Abstract
Casparian strip membrane domain protein-like (CASPL) genes are key genes for the formation and regulation of the Casparian strip and play an important role in plant abiotic stress. However, little research has focused on the members, characteristics, and biological functions of the patchouli PatCASPL gene family. In this study, 156 PatCASPL genes were identified at the whole-genome level. Subcellular localization predicted that 75.6% of PatCASPL proteins reside on the cell membrane. A phylogenetic analysis categorized PatCASPL genes into five subclusters alongside Arabidopsis CASPL genes. In a cis-acting element analysis, a total of 16 different cis-elements were identified, among which the photo-responsive element was the most common in the CASPL gene family. A transcriptome analysis showed that p-hydroxybenzoic acid, an allelopathic autotoxic substance, affected the expression pattern of PatCASPLs, including a total of 27 upregulated genes and 30 down-regulated genes, suggesting that these PatCASPLs may play an important role in the regulation of patchouli continuous cropping obstacles by affecting the formation and integrity of Casparian strip bands. These results provided a theoretical basis for exploring and verifying the function of the patchouli PatCASPL gene family and its role in continuous cropping obstacles.
1. Introduction
Pogostemon cablin (Blanco) Benth is a perennial herb or semi-shrub plant of Labiatae, mainly distributed in the tropical and subtropical regions of Asia [1], such as India, Sri Lanka, Malaysia, Indonesia, and the Philippines [2]. In China, P. cablin, also called ‘Guanghuoxiang’, is mainly distributed in Hainan and Guangdong provinces [3,4]. According to the cultivation and production areas, it can be divided into four cultivation types, ‘Hainan Guanghuoxiang’ (Nanxiang), ‘Zhanjiang Guanghuoxiang’ (Zhanxiang), ‘Shipai Guanghuoxiang’ (Paixiang), and ‘Zhaoqing Guanghuoxiang’ (Zhaoxiang) [5,6]. As one of the ‘Ten Southern Medicines’ and a traditional Chinese medicine, it has been used for aromatic dampness, clearing heat, and as an antiemetic [7,8], and shows important medicinal and economic value [9]. However, continuous cropping obstacles are a key problem in the cultivation and production of P. cablin, which seriously affects its yield and quality [10,11]. Previous studies have found that the deterioration of soil physiochemical properties, the accumulation of allelochemicals, and the imbalance of microbial communities are the main causes of continuous cropping obstacles [12,13,14]. Among them, p-hydroxybenzoic acid (p-HBA) has been proved as a key allelochemical inducing the occurrence of P. cablin continuous cropping obstacles [15,16]. However, how does p-HBA induce the occurrence of P. cablin continuous cropping obstacles? Which signaling pathways are involved in the regulation process? These issues need to be further studied.
The Casparian strip is a bolted and lignified band-thickening wall structure surrounded by the radial wall and transverse wall of the endothelial cells, which plays an important role in screening, blocking, and cutting off unwanted ions or macromolecules into the vascular column [17,18]. It has been found that the formation and regulation of the Casparian strip involve multiple genes and signaling pathways [19,20]. It mainly contains the following key genes: Casparian strip membrane domain proteins (CASPs) [21], leucine receptor kinase (GSO1/SGN3) [22], enhanced suberin 1 (ESB1) [23], MYB domain protein 36 (MYB36) [24], and endodermis Casparian strip integrity factors 1/2 (CIF1/2) [25]. Among them, CASPLs are pivotal membrane proteins specifically expressed in the Casparian strip formation region. These proteins contain a four-transmembrane domain with cytoplasmic amino and carboxyl ends and conserved extracellular loops, which play a key role in the formation of the Casparian strip [26,27]. At present, the number of CASPL family members varies among different plant species, with 39 identified in the Arabidopsis genome, 19 in rice [28], 48 in cotton [29], 61 in banana [30], and 33 in litchi [31] genomes, implying the diversity of CASPL family composition across species. Moreover, it has been found that CASPL family genes play an important role in abiotic stress, including salt tolerance [32,33,34] and cold tolerance [35,36]. However, the research on the function of plant CASPL genes is mainly focused on model plants such as Arabidopsis thaliana and rice, and there is no research reported on PatCASPL genes in P. cablin. Previous transcriptome data showed that PatCASPLs can respond to the allelochemical p-HBA, suggesting that the Casparian strip may play an important role in continuous cropping obstacles [16]. How many PatCASPL family genes are there in P. cablin? Which CASPL genes are involved in the continuous cropping obstacles of P. cablin? These questions need to be further answered.
In this study, based on the whole-genome data of P. cablin, a total of 156 PatCASPL family members were screened and identified through a bioinformatics analysis, and their protein physicochemical properties, chromosome distribution, promoter cis-acting elements, and evolutionary expression characteristics were analyzed in detail. Furthermore, 57 key CASPL candidate genes involved in the continuous cropping obstacles of P. cablin were screened and identified, via an analysis of the transcriptome data of P. cablin treated with p-HBA. The results of this study provide a theoretical basis for exploring the functions of the PatCASPL gene family and its role in continuous cropping obstacles.
2. Results
2.1. Identification of PatCASPL Gene Family Members and Analysis of Protein Physicochemical Properties in P. cablin
According to the 39 CASPL protein sequences of A. thaliana, 176 and 168 CASPL candidate genes were screened preliminarily in the P. cablin genome database by using the BLASTP [37] and HMM [38] alignment search methods, respectively. After further comparison and analysis using the SMART online program on the NCBI website (https://blast.ncbi.gov/ accessed on 10 May 2023), a total of 156 members of the PatCASPL gene family were finally identified. The physicochemical properties of the 156 CASPL protein sequences were analyzed, and the isoelectric point and molecular weight of the PatCASPL protein were predicted using the ExPASy [39] online analysis tool (Table 1). The results showed that the number of amino acids of PatCASPL protein varied from 102 (PatCASPL5C8) to 365 (PatCASPL1A6). The molecular weight of PatCASPL protein was between 11,043.99 and 39,579.28 Da, including 43 acidic proteins (pI ≤ 7) and 113 basic proteins (pI ≥ 7). The aliphatic index ranged from 64.93 to 127.1. The theoretical pI of the 156 CASPL proteins ranged from 4.93 to 10.14. In the CASPL gene family, the instability index of 40 CASPL genes is greater than 40, which highlights an unstable protein; the instability index of 116 CASPL genes was less than 40, which highlights stable proteins. The grand average of hydropathicity (GRAVY) of the PatCASPL2B4, PatCASPL2B2, PatCASPL1F10, PatCASPL2B5, PatCASPL4D12, PatCASPL1A8, PatCASPL1F11, PatCASPL2B8, PatCASPL4D3, PatCASPL2B1, PatCASPL1F9, PatCASPL1A7, PatCASPL4D4, PatCASPL1A9, PatCASPL2B3, PatCASPL1B1, and PatCASPL3A3 proteins was less than zero, implying that they are hydrophilic proteins; the average hydrophilicity of the remaining 139 CASPL genes was greater than zero, indicating that they were hydrophobic proteins.

Table 1.
Physicochemical properties of CASPL gene family proteins in Pogostemon cablin.
A secondary structure analysis of the PatCASPL protein in P. cablin using the SOPMA online tool [40] showed that the PatCASPL protein contained four types of structures: Alpha helix (α-helix, 22.09~71.31%), Beta turn (β-turn, 3.88~53.44%), random coil (1.06~10.4%), and extended strand (11.48~53.91%) (Table 2).

Table 2.
Subcellular localization and protein secondary structure analysis of CASPL gene family proteins in P. cablin.
A subcellular localization prediction analysis showed that the PatCASPL protein was mainly distributed on the cell membrane, accounting for 75%, and the possibility of prediction analysis on the cell membrane was between 50.31% and 100%. Among them, PatCASPL1C9, PatCASPL4D3, PatCASPL4D6, PatCASPL1C2, PatCASPL1C1, PatCASPL1C7, PatCASPL1C5, and PatCASPL4D4 gene distribution predicted the possibility of distribution on the cell membrane as high as 100%. In addition, 18 PatCASPL proteins were only distributed in the nucleus, with a probability of 51.39% to 98.88%. Eight gene proteins (PatCASPL4D11, PatCASPL4D7, PatCASPL1A6, PatCASPL5B3, PatCASPL4D8, PatCASPL1B2, PatCASPL1A8, PatCASPL5B4) were distributed on the chloroplast, and the probability ranged from 49.45% to 86.61%. PatCASPL2B5, PatCASPL2B7, and PatCASPL2B8 were distributed on peroxisomes, with a probability of 49.45% to 95%. The different subcellular localization of PatCASPL family proteins indicates that the gene family may play different biological functions and mainly act on the cell membrane, which is consistent with the characteristics of membrane proteins. These prediction results provide a reference for subsequent experiments.
2.2. Genetic Characterization and Phylogenetic Analysis of PatCASPL Gene Family Members in P. cablin
Based on the sequences of the PatCASPL gene family of P. cablin, the introns/exons and conserved motifs [41] were analyzed (Figure 1). A gene structure analysis showed that PatCASPL gene family members contained two to seven exons and two to eight introns. The members of the same subcluster have the same exons/introns, and this highly conserved gene structure affects the phylogenetic evolution relationship. Pfam (http://pfam.xfam.org/ accessed on 31 March 2023) and Batch CD-Search (https://www.ncbi.nlm.nih.gov/ accessed on 17 May 2023) were used to test whether the PatCASPL genes contained a complete domain, and the results showed that 156 PatCASPL gene family members contained DUF588 or MARVEL conserved domains. The two domains are selectively distributed in specific phylogenetic tree branches, showing the structural similarity between proteins in the same group. PatCASP10, PatCASPL1A3, PatCASPL1C9, PatCASPL1F2, and the other 34 genes containing the conserved domain of MARVEL were distributed in the E-subcluster (Group E) of the phylogenetic tree, and the remaining 122 genes containing the conserved domain of DUF588 were distributed in other subclusters of the phylogenetic tree (Figure 2). A subcellular localization prediction analysis showed that 25 genes, including PatCASPL1C1, PatCASPL1E3, PatCASPL1F2, and PatCASPL1D6, in 34 genes containing the MARVEL conserved domain were distributed on the cell membrane, and eight genes, including PatCASPL1A5, PatCASPL1A8, PatCASPL1B4, and PatCASPL1B2, were distributed on the chloroplast.
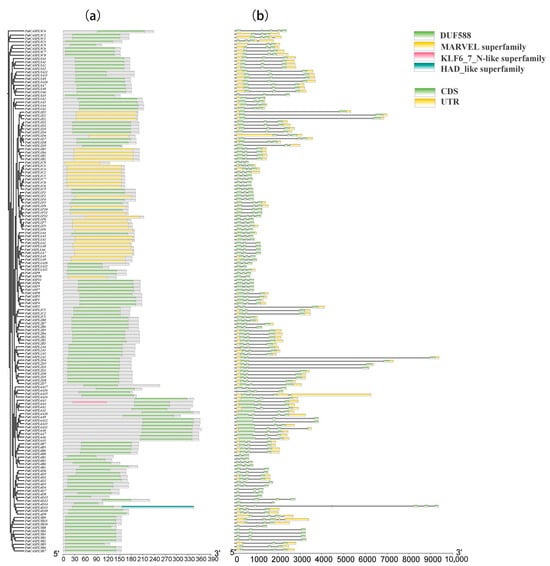
Figure 1.
Conserved domain (a) and gene structure (b) of CASPL family members in Pogostemon cablin.
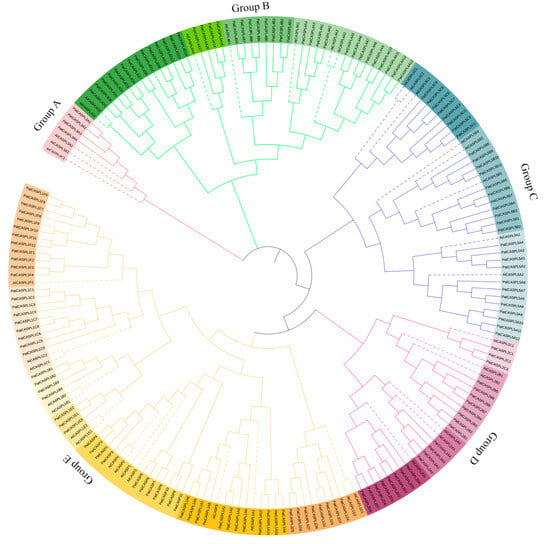
Figure 2.
Phylogenetic trees of CASPL genes in P. cablin (Pat) and Arabidopsis (At). The phylogenetic tree of CASPL protein in P. cablin and A. thaliana was constructed with MEGA11 software using maximum likelihood method, and then visualized with the ITOL online tool. Different subfamilies are represented by branches and frames of different colors.
In addition to the DUF588 structure, PatCASPL4A4 and PatCASPL4D11 also contain the KLF6_7_N-like superfamily and HAD_like superfamily, respectively, suggesting that these two genes may have multiple functions. The analysis found that the function of the conserved domain of DUF588 has not been identified, containing a conserved arginine and aspartic acid, which constitutes a site that may have catalytic activity [42]. It has been reported that some scholars have extended phylogenetic analysis beyond the plant kingdom and found that there is conservation between the CASPL and MARVEL protein families, and conserved residues are located in transmembrane domains, indicating that these domains are involved in the localization of CASPL [21].
In order to explore the phylogenetic relationship of the PatCASPL gene family in P. cablin, the phylogenetic tree of 195 CASPL protein sequences in P. cablin and A. thaliana was constructed using the MEGA11 software [43]. The results showed that 195 PatCASPL and AtCASPL members could be divided into five subclusters, among which the A-subcluster (Group A) had the largest number of members, containing 15 AtCASPL members and 59 PatCASPL members. The B-Subcluster (Group B) contains six AtCASPL members and 23 PatCASPL members. The C-subcluster (Group C) contains seven AtCASPL members and 39 PatCASPL members. The number of members in the D-subcluster (Group D) is the lowest, containing only four AtCASPL members and three PatCASPL members. The E-subcluster (Group E) contains eight AtCASPL members and 31 PatCASPL members (Figure 2). The proportion of AtCASPL members to PatCASPL members in each cluster was 1:1~1:3.9, indicating that the CASPL genes in the same subcluster of P. cablin and A. thaliana may be derived from the same ancestor, and the chromosome doubling event of P. cablin led to the expansion of the number of PatCASPL family genes in P. cablin.
The E-subcluster (Group E) contains 11 genes with the highest homology with AtCASP. AtCASP1-5 was identified as a gene related to the formation of Arabidopsis Casparian strip in A. thaliana. The function of AtCASPL protein in other subclusters has not been reported in depth. The genes distributed in the B-subcluster (Group B) may be involved in the response to abiotic stress. At3g55390 (AtCASPL4C1) has been reported to be induced by low-temperature and negatively regulates plant growth [35]. Therefore, it is speculated that PatCASPLs with high homology may have similar functions. Up to now, the biological function of most PatCASPL genes in P. cablin is still unclear. However, more and more CASPL genes of Arabidopsis have been functionally characterized. Therefore, the clustering and comparison of PatCASPL proteins and AtCASP proteins can predict their functions through a homologous analysis.
2.3. Analysis of Cis-Acting Elements of PatCASPL Gene Family in P. cablin
In order to explore the PatCASPL-involved signal regulation pathways, the cis-acting elements of the PatCASPL gene were analyzed [44]. A variety of different cis-elements were identified in the 2000 bp sequence upstream of the initiation codon of 156 PatCASPL gene families in P. cablin. Among them, the light response element is the most frequent in the CASPL gene, and 138 of the 156 genes contain light response elements, which is the most in the PatCASPL gene family, suggesting that the PatCASPLs may be involved in regulating the photomorphogenesis of P. cablin (Figure 3). Among the 156 members of the PatCASPL family, 152 members contained hormone-responsive elements, including 135 for abscisic acid, 88 for methyl jasmonate, 80 for gibberellin, 61 for auxin, and 61 for salicylic acid. There are 70 and 56 PatCASPL genes containing drought and low-temperature response elements, respectively, indicating that the PatCASPLs may play an important role in hormone regulation and the mitigation of stress. The cis-elements of different genes in the same subcluster of the PatCASPL gene family are not the same, suggesting that different PatCASPLs may play different functions in different growth, development, and stress response processes of P. cablin.

Figure 3.
Cis-acting elements of CASPL family members in P. cablin. The 2000 bp promoter sequences of P. cablin CASPL genes contain a variety of cis-acting elements, including photo responsive elements, hormone responsive elements, drought, low-temperature, anaerobic, wound and other response elements, as well as specific elements of meristem, seed, and endosperm.
2.4. Chromosome Localization of PatCASPL Gene Family in P. cablin
A phylogenetic analysis showed that the PatCASPL gene family members were distributed in five subclusters, and their distribution showed an apparent chromosome preference, essentially distributed at both ends of the chromosome. Gene density information was obtained and analyzed using a gene density profile tool. The results of the gene density analysis (Figure 4) showed that the gene density of the 21 chromosome front segments of the CASPL gene was low, and the gene density of the back end was high. The gene density of the front and back ends of the remaining chromosomes was high, and the gene density of the middle was low. Twelve genes such as PatCASPL4A15, PatCASPL4A1, PatCASPL2D1, and PatCASPL2D3 were distributed in the low-gene-density region, and 92% of the genes were located in the high-gene-density region.
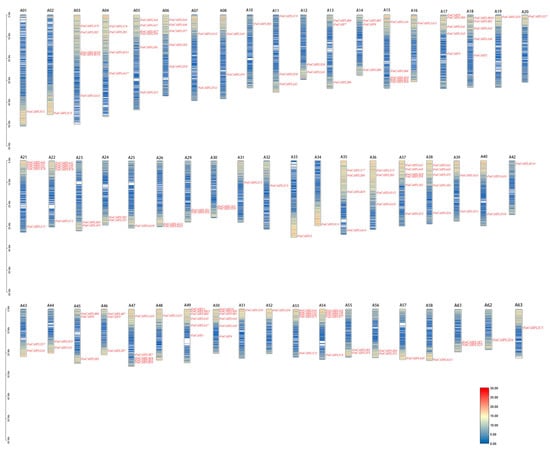
Figure 4.
Chromosomal mapping of CASPL family genes in P. cablin. The leftmost scales represent the chromosome length; A01–A63 represent the names of 57 chromosomes of mustard. There is no PatCASPL gene on chromosomes 9, 27, 28, 41, 59, and 60. Blue indicates low gene density and red indicates high gene density.
A total of 156 CASPL genes identified in the whole genome of P. cablin were distributed on 57 of 63 chromosomes of P. cablin (Figure 4). The number of CASPL genes on each chromosome ranged from zero to six (Table 3). Among them, chromosomes 5, 37, 49, and 50 were the most distributed members, with six CASPL genes; five CASPL genes were distributed on chromosomes 6, 15, 17, 18, 21, 22, 38, 47, 53, and 54, respectively. Four CASPL genes were distributed on chromosomes 4, 35, and 36. There are one to three CASPL genes on another 40 chromosomes, while there is no PatCASPL gene on chromosomes 9, 27, 28, 41, 59, and 60. It is speculated that the number of CASPL members on each chromosome is not related to chromosome size. In addition, no tandem duplication was found in the PatCASPL gene family of P. cablin.

Table 3.
Gene number per chromosome of CASPL gene family proteins in P. cablin.
2.5. Expression Analysis of PatCASPL Gene Family in P. cablin
In order to screen and identify the potential candidate genes of P. cablin in response to continuous cropping obstacles, the root transcriptome database (NCBI accession number PRJNA850618) of different periods (0 h, 6 h, 12 h, 24 h, 48 h, and 96 h) of p-HBA treatment was constructed in the previous study [16]. Cluster heat maps of 57 CASPL gene family transcripts were screened and constructed using the FPKM value of transcriptome data (Figure 5). The results showed that the CASPL genes in response to p-HBA could be divided into two types, namely p-HBA inhibited expression, and p-HBA promoted expression. In the first type, compared with the control (0 h), the expression levels of genes (PatCASPL1A2, PatCASPL1F2, PatCASPL2A2, PatCASPL4A10, PatCASPL4A11, PatCASPL4D9, PatCASPL5A5, PatCASPL5A7, PatCASPL5A10, and PatCASP6) were significantly down-regulated at 6 h, 12 h, and 24 h, while the expression levels of some genes (PatCASPL1A10, PatCASPL1D5, PatCASPL2D4, PatCASPL5C1, PatCASPL5A1, and PatCASPL5A3) were up-regulated at 48 h or 96 h, indicating that p-HBA inhibited the expression of these genes in the early stage, and the inhibitory effect gradually weakened over time or p-HBA consumption. In the second type, p-HBA treatment can promote the expression of CASPL genes such as PatCASPL1A3, PatCASPL1A4, PatCASPL1A5, PatCASPL1C3, PatCASPL1D3, PatCASPL4A5, PatCASPL4B5, PatCASPL4A8, and PatCASPL5A4, although the response time is different. Specifically, the gene expression levels of PatCASPL1D3 and PatCASPL4A8 peaked at 6 h and then decreased. The gene expression levels of PatCASPL5B7, PatCASPL4D3, and PatCASPL1C3 were the highest at the 12 h treatment period. The gene expression levels of PatCASPL4B5, PatCASPL1A4, PatCASPL1A3, PatCASPL1A7, and PatCASPL4A5 were the most significantly up-regulated at 24 h, while the gene expression levels of PatCASPL2D4, PatCASPL5A4, and PatCASPL1A5 were at the maximum at 48 h. The gene expression of PatCASPL4D9, PatCASPL1A2, PatCASPL1F1, PatCASP6, PatCASPL2A4, PatCASPL5A5, PatCASPL2B3, PatCASPL2A3, PatCASPL5A10, PatCASPL5A7, PatCASPL4A10, PatCASPL4A11, PatCASPL1F2, PatCASPL1F3, and PatCASPL1E3 showed a downward trend. The gene expression of PatCASPL4B3 was most significantly down-regulated at 6 h, while the gene expression levels of PatCASPL4D4 and PatCASPL5A12 were significantly down-regulated at 96 h. The above 57 genes were highly expressed in their corresponding periods.
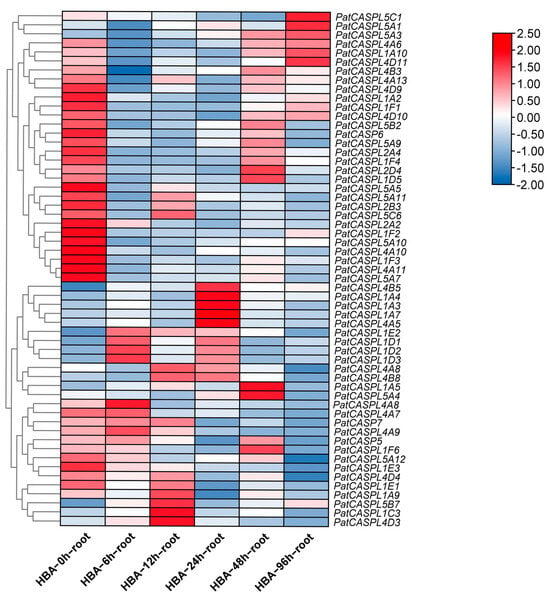
Figure 5.
Expression of PatCASPL gene in P. cablin at different treatment stages of p-HBA.
In the first type, the promoter regions of the significantly down-regulated genes at 6 h, 12 h, and 24 h contained abscisic acid-responsive elements, suggesting that their response to p-HBA may involve ABA signaling. Except PatCASPL5A3, which is distributed on chloroplasts, other genes are distributed on cell membranes, PatCASPL1A2 and PatCASPL1A10 contain MARVEL conserved domains, and other genes contain DUF588 conserved domains. In the second type, the promoter regions of PatCASPL1A9, PatCASPL1E1, PatCASPL5A12, PatCASPL4D4, and PatCASPL4A8, which were down-regulated in expression, contained drought response elements; PatCASPL4A8 was distributed in the nucleus, and the other genes were distributed on the cell membrane. In addition to PatCASPL1E1 containing the MARVEL conserved structure, other genes contained the DUF588 conserved domain. In summary, 34 genes containing the MARVEL conserved domain were more likely to be distributed on the cell membrane and contained a large number of MYB binding sites. It is speculated that they may indirectly affect the structure of the Casparian strip. In addition, the PatCASPL gene family of Pogostemon cablin also contains many other types of cis-acting elements, among which the number of light-responsive elements is the largest. It can be judged that the CASPL family genes may have biological functions on the regulation of circadian rhythm and photomorphogenesis.
The expression of PatCASPLs changed after p-HBA treatment, suggesting that it may play an important role in the response of P. cablin to p-HBA treatment. It can be used as a candidate in gene screening for further resistance research and functional analysis. It is speculated that p-HBA may affect the expression of these genes, thereby affecting the formation and integrity of the Casparian strip, reducing the tolerance of P. cablin to stress, and ultimately leading to continuous cropping obstacles. The specific functions of these genes need to be further verified.
3. Discussion
In this study, 156 CASPL genes were identified via a bioinformatics analysis based on the genome data of P. cablin. The number of PatCASPLs is much larger than those of A. thaliana (39) [21], rice (19) [28], cotton (48) [29], banana (61) [30], and litchi (33) [31]. The number of gene family members among species is affected by the number of genome doublings, tandem repeats, and natural selection [45]. Long-terminal repeat retrotransposons (LTR-RTs) can proliferate rapidly in the host, thus affecting the expression of genes. It has been reported that the transposition and amplification of LTR-RTs is an important factor in the expansion of the plant genome [46,47]. Previous studies have found that the genome of P. cablin had a chromosome doubling event (3.3 million years ago) and an LTR-RT insertion event (1.1 million years ago) [48]. This also explains why the number of PatCASPL family members in P. cablin is far more than that of other species and is consistent with the results that the number of AtCASPL members and PatCASPL members in each cluster is 1:1~1:3.9. The conserved domains of the CASPL gene family are also various in different species. The MARVEL domain is present in the CASPL gene family of A. thaliana, banana, and litchi [21]; however, it has not been reported in rice, indicating that the number and domain of CASPL gene family members in different species are also diverse.
In this research, the cis-elements analysis showed that the PatCASPL gene family has more hormone response and stress response elements, and it is speculated that the gene family may play an important role in regulating growth and development and stress tolerance. Previous studies have found that the expression of TaCASPLs can be induced in response to salt stress, osmotic stress, and calcium ion stress, and their expression is inhibited under low-temperature stress [49]. Moreover, Liu et al. [32] found that the SbCASP-LP1C1 gene was involved in the formation of an extracellular barrier and improved the salt tolerance of sweet sorghum. Similarly, Rushil et al. [50] showed that the OsCASPL1 protein has a certain role in salt stress tolerance. Yang et al. [35] identified a cold-inducible protein ClCASPL gene in watermelon. In addition, AtCASPL4C1, the homologous gene in A. thaliana, plays an important role in cold tolerance [51]. The above results suggest that the PatCASPL gene may also play a key role in abiotic stress responses.
In agricultural practice, the continuous planting of P. cablin on the same land will lead to changes in the soil’s physical and chemical properties [52,53], the microbial community structure, the aggravation of soil diseases, and more serious allelopathic autotoxicity, resulting in serious continuous cropping obstacles [54,55,56,57]. Previous studies have found that p-HBA is the main allelochemical that induces continuous cropping obstacles of P. cablin [16,58]. In this study, 30 down-regulated candidate genes and 27 up-regulated candidate genes were identified by analyzing the transcriptome data of P. cablin roots treated with p-HBA. The down-regulated expression of candidate genes may contribute to the incomplete Casparian strip, and then to the diffusion of the allelochemicals through the incomplete Casparian strip and long-distance transportation in a vascular bundle. These allelochemicals cause plant metabolic disorder, affecting plant growth and development, finally leading to the occurrence of P. cablin continuous cropping obstacles. The up-regulated expression of some genes may partially compensate for the incomplete Casparian strip and slow down the entry of allelochemicals into plant vascular bundles. The functions of these candidate genes need to be further verified.
4. Materials and Methods
4.1. Genome-Wide Identification of CASPL from P. cablin
The Arabidopsis genome data were downloaded from the TAIR website (https://www.arabidopsis.org/ accessed on 24 March 2023.), and the patchouli genome sequence file, protein sequence file, and gene structure annotation file were downloaded from the GSA website (https://ngdc.cncb.ac.cn/gsa/ accessed on 24 April 2023). The amino acid sequences of 39 CASPL proteins in A. thaliana were extracted using the TBTools (v1.120) software [59]. To obtain homologous sequences, the Arabidopsis CASPL gene protein sequences were aligned with the patchouli protein data (e-value < 1× 10−5) using BLASTP [60], followed by dereplication. At the same time, the Pfam (number PF04535) of CASPL was obtained in the InterPro [61] database (www.ebi.ac.uk/interpro accessed on 24 March 2023), and its hidden Markov model (HMM) file was downloaded. The HMM [62] file of CASPL was used as input and compared in the TBtools software to obtain candidate gene IDs and extract candidate gene protein sequences. The amino acid sequences of the candidate proteins obtained via the two alignment methods were merged and the repeat sequences were deleted. The conserved domain of the CASPL gene was further analyzed, the candidate genes with an incomplete or missing CASPL domain were deleted, and the PatCASPL genes were finally obtained. The Expasy [63] website (https://www.expasy.org// accessed on 19 July 2023) was used to predict the number of amino acids and the protein molecular weight of CASPL family members. The subcellular localization of CASPL family proteins was predicted using the Plant-mPLoc [64,65] online software (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc2/ accessed on 11 September 2023) and the YLoc [66] online software (https://abi-services.cs.uni-tuebingen.de/yloc/webloc.cgi accessed on 12 November 2023), and the secondary structure of the CASPL family proteins was predicted and analyzed using the SOPMA [40] online software (https://npsa-prabi.ibcp.fr// accessed on 11 September 2023).
4.2. Genetic Characteristics and Phylogenetic Analysis
The position information of the exons, introns, and UTR of CASPL family genes was obtained from the patchouli genome annotation information (gff) file, and the CASPL gene structure map was drawn using the TBtools software. We used MEME [67] online (http://meme-suite.org/ accessed on 17 May 2023) to analyze the conserved motifs of CASPL proteins. The maximum number and the length of motifs was set at eight and 6–50 amino acids, respectively. The TBtools software was used for a visual analysis. Using the original sequence, the PF04535.15 domain was found in the Pfam [68] database (http://pfam.xfam.org/ accessed on 31 March 2023). The domain information was extracted and the CASPL protein sequence from P. cablin and A. thaliana were compared using the muscle program [69]. The results were compared with the ProtTest to predict the optimal model [70]. The phylogenetic tree of the CASPL protein in P. cablin and A. thaliana was constructed with the MEGA11 software using the maximum likelihood method, and then visualized with the ITOL online tool (itol.embl.de/). The promoter sequences of the CASPL gene family members in P. cablin were extracted with TBtools, and the promoter cis-elements were predicted and analyzed with the PlantCARE [71] online program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ accessed on 8 August 2023).
4.3. Chromosomal Localization Analysis
Based on the patchouli genome annotation file, the length information of 63 chromosomes and the location information of all PatCASPL genes on chromosomes were obtained, followed by visualization. The gene density profile tool of the Tbtools software was used to obtain the gene density information of the PatCASPL gene. And the location information and distance relationship of all patchouli PatCASPL gene family members were marked on chromosomes via the TBtools software.
4.4. Expression Analysis
According to the previous transcriptome study [6,72] of P. cablin roots, the transcriptome data of P. cablin roots at different time points (0 h, 6 h, 12 h, 24 h, 48 h, and 96 h) under p-hydroxybenzoic acid (p-HBA) treatment were obtained. The above raw data have been uploaded to the NCBI (https://www.ncbi.nlm.nih.gov/ accessed on 28 July 2023) sequence read file (SRA) (accession number PRJNA850618) [11]. The transcript expression of CASPL genes were extracted from the above transcriptome data, and were preliminarily screened using the Excel software of Microsoft office 2019. After that, the selected data were further processed with the TBtools (v1.120) software and the clustering heat map was drawn.
5. Conclusions
In summary, in order to explore the biological function of the PatCASPL gene family and its role in continuous cropping obstacles, the members of the PatCASPL family were identified and analyzed at the genome-wide level. The composition, physicochemical properties, evolutionary relationship, and potential biological function of PatCASPL family members were characterized and figured out. These results provide an important theoretical basis for further exploring the biological function of the PatCASPL gene family and its role in continuous cropping obstacles.
Author Contributions
Conceptualization, Y.S. and Y.L. (Ya Liu); methodology, Y.S. and M.Z.U.H.; software, Y.S. and X.L.; data curation, Y.S., Y.L. (Yang Li), and M.Z.U.H.; visualization, Y.S. and J.Y.; writing—original draft preparation, Y.S.; writing—review and editing, Y.S., X.L., Y.L. (Yang Li), M.Z.U.H., J.Y., D.Y. and Y.W.; supervision, Y.W. and Y.L. (Ya Liu); funding acquisition, Y.L. (Ya Liu) and Y.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (Nos. 82304658 and 82260737), Startup Funding from Hainan University (No. KYQD(ZR)23018), and the Key Research and Development Program of Hainan Province (No. ZDYF2021SHFZ075).
Conflicts of Interest
The authors declare no conflict of interest.
References
- He, Y.; Peng, F.; Deng, C.; Xiong, L.; Huang, Z.; Zhang, R.; Liu, M.; Peng, C. Building an octaploid genome and transcriptome of the medicinal plant Pogostemon cablin from Lamiales. Sci. Data 2018, 5, 180274. [Google Scholar] [CrossRef]
- Swamy, M.K.; Sinniah, U.R. Patchouli (Pogostemon cablin Benth.): Botany, agrotechnology and biotechnological aspects. Ind. Crop. Prod. 2016, 87, 161–176. [Google Scholar] [CrossRef]
- Wu, Y.G.; Guo, Q.S.; He, J.C.; Lin, Y.F.; Luo, L.J.; Liu, G.D. Genetic diversity analysis among and within populations of Pogostemon cablin from China with ISSR and SRAP markers. Biochem. Syst. Ecol. 2010, 38, 63–72. [Google Scholar] [CrossRef]
- Xie, B.; Wu, X.F.; Luo, H.T.; Huang, X.L.; Huang, F.; Zhang, Q.Y.; Zhou, X.; Wu, H.Q. Chemical profiling and quality evaluation of Pogostemon cablin Benth by liquid chromatography tandem mass spectrometry combined with multivariate statistical analysis. J. Pharm. Biomed. Anal. 2022, 209, 114526. [Google Scholar] [CrossRef] [PubMed]
- Uh, Y.R.; Jang, C.S. Establishing DNA markers to differentiate agastache rugosa and Pogostemon cablin, which are confusedly used as medicinal herbs, using real-time PCR. Food Sci. Biotechnol. 2023, 32, 239–247. [Google Scholar] [CrossRef]
- Yan, W.; Ye, Z.; Cao, S.; Yao, G.; Yu, J.; Yang, D.; Chen, P.; Zhang, J.; Wu, Y. Transcriptome analysis of two Pogostemon cablin chemotypes reveals genes related to patchouli alcohol biosynthesis. PeerJ 2021, 9, e12025. [Google Scholar] [CrossRef] [PubMed]
- Swamy, M.; Sinniah, U. A comprehensive review on the phytochemical constituents and pharmacological activities of Pogostemon cablin Benth.: An aromatic medicinal plant of industrial importance. Molecules 2015, 20, 8521–8547. [Google Scholar] [CrossRef]
- Yoon, S.C.; Je, I.G.; Cui, X.; Park, H.R.; Khang, D.; Park, J.S.; Kim, S.H.; Shin, T.Y. Anti-allergic and anti-inflammatory effects of aqueous extract of Pogostemon cablin. Int. J. Mol. Med. 2016, 37, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.J.; Xiong, Y.; Zhang, H.Y.; He, M.L. In vitro induction and morphological characteristics of octoploid plants in Pogostemon cablin. Breed. Sci. 2016, 66, 169–174. [Google Scholar] [CrossRef]
- Yan, W.; Liu, X.; Cao, S.; Yu, J.; Zhang, J.; Yao, G.; Yang, H.; Yang, D.; Wu, Y. Molecular basis of Pogostemon cablin responding to continuous cropping obstacles revealed by integrated transcriptomic, miRNA and metabolomic analyses. Ind. Crop. Prod. 2023, 200, 116862. [Google Scholar] [CrossRef]
- Yan, W.; Cao, S.; Wu, Y.; Ye, Z.; Zhang, C.; Yao, G.; Yu, J.; Yang, D.; Zhang, J. Integrated analysis of physiological, mRNA sequencing, and miRNA sequencing data reveals a specific mechanism for the response to continuous cropping obstacles in Pogostemon cablin roots. Front. Plant Sci. 2022, 13, 853110. [Google Scholar] [CrossRef]
- Zhou, X.; Li, C.; Liu, L.; Zhao, J.; Zhang, J.; Cai, Z.; Huang, X. Control of fusarium wilt of Lisianthus by reassembling the microbial community in infested soil through reductive soil disinfestation. Microbiol. Res. 2019, 220, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Xu, J.; Zhang, L.; Cheng, R.; Wei, G.; Su, H.; Yang, J.; Qian, J.; Xu, R.; Chen, S. Rhizospheric microbial communities are driven by Panax ginseng at different growth stages and biocontrol bacteria alleviates replanting mortality. Acta Pharm. Sin. B 2018, 8, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Qin, X.; Tian, X.; Yang, T.; Deng, R.; Huang, J. Effects of continuous cropping of Pinellia Ternata (Thunb.) Breit. on soil physicochemical properties, enzyme activities, microbial communities and functional genes. Chem. Biol. Technol. Agric. 2021, 8, 43. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, Y.G.; Chen, Y.; Zhang, J.F.; Song, X.Q.; Zhu, G.P.; Hu, X.W. Autotoxicity in Pogostemon cablin and their allelochemicals. Rev. Bras. Farmacogn. 2015, 25, 117–123. [Google Scholar] [CrossRef]
- Yan, W.; Cao, S.; Liu, X.; Yao, G.; Yu, J.; Zhang, J.; Bian, T.; Yu, W.; Wu, Y. Combined physiological and transcriptome analysis revealed the response mechanism of Pogostemon cablin roots to p-Hydroxybenzoic Acid. Front. Plant Sci. 2022, 13, 980745. [Google Scholar] [CrossRef] [PubMed]
- Geldner, N. Casparian strips. Curr. Biol. 2013, 23, R1025–R1026. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, I.C.R.; Rojas-Murcia, N.; Geldner, N. The Casparian strip—One ring to bring cell biology to lignification? Curr. Opin. Biotechnol. 2019, 56, 121–129. [Google Scholar] [CrossRef] [PubMed]
- De Bang, T.C.; Lay, K.S.; Scheible, W.R.; Takahashi, H. Small peptide signaling pathways modulating macronutrient utilization in plants. Curr. Opin. Plant Biol. 2017, 39, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Murcia, N.; Hématy, K.; Lee, Y.; Emonet, A.; Ursache, R.; Fujita, S.; De Bellis, D.; Geldner, N. High-order mutants reveal an essential requirement for peroxidases but not laccases in Casparian strip lignification. Proc. Natl. Acad. Sci. USA 2020, 117, 29166–29177. [Google Scholar] [CrossRef]
- Roppolo, D.; Boeckmann, B.; Pfister, A.; Boutet, E.; Rubio, M.C.; Dénervaud-Tendon, V.; Vermeer, J.E.M.; Gheyselinck, J.; Xenarios, I.; Geldner, N. Functional and evolutionary analysis of the CASPARIAN STRIP MEMBRANE DOMAIN PROTEIN Family. Plant Physiol. 2014, 165, 1709–1722. [Google Scholar] [CrossRef] [PubMed]
- Pfister, A.; Barberon, M.; Alassimone, J.; Kalmbach, L.; Lee, Y.; Vermeer, J.E.; Yamazaki, M.; Li, G.; Maurel, C.; Takano, J.; et al. A Receptor-like kinase kutant with absent endodermal diffusion barrier displays selective nutrient homeostasis defects. eLife 2014, 3, e03115. [Google Scholar] [CrossRef] [PubMed]
- Hosmani, P.S.; Kamiya, T.; Danku, J.; Naseer, S.; Geldner, N.; Guerinot, M.L.; Salt, D.E. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc. Natl. Acad. Sci. USA 2013, 110, 14498–14503. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, T.; Borghi, M.; Wang, P.; Danku, J.M.C.; Kalmbach, L.; Hosmani, P.S.; Naseer, S.; Fujiwara, T.; Geldner, N.; Salt, D.E. The MYB36 transcription factor orchestrates Casparian strip formation. Proc. Natl. Acad. Sci. USA 2015, 112, 10533–10538. [Google Scholar] [CrossRef]
- Fujita, S. CASPARIAN STRIP INTEGRITY FACTOR (CIF) family peptides—Regulator of plant extracellular barriers. Peptides 2021, 143, 170599. [Google Scholar] [CrossRef]
- Kim, Y.Y.; Kim, D.Y.; Shim, D.; Song, W.Y.; Lee, J.; Schroeder, J.I.; Kim, S.; Moran, N.; Lee, Y. Expression of the novel wheat gene TM20 confers enhanced cadmium tolerance to bakers’ yeast. J. Biol. Chem. 2008, 283, 15893–15902. [Google Scholar] [CrossRef]
- Barbosa, I.C.R.; De Bellis, D.; Flückiger, I.; Bellani, E.; Grangé-Guerment, M.; Hématy, K.; Geldner, N. Directed growth and fusion of membrane-wall microdomains requires CASP-mediated inhibition and displacement of secretory foci. Nat. Commun. 2023, 14, 1626. [Google Scholar] [CrossRef]
- Roppolo, D.; De Rybel, B.; Tendon, V.D.; Pfister, A.; Alassimone, J.; Vermeer, J.E.M.; Yamazaki, M.; Stierhof, Y.-D.; Beeckman, T.; Geldner, N. A novel protein family mediates Casparian strip formation in the endodermis. Nature 2011, 473, 380–383. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Wang, L.; Pan, Z.; He, S.; Gao, Q.; Chen, B.; Gong, W.; Du, X. Casparian strip membrane domain proteins in Gossypium Arboreum: Genome-wide identification and negative regulation of lateral root growth. BMC Genom. 2020, 21, 340. [Google Scholar] [CrossRef]
- Ji, Y.; Zhang, C.; Li, D.; Lai, Z. Genome-wide identification and analysis of CASP gene family in banana. Subtrop. Agric. Res. 2021, 17, 192–199. [Google Scholar]
- Chen, T.; Liao, X. Identification and analysis of whole genome of Litchi CASP family. Mol. Plant Breed. 2022, 1–10. [Google Scholar]
- Liu, L.; Wei, X.; Yang, Z.; Yuan, F.; Han, G.; Guo, J.; Wang, B. SbCASP-LP1C1 improves salt exclusion by enhancing the root apoplastic barrier. Plant Mol. Biol. 2023, 111, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Fang, P.; Peng, Y.; Zeng, W.; Zhao, X.; Ding, Y.; Zhuang, Z.; Gao, Q.; Ren, B. comparative proteomics of salt-tolerant and salt-sensitive Maize inbred lines to reveal the molecular mechanism of salt tolerance. Int. J. Mol. Sci. 2019, 20, 4725. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Cai, X.; Wu, X.; Karahara, I.; Schreiber, L.; Lin, J. Casparian strip development and its potential function in salt tolerance. Plant Signal. Behav. 2011, 6, 1499–1502. [Google Scholar] [CrossRef]
- Yang, J.; Ding, C.; Xu, B.; Chen, C.; Narsai, R.; Whelan, J.; Hu, Z.; Zhang, M. A Casparian strip domain-like gene, CASPL, negatively alters growth and cold tolerance. Sci. Rep. 2015, 5, 14299. [Google Scholar] [CrossRef]
- Lee, M.; Jeon, H.S.; Kim, S.H.; Chung, J.H.; Roppolo, D.; Lee, H.; Cho, H.J.; Tobimatsu, Y.; Ralph, J.; Park, O.K. Lignin-based barrier restricts pathogens to the infection site and confers resistance in plants. EMBO J. 2019, 38, e101948. [Google Scholar] [CrossRef]
- McGinnis, S.; Madden, T.L. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004, 32, W20–W25. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by Consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Yang, Y.; Chen, M. Genome-wide survey of the amino acid transporter gene family in wheat (Triticum Aestivum L.): Identification, expression analysis and response to abiotic stress. J. Biol. Macromol. 2020, 162, 1372–1387. [Google Scholar] [CrossRef]
- Piegu, B.; Guyot, R.; Picault, N.; Roulin, A.; Sanyal, A.; Kim, H.; Collura, K.; Brar, D.S.; Jackson, S.; Wing, R.A.; et al. Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006, 16, 1262–1269. [Google Scholar] [CrossRef]
- Zhao, M.; Ma, J. Co-evolution of plant LTR-retrotransposons and their host genomes. Protein Cell 2013, 4, 493–501. [Google Scholar] [CrossRef]
- Shen, Y.; Li, W.; Zeng, Y.; Li, Z.; Chen, Y.; Zhang, J.; Zhao, H.; Feng, L.; Ma, D.; Mo, X.; et al. Chromosome-level and haplotype-resolved genome provides insight into the tetraploid hybrid origin of patchouli. Nat. Commun. 2022, 13, 3511. [Google Scholar] [CrossRef]
- Fei, Z. Analysis and Functional Identification of CASP Family Members in Wheat; College of Plant Science and Technology of Huazhong Agricultural University: Wuhan, China, 2021. [Google Scholar]
- Rushil, M. Cloning and in silico analysis of Casparian strip membrane domain protein (CASP) from rice. Madras Agric. J. 2019, 106, 155–159. [Google Scholar] [CrossRef]
- Zhang, W.; Sun, P.; He, Q.; Shu, F.; Deng, H. Transcriptome analysis of near-isogenic line provides novel insights into genes associated with panicle traits regulation in rice. PLoS ONE 2018, 13, e0199077. [Google Scholar] [CrossRef]
- Zhang, J.; He, L.; Wu, Y.; Ma, W.; Chen, H.; Ye, Z. Comparative proteomic analysis of Pogostemon cablin leaves after continuous cropping. Protein Expr. Purif. 2018, 152, 13–22. [Google Scholar] [CrossRef]
- Zeng, J.; Liu, J.; Lu, C.; Ou, X.; Luo, K.; Li, C.; He, M.; Zhang, H.; Yan, H. Intercropping with turmeric or ginger reduce the continuous cropping obstacles that affect Pogostemon cablin (patchouli). Front. Microbiol. 2020, 11, 579719. [Google Scholar] [CrossRef]
- Liu, Q.; Li, K.; Guo, X.; Ma, L.; Guo, Y.; Liu, Z. Developmental characteristics of grapevine seedlings root border cells and their response to p-Hydroxybenzoic acid. Plant Soil 2019, 443, 199–218. [Google Scholar] [CrossRef]
- Tan, G.; Liu, Y.; Peng, S.; Yin, H.; Meng, D.; Tao, J.; Gu, Y.; Li, J.; Yang, S.; Xiao, N.; et al. Soil potentials to resist continuous cropping obstacle: Three field cases. Environ. Res. 2021, 200, 111319. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Du, J.; Li, Y.; Tang, H.; Yin, Z.; Yang, L.; Ding, X. Evolutions and managements of soil microbial community structure drove by continuous cropping. Front. Microbiol. 2022, 13, 839494. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, Z.; Niu, J.; Dang, K.; Zhang, S.; Wang, S.; Wang, Z. Changes in physicochemical properties, enzymatic activities, and the microbial community of soil significantly influence the continuous cropping of Panax quinquefolius L. (American Ginseng). Plant Soil 2021, 463, 427–446. [Google Scholar] [CrossRef]
- Zeeshan Ul Haq, M.; Yu, J.; Yao, G.; Yang, H.; Iqbal, H.A.; Tahir, H.; Cui, H.; Liu, Y.; Wu, Y. A systematic review on the continuous cropping obstacles and control strategies in medicinal plants. Int. J. Mol. Sci. 2023, 24, 12470. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Sardi, G.; De Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A Top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- Xiong, E.; Zheng, C.; Wu, X.; Wang, W. Protein subcellular location: The gap between prediction and experimentation. Plant Mol. Biol. Rep. 2016, 34, 52–61. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Briesemeister, S.; Rahnenführer, J.; Kohlbacher, O. YLoc—An interpretable web server for predicting subcellular localization. Nucleic Acids Res. 2010, 38 (Suppl. S2), W497–W502. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Rombauts, S.; Dehais, P.; Van Montagu, M.; Rouze, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, W.; Lu, C.; He, M.; Yan, H. SMRT Sequencing of full-length transcriptome and gene expression analysis in two chemical types of Pogostemon cablin (Blanco) Benth. PeerJ 2022, 10, e12940. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).