Abstract
Cannabis (Cannabis sativa L.) is one of the earliest cultivated crops, valued for producing a broad spectrum of compounds used in medicinal products and being a source of food and fibre. Despite the availability of its genome sequences, few studies explore the molecular mechanisms involved in pathogen defense, and the underlying biological pathways are poorly defined in places. Here, we provide an overview of Cannabis defence responses against common pathogens, such as Golovinomyces spp., Fusarium spp., Botrytis cinerea and Pythium spp. For each of these pathogens, after a summary of their characteristics and symptoms, we explore studies identifying genes involved in Cannabis resistance mechanisms. Many studies focus on the potential involvement of disease-resistance genes, while others refer to other plants however whose results may be of use for Cannabis research. Omics investigations allowing the identification of candidate defence genes are highlighted, and genome editing approaches to generate resistant Cannabis species based on CRISPR/Cas9 technology are discussed. According to the emerging results, a potential defence model including both immune and defence mechanisms in Cannabis plant–pathogen interactions is finally proposed. To our knowledge, this is the first review of the molecular mechanisms underlying pathogen resistance in Cannabis.
1. Introduction
Cannabis (Cannabis sativa L.) is a dicotyledonous angiosperm originating from Central Asia but is cultivated across many parts of the world due to its ability to grow in a wide range of habitats and environmental conditions [1].
Cannabis belongs to the Cannabaceae family and is considered one of the earliest cultivated crops, being of particular interest due to its multiple uses. Cannabinoids are responsible for the pharmacological and psychoactive properties of this crop, and these therapeutic characteristics have drawn the attention of researchers from all over the world. Additionally, hemp, a Cannabis variety containing less than 0.3% of tetrahydrocannabinol (THC), is cultivated for biomass and fibre, which constitute feedstock for industrial uses. Conversely, medicinal Cannabis contains a greater amount of THC, which has been increasing in recent years, reaching 17–28% of the dry weight in some varieties [2], or even exceeding 30% in others [3].
Among the ~130 secondary metabolites identified in Cannabis [4], THC, along with cannabidiol (CBD), constitute the most relevant compounds produced by this crop and are the main focus of Cannabis breeding programs.
Breeding efforts to produce Cannabis with unique fragrance and flavour characteristics are also of interest. Consequently, the profile of terpenoids, which are highly abundant and largely responsible for the characteristic aroma of Cannabis, is of importance, with isoprenes, monoterpenes, and sesquiterpenes being the predominant classes [5,6].
Due to the legislation regulating Cannabis and related breeding programs, research into the cannabinoid biosynthetic pathway is underrepresented, and it has not been sufficiently characterised, especially at the molecular level [5,7]. Many other major crops have already been widely investigated from this perspective, especially after the advent of Next Generation Sequencing (NGS) technologies [7]. However, the recent modifications in legislation and less stringent regulations [8], as well as the availability of the Cannabis genomic sequence [9], have broadened research in this crop, with the aim also to improve its biomass quality, in the context of sustainable agriculture [10,11].
These legislative changes have also resulted in increased Cannabis production and, with it, a growth of the incidence and severity of crop pathogens, along with the detection of previously unreported diseases. Among emerging pathogens of Cannabis recently reported there are Botrytis cinerea [12,13], Fusarium spp. [14,15], Pythium [16,17] Golovinomyces spp. [12,13], and Hop latent viroid [18], where hop (Humulus lupulus) is a member of Cannabaceae and is closely related to Cannabis [19]. These pathogens can be grouped according to the tissues they infect: root and crown (Fusarium oxysporum, Fusarium proliferatum, Fusarium solani, Pythium myriotylum, Pythium dissotocum, Pythium aphanidermatum), leaves (Golovinomyces spp.), buds (Hop latent viroid) [12]. Botrytis cinerea is often classified as a postharvest pathogen and can attack Cannabis seeds, leaves, and stalks [12]. Fusarium and Pythium species are the most destructive root pathogens, especially when the infection occurs during vegetative growth. Crop losses resulting from the attack of these two pathogens can reach 30% of the total yield [12]. Botryis and Fusarium species also are harmful, as well as other fungi, such as Golovinomyces species, causing powdery mildew (PM, a common term for several taxa of plant pathogenic fungi), and colonizing foliar and flower tissues through the production of spores. Furthermore, extensive infection by fungi such as Fusarium can lead to mycotoxin accumulation in the tissues, potentially harmful to human health [20]. Hop latent viroid leads to malformation of buds and can infect other parts of the crop [18].
The above-reported fungi, oomycetes and the mentioned viroid have been investigated in Cannabis, as well as in several other crops, but little is known about infection within the seed, even though there are harmful pathogens, such as Alternaria, which can start their attack in developing seedlings [12]. The lack of significant research results on Cannabis bacteria pathogen defence mechanisms has also been underlined [21].
On the other hand, research into the characterization and use of biocontrol agents has consistently improved in recent years [22]. The use of synthetic fungicides to control fungal diseases has limitations due to toxicological risks, and it is necessary to replace them with safer means, for human health and with reduced environmental risks. Omics methods and their applications in the biocontrol field were recently reviewed by Massart et al. [23]. A better understanding of the molecular mechanisms underlying pathogen plant resistance can only have positive effects in this field of research.
Many of the above-reported pathogens have been identified using methods based on Polymerase Chain Reaction (PCR) of parts of rDNA, such as Internal Transcribed Spacer (ITS) and Inter Generic Spacer (IGS) regions [24]. However, for Golovinomyces and Botrytis, additional molecular markers were necessary to differentiate between species [25,26].
Few omics studies have explored the molecular mechanisms involved in Cannabis pathogen defence, and the underlying biological pathways need further investigation. As Cannabis contains many lipophilic cannabinoids and terpenoids, along with numerous other metabolites, the metabolic and genetic analyses of this crop are challenging [27]. A detailed analysis of the mechanisms involved in pathogen defence, at both the chemical and molecular level, therefore requires the use of innovative and advanced approaches.
Here, we provide an overview of Cannabis defence responses against some relevant pathogens, focusing on Golovinomyces spp., Fusarium spp., Botrytis cinerea and Pythium. First, we illustrate a summary of the most important molecular mechanisms of resistance, including those associated with pattern-triggered immunity in Cannabis, as well as the subsequent effector-triggered response activated by disease resistance genes (R genes). Secondly, for each of the previously reported pathogens, significant results of studies aimed to identify and describe genes involved in Cannabis defence mechanisms are reported. Many studies focus on the potential involvement of disease-resistance genes, and others draw comparisons to better-studied plants. Studies based on the use of omics science, allowing the identification of Cannabis candidate resistance genes, are also highlighted. They provide a starting point for genome editing approaches to generate disease-resistant crops, which are finally discussed. According to the emerging results of these studies, a potential defence model including both immune and defence mechanisms in Cannabis plant–pathogen interactions is proposed.
Abbreviations used throughout the manuscript are listed in Table 1.

Table 1.
List of abbreviations used in this manuscript.
2. Overview of Cannabis Resistance Genes to Pathogens
Cannabis includes genotypes whose origins are geographically very different [28], and this genetic diversity leads us to believe the existence of naturally occurring genotypes characterised by resistance to specific pathogens. Indeed, among 12 Cannabis genotypes evaluated, it was found that seven displayed partial or complete resistance to PM [29]. Furthermore, a recent study provided insight on the variability of Cannabis cultivars on disease resistance and cannabinoid accumulation over the course of crop maturation [30]. Here, PM resistance was shown for ‘FL 58’ cultivar, on which PM was never observed, as well as ‘RN13a’, ‘Otto II’, and ‘AC/DC’, cultivars, showing very low levels of PM disease.
Studies on other crop species have investigated the molecular mechanisms of resistance to Fusarium and PM [31,32], providing insights for further research on disease resistance responses in Cannabis. Conversely, the search for Cannabis resistance traits to viral pathogens did not yield answers so quickly [33].
Several studies focused on non-host resistance (NHR), a resistance of plant species against all non-adapted pathogens, which is considered the most durable and efficient immune system of plants, as described in the review by Oh and Choi [34]. Most non-adapted pathogen attacks are stopped by an innate defence response based on the recognition of pathogen-associated molecular patterns (PAMPs) by the plant pattern recognition receptors (PRRs), which activates PAMP-triggered immunity (PTI), also induced by reactive oxygen species (ROS) production and mitogen-activated protein kinase (MAPK) [35]. Specific PAMPs, harpin and flg22, were analyzed to study the response to Pythium in Cannabis [36]. Results showed that harpin-enhanced hemp seedlings resistant to Pythium aphanidermatum, while flg22 did not contribute to the defence mechanism against P. aphanidermatum. The lack of comprehensive experimental evidence supporting the recognition of PAMPs in Cannabis opens a field of future research.
The salicylic acid (SA) or the jasmonic acid (JA)/ethylene (ET) signalling pathways, which are known to have an antagonistic interaction [37], are also involved in the activation of disease resistance mechanisms. SA is involved in several key components of plant defence through complex networks, generally activated by biotrophic pathogens, and JA/ET signalling pathways are usually required for the activation of plant defence against necrotrophic pathogens [37].
However, specialised pathogens can suppress PTI responses through effector proteins, which can, in turn, activate subsequent defence responses called effector-triggered immunity (ETI) in plants with immunity to a specialised pathogen. ETI is also activated by SA or JA/ET pathway [38]. An ETI response is generally able to control specific pathogen attacks [39]. The majority of disease resistance genes in plants encode the conserved nucleotide binding site-leucine-rich repeat (NBS-LRR) disease resistance proteins [40,41], which can identify specific effectors to trigger ETI [38,42].
Studies on Wall-Associated receptor Kinases (WAKs) and WAK-like (WAKLs) genes have underlined their role in pathogen resistance across a wide range of plants [43]. The Arabidopsis WAKL22 gene is the homolog of Cannabis WAK7 and was shown to be responsible for dominant resistance against several Fusarium strains [44]. The cotton WAK18 and WAK29 (homolog of CsWAK4 and CsWAK7, respectively), specifically expressed in flowers, showed pathogen resistance characteristics [45]. The Juglans regia WAK9, the homolog of CsWAK1, has been demonstrated to be involved in pathogen response [46]. A recent analysis of the WAK gene family in Cannabis sativa investigated some CsWAKs/CsWAKLs (CsWAK1, CsWAK4, CsWAK7, CsWAKL1, and CsWAKL7) in leaf tissues, showing how their expression differs from their homologs in other plants [47]. Furthermore, the hemp WAK1 gene is highly expressed under drought stress conditions, and its expression can be induced by phytohormones like salicylic acid, methyl jasmonate, and ethylene [48]. These findings put the bases for future research on the potential roles of CsWAK/CsWAKLs in response to hormone treatments and abiotic/biotic stresses, including pathogen attacks.
Biosynthesis of specific terpenes may affect biotic and abiotic stress plant response and disease resistance [49]. For instance, few studies showed that while phytoanticipins terpenes are constitutively secreted in the absence of plant pathogen infection, phytoalexins are produced in response to pathogenic microbes [49,50]. A whole genome resequencing data across diverse samples of feral and domesticated lineages of C. sativa, aimed to examine their population structure, also allowed the identification of 6 loci related to stress response and 1 gene potentially involved in disease resistance [51]. This gene was annotated as mevalonate kinase (MEV kinase) and is involved in sesquiterpenes biosynthesis via the mevalonic acid pathway, with sesquiterpenes known for their antifungal properties [52]. Despite successful breeding efforts to modify terpene profiles, plant pathogens still constitute a significant cause of crop loss in Cannabis production [53].
3. Powdery Mildew Pathogens—Golovinomyces spp.
Cannabis is susceptible to the common PM pathogen Golovinomyces spp. [12]. Symptoms first appear as white circular patches of epiphytic mycelia and conidia on the leaf surface, which progress to cover the entire surface, and spread to the flowers and buds. Thaumatin-like proteins (TLPs), whose antifungal properties are thought to result from their β-1,3-glucanase activity [54], have also been associated with PM resistance in hops [55]. However, the antifungal activity of Endochitinase genes has not been widely studied [56]. The opposite occurs in the mildew loci O (MLO) gene family, which encodes plant proteins in conserved clades, of which clades IV and V are known for their susceptibility to PM [57]. In a recent study [58], the expression analysis of CsMLOs belonging to clade V, (CsMLO1 and CsMLO4 genes) revealed that these genes were significantly upregulated under Golovinomyces ambrosiae infection, confirming their possible involvement in PM susceptibility and as negative regulatory in immune system.
Subsequently, evidence was provided for the first R gene in Cannabis, represented by a single dominant locus able to confer complete resistance to an isolate of the PM pathogen G. ambrosiae [59]. Here, by using the “CBDRx” genome and linkage mapping with ~10,000 single nucleotide polymorphism (SNP) markers, 10 candidate genes of a single dominant R gene type, designed PM1, were detected, and this gene was mapped to a region rich in genes containing NBS and LRR domains. Specifically, a cluster of these putative disease resistance proteins contained N-terminal coiled-coil (CC) and nucleotide-binding arc (NB-ARC) domains, and two genes also contained LRR characteristics. Three annotations were also observed for tetratricopeptide repeat-containing proteins. Overall, the study identifies a key area for further research into the genetic basis for Cannabis resistance to G.ambrosiae.
NBS-LRR are involved in resistance to PM in several other species, like Vitis vinifera [60] and Triticum aestivum [61], and NBS proteins have been associated with candidate resistance genes to PM in hops [62]. According to these results and Mihalyov and Garfinkel’s [59] findings, the NBS-LRR resistance may be hypothesised for Cannabis.
4. Fusarium spp.
Pathogens in the genus Fusarium are among the most destructive in Cannabis, especially when affecting the roots or when infection occurs in the vegetative growth phase [10,63]. Sixteen species of Fusarium were reported as associated to Cannabis and classified into seven species complexes: Fusarium oxysporum, F. solani, F. incarnatum-equiseti, F.sambucinum, F. tricinctum, F. graminearum and F. fujikuroi [63,64]. The most evident Fusarium symptoms include yellowing of foliage and stem necrosis, and the related disease pathology is typically vascular wilt, but several Fusarium species can also result in seedling damping-off, crown rot, and reduced growth of stems and roots [12,63]. The main causative species in root and stem rots are F. solani and F. oxysporum [12].
F. oxysporum, a soilborne pathogen, can cause devastating vascular wilt in more than 100 plant species, and most of these fungi are formae speciales (f. sp.), an informal taxonomic group only infecting a single host plant species [65]. Two forms are reported as causal agents of Fusarium wilt in Cannabis, F. oxysporum f. sp. Cannabis (FoxC) and F. oxysporum f. sp. vasinfectum (FoxV). No host other than Cannabis has been reported for FoxC [14]. There are few studies on plant resistance to diseases caused by Fusarium and by F. oxysporum in Cannabis [66]. However, some resistance mechanisms and related gene families, common to a large set of plants and not specific to Cannabis, have been investigated [66].
The plant cell wall is the first barrier that F. oxysporum encounters during an attack, and this barrier defines the primary resource to fight the pathogen. Genes, which are reported to strengthen the plant cell walls, such as genes encoding 4-coumarate-CoA ligase, polyphenol oxidase and cellulose synthase, resulted upregulated in resistant Cavendish banana roots [67]. These findings suggest that the strengthened cell walls possibly confer enhanced pathogen resistance, which could also be the case in Cannabis.
Other gene families conferring F. oxysporum resistance include those involved in biosynthesis of JA and encoding P450 proteins [68]. Genes encoding dirigent-like proteins, CAP family proteins (cysteine-rich secretory proteins) and wound-responsive family proteins have also been demonstrated to be overexpressed during F. oxysporum infection in Arabidopsis [66]. NADPH (nicotinamide adenine dinucleotide phosphate) oxidases (or Respiratory burst oxidase) are also upregulated in several plants infected by F. oxysporum, such as wheat, cotton, and cucumber [67]. However, how these oxidases confer basal resistance to the pathogen is still unclear.
A large number of genes responsive to F. oxysporum Arabidopsis infection were detected in Zhu et al. [69]. Results confirmed that the ET, JA, auxin and SA pathways are all activated in response to infection by F. oxysporum. WAK gene upregulation was demonstrated to be induced upon F. oxysporum attack, and it seems that plants use these receptors to detect elicitors released by this fungus [69]. The induction of a number of genes encoding receptor-like kinases (RLKs) by F. oxysporum infection was also reported, and similar to what happens in the PM response, NBS-LRR-encoding genes also showed F. oxysporum resistance properties [69]. Transcription factors (TFs) play an important role as positive and negative regulators of antimicrobial compounds during the pathogenesis, and few of them belonging to the WRKY, ERF, MYB, and NAC gene families resulted constitutively up-regulated during the fungal infection [69].
Furthermore, the overexpression in Arabidopsis of genes belonging to the ERF family was related to the resistance to F. oxysporum [66]. Indeed, overexpression of ERF1, ERF2, and ERF14, which encode proteins belonging to the APETALA/ethylene (ET)-responsive-element binding protein (EREBP) family, provide resistance against this fungus. Conversely, overexpression of ERF4 leads to decreased resistance against F. oxysporum.
5. Botrytis cinerea
Botrytis cinerea attacks over 1000 crops, including legumes, berries, and some ornamental plants [70]. Because of this wide host range, it is the most studied necrotrophic pathogen. Cannabis is also susceptible to the gray mold disease caused by this fungus [12,13]. This air-borne necrotrophic pathogen can attack Cannabis seeds, leaves, inflorescences and stalks, causing lesions covered by a conidia grey layer and often leading to broader crop decay [71]. Its pathogenic strategy consists of the secretion of enzymes to digest the plant surface, and in the synthesis of phytotoxic metabolites leading to the host cell death [72]. Generally, the infection strategies rely on several virulence factors, like toxins and plant cell wall degrading enzymes (PCWDEs), transporter proteins and enzymes that protect B. cinerea from oxidative stress [73].
To fight this disease, plants activate a complex network of defence pathways, which allow them to respond to the pathogen. Plant cells recognise B. cinerea rapidly and activate the production of pathogenesis-related proteins (PRs), and increase the production of hormones such as SA, JA, ET, abscisic acid (ABA) and brassinosteroids (BR), which have been known to play a key role in defence against this pathogen [74]. ET and JA have synergistic effects in plant B. cinerea resistance. JA targets the JA-Zim (JAZ) repressor for degradation, activates JA/ET-related defence genes, and SA negatively regulates this transcriptional cascade. ABA decreases resistance to B. cinerea through the reduction of nitric oxide (NO) formation and suppresses both ROS and ET production. BRs also regulate plant immunity mainly by interacting with TFs, playing a key role for pathway crosstalk and signal integration, and allowing regulation of plant growth and defence [74]. Findings suggest that these defence mechanisms involved in the B. cinerea infection process are common to several plant hosts [75].
In a recent study [76], Cannabis defence responses against B. cinerea were explored at the molecular level. Symptoms were monitored, and the expression of putative defence genes was verified in leaves by quantitative reverse transcription PCR (RT-qPCR). Five putative defence genes, involved in JA/ET-pathway (ethylene response factor 1 (ERF1), encoding hevein-like protein (HEL) and phenylalanine ammonia-lyase (PAL) proteins), and in SA-pathway (pathogenesis-related protein 1 (PR1) and pathogenesis-related protein 2 (PR2), were identified, showing upregulation during all infection phases and thus strongly induced by B. cinerea pathogen.
6. Pythium
Pythium sp. are classified as oomycetes and are soil-borne plant pathogens commonly referred to as water molds [77]. They cause seed death, seedling damping-off, root browning and stunting, decay of fruits and vegetables during cultivation, culminating in serious damage to a wide range of crops, like beans, opium, spinach, strawberry, soybean and tobacco [17]. P. aphanidermatum, typical of plants in warm regions, appeared to be the most aggressive, especially towards plants in their germination and seedling stages [17]. Together with P. ultimum it was reported to cause damping-off of hemp seedlings [78] and was also recently shown to cause crown and root rot on hemp [79]. Furthermore, under greenhouse hydroponic conditions, it can cause the death of mature plants [17]. Moreover, two Pythium species, P. dissotocum Drechsler and P. myriotylum Drechsler are able to produce zoospores at 24–27 °C, and were shown to cause root damage in Cannabis, resulting in browning and stunting [17]. Neither of these two species has been previously reported to infect Cannabis [17].
In a recent study, two PAMPs, harpin and flg22, were shown to activate immune responses in various plant species [36]. Harpin has been considered to increase Cannabis growth and to help its disease resistance; however, there have been no scientific studies supporting this statement. In any case, it is known that harpin induces insect defence and activates the ethylene signalling pathway [80]. Flg22 is known to induce PTI in plants, resulting in ethylene biosynthesis and activation of MAPK cascades [81].
In hemp, pretreatment with harpin was shown to enhance seedling resistance to P. aphanidermatum PAMPs; however, flg22 did not induce the same defence mechanism towards this pathogen species [36]. Both harpin and flg22 pretreatment induced ethylene-responsive genes, but harpin-treated seedlings showed a significant increase in CsERF1 expression, while flg22 treatment did not affect the expression of this gene. Furthermore, both harpin and flg22-induced CsFRK1 (FLG22-induced receptor-like kinase1) and CsPR1, two marker genes associated with plant innate immunity in uninfected plants
7. Genome Editing to Generate Disease-Resistant Cannabis Varieties
Omics approaches are comprehensive methods for investigating defence response pathways and have been used broadly in medicinal plants [82,83]. Furthermore, by identifying candidate resistance genes and yielding an in-depth knowledge of the underlying molecular mechanism, they provide a strong basis for genome editing studies to generate disease-resistant Cannabis varieties [83].
The use of genetic engineering methods in Cannabis to enhance its resistance to pathogens and to improve desirable traits is a subject of investigation in several research projects [84]. However, it is challenging to regenerate fully developed Cannabis transgenic plants [85], and, despite some candidate genes involved in pathogen resistance having been identified, functions of these genes are not yet fully validated, and only a few studies report stable transformation for Cannabis tissues [44].
The first edited Cannabis line was developed by Agrobacterium-mediated transformation [86], in which overexpressing the Cannabis developmental regulator chimera in the embryo hypocotyls of unripe grains increased the regeneration efficiency. By applying this method, the development of transgenic callus from Cannabis has been achieved [87]. Evidence suggests that the overexpression of Non-expressor of Pathogenesis-Related genes-1 (NPR1) in Arabidopsis can confer disease resistance to different pathogens in various plants, such as cotton [88] and Brassica juncea [89]. The AtNPR1 gene has been introduced into C. sativa and confirmed by PCR and RT-PCR, showing that Cannabis can be transformed to generate disease-resistant varieties [89].
A recent mini-review on hemp genome editing [90] discusses the opportunity offered by next-generation genome editing technology. The direct delivery of CRISPR/Cas (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-Associated Protein) ribonucleoprotein complexes into plant tissue overcomes the drawback of Agrobacterium-mediated transformation, by which external plasmid DNA is introduced into the crop genome. CRISPR/Cas technology, which is still less commonly used in Cannabis, can be applied to introduce a specific DNA fragment to a precise location in the genome. It could have broad applications in Cannabis breeding, modifying gene regulation and developing pathogen-resistant plants, as already performed in other recalcitrant plants, such as grapes [90]. For instance, a protocol for this type of transformation in C. sativa was developed, and genome-edited Cannabis was produced by CRISPR/Cas9 approach [90].
By using CRISPR/Cas9, the previously discussed results of the study of Mihalyov and Garfinkel [59], consisting of a set of R candidate genes, could be used as target genes to improve PM resistance in the crop.
Furthermore, results reported in other plants could provide useful inputs for Cannabis gene editing. For instance, the genetic transformation of wheat with TLP and glucanases resulted in enhanced resistance to Fusarium [91], and MLO-7 was used as a host susceptibility (S) gene to improve grapevine and apple disease resistance to PM [92].
Overall, this advanced genome editing approach, based on a transgene-free framework, can address many problems associated with transgenic-based approaches and could be applied to produce improved non-transgenic Cannabis, with the most industrially desirable traits, including pathogen resistance traits.
Another alternative to Agrobacterium transformation protocol is represented by the use of a nanoparticle-based transient gene; through this method, multiple gene plasmids were expressed simultaneously in Cannabis leaf cells [93]. However, the study of disease resistance through this method is still in its infancy. It offers promising new perspectives in regulating the content of secondary metabolites, inducing pathogen resistance genes, and obtaining transgenic disease-resistant plants [94,95].
On this basis, there is a real possibility to improve Cannabis disease resistance by acting on targeted R genes or on S genes. A deep understanding of the underlying molecular mechanisms in which they are involved, as well as of plant-pathogen interactions, and the application of innovative molecular techniques is leading to innovations in the development of pathogen-resistant plants [96].
To date, it is still challenging to produce transgenic or gene-edited Cannabis, but the previously reported studies, and several gene editing approaches applied in other plant species, constitute good reference points for further Cannabis resistance research.
8. Conclusions
With the recent easing of legislation regulating Cannabis cultivation, the production and the consequent disease spread of this crop are growing rapidly. However, with this also comes a renewed interest in researching this plant from an agronomic perspective, and innovative approaches to crop improvement are being developed. Simultaneously, NGS technologies and recent advances in biotechnology have made it possible to greatly improve our understanding of Cannabis genetics and metabolomics, and several candidate genes involved in pathogen and resistance response have been elucidated. Figure 1 shows a potential model of immune and defence Cannabis responses in plant–pathogen interactions, referred in particular to PM, Fusarium spp., B. cinerea and Pythium, extrapolated, taking into account the studies discussed in this review. It summarises the initial plant pathogen response generally shared among plants and previously discussed in Section 2 and shows the consequent Cannabis defence response, detailing some genes/genes family or proteins associated with the enhancing cell walls, the activation of SA response, the activation of JA/ET response, the secondary metabolites involvement, and the overall reducing of pathogenicity, according to results discussed in Section 3, Section 4, Section 5 and Section 6.
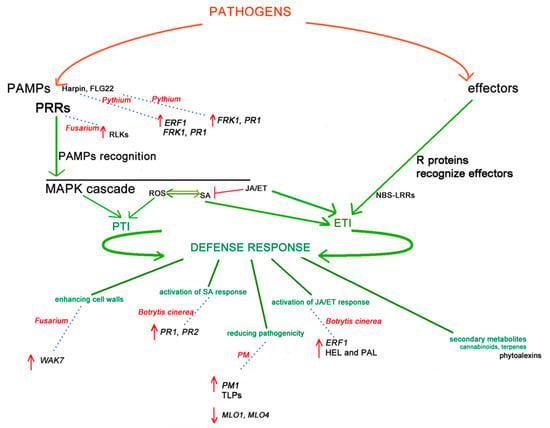
Figure 1.
A model of Cannabis immune and defence responses to PM, Fusarium spp., Botrytis cinerea and Pythium interactions. The initial defence response, shared among plants, starts with two types of molecules which are derived from pathogens. Pathogen-associated molecular patterns (PAMPs), recognised by the plant pattern recognition receptors (PRRs), activate the PAMP-triggered immunity (PTI) and initiate the first plant defence response, giving a basic level of resistance to most non-specialised pathogens. Two PAMPs, Harpin and Flg22, are involved in the Pythium defence response, both induce FRK1 and PR1 and only Harpin induces ERF1. While PRRs, receptor-like kinases (RLKs), are involved in the Fusarium defence response. Reactive oxygen species (ROS) production and mitogen-activated protein kinase (MAPK) also induce PTI. Effector proteins are the other type of initiators. Effector-triggered immunity (ETI) induced by the interactions of R proteins (e.g., nucleotide binding site-leucine-rich repeat (NBS-LRR) proteins) and pathogen effectors can start the second line of host-induced defence responses. The salicylic acid (SA) and jasmonic acid (JA)/ethylene (ET) signalling pathways are involved in PTI and ETI activation and the resistance response to pathogen infections, stimulating downstream transcription factors and plant defence responses (e.g., enhancing cell walls, the activation of SA response, the reduction of pathogenicity, the activation of JA/ET response and the secondary metabolites involvement). Genes reported to strengthen the plant cell walls, such as the WAK7 gene, are upregulated (red arrow in the model) in Fusarium Cannabis response. Genes involved in SA response, such as PR1, PR2 (Botrytis cinerea response), and JA/ET response, such as ERF1, encoding HEL and PAL (Botrytis cinerea response) are all upregulated during infection. Other genes are reported to reduce virulence, such as the PM1 gene and MLO1 and MLO4 (Powdery Mildew (PM) response), which are upregulated during PM infection. The phytoalexins involved in the biosynthesis of specific terpenes may also be involved in pathogen response. Arrows (in green colour) indicate positive regulation, and open blocks (in red colour) indicate negative regulation. Green lines indicate defence response mechanisms associated with different pathways. Blue pointing lines and the associated red pathogen name indicate genes/gene families involved in the defence response.
To date, few Cannabis omics studies are focused on its defence mechanisms against pathogens and the associated resistance genes. However, these studies, along with omics investigations of disease resistance molecular mechanisms in other crops (see Table 2), could constitute a suitable starting point for further Cannabis research in this field, especially if combined with gene editing approaches which have recently made significant progress, opening new perspectives in regulating the content of secondary metabolites and inducing pathogen resistance genes.

Table 2.
Table summarizing the main studies examined in this review.
Author Contributions
Conceptualization, T.M.S. and N.D.S.; writing preparation, T.M.S., R.A.L. and N.D.S.; supervision, N.D.S. and T.M.S. All authors have read and agreed to the published version of the manuscript.
Funding
Natasha D. Spadafora benefits from funding of the program PON “Research and Innovation” 2014–2020 (PON R&I), Action IV.6 “Contratti di ricerca su tematiche Green”.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Clarke, R.C.; Merlin, M.D. Cannabis domestication, breeding history, present day genetic diversity, and future prospects. Crit. Rev. Plant Sci. 2016, 35, 293–327. [Google Scholar] [CrossRef]
- Stuyt, E. The problem with the current high potency THC marijuana from the perspective of an addiction psychiatrist. Mo. Med. 2018, 115, 482–486. [Google Scholar] [PubMed]
- Schwabe, A.L.; Johnson, V.; Harrelson, J.; McGlaughlin, M.E. Uncomfortably high: Testing reveals inflated THC potency on retail Cannabis labels. PLoS ONE 2023, 18, e0282396. [Google Scholar] [CrossRef]
- Morales, P.; Hurst, D.P.; Reggio, P.H. Molecular targets of the phytocannabinoids: A complex picture. Prog. Chem. Org. Nat. Prod. 2017, 103, 103–131. [Google Scholar] [PubMed]
- Sirangelo, T.M.; Ludlow, R.A.; Spadafora, N.D. Multi-Omics approaches to study molecular mechanisms in Cannabis sativa. Plants 2022, 11, 2182. [Google Scholar] [CrossRef]
- Chandra, S.; Lata, H.; Khan, I.A.; Elsohly, M.A. Cannabis sativa L.: Botany and horticulture. In Cannabis sativa L.—Botany and Biotechnology; Chandra, S., Lata, H., Elsohly, M.A., Eds.; Springer: Cham, Switzerland, 2017; pp. 79–100. [Google Scholar]
- Hurgobin, B.; Tamiru-Oli, M.; Welling, M.T.; Doblin, M.S.; Bacic, A.; Whelan, J.A.; Lewsey, M.G. Recent advances in Cannabis sativa genomics research. N. Phytol. 2020, 230, 73–89. [Google Scholar] [CrossRef]
- Pacula, R.L.; Smart, R. Medical marijuana and marijuana legalization. Ann. Rev. Clin. Psychol. 2017, 13, 397–419. [Google Scholar] [CrossRef]
- Gao, S.; Wang, B.; Xie, S.; Xu, X.; Zhang, J.; Pei, L.; Yu, Y.; Yang, W.; Zhang, Y. A high quality reference genome of wild Cannabis sativa. Hortic. Res. 2020, 7, 73. [Google Scholar] [CrossRef]
- Petit, J.; Salentijn, E.M.J.; Paulo, M.J.; Denneboom, C.; van Loo, E.N.; Trindade, L.M. Elucidating the genetic architecture of fiber quality in hemp (Cannabis sativa L.) using a Genome-Wide Association study. Front. Genet. 2020, 11, 566314. [Google Scholar] [CrossRef]
- Sirangelo, T.M.; Ludlow, R.A.; Chenet, T.; Pasti, L.; Spadafora, N.D. Multi-Omics and genome editing studies on plant cell walls to improve biomass quality. Agriculture 2023, 13, 752. [Google Scholar] [CrossRef]
- Punja, Z.K. Emerging diseases of Cannabis sativa and sustainable management. Pest Manag. Sci. 2021, 77, 3857–3870. [Google Scholar] [CrossRef]
- Punja, Z.K.; Collyer, D.; Scott, C.; Lung, S.; Holmes, J.; Sutton, D. Pathogens and molds affecting production and quality of Cannabis sativa L. Front. Plant Sci. 2019, 10, 1120. [Google Scholar] [CrossRef]
- McCain, A.H.; Noviello, C. Biological control of Cannabis sativa. In Proceedings of the VI International Symposium on Biological Control of Weeds, Vancouver, WA, Canada, 19–25 August 1984; Delfosse, E.S., Ed.; Agricultural Canada: Ottawa, ON, Canada, 1985; pp. 635–642. [Google Scholar]
- Punja, Z.K. Epidemiology of Fusarium oxysporum causing root and crown rot of Cannabis (Cannabis sativa L., marijuana) plants in commercial greenhouse production. Can. J. Plant Pathol. 2020, 43, 216–235. [Google Scholar] [CrossRef]
- Cheng, Y.; Tang, X.; Gao, C.; Li, Z.; Chen, J.; Guo, L.; Wang, T.; Xu, J. Molecular diagnostics and pathogenesis of fungal pathogens on bast fiber crops. Pathogens 2020, 9, 223. [Google Scholar] [CrossRef]
- Punja, Z.K.; Rodriguez, G. Fusarium and Pythium species infecting roots of hydroponically grown marijuana (Cannabis sativa L.) plants. Can. J. Plant Pathol. 2018, 40, 498–513. [Google Scholar] [CrossRef]
- Warren, J.G.; Mercado, J.; Grace, D. Occurrence of hop latent viroid causing disease in Cannabis sativa in California. Plant Dis. 2019, 103, 2699. [Google Scholar] [CrossRef]
- Wolfenbarger, S.N.; Massie, S.T.; Ocamb, C.; Eck, E.B.; Grove, G.G.; Nelson, M.E.; Probst, C.; Twomey, M.C.; Gent, D.H. Distribution and characterization of Podosphaera macularis virulent on hop cultivars possessing R6-Based resistance to Powdery Mildew. Plant Dis. 2016, 100, 1212–1221. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Desjardins, A.E. Fusarium Mycotoxins: Chemistry, Genetics, and Biology; APS Press: St. Paul, MN, USA, 2006. [Google Scholar]
- Vujanovic, V.; Korber, D.R.; Vujanovic, S.; Vujanovic, J.; Jabaji, S. Scientific prospects for cannabis-microbiome research to ensure quality and safety of products. Microorganisms 2020, 8, 290. [Google Scholar] [CrossRef]
- Palmieri, D.; Ianiri, G.; Del Grosso, C.; Barone, G.; De Curtis, F.; Castoria, R.; Lima, G. Advances and perspectives in the use of biocontrol agents against fungal plant diseases. Horticulturae 2022, 8, 577. [Google Scholar] [CrossRef]
- Massart, S.; Martinez-Medina, M.; Jijakli, M.H. Biological control in the microbiome era: Challenges and opportunities. Biol. Control 2015, 89, 98–108. [Google Scholar] [CrossRef]
- Op De Beeck, M.; Lievens, B.; Busschaert, P.; Declerck, S.; Vangronsveld, J.; Colpaert, J.V. Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS ONE 2014, 9, e97629. [Google Scholar] [CrossRef]
- Pépin, N.; Punja, Z.K.; Joly, D.L. Occurrence of powdery mildew caused by Golovinomyces chicoracearum sensu lato on Cannabis sativa in Canada. Plant Dis. 2018, 102, 2644. [Google Scholar] [CrossRef]
- Garfinkel, A.R. Three Botrytis species found causing gray mold on industrial hemp (Cannabis sativa) in Oregon. Plant Dis. 2020, 104, 2026. [Google Scholar] [CrossRef]
- Aliferis, K.A.; Bernard-Perron, D. Cannabinomics: Application of metabolomics in Cannabis (Cannabis sativa L.). Research and development. Front. Plant. Sci. 2020, 11, 554. [Google Scholar] [CrossRef] [PubMed]
- Punja, Z.K.; Rodriguez, G.; Chen, S. Assessing genetic diversity in Cannabis sativa using molecular approaches. In Cannabis sativa L.-Botany and Biotechnology; Chandra, S., Lata, L., ElSohly, M.A., Eds.; Springer: Berlin, Germany, 2017; pp. 395–418. [Google Scholar]
- Scott, C.; Punja, Z.K. Evaluation of disease management approaches for powdery mildew on Cannabis sativa L. (marijuana) plants. Can. J. Plant Pathol. 2021, 34, 394–412. [Google Scholar] [CrossRef]
- Stack, G.M.; Toth, J.A.; Carlson, C.H.; Cala, A.R.; Marrero-González, M.I.; Wilk, R.L.; Gentner, D.R.; Crawford, J.L.; Philippe, G.; Rose, J.K.C. Season-long characterization of high-cannabinoid hemp (Cannabis sativa L.) reveals variation in cannabinoid accumulation, flowering time, and disease resistance. GCB Bioenergy 2021, 13, 546–561. [Google Scholar] [CrossRef]
- Bai, Y.; Huang, C.C.; Hulst, R.V.D.; Meijer-Dekens, F.; Bonnema, G.; Lindhout, P. QTLs for tomato powdery mildew resistance (Oidium lycopersici) in Lycopersicon parviflorum G1.1601 localize with two qualitative powdery mildew resistance genes. Mol. Plant Microbe Interact. 2003, 16, 169–176. [Google Scholar] [CrossRef]
- Tassone, M.R.; Bagnaresi, P.; Desiderio, F.; Bassolino, L.; Barchi, L.; Florio, F.E.; Sunseri, F.; Sirangelo, T.M.; Rotino, G.L.; Toppino, L. A Genomic BSAseq Approach for the characterization of QTLs underlying resistance to Fusarium oxysporum in eggplant. Cells 2022, 11, 2548. [Google Scholar] [CrossRef]
- Nachappa, P.; Fulladolsa, A.C.; Stenglein, M. Wild wild west: Emerging viruses and viroids of hemp. Outlooks Pest Manag. 2020, 31, 175–179. [Google Scholar] [CrossRef]
- Oh, S.; Choi, D. Receptor-mediated nonhost resistance in plants. Essays Biochem. 2022, 66, 435–445. [Google Scholar]
- Lee, H.A.; Lee, H.Y.; Seo, E.; Lee, J.; Kim, S.B.; Oh, S.; Choi, E.; Choi, E.; Lee, S.E.; Choi, D. Current understandings of plant nonhost resistance. Mol. Plant-Microbe Interact. 2017, 30, 5–15. [Google Scholar] [CrossRef]
- Sands, L.B.; Cheek, T.; Reynolds, J.; Ma, Y.; Berkowitz, G.A. Effects of Harpin and Flg22 on growth enhancement and pathogen defense in Cannabis sativa seedlings. Plants 2022, 11, 1178. [Google Scholar] [CrossRef]
- Benjamin, G.; Pandharikar, G.; Frendo, P. Salicylic acid in plant symbioses: Beyond plant pathogen interactions. Biology 2022, 11, 861. [Google Scholar] [CrossRef]
- Ding, L.N.; Li, Y.T.; Wu, Y.Z.; Li, T.; Geng, R.; Cao, J.; Zhang, W.; Tan, X.-L. Plant disease resistance-related signaling pathways: Recent progress and future prospects. Int. J. Mol. Sci. 2022, 23, 16200. [Google Scholar] [CrossRef] [PubMed]
- Boba, A.; Kostyn, K.; Kozak, B.; Zalewski, I.; Szopa, J.; Kulma, A. Transcriptomic profiling of susceptible and resistant flax seedlings after Fusarium oxysporum lini infection. PLoS ONE 2021, 16, e0246052. [Google Scholar] [CrossRef] [PubMed]
- McHale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 2006, 7, 212. [Google Scholar] [CrossRef] [PubMed]
- Andolfo, G.; Dohm, J.C.; Himmelbauer, H. Prediction of NB-LRR resistance genes based on full-length sequence homology. Plant J. 2022, 110, 1592–1602. [Google Scholar] [CrossRef]
- Xie, S.S.; Duan, C.G. Epigenetic regulation of plant immunity: From chromatin codes to plant disease resistance. aBIOTECH 2023, 1–16. [Google Scholar] [CrossRef]
- He, Z.H.; He, D.; Kohorn, B.D. Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 1998, 14, 55–63. [Google Scholar] [CrossRef]
- Diener, A.C.; Ausubel, F.M. Resistance to Fusarium oxysporum 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 2005, 171, 305–321. [Google Scholar] [CrossRef]
- Yang, J.; Xie, M.; Wang, X.; Wang, G.; Zhang, Y.; Li, Z.; Ma, Z. Identification of cell wall-associated kinases as important regulators involved in Gossypium hirsutum resistance to Verticillium dahliae. BMC Plant Biol. 2021, 21, 220. [Google Scholar] [CrossRef]
- Li, L.; Yu, S.; Chen, J.; Cheng, C.; Sun, J.; Xu, Y.; Deng, C.; Dai, Z.; Yang, Z.; Chen, X.; et al. Releasing the full potential of Cannabis through biotechnology. Agronomy 2022, 12, 2439. [Google Scholar] [CrossRef]
- Sipahi, H.; Whyte, T.D.; Ma, G.; Berkowitz, G. Genome-Wide identification and expression analysis of Wall-Associated Kinase (WAK) gene family in Cannabis sativa L. Plants 2022, 11, 2703. [Google Scholar] [CrossRef] [PubMed]
- Maravaneh, H.; Davarpanah, S.J. Study of cannabinoids biosynthesis-related genes in hemp (Cannabis sativa L.) under drought stress by in vitro and in silico tools. J. Appl. Biotechnol. Rep. 2022, 9, 504–510. [Google Scholar]
- Ninkuu, V.; Zhang, L.; Yan, J.; Fu, Z.; Yang, T.; Zeng, H. Biochemistry of terpenes and recent advances in plant protection. Int. J. Mol. Sci. 2021, 22, 5710. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.N.; Zimmer, K.R.; Macedo, A.J.; Trentin, D.S. Plant natural products targeting bacterial virulence factors. Chem. Rev. 2016, 116, 9162–9236. [Google Scholar] [CrossRef]
- Woods, W.; Price, N.; Matthews, P.; McKay, J.K. Genome-wide polymorphism and genic selection in feral and domesticated lineages of Cannabis sativa. G3 2023, 13, jkac209. [Google Scholar] [CrossRef] [PubMed]
- Sommano, S.R.; Chittasupho, C.; Ruksiriwanich, W.; Jantrawut, P. The Cannabis terpenes. Molecules 2020, 25, 5792. [Google Scholar] [CrossRef] [PubMed]
- Backer, R.; Schwinghamer, T.; Rosenbaum, P.; McCarty, V.; Eichhorn Bilodeau, S.; Lyu, D.; Ahmed, M.B.; Robinson, G.; Lefsrud, M.; Wilkins, O.; et al. Closing the yield gap for Cannabis: A meta-analysis of factors determining Cannabis yield. Front. Plant Sci. 2019, 10, 495. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.J. Fungal cell wall chitinases and glucanases. Microbiology 2004, 150, 2029–2035. [Google Scholar] [CrossRef]
- Kappagantu, M.; Bullock, J.M.; Nelson, M.E.; Eastwell, K.C. Hop stunt viroid: Effect on host (Humulus lupulus) Transcriptome and its interactions with hop Powdery Mildew (Podospheara macularis). Mol. Plant-Microbe Interact. 2017, 30, 842–851. [Google Scholar] [CrossRef]
- Viterbo, A.; Haran, S.; Friesem, D.; Ramot, O.; Chet, I. Antifungal activity of a novel endochitinase gene (chit36) from Trichoderma harzianum Rifai TM. FEMS Microbiol. Lett. 2001, 200, 169–174. [Google Scholar] [CrossRef]
- Acevedo-Garcia, J.; Kusch, S.; Panstruga, R. Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. 2014, 204, 273–281. [Google Scholar] [CrossRef]
- Pépin, N.; Hebert, F.O.; Joly, D.L. Genome-Wide characterization of the MLO gene family in Cannabis sativa reveal two genes as strong candidates for Powdery Mildew susceptibility. Front. Plant Sci. 2021, 12, 729261. [Google Scholar] [CrossRef]
- Mihalyov, P.D.; Garfinkel, A.R. Discovery and genetic mapping of PM1, a powdery mildew resistance gene in Cannabis sativa L. Front. Agron. 2021, 3, 720215. [Google Scholar] [CrossRef]
- Goyal, N.; Bhatia, G.; Sharma, S.; Garewal, N.; Upadhyay, A.; Upadhyay, S.K.; Singh, K. Genome-wide characterization revealed role of NBS-LRR genes during powdery mildew infection in Vitis vinifera. Genomics 2020, 112, 312–322. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Zhu, S.; Zhao, R.; Jiang, Z.; Ji, Y.; Ji, J.; Qiu, D.; Li, H.; Bie, T. Pm21, encoding a typical CC-NBS-LRR protein, confers broad spectrum resistance to wheat powdery mildew disease. Mol. Plant. 2018, 11, 879–882. [Google Scholar] [CrossRef] [PubMed]
- Padgitt-Cobb, L.K.; Kingan, S.B.; Henning, J.A. Genomic analysis of powdery mildew resistance in a hop (Humulus lupulus L.) bi-parental population segregating for “R6-locus”. Euphytica 2019, 216, 10. [Google Scholar]
- Gwinn, K.D.; Hansen, Z.; Kelly, H.; Ownley, B.H. Diseases of Cannabis sativa caused by diverse Fusarium Species. Front. Agron. 2022, 3, 796062. [Google Scholar] [CrossRef]
- Yulfo-Soto, G.E.; Smith, H.; Szarka, D.; Dixon, E.; Vaillancourt, L.J.; Gauthier, N. First report of Fusarium graminearum causing flower blight on hemp (Cannabis sativa) in Kentucky. Plant Dis. 2022, 106, 334. [Google Scholar] [CrossRef] [PubMed]
- Agrios, G.N. (Ed.) Plant Pathology, 5th ed.; Elsevier Academic Press: Burlington, MA, USA, 2005. [Google Scholar]
- Husaini, A.M.; Sakina, A.; Cambay, S.R. Host-Pathogen Interaction in Fusarium oxysporum infections: Where do we stand? Mol. Plant-Microbe Interact. 2018, 31, 889–898. [Google Scholar] [CrossRef]
- Li, C.Y.; Deng, G.M.; Yang, J.; Viljoen, A.; Jin, Y.; Kuang, R.B.; Zuo, C.W.; Lv, Z.C.; Yang, Q.S.; Sheng, O.; et al. Transcriptome profiling of resistant and susceptible Cavendish banana roots following inoculati on with Fusarium oxysporum f. sp. cubense tropical race 4. BMC Genom. 2012, 13, 374. [Google Scholar] [CrossRef]
- Liu, F.; Jiang, H.; Ye, S.; Chen, W.P.; Liang, W.; Xu, Y.; Sun, B.; Sun, J.; Wang, Q.; Cohen, J.D.; et al. The Arabidopsis P450 protein CYP82C2 modulates jasmonate-induced root growth inhibition, defense gene expression and indole glucosinolate biosynthesis. Celi. Res. 2010, 20, 539–552. [Google Scholar] [CrossRef]
- Zhu, Q.H.; Stephen, S.; Kazan, K.J.; In, G.; Fan, L.; Taylor, J.; Dennis, E.S.; Helliwell, C.A.; Wang, M.B. Characterization of the defense transcriptome responsive to Fusarium oxysporum-infection in Arabidopsis using RNA-seq. Gene 2013, 512, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, M.; BenRejeb, I.; Punja, Z.K.; Buirs, L.; Jabaji, S. Review: Understanding bud rot development, caused by Botrytis cinerea, on cannabis (Cannabis sativa L.) plants grown under greenhouse conditions. Botany 2023, 101, 200–231. [Google Scholar] [CrossRef]
- McPartland, J.M.; Clarke, R.C.; Watson, D.P. Hemp Diseases and Pests Management and Biological Control; CABI Publishing: Trowbridge, UK, 2000. [Google Scholar]
- van Kan, J.A. Licensed to kill: The lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 2006, 11, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Choquer, M.; Fournier, E.; Kunz, C.; Levis, C.; Pradier, J.M.; Simon, A.; Viaud, M. Botrytis cinerea virulence factors: New insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett. 2007, 277, 1–10. [Google Scholar] [CrossRef]
- AbuQamar, S.F.; Moustafa, K.; Tran, L.S. Mechanisms and strategies of plant defense against Botrytis cinerea. Crit. Rev. Biotechnol. 2017, 37, 262–274. [Google Scholar] [CrossRef]
- Reboledo, G.; Agorio, A.; Vignale, L.; Batista-García, R.A.; Ponce De León, I. Botrytis cinerea transcriptome during the infection process of the Bryophyte Physcomitrium patens and angiosperms. J. Fungi 2021, 7, 11. [Google Scholar] [CrossRef]
- Balthazar, C.; Cantin, G.; Novinscak, A.; Joly, D.L.; Filion, M. Expression of putative defense responses in Cannabis primed by Pseudomonas and/or Bacillus strains and infected by Botrytis cinerea. Front. Plant Sci. 2020, 11, 572112. [Google Scholar] [CrossRef] [PubMed]
- El Ghaouth, A.; Arul, J.; Grenier, J.; Benhamou, N.; Asselin, A.; Belanger, R. Effect of chitosan on cucumber plants: Suppression of Pythium aphanidermatum and induction of defense reactions. Am. Phytophatological. Soc. 1993, 84, 313–320. [Google Scholar] [CrossRef]
- McPartland, J.M. A review of Cannabis diseases. J. Int. Hemp. Assoc. 1996, 3, 19–23. [Google Scholar]
- Beckerman, J.; Nisonson, H.; Albright, N.; Creswell, T. First report of Pythium aphanidermatum causing crown and root rot of industrial hemp in the United States. Plant Dis. 2017, 101, 1038. [Google Scholar] [CrossRef]
- Li, X.; Han, B.; Xu, M.; Han, L.; Zhao, Y.; Liu, Z.; Dong, H.; Zhang, C. Plant growth enhancement and associated physiological responses are coregulated by ethylene and gibberellin in response to harpin protein Hpa1. Planta 2014, 239, 831–846. [Google Scholar] [CrossRef][Green Version]
- Mersmann, S.; Bourdais, G.; Rietz, S.; Robatzek, S. Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 2010, 154, 391–400. [Google Scholar] [CrossRef]
- Amer, B.; Baidoo, E.E.K. Omics-driven biotechnology for industrial applications. Front. Bioeng. Biotechnol. 2021, 9, 613307. [Google Scholar] [CrossRef]
- Yang, L.; Yang, Y.; Huang, L.; Cui, X.; Liu, Y. From single- to multi-omics: Future research trends in medicinal plants. Brief. Bioinform. 2023, 24, bbac485. [Google Scholar] [CrossRef] [PubMed]
- Punja, Z.K.; Holmes, J.; Collyer, D.; Lung, S. Development of tissue culture methods for marijuana (Cannabis sativa L.) strains to achieve Agrobacterium-mediated transformation to enhance disease resistance. Vitro Cell. Dev. Biol. Anim. 2019, 55, 523. [Google Scholar]
- Feeney, M.; Punja, Z.K. The role of Agrobacterium-mediated and other gene-transfer technologies in Cannabis research and product development, in Cannabis sativa L. In Botany and Biotechnology; Chandra, S., Lata, L., ElSohly, M.A., Eds.; Springer: Berlin, Germany, 2017; pp. 343–363. [Google Scholar]
- Zhang, X.; Xu, G.; Cheng, C.; Lei, L.; Sun, J.; Xu, Y.; Deng, C.; Dai, Z.; Yang, Z.; Chen, X.; et al. Establishment of an Agrobacterium mediated genetic transformation and CRISPR/Cas9-mediated targeted mutagenesis in Hemp (Cannabis sativa L.). Plant Biotechnol. J. 2021, 19, 1979–1987. [Google Scholar] [CrossRef] [PubMed]
- Holmes, J.E.; Punja, Z.K. Agrobacterium-mediated transformation of THC-containing Cannabis sativa L. yields a high frequency of transgenic calli expressing bialaphos resistance and non-expressor of PR1 (NPR1) genes. Botany, 2023; in press. [Google Scholar] [CrossRef]
- Kumar, V.; Joshi, S.G.; Bell, A.A.; Rathore, K.S. Enhanced resistance against Thielaviopsis basicola in transgenic cotton plants expressing Arabidopsis NPR1 gene. Transgenic Res. 2013, 22, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Mir Zahoor, A.; Tyagi, A.; Mehari, H.; Meena, R.P.; Bhat, J.A.; Yadav, P.; Papalou, P.; Rawat, S.; Grover, A. Overexpression of NPR1 in Brassica juncea confers broad spectrum resistance to fungal pathogens. Front. Plant Sci. 2017, 8, 1693. [Google Scholar] [CrossRef] [PubMed]
- Shiels, D.; Prestwich, B.D.; Koo, O.; Kanchiswamy, C.N.; O’Halloran, R.; Badmi, R. Hemp genome editing—Challenges and opportunities. Front. Genome 2022, 4, 823486. [Google Scholar] [CrossRef]
- Mackintosh, C.A.; Lewis, J.; Radmer, L.E.; Shin, S.; Heinen, S.J.; Smith, L.A.; Wyckoff, M.N.; Dill-Macky, R.; Evans, C.K.; Kravchenko, S.; et al. Overexpression of defense response genes in transgenic wheat enhances resistance to Fusarium head blight. Plant Cell Rep. 2007, 26, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Malnoy, M.; Viola, R.; Jung, M.H.; Koo, O.J.; Kim, S.; Kim, J.S.; Velasco, R.; Nagamangala Kanchiswamy, C. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef]
- Ahmed, S.; Gao, X.; Jahan, M.A.; Adams, M.; Wu, N.; Kovinich, N. Nanoparticle-based genetic transformation of Cannabis sativa. J. Biotechnol. 2021, 326, 48–51. [Google Scholar] [CrossRef]
- Dolgin, E. The bioengineering of Cannabis. Nature 2019, 572, 5–7. [Google Scholar] [CrossRef]
- Dong, B.R.; Jiang, R.; Chen, J.F.; Xiao, Y.; Lv, Z.Y.; Chen, W.S. Strategic nanoparticle-mediated plant disease resistance. Crit. Rev. Biotechnol. 2022, 43, 22–37. [Google Scholar] [CrossRef]
- Tyagi, S.; Kumar, R.; Kumar, V.; Won, S.Y.; Shukla, P. Engineering disease resistant plants through CRISPR-Cas9 technology. GM Crops Food 2021, 12, 125–144. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).