Free Amino Acids and Biogenic Amines Profiling and Variation in Wild and Sub-Endemic Cardueae Species from Sardinia and Corse
Abstract
1. Introduction
2. Results and Discussion
2.1. Phytochemical Analyses
2.1.1. Optimization of the Extraction Procedure
2.1.2. DEEMM Derivatization-Targeted Characterization by HPLC-MS/MS
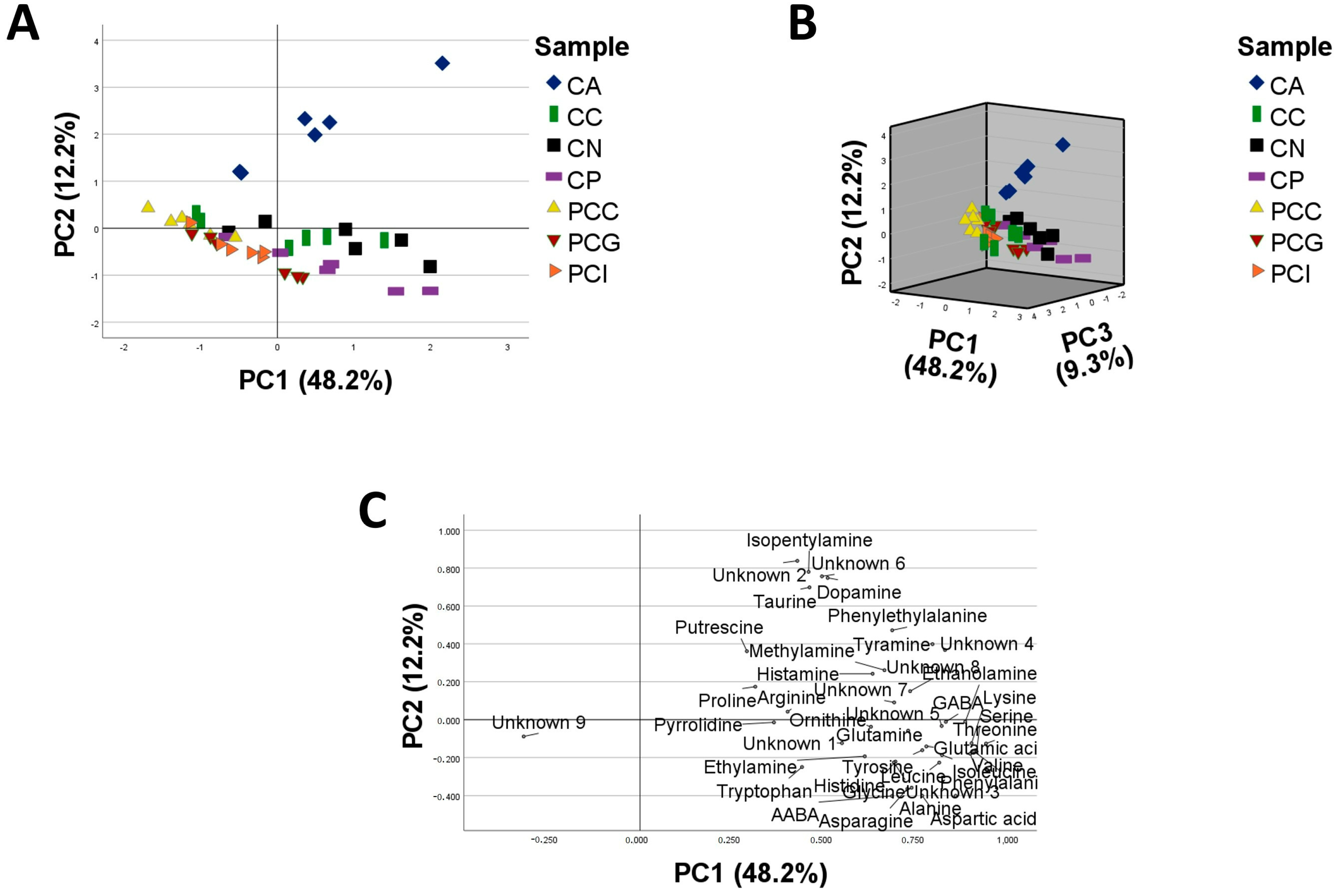
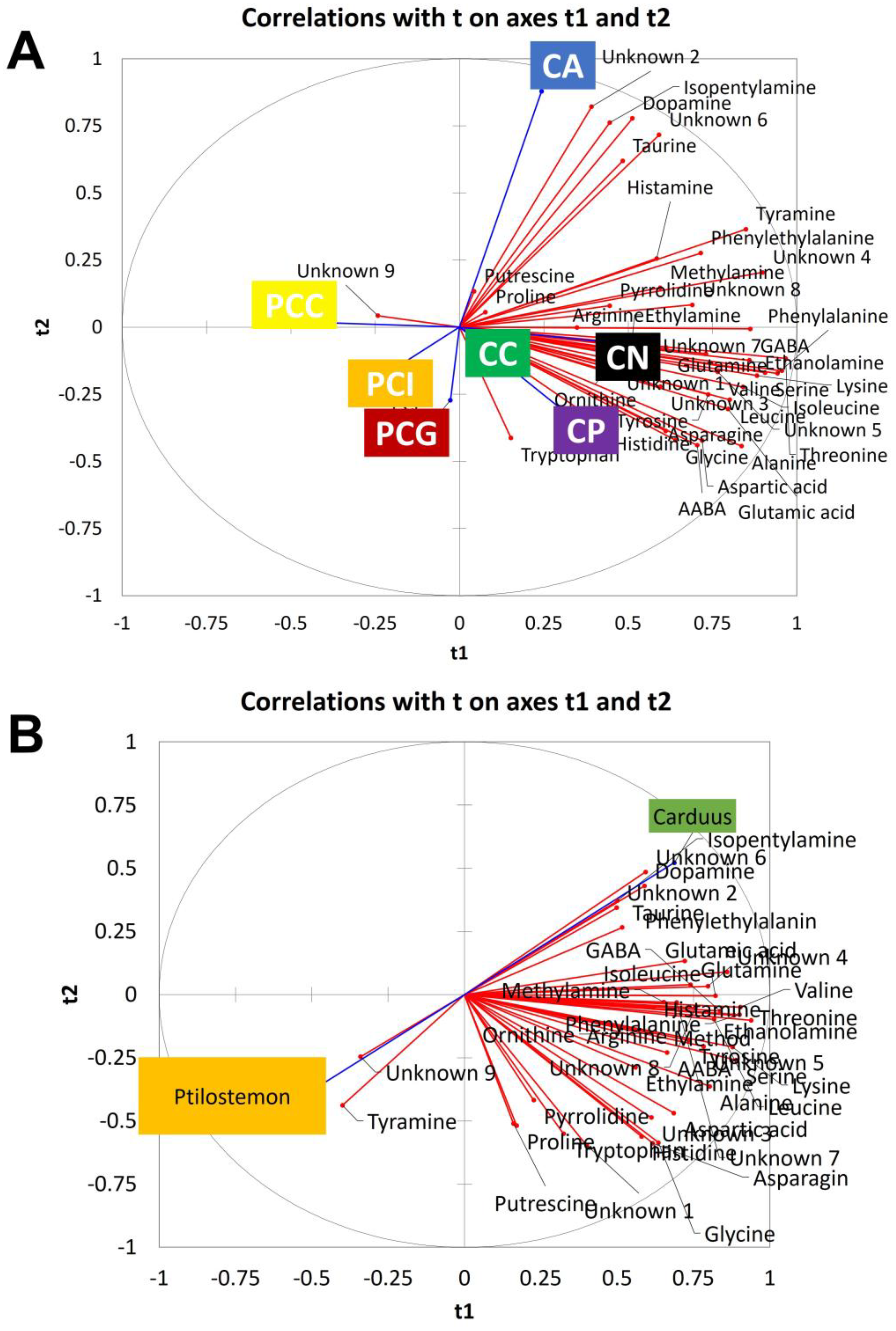
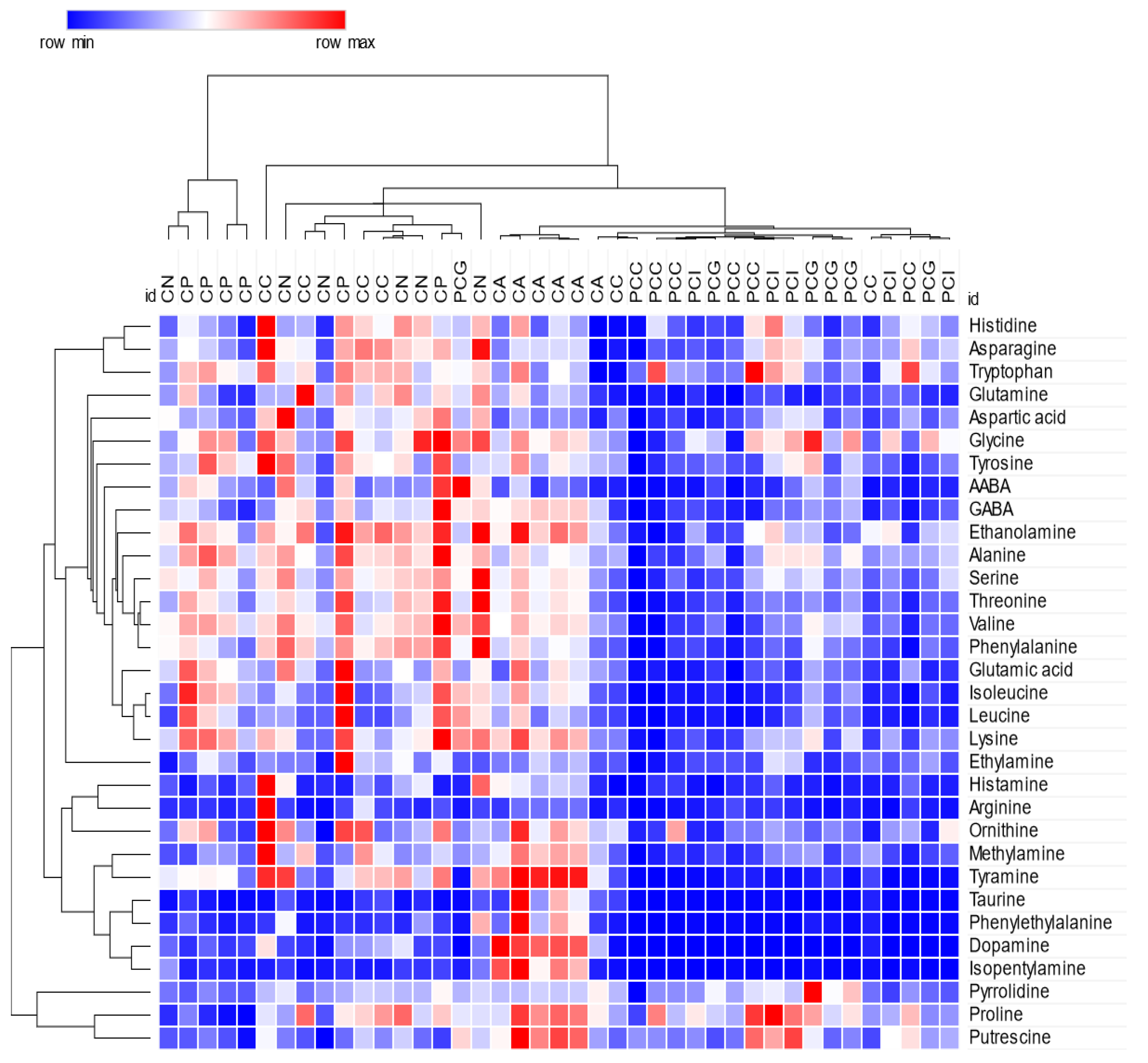
2.2. Statistical Analysis
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Preparation of the Extracts and Pre-Column Derivatization
3.4. HPLC-MS/MS Analysis
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pignatti, S. La Flora d’Italia; Edagricole: Milano, Italy, 1982. [Google Scholar]
- Tutin, T.G.; Heywood, V.H.; Burges, N.A.; Valentine, D.H.; Walters, S.M.; Webb, D.A. Flora Europaea; Cambridge University Press: Cambridge, UK, 1968. [Google Scholar]
- Barres, L.; Sanmartín, I.; Anderson, C.L.; Susanna, A.; Buerki, S.; Galbany-Casals, M.; Vilatersana, R. Reconstructing the evolution and biogeographic history of tribe Cardueae (compositae). Am. J. Bot. 2013, 100, 867–882. [Google Scholar] [CrossRef]
- Ackerfield, J.; Susanna, A.; Funk, V.; Kelch, D.; Park, D.S.; Thornhill, A.H.; Yildiz, B.; Arabaci, T.; Dirmenci, T. A prickly puzzle: Generic delimitations in the Carduus-Cirsium group (Compositae: Cardueae: Carduinae). Taxon 2020, 69, 715–738. [Google Scholar] [CrossRef]
- Mattalia, G.; Sõukand, R.; Corvo, P.; Pieroni, A. Wild food thistle gathering and pastoralism: An inextricable link in the biocultural landscape of Barbagia, central Sardinia (Italy). Sustain. 2020, 12, 5105. [Google Scholar] [CrossRef]
- Guarrera, P.M.; Savo, V. Wild food plants used in traditional vegetable mixtures in Italy. J. Ethnopharmacol. 2016, 185, 202–234. [Google Scholar] [CrossRef]
- Esteban Hernandez Bermejo, J.; Delucchi, G.; Charra, G.; Pochettino, M.L.; Hurrell, J.A. “Cardos” of two worlds: Transfer and resignification of the uses of thistles from the Iberian Peninsula to Argentina. Ethnobiol. Conserv. 2019, 8, 1–11. [Google Scholar] [CrossRef]
- Marengo, A.; Fumagalli, M.; Sanna, C.; Maxia, A.; Piazza, S.; Cagliero, C.; Rubiolo, P.; Sangiovanni, E.; Dell’Agli, M. The hydro-alcoholic extracts of Sardinian wild thistles (Onopordum spp.) inhibit TNFα-induced IL-8 secretion and NF-κB pathway in human gastric epithelial AGS cells. J. Ethnopharmacol. 2018, 210, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Kurt, A.; Ozcan, M.; Colak, N.; Ozogul, Y.; Glew, R.; Ozogul, F.; Ayaz, F.A. Fatty acids of oil and antioxidant capacity of phenolics from fruits of 11 Cardueae (Carduoideae, Asteraceae) taxa from northeast Anatolia (Turkey). Bot. Serbica 2019, 43, 31–45. [Google Scholar] [CrossRef]
- Marengo, A.; Maxia, A.; Sanna, C.; Mandrone, M.; Bertea, C.M.; Bicchi, C.; Sgorbini, B.; Cagliero, C.; Rubiolo, P. Intra-specific variation in the little-known Mediterranean plant Ptilostemon casabonae (L.) Greuter analysed through phytochemical and biomolecular markers. Phytochemistry 2019, 161, 21–27. [Google Scholar] [CrossRef]
- Marengo, A.; Maxia, A.; Sanna, C.; Bertea, C.M.; Bicchi, C.; Ballero, M.; Cagliero, C.; Rubiolo, P. Characterization of four wild edible Carduus species from the Mediterranean region via phytochemical and biomolecular analyses. Food Res. Int. 2017, 100, 822–831. [Google Scholar] [CrossRef]
- Atzei, A.D. Le Piante Nella Tradizione Popolare Della Sardegna; Carlo Delfino Editore: Sassari, Italy, 2003. [Google Scholar]
- Paulis, G. I nomi popolari delle piante in Sardegna. Etimologia storia e tradizioni; Carlo Delfino Editore: Sassari, Italy, 1992. [Google Scholar]
- Marengo, A.; Fenu, G.; Gennai, M.; Cogoni, D.; Fois, M.; Bacchetta, G. Schede per una Lista Rossa della Flora vascolare e crittogamica italiana. Ptilostemon casabonae (L.). Greuter. Inf. Bot. Ital. 2015, 47, 245–289. [Google Scholar]
- Sanna, C.; Maxia, A.; Fenu, G.; Loi, M.C. So uncommon and so singular, but underexplored: An updated overview on ethnobotanical uses, biological properties and phytoconstituents of sardinian endemic plants. Plants 2020, 9, 958. [Google Scholar] [CrossRef]
- Fois, M.; Farris, E.; Calvia, G.; Campus, G.; Fenu, G.; Porceddu, M.; Bacchetta, G. The Endemic Vascular Flora of Sardinia: A Dynamic Checklist with an Overview of Biogeography and Conservation Status. Plants 2022, 11, 601. [Google Scholar] [CrossRef]
- Kozyra, M.; Komsta, Ł.; Wojtanowski, K. Analysis of phenolic compounds and antioxidant activity of methanolic extracts from inflorescences of Carduus sp. Phytochem. Lett. 2019, 31, 256–262. [Google Scholar] [CrossRef]
- Al-Shammari, L.A.; Hassan, W.H.B.; Al-Youssef, H.M. Phytochemical and biological studies of Carduus pycnocephalus L. J. Saudi Chem. Soc. 2015, 19, 410–416. [Google Scholar] [CrossRef]
- de Falco, B.; Incerti, G.; Pepe, R.; Amato, M.; Lanzotti, V. Metabolomic Fingerprinting of Romaneschi Globe Artichokes by NMR Spectroscopy and Multivariate Data Analysis. Phytochem. Anal. 2016, 27, 304–314. [Google Scholar] [CrossRef]
- Rotaru, L.T.; Varut, R. Carduus Acanthoides Folium Tincture: Chemical Composition, Antibacterial Activity and Synergistic / Antagonist Effect in Combination With Antibiotics. J. Sci. Arts 2020, 20, 467–474. [Google Scholar]
- Albergamo, A.; Rotondo, A.; Salvo, A.; Pellizzeri, V.; Bua, D.G.; Maggio, A.; Cicero, N.; Dugo, G. Metabolite and mineral profiling of “Violetto di Niscemi” and “Spinoso di Menfi” globe artichokes by1H-NMR and ICP-MS. Nat. Prod. Res. 2017, 31, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Maciel, L.S.; Marengo, A.; Rubiolo, P.; Leito, I.; Herodes, K. Derivatization-targeted analysis of amino compounds in plant extracts in neutral loss acquisition mode by liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2021, 1656, 462555. [Google Scholar] [CrossRef]
- Liu, P.; Weng, R.; Sheng, X.; Wang, X.; Zhang, W.; Qian, Y.; Qiu, J. Profiling of organosulfur compounds and amino acids in garlic from different regions of China. Food Chem. 2020, 305, 125499. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, A.A.G.; Wrobel, K.; Escobosa, A.R.C.; Elguera, J.C.T.; Garay-Sevilla, M.E.; Wrobel, K. Determination of putrescine, cadaverine, spermidine and spermine in different chemical matrices by high performance liquid chromatography-electrospray ionization-ion trap tandem mass spectrometry (HPLC-ESI-ITMS/MS). J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2015, 1002, 176–184. [Google Scholar] [CrossRef]
- Wu, Y.J.; Cai, X.Y.; Jiang, S.H. Simultaneous identification and determination of biogenic amines in Pueraria lobata by ultra-high performance liquid chromatography. SN Appl. Sci. 2020, 2, 1–8. [Google Scholar] [CrossRef]
- Priscila del Campo, C.; Garde-Cerdán, T.; Sánchez, A.M.; Maggi, L.; Carmona, M.; Alonso, G.L. Determination of free amino acids and ammonium ion in saffron (Crocus sativus L.) from different geographical origins. Food Chem. 2009, 114, 1542–1548. [Google Scholar] [CrossRef]
- Redruello, B.; Ladero, V.; Cuesta, I.; Álvarez-Buylla, J.R.; Martín, M.C.; Fernández, M.; Alvarez, M.A. A fast, reliable, ultra high performance liquid chromatography method for the simultaneous determination of amino acids, biogenic amines and ammonium ions in cheese, using diethyl ethoxymethylenemalonate as a derivatising agent. Food Chem. 2013, 139, 1029–1035. [Google Scholar] [CrossRef]
- Fernández, M.; Linares, D.M.; Del Río, B.; Ladero, V.; Alvarez, M.A. HPLC quantification of biogenic amines in cheeses: Correlation with PCR-detection of tyramine-producing microorganisms. J. Dairy Res. 2007, 74, 276–282. [Google Scholar] [CrossRef]
- Acquadro, S.; Appleton, S.; Marengo, A.; Bicchi, C.; Sgorbini, B.; Mandrone, M.; Gai, F.; Peiretti, P.G.; Cagliero, C.; Rubiolo, P. Grapevine green pruning residues as a promising and sustainable source of bioactive phenolic compounds. Molecules 2020, 25, 464. [Google Scholar] [CrossRef]
- Benedetti, B.; Caponigro, V.; Ardini, F. Experimental Design Step by Step: A Practical Guide for Beginners. Crit. Rev. Anal. Chem. 2020, 52, 1015–1028. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Xiang, Z.; Li, H.; Zhao, H.; Lin, Y.; Pan, W.; Lei, B. Free amino acids, biogenic amines, and ammonium profiling in tobacco from different geographical origins using microwave-assisted extraction followed by ultra high performance liquid chromatography. J. Sep. Sci. 2017, 40, 4571–4582. [Google Scholar] [CrossRef] [PubMed]
- McCusker, S.; Buff, P.R.; Yu, Z.; Fascetti, A.J. Amino acid content of selected plant, algae and insect species: A search for alternative protein sources for use in pet foods. J. Nutr. Sci. 2014, 3, 1–5. [Google Scholar] [CrossRef]
- Redruello, B.; Ladero, V.; del Rio, B.; Fernández, M.; Martin, M.C.; Alvarez, M.A. A UHPLC method for the simultaneous analysis of biogenic amines, amino acids and ammonium ions in beer. Food Chem. 2017, 217, 117–124. [Google Scholar] [CrossRef]
- Bouchereau, A.; Guénot, P.; Larher, F. Analysis of amines in plant materials. J. Chromatogr. B Biomed. Sci. Appl. 2000, 747, 49–67. [Google Scholar] [CrossRef]
- Thevenet, D.; Pastor, V.; Baccelli, I.; Balmer, A.; Vallat, A.; Neier, R.; Glauser, G.; Mauch-Mani, B. The priming molecule β-aminobutyric acid is naturally present in plants and is induced by stress. New Phytol. 2017, 213, 552–559. [Google Scholar] [CrossRef]
- Liu, Q.; Gao, T.; Liu, W.; Liu, Y.; Zhao, Y.; Liu, Y.; Li, W.; Ding, K.; Ma, F.; Li, C. Functions of dopamine in plants: A review. Plant Signal. Behav. 2020, 15, 1827782. [Google Scholar] [CrossRef]
- Boonen, J.; Bronselaer, A.; Nielandt, J.; Veryser, L.; De Tré, G.; De Spiegeleer, B. Alkamid database: Chemistry, occurrence and functionality of plant N-alkylamides. J. Ethnopharmacol. 2012, 142, 563–590. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Sharma, A.; Kohli, S.K.; Yadav, P.; Bali, S.; Bakshi, P.; Parihar, R.D.; Yuan, H.; Yan, D.; He, Y.; et al. Amino acids distribution in economical important plants: A review. Biotechnol. Res. Innov. 2019, 3, 197–207. [Google Scholar] [CrossRef]
- Qiu, X.; Reynolds, R.; Johanningsmeier, S.; Truong, V. Den Determination of free amino acids in five commercial sweetpotato cultivars by hydrophilic interaction liquid chromatography-mass spectrometry. J. Food Compos. Anal. 2020, 92, 103522. [Google Scholar] [CrossRef]
- Wistaff, E.A.; Beller, S.; Schmid, A.; Neville, J.J.; Nietner, T. Chemometric analysis of amino acid profiles for detection of fruit juice adulterations—Application to verify authenticity of blood orange juice. Food Chem. 2021, 343, 128452. [Google Scholar] [CrossRef]
- Lähdesmäki, P. Determination of taurine and other acidic amino acids in plants. Phytochemistry 1986, 25, 2409–2411. [Google Scholar] [CrossRef]
- Kotzamanis, Y.; Tsironi, T.; Brezas, A.; Grigorakis, K.; Ilia, V.; Vatsos, I.; Romano, N.; van Eys, J.; Kumar, V. High taurine supplementation in plant protein-based diets improves growth and organoleptic characteristics of European seabass (Dicentrarchus labrax). Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Violi, J.P.; Bishop, D.P.; Padula, M.P.; Steele, J.R.; Rodgers, K.J. Considerations for amino acid analysis by liquid chromatography-tandem mass spectrometry: A tutorial review. TrAC Trends Anal. Chem. 2020, 131, 116018. [Google Scholar] [CrossRef]
- Farrés, M.; Platikanov, S.; Tsakovski, S.; Tauler, R. Comparison of the variable importance in projection (VIP) and of the selectivity ratio (SR) methods for variable selection and interpretation. J. Chemom. 2015, 29, 528–536. [Google Scholar] [CrossRef]
- Carratù, B.; Boniglia, C.; Giammarioli, S.; Mosca, M.; Sanzini, E. Free amino acids in botanicals and botanical preparations. J. Food Sci. 2008, 73, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Kahsay, B.N.; Ziegler, J.; Imming, P.; Gebre-Mariam, T.; Neubert, R.H.H.; Moeller, L. Free amino acid contents of selected Ethiopian plant and fungi species: A search for alternative natural free amino acid sources for cosmeceutical applications. Amino Acids 2021, 53, 1105–1122. [Google Scholar] [CrossRef]
- Sánchez-Pérez, S.; Comas-Basté, O.; Rabell-González, J.; Veciana-Nogués, M.T.; Latorre-Moratalla, M.L.; Vidal-Carou, M.C. Biogenic amines in plant-origin foods: Are they frequently underestimated in low-histamine diets? Foods 2018, 7, 205. [Google Scholar] [CrossRef] [PubMed]
- Yılmaz, C.; Özdemir, F.; Gökmen, V. Investigation of free amino acids, bioactive and neuroactive compounds in different types of tea and effect of black tea processing. Lwt 2020, 117, 108655. [Google Scholar] [CrossRef]
- Leardi, R.; Melzi, C.; Polotti, G. CAT (Chemometric Agile Tool). Available online: Http://Gruppochemiometria.It/Index.Php/Software; http://gruppochemiometria.it/index.php/software (accessed on 20 December 2022).
- Broad Institute. Morpheus. Available online: https://software.broadinstitute.org/morpheus (accessed on 20 December 2022).
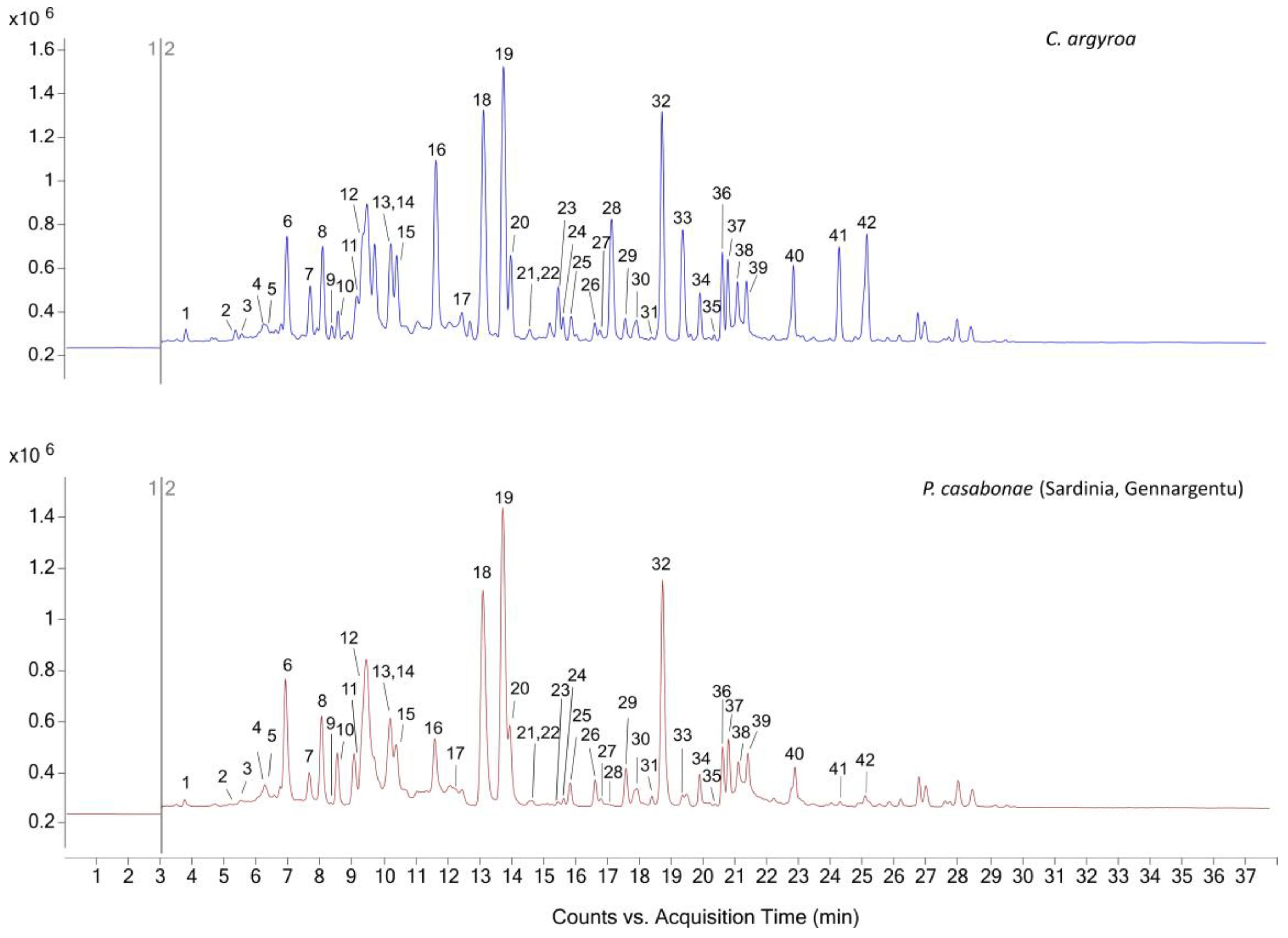
| N° | Retention Time (min) | ESI+ Protonated Compound (m/z) | Derivative Nominal Mass (g/mol) | Nominal Mass (g/mol) | Compound Name | Reference |
|---|---|---|---|---|---|---|
| 1 | 3.78 | 326 | 325 | 155 | Histidine | [31] |
| 2 | 5.35 | 296 | 295 | 125 | Taurine | [32] |
| 3 | 5.54 | 282 | 281 | 111 | Histamine | [33] |
| 4 | 6.24 | 345 | 344 | 174 | Arginine | [31] |
| 5 | 6.32 | 365 | 364/342 1 | 194/172 1 | Unknown 1 | |
| 6 | 6.97 | 303 | 302 | 132 | Asparagine | [31] |
| 7 | 7.70 | 317 | 316 | 146 | Glutamine | [31] |
| 8 | 8.10 | 276 | 275 | 105 | Serine | [31] |
| 9 | 8.40 | 301 | 300/278 1 | 130/108 1 | Unknown 2 | |
| 10 | 8.59 | 259 | 258/236 1 | 88/66 1 | Unknown 3 | |
| 11 | 9.20 | 304 | 303 | 133 | Aspartic acid | [31] |
| 12 | 9.37 | 232 | 231 | 61 | Ethanolamine | |
| 13 | 10.24 | 246 | 245 | 75 | Glycine | [31] |
| 14 | 10.28 | 318 | 317 | 147 | Glutamic acid | [31] |
| 15 | 10.45 | 290 | 289 | 119 | Threonine | [31] |
| 16 | 11.68 | 260 | 259/237 1 | 89/671 | Unknown 4 | |
| 17 | 12.51 | 202 | 201 | 31 | Methylamine | [34] |
| 18 | 13.20 | 274 | 273 | 103 | γ-aminobutyric acid (GABA) | [31] |
| 19 | 13.83 | 260 | 259 | 89 | Alanine | [31] |
| 20 | 14.07 | 286 | 285 | 115 | Proline | [31] |
| 21 | 14.66 | 274 | 273/251 1 | 103/811 | Unknown 5 | |
| 22 | 14.70 | 274 | 273 | 103 | β-aminobutyric acid (BABA) | [35] |
| 23 | 15.57 | 288 | 287/265 1 | 117/95 1 | Unknown 6 | |
| 24 | 15.73 | 440 | 439 | 269 | Unknown 7 | |
| 25 | 15.97 | 352 | 351 | 181 | Tyrosine | [31] |
| 26 | 16.73 | 274 | 273 | 103 | α-aminobutyric acid (AABA) | [35] |
| 27 | 16.89 | 216 | 215 | 45 | Ethylamine | [34] |
| 28 | 17.26 | 324 | 323 | 153 | Dopamine | [36] |
| 29 | 17.70 | 242 | 241 | 71 | Pyrrolidine | [37] |
| 30 | 18.06 | 288 | 287/265 1 | 117/95 1 | Unknown 8 | |
| 31 | 18.51 | 301 | 300/278 1 | 130/108 1 | Unknown 9 | |
| 32 | 18.86 | 288 | 287 | 117 | Valine | [31] |
| 33 | 19.47 | 308 | 307 | 137 | Tyramine | [31] |
| 34 | 20.05 | 375 | 374 | 204 | Tryptophan | [31] |
| 35 | 20.72 | 4272 | 472 | 132 | Ornithine | [31] |
| 36 | 20.76 | 336 | 335 | 165 | Phenylalanine | [31] |
| 37 | 20.93 | 302 | 301 | 131 | Isoleucine | [31] |
| 38 | 21.24 | 302 | 301 | 131 | Leucine | [31] |
| 39 | 21.54 | 441 2/509 3 | 440 | 270 | Lysine | [31] |
| 40 | 23.02 | 383 | 382 | 212 | Putrescine | [31] |
| 41 | 24.47 | 292 | 291 | 121 | Phenylethylamine | [31] |
| 42 | 25.35 | 258 | 257 | 87 | Isopentylamine | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marengo, A.; Maciel, L.S.; Cagliero, C.; Rubiolo, P.; Herodes, K. Free Amino Acids and Biogenic Amines Profiling and Variation in Wild and Sub-Endemic Cardueae Species from Sardinia and Corse. Plants 2023, 12, 319. https://doi.org/10.3390/plants12020319
Marengo A, Maciel LS, Cagliero C, Rubiolo P, Herodes K. Free Amino Acids and Biogenic Amines Profiling and Variation in Wild and Sub-Endemic Cardueae Species from Sardinia and Corse. Plants. 2023; 12(2):319. https://doi.org/10.3390/plants12020319
Chicago/Turabian StyleMarengo, Arianna, Larissa Silva Maciel, Cecilia Cagliero, Patrizia Rubiolo, and Koit Herodes. 2023. "Free Amino Acids and Biogenic Amines Profiling and Variation in Wild and Sub-Endemic Cardueae Species from Sardinia and Corse" Plants 12, no. 2: 319. https://doi.org/10.3390/plants12020319
APA StyleMarengo, A., Maciel, L. S., Cagliero, C., Rubiolo, P., & Herodes, K. (2023). Free Amino Acids and Biogenic Amines Profiling and Variation in Wild and Sub-Endemic Cardueae Species from Sardinia and Corse. Plants, 12(2), 319. https://doi.org/10.3390/plants12020319








