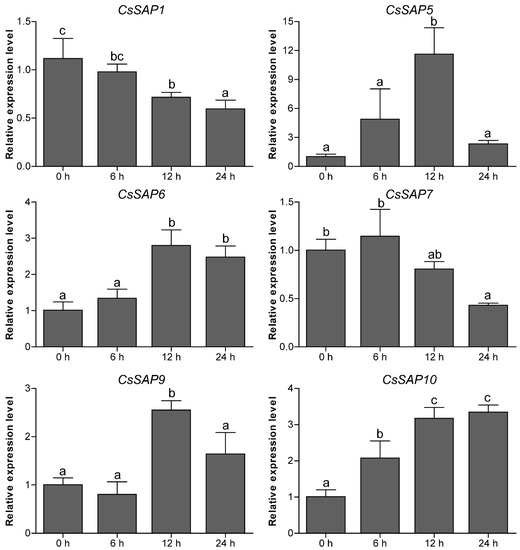
Figure 7.
RT-qPCR analysis of six selected CsSAP genes under cold treatment. Error bars represent the standard error from three replicates, and different letters above the bars represent significant differences of the data at P < 0.05 (Tukey’s test).
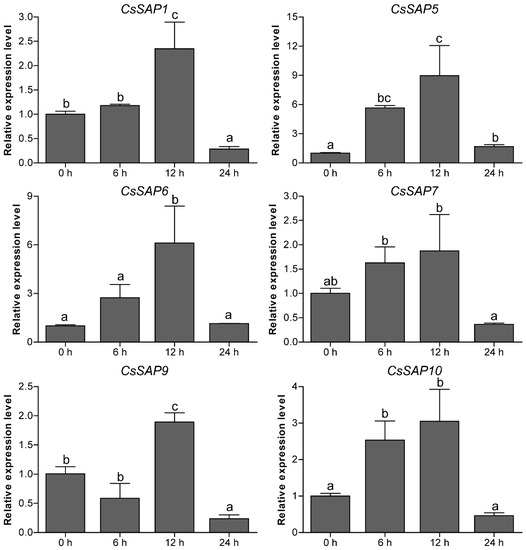
Figure 8.
Expression profiles of six selected CsSAP genes under drought treatment as revealed by RT-qPCR. Error bars represent the standard error from three replicates, and different letters above the bars represent significant differences of the data at P < 0.05 (Tukey’s test).
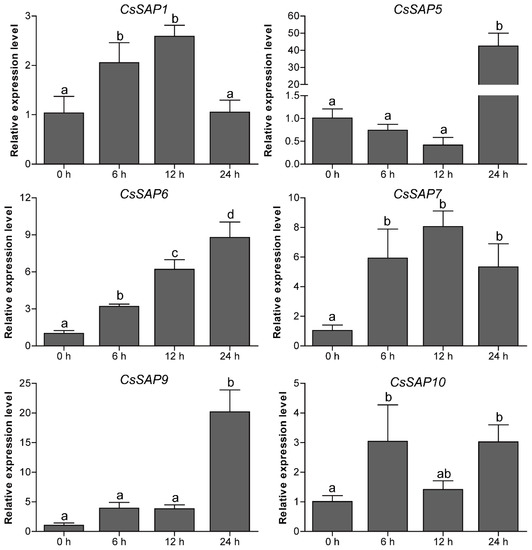
Figure 9.
Expression profiles of six selected CsSAP genes under salt treatment as revealed by RT-qPCR. Error bars represent the standard error from three replicates, and different letters above the bars represent significant differences of the data at P < 0.05 (Tukey’s test).
3. Discussion
In the present study, a total of 12 SAP gene members were identified and characterized in cucumber (Table 1). Genome-wide survey of SAP gene family composition has been performed in various plant species and there is a high variability in terms of the number of gene members. For example, 27 SAP genes were reported in Glycine max [5], 57 in Brassica napus [6], 16 in Artemisia annua [7], 30 in Malus domestica [8], 17 in Medicago truncatula [3], 37 in Gossypium hirsutum [9], 13 in Solanum lycopersicum [1], 18 in Oryza sativa, and 14 in Arabidopsis thaliana [10]. It turns out that there are fewer SAP family genes in cucumber than in these plants, possibly because of the tandem and segmental duplications of the SAP gene family. In a previous report, 36 out of the 37 GhSAP genes were found to form 18 pairs of putatively duplicated genes in cotton [9]. In apple, a total of 17 and two MdSAP genes have undergone tandem and segmental duplication events, respectively [8]. In the present study, only two segmental duplications and one tandem duplication were observed among the SAP genes in cucumber (Figure 1). In addition, a majority of CsSAP proteins (9 out of 12) harbor one A20 and one AN1 zinc finger domain, while the remaining three SAPs have no A20 domain (Table 1; Figure 3). However, SAP proteins with a single A20 domain were observed in some plants. For example, a total of seven BnaSAPs have a single A20 domain [6], and MdSAP22 and GmSAP23 also possess only one single A20 domain in apple and soybean, respectively [5,8].
Analysis of the evolutionary relationships of SAP proteins among cucumber and other four plants indicated that these SAPs can be divided into seven groups, and most proteins in the same group have the same arrangement of AN1 and A20 domains (Figure 2). We further examined the conserved motif distributions of cucumber SAP proteins according to the evolutionary relationship. A total of 10 conserved motifs were identified, and their distributions exhibited strong evolutionary conservation (Figure 3). Gene structure can also effectively reveal the evolutionary relationships among gene families. In this work, a large proportion of CsSAP genes (9 out of 12) were found to be intronless, which is in accordance with the previous reports [3,5,8,9]. The intronless genes were suggested to be able to reduce post-transcriptional processing for immediate responses to abiotic stresses [3]. These findings suggest that SAP genes are highly evolutionarily conserved in plants. Moreover, some CsSAP genes clustered together exhibited similar tissue expression patterns (Figure 2 and Figure 6). For example, CsSAP1, CsSAP7, and CsSAP11 showed a board spectrum of expression in different tissues, and all displayed the highest expression in unfertilized ovaries (Figure 6A).
Previous reports have concluded that plant SAP genes play crucial roles in regulating a variety of abiotic and biotic stresses [22,30,31,41]. In the present study, a series of stress-responsive cis-elements were observed in the promoters of CsSAP genes (Figure 5), implying that the CsSAP genes may participate in stress responses. Additionally, RT-qPCR assay of the selected CsSAP genes under cold, drought, and salt stresses revealed that the transcript levels of these genes were obviously changed under the three stress conditions (Figure 7, Figure 8 and Figure 9). For instance, the transcript levels of CsSAP5, CsSAP6, CsSAP9, and CsSAP10 were obviously elevated at certain time points under all of the three stresses, suggesting their potential roles in stress response. Both OsiSAP1 and OsiSAP8 are multiple stress-inducible genes in rice, and transgenic plants overexpressing either of them displayed tolerance to various abiotic stresses [12,20,21,22]. Similarly, AtSAP5 was significantly up-regulated under salt, osmotic, drought, and cold stress conditions, and overexpression of AtSAP5 conferred tolerance to a series of abiotic stresses in transgenic Arabidopsis plants [17,41]. Notably, CsSAP5 displayed much more observably up-regulated expression than other CsSAP genes under the three stress conditions (Figure 7, Figure 8 and Figure 9), suggesting that it might contribute greatly to the resistance against stress in cucumber. It should be noted that CsSAP1 and CsSAP7 were suppressed by cold stress (Figure 7). They have closer evolutionary relationships than other CsSAP genes, and their encoded proteins display similar conserved motif arrangements (Figure 2 and Figure 3). These findings suggest that CsSAP1 and CsSAP7 may play negative roles in response to cold stress. However, no low-temperature-responsive (LTR) cis-elements were found in the promoter regions of CsSAP1 and CsSAP7 (Figure 5), implying that their promoters may harbor other unknown cold-responsive cis-elements. Some genes playing negative roles in stress tolerance were also observed in different plant species. For instance, ZFP185/OsSAP4 was found to have negative functions in stress tolerance and overexpression of ZFP185 in rice increased its sensitivity to drought, cold, and salt stresses [42]. Down-regulation of PagSAP1 in poplar resulted in significantly enhanced tolerance to salt stress with an increased K+/Na+ ratio in root and altered expression of genes associated with cellular ionic homeostasis [43]. However, the expression levels of CsSAP1 and CsSAP7 showed observable increases under drought and salt treatments at certain time points (Figure 8 and Figure 9). These findings indicate that the CsSAP genes may have vital regulatory functions in response to a variety of abiotic stresses, and cucumber may have developed diverse regulatory mechanisms when suffering from different environmental stimuli [2]. In a previous study, overexpression of ZmAN13 in Arabidopsis plants led to enhanced tolerance to cold stress but increased sensitivity to salt and drought stress [15].
4. Materials and Methods
4.1. Identification of the SAP Family Members in Cucumber
The cucumber v2 protein sequences were acquired from Cucurbit Genomics Database (CuGenDB, http://cucurbitgenomics.org/). The A20 domain (PF01754) and AN1 domain (PF01428) were obtained from the Pfam server (http://pfam.xfam.org/), and employed for the identification of cucumber SAP members using HMMER3.0. In addition, the protein sequences of SAP family members of Arabidopsis and rice were retrieved from a previous report [10], and then used as queries to conduct an extensive BLASTp search against the cucumber v2 proteome. After removing the redundant sequences, each SAP candidate was checked via Pfam and SMART (http://smart.embl-heidelberg.de/) to verify that they had the A20 domain and/or the AN1 domain.
4.2. Protein Property and Sequence Analysis of Cucumber SAP Proteins
The ProtParam tool on the ExPASy server (http://web.expasy.org/protparam) was employed to predict the molecular weight (MW), isoelectric points (pI), and grand average of hydropathicity (GRAVY) of each cucumber putative SAP protein. The conserved motifs of cucumber SAP proteins were examined with MEME Version 5.1.0 (http://meme-suite.org/tools/meme) using the minimum and maximum width of the motif set to 6 and 50, and the number of different motifs was set to 10. The MEME results were illustrated with the TBtools software [44]. The distributions of A20, AN1, and C2H2 domains were visualized with the Illustrator for Biological Sequences (IBS) tool (http://ibs.biocuckoo.org/index.php). Subcellular localization analysis of each cucumber SAP protein was conducted with the Plant-mPLoc server (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) [45]. For multiple sequence alignment, the full-length SAP amino acid sequences from cucumber and other plant species were aligned using the MAFFT server (https://www.ebi.ac.uk/Tools/msa/mafft/) with default settings. Subsequently, the alignment was imported into the MEGA 7.0 software to create a phylogenetic tree by the neighbor-joining (NJ) method with a bootstrap option of 1000 replications.
4.3. Chromosomal Distribution, Gene Structure, and Cis-Element Analyses of Cucumber SAP Genes
The information about chromosomal distribution, coding sequence (CDS), and genomic DNA (gDNA) sequence was retrieved from the cucumber genome database (http://cucurbitgenomics.org/organism/2). The chromosomal location of cucumber SAP genes was drawn with the MapInspect software following the procedure previously described [2]. For gene duplication analysis, the MCScanX software was employed to determine the tandem and segmental duplication events using the criteria from a previous study [46]. Gene structure was determined with the GSDS tool (http://gsds.cbi.pku.edu.cn/index.php) by comparison of CDS and respective gDNA sequence of each cucumber SAP gene. For identification of cis-elements located at the promoter regions of SAP genes, the 2.0-kb upstream sequence of the start codon (ATG) of each CsSAP gene was downloaded from the cucumber genome database. A search was performed by using the PlantCARE server (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
4.4. Transcript Analysis of CsSAP Genes in Different Plant Tissues and during Fruit Ripening
To assess the tissue-specific expression profiles of CsSAP genes, the RNA-seq reads were downloaded in NCBI SRA database (https://www.ncbi.nlm.nih.gov/sra/?term=sra046916) and the expression profiles of CsSAP genes in flowers, leaves, ovaries, roots, stems, and tendrils were analyzed based on a previous study [47]. To gain insights into the possible roles of CsSAP genes during fruit development, their digital gene expression was derived from the fruitENCODE database (http://www.epigenome.cuhk.edu.hk/encode.html). All of the expression profiles of CsSAP genes were displayed as the reads per kilobase per million (RPKM) values, and the heat maps were illustrated with TBtools [44].
4.5. Transcript Analysis of CsSAP Genes under Different Abiotic Stresses
Cucumber (Cucumis sativus var. sativus line 9930) seedlings were grown in poly trays containing peat, sand, and pumice (1:1:1, v/v/v) within a growth chamber under a 16 h photoperiod, a night temperature of 18 °C and day temperature of 24 °C, and a relative humidity of 60–70%. Two-week-old cucumber seedlings were treated with different stress conditions including 4 °C (cold), NaCl (salt), and PEG (drought) as described in our previous reports [2,34]. Briefly, seedlings in the trays were transferred to 4 °C for cold stress. For salt and drought stress, 2-week-old seedlings were grown in liquid Murashige and Skoog (MS) medium containing 200 mM NaCl and 10% PEG-6000 (w/v), respectively. Samples were taken at 0, 6, 12, and 24 h after plant exposure to each stress condition. Each sample consisted of leaves taken from at least ten independent plants. Three biological repetitions were considered for each time point. All samples were frozen immediately in liquid nitrogen and stored at −80 °C until further processing.
Total RNA was isolated using the Eastep Super Total RNA Extraction Kit (Promega, Madison, WA, USA), and then about 3 μg RNA was reverse-transcribed into cDNA using the M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturers’ protocols as previously described [47]. The RT-qPCR was conducted with the Roche Lightcyler 480II PCR System using the TB Green Premix Ex TaqII Kit (TaKaRa, Dalian, China). All reactions were carried out with three technical replicates, and the RT-qPCR parameters were used as described previously [2]. CsAct3 was selected as a housekeeping gene [2], and the 2−ΔΔCt method was used to calculate the relative expression levels of CsSAP genes, which are shown relative to those at 0 h post stress exposure (hps). Gene-specific primers (Table S3) were designed by Primer Express software (Applied Biosystems, Foster City, CA, USA), and primer specificity was verified by Primer-BLAST at NCBI databases (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). A melting curve analysis was conducted to ensure the specificity in amplification. All data were subjected to one-way analysis of variance (ANOVA) with Tukey’s test.
5. Conclusions
In the present study, we carried out the systematic identification and characterization of SAP gene family members in cucumber. Their characteristics, including genomic locations, gene duplications, evolutionary relationships, conserved domains and motifs, gene structures, and cis-elements in promoter regions, were analyzed based on bioinformatics methods. In addition, the expression patterns of CsSAP genes in different tissues and fruit development stages were investigated based on RNA-seq. Moreover, RT-qPCR assays showed that all six selected CsSAP genes were regulated by three stress treatments (cold, drought, and salt), suggesting that the CsSAP genes may play certain roles in stress responses. This study provides essential information for elucidating the potential roles of cucumber SAP genes in stress responses and may help to further promote the stress resistance of cucumber and other plants.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/3/400/s1, Table S1: Accession numbers of SAPs for phylogenetic analysis. Table S2: Sequences and lengths of motifs among cucumber SAP proteins. Table S3: Gene-specific primers used for RT-qPCR.
Author Contributions
Data curation, W.L. and Y.Z.; funding acquisition, Y.Y. and S.L.; investigation, W.L., Y.Z., R.P., L.L., J.H., and H.L.; methodology, R.P., L.L., J.H., Y.Y., and S.L.; resources, L.L., H.L., and S.L.; software, W.L., Y.Z., and R.P.; validation, W.L. and Y.Z.; visualization, Y.Y.; writing—original draft, W.L., Y.Z., and S.L.; writing—review and editing, Y.Z., and S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Key Project of Science Foundation of Jiangxi Province (20171ACB21025 and 20181ACB20012), and the National Natural Science Foundation of China (31660578).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Solanke, A.U.; Sharma, M.K.; Tyagi, A.K.; Sharma, A.K. Characterization and phylogenetic analysis of environmental stress-responsive SAP gene family encoding A20/AN1 zinc finger proteins in tomato. Mol. Genet. Genom. 2009, 282, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Hu, L.; Xu, S.; Jiang, L.; Liu, S. Identification and transcriptional analysis of dehydrin gene family in cucumber (Cucumis sativus). Acta Physiol. Plant 2018, 40, 144. [Google Scholar] [CrossRef]
- Zhou, Y.; Zeng, L.; Chen, R.; Wang, Y.; Song, J. Genome-wide identification and characterization of the stress associated protein (SAP) gene family encoding A20/AN1 zinc finger proteins in Medicago truncatula. Arch. Biol. Sci. 2018, 70, 87–98. [Google Scholar] [CrossRef]
- Giri, J.; Dansana, P.K.; Kothari, K.S.; Sharma, G.; Vij, S.; Tyagi, A.K. SAPs as novel regulators of abiotic stress response in plants. Bioessays 2013, 35, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Z.; Zheng, W.J.; Cao, X.Y.; Cui, X.Y.; Zhao, S.P.; Yu, T.F.; Chen, J.; Zhou, Y.B.; Chen, M.; Chai, S.C.; et al. Genomic analysis of stress associated proteins in soybean and the role of GmSAP16 in abiotic stress responses in Arabidopsis and soybean. Front. Plant Sci. 2019, 10, 1453. [Google Scholar] [CrossRef]
- He, X.; Xie, S.; Xie, P.; Yao, M.; Liu, W.; Qin, L.; Liu, Z.; Zheng, M.; Liu, H.; Guan, M.; et al. Genome-wide identification of stress-associated proteins (SAP) with A20/AN1 zinc finger domains associated with abiotic stresses responses in Brassica napus. Environ. Exp. Bot. 2019, 165, 108–119. [Google Scholar] [CrossRef]
- Wang, Y.; Fu, X.; Xie, L.; Qin, W.; Li, L.; Sun, X.; Xing, S.; Tang, K. Stress associated protein 1 regulates the development of glandular trichomes in Artemisia annua. Plant Cell Tiss. Organ. Cult. 2019, 139, 249–259. [Google Scholar] [CrossRef]
- Dong, Q.; Duan, D.; Zhao, S.; Xu, B.; Luo, J.; Wang, Q.; Huang, D.; Liu, C.; Li, C.; Gong, X.; et al. Genome-wide analysis and cloning of the apple stress-associated protein gene family reveals MdSAP15, which confers tolerance to drought and osmotic stresses in transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2478. [Google Scholar] [CrossRef]
- Gao, W.; Long, L.; Tian, X.; Jin, J.; Liu, H.; Zhang, H.; Xu, F.; Song, C. Genome-wide identification and expression analysis of stress-associated proteins (SAPs) containing A20/AN1 zinc finger in cotton. Mol. Genet. Genom. 2016, 291, 2199–2213. [Google Scholar] [CrossRef]
- Vij, S.; Tyagi, A.K. Genome-wide analysis of the stress associated protein (SAP) gene family containing A20/AN1 zinc-finger(s) in rice and their phylogenetic relationship with Arabidopsis. Mol. Genet. Genom. 2006, 276, 565–575. [Google Scholar] [CrossRef]
- Li, J.; Sun, P.; Xia, Y.; Zheng, G.; Sun, J.; Jia, H. A stress-associated protein, PtSAP13, from Populus trichocarpa provides tolerance to salt stress. Int. J. Mol. Sci. 2019, 20, 5782. [Google Scholar] [CrossRef] [PubMed]
- Kothari, K.S.; Dansana, P.K.; Giri, J.; Tyagi, A.K. Rice stress associated protein 1 (OsSAP1) interacts with aminotransferase (OsAMTR1) and pathogenesis-related 1a protein (OsSCP) and regulates abiotic stress responses. Front. Plant Sci. 2016, 7, 1057. [Google Scholar] [CrossRef] [PubMed]
- Ben Saad, R.; Safi, H.; Ben Hsouna, A.; Brini, F.; Ben Romdhane, W. Functional domain analysis of LmSAP protein reveals the crucial role of the zinc-finger A20 domain in abiotic stress tolerance. Protoplasma 2019, 256, 1333–1344. [Google Scholar] [CrossRef] [PubMed]
- Giri, J.; Vij, S.; Dansana, P.K.; Tyagi, A.K. Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol. 2011, 191, 721–732. [Google Scholar] [CrossRef]
- Xuan, N.; Jin, Y.; Zhang, H.; Xie, Y.; Liu, Y.; Wang, G. A putative maize zinc-finger protein gene, ZmAN13, participates in abiotic stress response. Plant Cell Tiss. Organ. Cult. 2011, 107, 101. [Google Scholar] [CrossRef]
- Chang, L.; Chang, H.H.; Chang, J.C.; Lu, H.C.; Wang, T.T.; Hsu, D.W.; Tzean, Y.; Cheng, A.P.; Chiu, Y.S.; Yeh, H.H. Plant A20/AN1 protein serves as the important hub to mediate antiviral immunity. PLoS Pathog. 2018, 14, e1007288. [Google Scholar] [CrossRef]
- Kang, M.; Fokar, M.; Abdelmageed, H.; Allen, R.D. Arabidopsis SAP5 functions as a positive regulator of stress responses and exhibits E3 ubiquitin ligase activity. Plant Mol. Biol. 2011, 75, 451–466. [Google Scholar] [CrossRef]
- Kang, M.; Abdelmageed, H.; Lee, S.; Reichert, A.; Mysore, K.S.; Allen, R.D. AtMBP-1, an alternative translation product of LOS2, affects abscisic acid responses and is modulated by the E3 ubiquitin ligase AtSAP5. Plant J. 2013, 76, 481–493. [Google Scholar] [CrossRef]
- Dixit, A.; Tomar, P.; Vaine, E.; Abdullah, H.; Hazen, S.; Dhankher, O.P. A stress-associated protein, AtSAP13, from Arabidopsis thaliana provides tolerance to multiple abiotic stresses. Plant Cell Environ. 2018, 41, 1171–1185. [Google Scholar] [CrossRef]
- Dansana, P.K.; Kothari, K.S.; Vij, S.; Tyagi, A.K. OsiSAP1 overexpression improves water-deficit stress tolerance in transgenic rice by affecting expression of endogenous stress-related genes. Plant Cell Rep. 2014, 33, 1425–1440. [Google Scholar] [CrossRef]
- Mukhopadhyay, A.; Vij, S.; Tyagi, A.K. Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. USA 2004, 101, 6309–6314. [Google Scholar] [CrossRef] [PubMed]
- Kanneganti, V.; Gupta, A.K. Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol. Biol. 2008, 66, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Ben Saad, R.; Zouari, N.; Ben Ramdhan, W.; Azaza, J.; Meynard, D.; Guiderdoni, E.; Hassairi, A. Improved drought and salt stress tolerance in transgenic tobacco overexpressing a novel A20/AN1 zinc-finger “AlSAP” gene isolated from the halophyte grass Aeluropus littoralis. Plant Mol. Biol. 2010, 72, 171–190. [Google Scholar] [CrossRef] [PubMed]
- Ben Saad, R.; Fabre, D.; Mieulet, D.; Meynard, D.; Dingkuhn, M.; Al-Doss, A.; Guiderdoni, E.; Hassairi, A. Expression of the Aeluropus littoralis AlSAP gene in rice confers broad tolerance to abiotic stresses through maintenance of photosynthesis. Plant Cell Environ. 2012, 35, 626–643. [Google Scholar] [CrossRef] [PubMed]
- Ben Saad, R.; Ben Hsouna, A.; Saibi, W.; Ben Hamed, K.; Brini, F.; Ghneim-Herrera, T. A stress-associated protein, LmSAP, from the halophyte Lobularia maritima provides tolerance to heavy metals in tobacco through increased ROS scavenging and metal detoxification processes. J. Plant Physiol. 2018, 231, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Ben Saad, R.; Farhat-Khemekhem, A.; Ben Halima, N.; Ben Hamed, K.; Brini, F.; Saibi, W. The LmSAP gene isolated from the halotolerant Lobularia maritima improves salt and ionic tolerance in transgenic tobacco lines. Funct. Plant Biol. 2018, 45, 378–391. [Google Scholar] [CrossRef]
- Kang, M.; Lee, S.; Abdelmageed, H.; Reichert, A.; Lee, H.K.; Fokar, M.; Mysore, K.S.; Allen, R.D. Arabidopsis stress associated protein 9 mediates biotic and abiotic stress responsive ABA signaling via the proteasome pathway. Plant Cell Environ. 2017, 40, 702–716. [Google Scholar] [CrossRef]
- Xu, Q.; Mao, X.; Wang, Y.; Wang, J.; Xi, Y.; Jing, R. A wheat gene TaSAP17-D encoding an AN1/AN1 zinc finger protein improves salt stress tolerance in transgenic Arabidopsis. J. Integr. Agric. 2018, 17, 507–516. [Google Scholar] [CrossRef]
- Tyagi, H.; Jha, S.; Sharma, M.; Giri, J.; Tyagi, A.K. Rice SAPs are responsive to multiple biotic stresses and overexpression of OsSAP1, an A20/AN1 zinc-finger protein, enhances the basal resistance against pathogen infection in tobacco. Plant Sci. 2014, 225, 68–76. [Google Scholar] [CrossRef]
- Liu, S.; Yuan, X.; Wang, Y.; Wang, H.; Wang, J.; Shen, Z.; Gao, Y.; Cai, J.; Li, D.; Song, F. Tomato stress-associated protein 4 contributes positively to immunity against necrotrophic fungus Botrytis cinerea. Mol. Plant Microbe Interact. 2019, 32, 566–582. [Google Scholar] [CrossRef]
- Liu, S.; Wang, J.; Jiang, S.; Wang, H.; Gao, Y.; Zhang, H.; Li, D.; Song, F. Tomato SlSAP3, a member of the stress-associated protein family, is a positive regulator of immunity against Pseudomonas syringae pv. tomato DC3000. Mol. Plant Pathol. 2019, 20, 815–830. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, Y.; Xiao, J.; Ma, Q.; Li, D.; Xue, Z.; Chong, K. OsDOG, a gibberellin-induced A20/AN1 zinc-finger protein, negatively regulates gibberellin-mediated cell elongation in rice. J. Plant Physiol. 2011, 168, 1098–1105. [Google Scholar] [CrossRef] [PubMed]
- Lloret, A.; Conejero, A.; Leida, C.; Petri, C.; Gil-Munoz, F.; Burgos, L.; Badenes, M.L.; Rios, G. Dual regulation of water retention and cell growth by a stress-associated protein (SAP) gene in Prunus. Sci. Rep. 2017, 7, 332. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, S.; Yang, Z.; Yang, Y.; Jiang, L.; Hu, L. CsCAT3, a catalase gene from Cucumis sativus, confers resistance to a variety of stresses to Escherichia coli. Biotechnol. Biotechnol. Equip. 2017, 31, 886–896. [Google Scholar] [CrossRef]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Huang, S.; Xie, B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011, 12, 471. [Google Scholar] [CrossRef]
- Baloglu, M.C.; Eldem, V.; Hajyzadeh, M.; Unver, T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE 2014, 9, e96014. [Google Scholar] [CrossRef]
- Xu, X.; Liu, M.; Lu, L.; He, M.; Qu, W.; Xu, Q.; Qi, X.; Chen, X. Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Mol. Genet. Genom. 2015, 290, 1403–1414. [Google Scholar] [CrossRef]
- Zhang, X.M.; Yu, H.J.; Sun, C.; Deng, J.; Zhang, X.; Liu, P.; Li, Y.Y.; Li, Q.; Jiang, W.J. Genome-wide characterization and expression profiling of the NAC genes under abiotic stresses in Cucumis sativus. Plant Physiol. Biochem. 2017, 113, 98–109. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Z.; Yan, P.; Huang, S.; Fei, Z.; Lin, K. RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genom. 2011, 12, 540. [Google Scholar] [CrossRef]
- Cheng, Y.; He, P.; Jiang, L.; Liu, S.; Zhou, Y. Identification and characterization of a SEPALLATA-like MADS-box gene from cucumber (Cucumis sativus L.). Not. Bot. Horti Agrobot. 2019, 47, 1168–1177. [Google Scholar] [CrossRef]
- Hozain, M.D.; Abdelmageed, H.; Lee, J.; Kang, M.; Fokar, M.; Allen, R.D.; Holaday, A.S. Expression of AtSAP5 in cotton up-regulates putative stress-responsive genes and improves the tolerance to rapidly developing water deficit and moderate heat stress. J. Plant Physiol. 2012, 169, 1261–1270. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lan, H.; Shao, Q.; Wang, R.; Chen, H.; Tang, H.; Zhang, H.; Huang, J. An A20/AN1-type zinc finger protein modulates gibberellins and abscisic acid contents and increases sensitivity to abiotic stress in rice (Oryza sativa). J. Exp. Bot. 2016, 67, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.K.; Bae, E.K.; Lee, H.; Choi, Y.I.; Han, M.; Choi, H.; Kang, K.S.; Park, E.J. Downregulation of stress-associated protein 1 (PagSAP1) increases salt stress tolerance in poplar (Populus alba × P. glandulosa). Trees 2018, 32, 823–833. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; He, Y.; Xia, R. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. bioRxiv 2018. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- You, J.; Wang, Y.; Zhang, Y.; Dossa, K.; Li, D.; Zhou, R.; Wang, L.; Zhang, X. Genome-wide identification and expression analyses of genes involved in raffinose accumulation in sesame. Sci. Rep. 2018, 8, 4331. [Google Scholar] [CrossRef]
- Chen, L.; Zhou, Y.; Lai, W.; Hu, L.; Jiang, L.; Liu, S. In silico identification and expression analysis of nuclear factor Y (NF-Y) transcription factors in cucumber. Agronomy 2020, 10, 236. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).