Abstract
N6-methyldeoxyadenosine (6mA) is a recently discovered DNA modification involved in regulating plant adaptation to abiotic stresses. However, the mechanisms and changes of 6mA under cold stress in plants are not yet fully understood. Here, we conducted a genome-wide analysis of 6mA and observed that 6mA peaks were predominantly present within the gene body regions under both normal and cold conditions. In addition, the global level of 6mA increased both in Arabidopsis and rice after the cold treatment. The genes that exhibited an up-methylation showed enrichment in various biological processes, whereas there was no significant enrichment observed among the down-methylated genes. The association analysis revealed a positive correlation between the 6mA level and the gene expression level. Joint analysis of the 6mA methylome and transcriptome of Arabidopsis and rice unraveled that fluctuations in 6mA levels caused by cold exposure were not correlated to changes in transcript levels. Furthermore, we discovered that orthologous genes modified by 6mA showed high expression levels; however, only a minor amount of differentially 6mA-methylated orthologous genes were shared between Arabidopsis and rice under low-temperature conditions. In conclusion, our study provides information on the role of 6mA in response to cold stress and reveals its potential for regulating the expression of stress-related genes.
1. Introduction
N6-methyldeoxyadenosine (6mA) is a prevalent DNA modification commonly found in lower organisms, such as bacteria, archaea, protozoa, and fungi. Its involvement in diverse biological processes includes gene transcription, DNA–protein interactions, DNA repair, and transposon insertion [,,]. While 5mC is the primary DNA modification in higher eukaryotes, 6mA is present in trace amounts [,]. Advances in research have led to the increased recognition of 6mA’s features and roles in higher organisms. Recent studies using various techniques, such as liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS), dot blot assays, 6mA-IP-seq, Single-molecule real-time (SMRT) sequencing, and Oxford Nanopore long-read sequencing, have revealed the widespread presence of 6mA in higher plant and animal species [,,,,,,,]. In Chlamydomonas and Tetrahymena, 6mA contributes to the localization of nucleosomes near the (transcription start site) TSS and appears to mark active genes [,]. In Drosophila, 6mA is enriched in transposons and affects their expression []. In Arabidopsis and rice, 6mA is widespread throughout the genome, and it is positively associated with the gene expression level when it marks in gene bodies [,,]. Moreover, the roles of 6mA extend to various physiological processes, including fertilized egg development and human malignant glioma development [,,]. Collectively, these findings indicate that 6mA plays versatile and essential roles in biological functions.
Stress-induced changes in DNA methylation and histone modifications have been shown to regulate stress responsive gene expression. For example, 5mC methylation of the ALN promoter is stimulated by cold, and this leads to the suppression of ALN expression and further promotion of seed dormancy []. Methylation of CHH in the RhAG promoter region regulates the expression of the RhAG gene, which impacts the development of rose petals under low temperatures []. Furthermore, recent studies have linked 6mA with environmental stress responses. The overall 6mA levels are significantly elevated upon stress in mouse brains, and 6mA dynamic changes were significantly associated to functional genes involved in learning, action, and neurogenesis []. For instance, heat stress response genes were marked by 6mA and H3K9me3 modification, resulting in high expression and extended lifespan in the progeny of Caenorhabditis elegans [,]. The 6mA level in mitochondrial DNA mediated by METTL14 was elevated under hypoxic stress in human cells []. These findings indicate a prevalent involvement of 6mA modification in response to environmental stresses, although the exact mechanisms underlying the regulation, especially in stress tolerance, are not fully understood.
Low temperature is a pervasive environmental stress that can negatively impact plant growth and development, ultimately reducing crop yields. Research over the past few decades has focused on the C-repeat binding factor/dehydration-responsive element-binding protein1 (CBF/DREB1)-dependent cold signaling pathway in Arabidopsis. The CBF proteins, CBF1–3, are transcription factors that activate the expression of COR (Cold-regulated) genes in response to cold by binding to conserved CRT/DRE motifs in their promoters [,,]. In rice, several genes associated with chilling tolerance, including Ctb1, qLTG3-1, COLD1, OsSAP16, and qCTS-9, have been mapped and functionally characterized [,,,,]. Despite the numerous advances in understanding responses to cold stress in plants, the role of 6mA in this process remains ambiguous.
Here, we profiled 6mA in the genomic DNA (gDNA) of Arabidopsis and rice under normal and cold conditions. We found 6mA was mostly located in gene body regions, and it positively correlated with gene expression. Cold treatment increased the overall level of 6mA methylation, and the differentially methylated genes (DMGs) exhibited enrichment in diverse biological processes. Joint analysis of the 6mA methylome and transcriptome of Arabidopsis and rice under normal and low temperatures revealed that 6mA-containing genes generally have higher expression levels than those without, and, while the overall level of 6mA increased under cold treatment, no correlation was found between its abundance and changes in expression level. Our results reveal that 6mA is a dynamic epigenetic mark in response to cold and participates in regulating of expression of stress responsive genes.
2. Results
2.1. The Distribution of 6mA and Its Association with Gene Expression in Arabidopsis and Rice under Normal Conditions
To investigate the distribution and association of the 6mA modification with gene expression in Arabidopsis and rice, we performed 6mA-IP (immunoprecipitation)-seq with two biological replicates for each species. We obtained more than 70 million unique mapped reads per 6mA-IP-seq sample for subsequent analysis (Table S1). High reproducibility of the sequence data was observed with high Pearson correlation coefficients (R > 0.97) in Arabidopsis and rice between the two biological replicates (Figure S1A,B). We identified 4065 high-confidence 6mA peaks in Arabidopsis (Figure 1A; Table S2) and 8412 high-confidence 6mA peaks in rice (Figure 1B, Table S3) after peak calling. To examine the epigenetic roles of 6mA, we investigated its location in the genome, including intergenic regions, promoter, gene bodies, and their subregions. A majority of the 6mA peaks were located in gene body regions in both Arabidopsis and rice. In Arabidopsis, 82% of the 6mA peaks were positioned in gene bodies, with half of the peaks occupying exons (Figure 1C). In rice, more than half of the peaks were located in gene bodies, especially in the intron regions (Figure 1D). These observations indicated a high enrichment of 6mA peaks on gene bodies in both species.
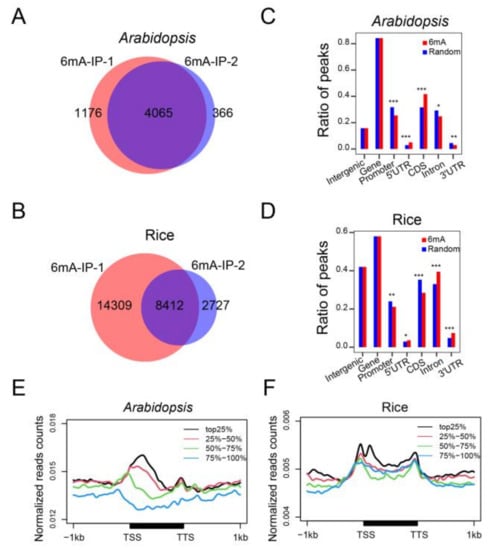
Figure 1.
Genome-wide mapping and distribution of 6mA in Arabidopsis and rice under normal conditions. (A,B) Venn diagrams showing the overlapped 6mA peaks between two biological repeats in Arabidopsis (A) and rice (B) under normal conditions. (C,D) Distributions (expressed as percentages) of 6mA peaks found among intergenic regions, promoters (within 1 kb upstream of the TSS), and gene bodies. Gene bodies were further divided into 5′ and 3′ UTRs (untranslated regions), CDS (coding sequences), and introns. (* p < 0.05; ** p < 0.01; *** p < 0.001, Fisher’s exact test). (E,F) Metaplot showing that 6mA is positively correlated with gene expression levels in Arabidopsis (E) and rice (F). All genes were divided into 4 groups, from high to low (top 25%, 25–50%, 50–75%, and 75–100% based on FPKM) expression levels. TSS, transcription start site; TTS, transcription terminal site.
We also investigated the relationship between 6mA abundance and gene expression level. To this end, we carried out RNA-seq with three biological replicates to analyze the gene expression level. The correlation between the individual gene expression levels demonstrated high Pearson correlation coefficients (R > 0.88) (Figure S2A,B). We divided all the protein-coding genes in the genome into four classes based on their expression level: top 25%, 25–50%, 50–75%, and 75–100%. Plotting of 6mA abundance in these four classes revealed that strongly expressed genes had a higher occupancy of 6mA, particularly at the TSS region, than weakly expressed genes in both Arabidopsis and rice (Figure 1E,F). Thus, the 6mA modification is positively correlated with gene expression.
2.2. Genome-Wide Mapping of 6mA under Low Temperature
To explore whether 6mA responds to cold stress, we examined the overall 6mA methylation level after cold treatment (4 °C). We performed dot-blot assays for rosette leaves of 3-week-old Arabidopsis and 2-week-old rice seedlings. The 6mA signals were detected to have increased 2.1- and 1.7-fold, respectively, following a 24 h cold treatment in Arabidopsis and a 6 day cold treatment in rice (Figure 2A,B). We then profiled 6mA distribution for Arabidopsis and rice plants under cold conditions using 6mA-IP-seq. Two biological replicates were performed for each species, and high Pearson correlation coefficients (R > 0.99) were observed between the two biological replicates (Figure S3A,B). Peak calling led to the identification of 7557 and 19,013 high-confidence 6mA peaks in Arabidopsis and rice under cold conditions, respectively (Figure S3C,D, Tables S2 and S3). We further analyzed the distribution of 6mA methylation within each gene by plotting the read number ratio of m6A-IP-seq along the gene body region (from 1kb 5′ of TSS to 1 kb 3′ of TTS). In both Arabidopsis and rice, 6mA methylation showed a higher enrichment on the gene body region under cold conditions compared to normal conditions (Figure 2C,D). Box plot analysis confirmed a significant increase in the overall enrichment of 6mA peaks after cold treatment in both Arabidopsis and rice (Figure 2E,F). These results demonstrate that the genome-wide 6mA methylation level is increased under low temperature conditions.
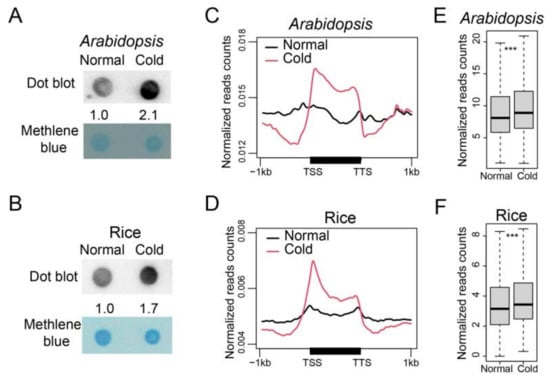
Figure 2.
The overall 6mA level increased under cold. (A,B) 6mA dot-blot of genomic DNA samples from 3-week-old Col-0 plants (A) and 2-week-old rice seedlings (B) with 4 °C cold treatment for 24 h and 6 d, respectively. An amount of 200 ng of genomic DNA was loaded. Marked numbers indicate the relative amount in cold-treated samples compared to non-treated sample quantified by Image J. (C,D) 6mA profiles of Arabidopsis (C) and rice (D) under normal and cold conditions in gene regions. The 1 kb upstream and downstream flanking coding regions were aligned for all genes. TSS, transcription start site; TTS, transcription terminal site. (E,F) Box plot of the enrichment of 6mA peaks in Arabidopsis (E) and rice (F) under normal and cold conditions. The p-values were calculated for significant differences between two groups by the Mann–Whitney U test. ***: p < 0.001.
2.3. Identification and Gene Ontology (GO) Analysis of Differentially Methylated Genes (DMGs) in Response to Cold Stress
In order to further examine 6mA alterations caused by chilling, we selected differentially methylated regions (DMRs) defined by a 1.5-fold or greater change in 6mA enrichment and with a significant test at q < 0.05. In Arabidopsis, we identified a total of 1391 up-regulated and 434 down-regulated differentially methylated regions (DMRs) (Figure 3A, Table S4), while in rice, we identified 3522 up-regulated and 598 down-regulated DMRs between cold and normal temperatures (Figure 3B, Table S5). We then analyzed the distribution of DMRs and found that down-regulated DMRs were primarily enriched in the intergenic and promoter regions, while up-regulated DMRs were mainly enriched in the coding sequence (CDS) and 5′ untranslated region (UTR) in both Arabidopsis and rice (Figure 3C,D). These up- and down-regulated DMRs corresponded to 1427 and 255 genes in Arabidopsis, which we defined as differentially methylated genes (DMGs), as shown in Table S4. The GO analysis of these DMGs revealed that up-DMGs were enriched in diverse terms such as plasmodesmata, ATP binding, response to temperature stimuli, and others (Figure 3E, Table S5). In rice, these up- and down-regulated DMRs corresponded to 2339 and 211 DMGs, respectively (Table S6). GO analysis of the up-regulated DMGs revealed enriched terms such as ATP binding, apoptosis, defense response, and others (Figure 3F, Table S7). No GO term was significantly enriched for the down-regulated DMRs in either Arabidopsis or rice. Furthermore, we discovered that the 6mA enrichment in key stress response genes increased after cold treatment. For instance, the Arabidopsis MYB family transcription factor MYB108/BOS1 and the rice bZIP transcription factor OsbZIP52/RISBZ5, known for their involvement in abiotic stress regulation, displayed substantially increased 6mA levels under cold stress (Figure 3G,H). These findings suggest that 6mA plays a role in the regulation of diverse biological processes in Arabidopsis and rice.
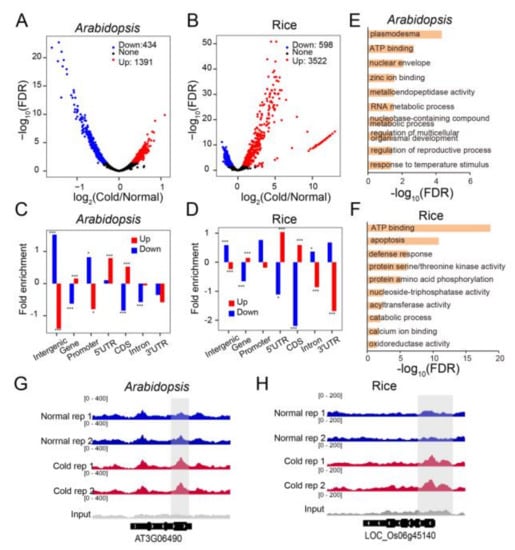
Figure 3.
Identification and gene ontology (GO) analysis of DMGs in Arabidopsis and rice upon cold treatment. (A,B) Volcano plot showing the changes of significantly up- and down-regulated enrichment of 6mA peaks after cold treatment in Arabidopsis (A) and rice (B). Each dot represents 6mA-modified region under normal and cold conditions. Fold changes in 6mA reads (Cold/Normal) are indicated on the x-axis, and -log10 (FDR) of each 6mA-modified region is indicated on the y-axis. (C,D) Barplot showing the changes of 6mA enrichment among intergenic regions, promoters (within 1 kb upstream of the TSS), and gene bodies in Arabidopsis (C) and rice (D) after cold treatment. Gene bodies were further divided into 5′ and 3′ UTRs, CDS (coding sequence), and introns. (* p < 0.05; *** p < 0.001, Fisher’s exact test). (E,F), GO analysis of up-DMGs in Arabidopsis (E) and rice (F) after cold treatment. (G,H) Genomic visualization of 6mA-IP-seq for MYB108 (AT3G06490) in Arabidopsis. (G) and bZIP52 (LOC_Os06g45140) in rice (H) under normal and cold conditions.
2.4. The Effect of 6mA Modification on Gene Expression in Response to Low Temperatures
To investigate the role of 6mA modification in regulating gene expression in response to low temperatures, we initially analyzed the global association of 6mA on gene expression. Individual gene expression was analyzed using RNA-seq with three biological replicates of Arabidopsis and rice, where high Pearson correlation coefficients (R > 0.95) were observed (Figure S2A,B). To ensure the accuracy of our expression profiling data, we conducted a comparison with published RNA-seq data in Arabidopsis available from NCBI. However, we were unable to find comparable expression profiling data in rice at the same growth stage and under equivalent cold treatment conditions. As an alternative, we employed RNA-seq data obtained by Jabre et al. for their cold-treated (SRR10553430, SRR10553431, and SRR10553432) and control (SRR10553433, SRR10553434, and SRR10553435) samples []. By analyzing this data, we identified 11,604 differentially expressed genes (DEGs), which is slightly less than the 14,293 DEGs found in our study. Importantly, over 74% of the DEGs were common between these independent studies, providing strong support for the reliability and validity of our data (Figure S2C).
The genes were subsequently divided into two classes, 6mA-containing genes and non-6mA-containing genes, to assess the association between 6mA modification and transcript abundance. Both Arabidopsis and rice showed that 6mA-containing genes had a higher average expression level than non-6mA-containing genes under cold conditions (Figure 4A,B). This indicates a positive role of 6mA modification in gene expression levels under low temperature.
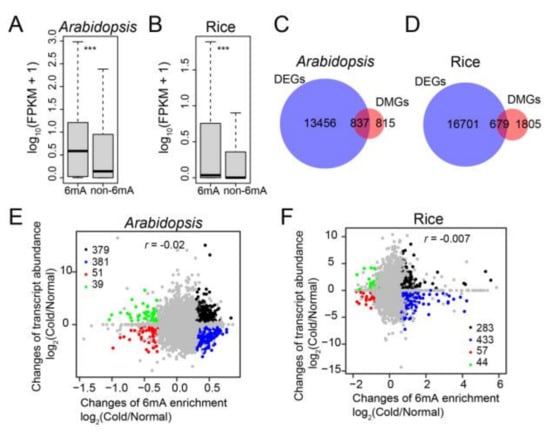
Figure 4.
Association analysis of DMGs and DEGs in Arabidopsis and rice upon cold treatment. (A,B) Box plot of gene expression levels (based on FPKM values) for genes with or without 6mA peaks (6mA and non-6mA) in Arabidopsis (A) and rice (B) under normal and cold conditions. The p values were calculated for significant differences between the two groups by the Mann–Whitney U test. ***: p < 0.001. (C,D) Venn diagrams showing the overlaps between DMGs and DEGs in Arabidopsis (C) and rice (D) after cold treatment. (E,F) Correlation of fold changes between 6mA modification level and the expression of 6mA-containing genes in Arabidopsis (E) and rice (F) in response to low temperature.
We investigated whether the cold-induced alterations of 6mA modification affected changes in gene expression. In Arabidopsis and rice, we identified a total of 14,293 (6804 up/7489 down) and 17,380 (8364 up/9572 down) differentially expressed genes (DEGs), respectively, when comparing 4 °C to 22 °C (Figure S4A,B, Table S8). We observed that 50.7% and 27.3% of DMGs overlapped with DEGs in Arabidopsis and rice, respectively (Figure 4C,D). Furthermore, these overlapped genes were significantly enriched in the response to stimulus (Figure S4C,D, Table S9). However, the DMGs of 4 °C versus 22 °C exhibited an increased or decreased level of expression at 4 °C versus 22 °C (Figure 4E,F). In Arabidopsis and rice, the Pearson correlations between the changes in 6mA modification and gene expression levels were −0.02 and −0.007, respectively (Figure 4E,F). This suggests that the fold changes in gene expression induced by cold were not correlated with 6mA enrichment in both Arabidopsis and rice.
2.5. The Response of 6mA-Modified Orthologous Genes between Arabidopsis and Rice to Low Temperature
To investigate the evolutionary conservation and functional significance of 6mA-modified orthologous genes between Arabidopsis and rice, we compared the sequence homology of genes in the two plants. We identified a total of 14,896 and 14,327 orthologs in Arabidopsis and rice, respectively, which we named Arabidopsis orthologous genes (AOGs) and rice orthologous genes (ROGs) (Figure S5A). Among these genes, 52.9% and 38.3% of 6mA-methylated genes overlapped with AOGs and ROGs, respectively, in Arabidopsis and rice (Figure 5A,B). Notably, the expression levels of 6mA-methylated orthologous genes were significantly higher than those of non-6mA-modified orthologous genes as well as 6mA-modified non-orthologous genes (Figure 5C,D). Furthermore, we identified about 370 common 6mA-methylated orthologous genes between Arabidopsis and rice, which exhibited higher expression levels than unique 6mA-methylated orthologous genes (Figure 5E–G). These results suggested that 6mA-methylated orthologous genes tend to be highly expressed. However, upon cold treatment, just over 40 differentially 6mA-methylated orthologous genes were shared between Arabidopsis and rice (Figure 5H), indicating that certain functions of 6mA-methylated orthologous genes may differ in response to cold. To investigate whether there are any common features among orthologous genes that exhibit changes in 6mA modifications between Arabidopsis and rice after cold treatment, we analyzed the trends in 6mA modification and expression level in response to low temperature. Our analysis revealed that over 70% of these genes display simultaneous upregulation in both Arabidopsis and rice, while only 26% exhibit similar alterations (both up or down) between the two species (Figure S5B,C). These findings are consistent with the global changes and relationship between 6mA modification and gene expression observed in both Arabidopsis and rice.
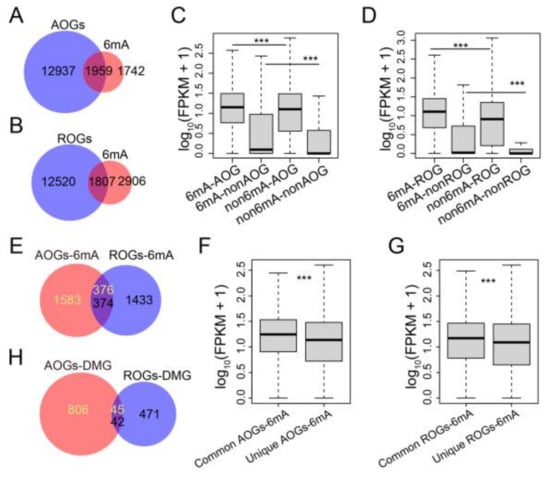
Figure 5.
The response of 6mA-methylated orthologous genes between Arabidopsis and rice upon cold treatment. (A,B) Venn diagram showing the 6mA-methylated orthologous genes in Arabidopsis (A) and rice (B). (C,D) Box plot of gene expression for AOGs (C) and ROGs (D) with or without 6mA peaks (6mA and non-6mA). The p values were calculated for significant differences between the two groups by the Mann–Whitney U test. ***: p < 0.001. (E) Venn diagram showing the overlaps of 6mA-methylated orthologous genes between Arabidopsis and rice. (F,G) Box plot of gene expression for the common and unique 6mA-methylated orthologous genes in Arabidopsis (F) and rice (G). The p values were calculated for significant differences between the two groups by the Mann–Whitney U test. ***: p < 0.001. (H) Venn diagram showing the overlapped differentially 6mA-methylated orthologous genes between Arabidopsis and rice upon cold treatment.
3. Discussion
6mA is a recently discovered DNA modification that plays a crucial role in numerous biological processes. Despite the low levels of 6mA in higher eukaryotes [,], recent research has confirmed its occurrence in animals and plants [,,,] and uncovered its function in various processes. This study focused on the biological implications of 6mA DNA modifications in Arabidopsis and rice under cold conditions. Our results revealed that there was a higher density of 6mA in gene bodies, which positively correlated with gene expression. Cold stress resulted in a higher abundance of 6mA, and up-DMGs exhibited involvement in various biological processes, but no GO term enrichment was observed for down-DMGs. Furthermore, we observed no correlation between changes in 6mA level and transcript level upon exposure to low temperature, indicating the intricate role that 6mA plays in regulating gene expression.
Under cold conditions, 6mA showed a significant increase in enrichment around the TSS region. The addition of a methyl group to the sixth position of adenine can potentially affect base-pairing energy and protein–DNA interactions and, in turn, influence transcription through the recruitment of transcription factors, RNA polymerases, histones, or other components within the surrounding chromatin context. Therefore, alterations in the position or density of 6mA in response to cold can potentially impact chromatin accessibility, nucleosome positioning, and gene expression level. Notably, other histone modifications, including histone acetylation (H3K9ac, H3K18ac, and H3K23ac) and histone methylation (H3K4me3 and H3K36me3), were also found to be present near the TSS [,,,], indicating that changes in 6mA due to low temperature likely interact with other histone modifications to modulate gene expression. Previous studies have demonstrated intricate interactions between DNA methylation and histone methylation. For example, past research has shown 5mC methylation to control histone H3K9 methylation, and a mutual enhancement between H3K9me2 and 5mC methylation has been observed in Arabidopsis [,,]. The mechanism behind the association between 6mA and other epigenetic marks should be addressed in future research.
Previous studies have shown that overall changes in 5mC under abiotic stress in Arabidopsis and rice do not correlate with changes in gene expression []; however, 5mC is known to substantially modulate the expression of some stress responsive genes [,,,]. In our study, we found the changes of 6mA enrichment were also not correlated with alterations of gene expression after cold shift. GO analysis of up-DMGs revealed enrichment in diverse processes including temperature stimulus, and some important stress response genes demonstrated changes in 6mA level, indicating that 6mA potentially participates in the regulation of stress response gene expression. In this study, it was observed that the 6mA enrichment in qLTG3-1, which is a significant quantitative trait locus responsible for controlling low-temperature germinability in rice [], increased when subjected to cold stress. Further research is necessary to investigate the significance of 6mA in adapting to environmental cues through the regulation of gene expression.
The distribution of 6mA and its association with transcription appear to differ between animals. For instance, 6mA sites are evenly distributed throughout the genome in worms [], but in flies, they are enriched in transposable elements and correlate with transposon expression []. However, in this study, we found that the genomic distribution and association of 6mA with transcription are conserved between Arabidopsis and rice. The 6mA peaks were predominantly present within gene body regions, and the density of the 6mA peaks positively correlated with gene expression levels in both species. Additionally, we observed that 52.9% and 38.3% of 6mA-methylated genes overlapped with AOGs and ROGs in Arabidopsis and rice, respectively (Figure 5A,B). Moreover, common 6mA-methylated orthologous genes between Arabidopsis and rice exhibited higher expression levels than unique 6mA-methylated orthologous genes (Figure 5E–G). The evolution of 6mA is an interesting topic that requires further investigation.
In conclusion, this study found that the overall 6mA levels increase under low temperatures and analyzed 6mA-modified regions under both normal and cold conditions. The changes in 6mA modification due to low temperature were primarily distributed in gene body regions and displayed differential correlations with gene expression levels. The DMGs were enriched in various biological processes, including stress response, and several key stress response genes revealed differential methylation levels of 6mA following cold treatment. The discovery of the roles of 6mA epi-modification in adjusting to cold temperatures not only enhances our understanding of environmental adaptation but also broadens the options available for the generation of stress-tolerant plants.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Arabidopsis thaliana Col-0 was grown in a growth chamber at 22 °C under long day conditions (16 h light/8 h dark) with a light intensity of 100 µmol m−2 s−1. Upon cold treatment, 3-week-old plants were transferred to 4 °C for 24 h. Rice (Oryza sativa, Nipponbare) was grown in a growth chamber at 28 °C under long-day conditions (16 h light/8 h dark) with a light intensity of 200 µmol m−2 s−1 for 2 weeks. For cold treatment, 2-week-old seedlings were transferred to 4 °C for 6 days.
4.2. Isolation of gDNA and 6mA-IP-Seq
gDNA of Arabidopsis and rice were extracted by using a DNAsecure Plant Kit (TIANGEN, Cat. DP320-03, Beijing, China) according to its procedures. Isolated gDNA was sonicated into 200–400 bp with Biorupter UCD-600, followed by end repair, 3′-adenylation, and adaptor ligation according to NEB Next Ultra II DNA Library Prep Kit for Illumina (E764S, NEB, Ipswich, MA, USA). The ligated DNA was denatured at 95 °C for 10 min and chilled on ice for 10 min. 10 µL of denatured DNA was saved as input. An amount of 1µg DNA was incubated with 3 µg of anti-m6A antibody (202-003, Synaptic Systems, Göttingen, Germany) at 4 °C for 6 h in 1 × IP buffer (10 mM Tris-HCl PH 7.4, 150 mM NaCl, 0.1% IGEPAL CA-630). Dynabeads Protein A (10001D, Invitrogen, Waltham, MA, USA) was washed twice with 1 × IP buffer and pre-blocked in 0.5 mL 1 × IP buffer with 20 µg/µL BSA for 2 h at 4 °C. After pre-blocked beads were washed twice with 1 × IP buffer, the DNA–antibody mixture was added to beads, rotating overnight at 4 °C. The beads were washed 4 times with 1 × IP buffer. Methylated DNA was eluted twice by 100 μL elution buffer containing N6-methyladenosine 5′-monophosphatesodium salt (M2780, Sigma, St. Louis, MO, USA) at 4 °C for 1 h. Eluted DNA was combined, then added to 20 μL of 3 M NaOAc (PH 5.3), 500 μL ethanol, and 0.5 μL glycogen. The mixture was frozen at −80 °C overnight and centrifuged. The precipitated DNA was dissolved in 20 μL ddH2O, followed by 8–10 cycles PCR amplification. The PCR product was purified by VAHTS DNA clean beads (N411-01, Vazyme, Nanjing, China). Sequencing was conducted by the Illumina NovaSeq6000 platform with 150-bp paired-end reads at Jiangbei New Area Biopharmaceutical Public Service Platform Co., Ltd. (Nanjing, China).
4.3. RNA Extraction and RNA-Seq
Total RNA of Arabidopsis and rice were isolated with TRIzol (15596026, Thermo Fisher, Waltham, MA, USA). Ribosomal RNA was removed by using an mRNA Miniprep Kit (Sigma, MRN10). An NEB Next Ultra II RNA Library Prep Kit for Illumina (NEB, E7770S) was used to construct the library, and sequencing was performed on an Illumina HiSeq 2000 platform with 100-bp paired-end reads at Jiangbei New Area Biopharmaceutical Public Service Platform Co., Ltd. (Nanjing, China).
4.4. Data Analyses
The reads of raw data were trimmed according to adapt sequence using cutadapt (v1.1) []. Then, the clean data were mapped to Arabidopsis genome (TAIR10) or rice genome (MSU7.0) using bowtie []; the parameter of maximum insert size was reset to 1000. Only unique reads were used to detect 6mA enrichment region by SCIER (v1.1) [] (using parameters redundancy threshold = 1 window size (bp) = 200 fragment size = 100 effective genome fraction = 0.74 gap size (bp) = 200 FDR = 0.01). Overlapping peaks (at least 1 bp overlap) between two biological replicates of a sample were retained for next analyses. DESeq2 (1.24.0) [] was used to compare all 6mA region from normal and cold stress with the consideration of sample variation. Differential regions with p value < 0.05 (false discovery rate < 0.05) were considered significant (DMRs).
The sequencing data were mapped to the Arabidopsis genome with matching annotation by tophat (v2.1.1) [] using default parameters. The expression value (FPKM) of each gene was calculated with cufflinks (v2.1.1) [], and cuffdiff (v2.1.1) [] was used to calculate the fold change of gene expression and FDR. Differential expression genes (DEGs) with false discovery rate < 0.05 were considered significant.
The agriGO v2.0 [] was used for GO analysis. The GO terms with FDR ≤ 0.05 were considered to be enriched. For GO terms with a hierarchical relationship, the child GO terms were kept and the parent terms were removed to avoid redundancy in this study. Orthofinder (v2.3.12) [] was run to search homologous genes between rice and Arabidopsis thaliana.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/plants12122373/s1. Figure S1. Pearson correlation analysis of 6mA-IP-seq read counts in Arabidopsis and rice under normal conditions; Figure S2. Pearson correlation analysis of RNA-seq data in Arabidopsis and rice under normal and cold conditions; Figure S3. Pearson correlation analysis of 6mA-IP-seq read counts in Arabidopsis and rice under cold conditions; Figure S4. The DEGs identified in Arabidopsis and rice after cold treatment; Figure S5. Characteristics of overlapping orthologous genes displaying changes in 6mA modifications between Arabidopsis and rice following exposure to cold treatment; Table S1. A summary of 4acC-IP-seq and RNA-seq information; Table S2. Identification of 6mA peaks under normal and cold conditions in Arabidopsis; Table S3. Identification of 6mA peaks under normal and cold conditions in rice; Table S4. Identification of DMPs and DMGs upon cold treatment in Arabidopsis; Table S5. GO analysis of up-DMGs in Arabidopsis after cold treatment; Table S6. Identification of DMPs and DMGs upon cold treatment in rice; Table S7. GO analysis of up-DMGs in rice after cold treatment; Table S8. The DEGs identified in Arabidopsis and rice after cold treatment; Table S9. GO analysis of the overlapped genes between DMGs and DEGs in Arabidopsis and rice upon cold treatment.
Author Contributions
Conceptualization, Y.W. and S.W.; methodology, F.M. and H.X.; software and validation, F.M., Y.S. and S.J.; investigation, F.M. and H.X.; formal analysis, F.M., H.X., Y.W. and S.W.; resources, F.M. and H.X.; data curation, F.M. and H.X.; writing—original draft preparation, F.M. and S.W.; writing—review and editing, Y.W. and S.W.; visualization, F.M. and H.X.; supervision, Y.W. and S.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the National Science Foundation of China (31671323; 32000207), the Innovative Project of the National Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, and the Jiangsu Collaborative Innovation Center for Modern Crop Production.
Data Availability Statement
All sequencing data generated from this study have been deposited into the NCBI GEO database under the accession number GSE230463 (Review token: efajkagklduppkt).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ratel, D.; Ravanat, J.-L.; Berger, F.; Wion, D. N6-methyladenine: The other methylated base of DNA. BioEssays 2006, 28, 309–315. [Google Scholar] [CrossRef]
- Mondo, S.J.; Dannebaum, R.O.; Kuo, R.C.; Louie, K.B.; Bewick, A.J.; LaButti, K.; Haridas, S.; Kuo, A.; Salamov, A.; Ahrendt, S.R.; et al. Widespread adenine N6-methylation of active genes in fungi. Nat. Genet. 2017, 49, 964–968. [Google Scholar] [CrossRef] [PubMed]
- Wion, D.; Casadesús, J. N6-methyl-adenine: An epigenetic signal for DNA–protein interactions. Nat. Rev. Microbiol. 2006, 4, 183–192. [Google Scholar] [CrossRef]
- Fang, G.; Munera, D.; Friedman, D.I.; Mandlik, A.; Chao, M.C.; Banerjee, O.; Feng, Z.; Losic, B.; Mahajan, M.C.; Jabado, O.J.; et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat. Biotechnol. 2012, 30, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Luo, G.Z.; Chen, K.; Deng, X.; Yu, M.; Han, D.; Hao, Z.; Liu, J.; Lu, X.; Dore, L.C.; et al. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 2015, 161, 879–892. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Blanco, M.A.; Gu, L.; Sendinc, E.; Liu, J.; Aristizabal-Corrales, D.; Hsu, C.H.; Aravind, L.; He, C.; Shi, Y. DNA Methylation on N6-Adenine in C. elegans. Cell 2015, 161, 868–878. [Google Scholar] [CrossRef]
- Zhang, G.; Huang, H.; Liu, D.; Cheng, Y.; Liu, X.; Zhang, W.; Yin, R.; Zhang, D.; Zhang, P.; Liu, J.; et al. N6-methyladenine DNA modification in Drosophila. Cell 2015, 161, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.Z.; Wang, F.; Weng, X.; Chen, K.; Hao, Z.; Yu, M.; Deng, X.; Liu, J.; He, C. Characterization of eukaryotic DNA N(6)-methyladenine by a highly sensitive restriction enzyme-assisted sequencing. Nat. Commun. 2016, 7, 11301. [Google Scholar] [CrossRef] [PubMed]
- O’Brown, Z.K.; Greer, E.L. N6-Methyladenine: A Conserved and Dynamic DNA Mark. Adv. Exp. Med. Biol. 2016, 945, 213–246. [Google Scholar] [CrossRef]
- Ye, P.; Luan, Y.; Chen, K.; Liu, Y.; Xiao, C.; Xie, Z. MethSMRT: An integrative database for DNA N6-methyladenine and N4-methylcytosine generated by single-molecular real-time sequencing. Nucleic Acids Res. 2017, 45, D85–D89. [Google Scholar] [CrossRef]
- Goh, W.S.S. Single-Nucleotide-Resolution Sequencing of N(6)-Methyldeoxyadenosine. Methods Mol. Biol. 2021, 2198, 369–377. [Google Scholar] [CrossRef]
- Liu, Q.; Fang, L.; Yu, G.; Wang, D.; Xiao, C.-L.; Wang, K. Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nat. Commun. 2019, 10, 2449. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, X.; Sheng, Y.; Liu, Y.; Gao, S. N6-adenine DNA methylation is associated with the linker DNA of H2A.Z-containing well-positioned nucleosomes in Pol II-transcribed genes in Tetrahymena. Nucleic Acids Res. 2017, 45, 11594–11606. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Wang, C.; Liu, H.; Zhou, Q.; Liu, Q.; Guo, Y.; Peng, T.; Song, J.; Zhang, J.; Chen, L.; et al. Identification and analysis of adenine N6-methylation sites in the rice genome. Nat. Plants 2018, 4, 554–563. [Google Scholar] [CrossRef]
- Liang, Z.; Shen, L.; Cui, X.; Bao, S.; Geng, Y.; Yu, G.; Liang, F.; Xie, S.; Lu, T.; Gu, X.; et al. DNA N6-Adenine Methylation in Arabidopsis thaliana. Dev. Cell 2018, 45, 406–416.e3. [Google Scholar] [CrossRef]
- Zhang, Q.; Liang, Z.; Cui, X.; Ji, C.; Li, Y.; Zhang, P.; Liu, J.; Riaz, A.; Yao, P.; Liu, M.; et al. N6-Methyladenine DNA Methylation in Japonica and Indica Rice Genomes and Its Association with Gene Expression, Plant Development, and Stress Responses. Mol. Plant 2018, 11, 1492–1508. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Zhang, G.; Wang, J.; Gao, Y.; Sun, R.; Cao, Z.; Chen, Z.; Zheng, X.; Yuan, J.; Luo, Y.; et al. 6mA-DNA-binding factor Jumu controls maternal-to-zygotic transition upstream of Zelda. Nat. Commun. 2019, 10, 2219. [Google Scholar] [CrossRef]
- Xie, Q.; Wu, T.P.; Gimple, R.C.; Li, Z.; Prager, B.C.; Wu, Q.; Yu, Y.; Wang, P.; Wang, Y.; Gorkin, D.U.; et al. N6-methyladenine DNA Modification in Glioblastoma. Cell 2018, 175, 1228–1243.e20. [Google Scholar] [CrossRef]
- Xiao, C.-L.; Zhu, S.; He, M.; Chen, D.; Zhang, Q.; Chen, Y.; Yu, G.; Liu, J.; Xie, S.-Q.; Luo, F.; et al. N6-Methyladenine DNA Modification in the Human Genome. Mol. Cell 2018, 71, 306–318.e7. [Google Scholar] [CrossRef]
- Iwasaki, M.; Hyvärinen, L.; Piskurewicz, U.; Lopez-Molina, L. Non-canonical RNA-directed DNA methylation participates in maternal and environmental control of seed dormancy. eLife 2019, 8, e37434. [Google Scholar] [CrossRef]
- Ma, N.; Chen, W.; Fan, T.; Tian, Y.; Zhang, S.; Zeng, D.; Li, Y. Low temperature-induced DNA hypermethylation attenuates expression of RhAG, an AGAMOUS homolog, and increases petal number in rose (Rosa hybrida). BMC Plant Biol. 2015, 15, 237. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Cheng, Y.; Wang, Z.; Li, Y.; Chen, L.; Huang, L.; Zhang, W.; Chen, D.; Wu, H.; Tang, B.; et al. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat. Commun. 2017, 8, 1122. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Niu, R.; Huang, T.; Shao, L.-W.; Peng, Y.; Ding, W.; Wang, Y.; Jia, G.; He, C.; Li, C.-Y.; et al. N6-methyldeoxyadenine is a transgenerational epigenetic signal for mitochondrial stress adaptation. Nat. Cell Biol. 2019, 21, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.-L.; Meng, X.; Dai, W.; Luo, Z.; Wang, C.; Fu, X.; Yang, J.; Ye, Q.; Zhou, Q. N6-methyldeoxyadenine and histone methylation mediate transgenerational survival advantages induced by hormetic heat stress. Sci. Adv. 2021, 7, eabc3026. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Wu, T.; Cui, X.; Zhu, P.; Tan, C.; Dou, X.; Hsu, K.-W.; Lin, Y.-T.; Peng, P.-H.; Zhang, L.-S.; et al. N6-Deoxyadenosine Methylation in Mammalian Mitochondrial DNA. Mol. Cell 2020, 78, 382–395.e8. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Zarka, D.G.; Stockinger, E.J.; Salazar, M.P.; Houghton, J.M.; Thomashow, M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. Cell Mol. Biol. 1998, 16, 433–442. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef]
- Medina, J.; Bargues, M.; Terol, J.; Perez-Alonso, M.; Salinas, J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression Is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 1999, 119, 463–470. [Google Scholar] [CrossRef]
- Fujino, K.; Sekiguchi, H.; Matsuda, Y.; Sugimoto, K.; Ono, K.; Yano, M. Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 12623–12628. [Google Scholar] [CrossRef]
- Wang, X.; Zou, B.; Shao, Q.; Cui, Y.; Lu, S.; Zhang, Y.; Huang, Q.; Huang, J.; Hua, J. Natural variation reveals that OsSAP16 controls low-temperature germination in rice. J. Exp. Bot. 2018, 69, 413–421. [Google Scholar] [CrossRef]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. COLD1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, S.; Dong, J.; Yang, T.; Mao, X.; Liu, Q.; Wang, X.; Liu, B. A novel functional gene associated with cold tolerance at the seedling stage in rice. Plant Biotechnol. J. 2017, 15, 1141–1148. [Google Scholar] [CrossRef]
- Saito, K.; Hayano-Saito, Y.; Kuroki, M.; Sato, Y.J.P.S. Map-based cloning of the rice cold tolerance gene Ctb1. Plant Sci. 2010, 179, 97–102. [Google Scholar] [CrossRef]
- Jabre, I.; Chaudhary, S.; Guo, W.; Kalyna, M.; Reddy, A.S.N.; Chen, W.; Zhang, R.; Wilson, C.; Syed, N.H. Differential nucleosome occupancy modulates alternative splicing in Arabidopsis thaliana. New Phytol. 2021, 229, 1937–1945. [Google Scholar] [CrossRef] [PubMed]
- Vanyushin, B.F.; Alexandrushkina, N.I.; Kirnos, M.D. N6-Methyladenine in mitochondrial DNA of higher plants. FEBS Lett. 1988, 233, 397–399. [Google Scholar] [CrossRef]
- Zhang, X.; Bernatavichute, Y.V.; Cokus, S.; Pellegrini, M.; Jacobsen, S.E. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009, 10, R62. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, X.; He, K.; Charron, J.B.; Elling, A.A.; Deng, X.W. Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol. Biol. 2010, 72, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Lam, E. ANCORP: A high-resolution approach that generates distinct chromatin state models from multiple genome-wide datasets. Plant J. Cell Mol. Biol. 2010, 63, 339–351. [Google Scholar] [CrossRef]
- Lu, L.; Chen, X.; Sanders, D.; Qian, S.; Zhong, X. High-resolution mapping of H4K16 and H3K23 acetylation reveals conserved and unique distribution patterns in Arabidopsis and rice. Epigenetics 2015, 10, 1044–1053. [Google Scholar] [CrossRef]
- Du, J.; Johnson, L.M.; Groth, M.; Feng, S.; Hale, C.J.; Li, S.; Vashisht, A.A.; Wohlschlegel, J.A.; Patel, D.J.; Jacobsen, S.E. Mechanism of DNA methylation-directed histone methylation by KRYPTONITE. Mol. Cell 2014, 55, 495–504. [Google Scholar] [CrossRef]
- Ebbs, M.L.; Bartee, L.; Bender, J. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol. Cell. Biol. 2005, 25, 10507–10515. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.P.; Lindroth, A.M.; Cao, X.; Jacobsen, S.E. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 2002, 416, 556–560. [Google Scholar] [CrossRef] [PubMed]
- Secco, D.; Wang, C.; Shou, H.; Schultz, M.D.; Chiarenza, S.; Nussaume, L.; Ecker, J.R.; Whelan, J.; Lister, R. Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife 2015, 4, e09343. [Google Scholar] [CrossRef]
- Xu, R.; Wang, Y.; Zheng, H.; Lu, W.; Wu, C.; Huang, J.; Yan, K.; Yang, G.; Zheng, C. Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J. Exp. Bot. 2015, 66, 5997–6008. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Jiang, J.; Chung, J.-S.; Wang, B.; Chen, J.; Xin, Z.; Shi, H. Regulated AtHKT1 Gene Expression by a Distal Enhancer Element and DNA Methylation in the Promoter Plays an Important Role in Salt Tolerance. Plant Cell Physiol. 2010, 52, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Sun, Y.; Cheng, B.; Xue, S.; Cheng, D.; Liu, L.; Meng, L.; Qiang, S. Variation in ICE1 Methylation Primarily Determines Phenotypic Variation in Freezing Tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2019, 60, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Xu, S.; Grullon, S.; Ge, K.; Peng, W. Spatial Clustering for Identification of ChIP-Enriched Regions (SICER) to Map Regions of Histone Methylation Patterns in Embryonic Stem Cells. In Stem Cell Transcriptional Networks: Methods and Protocols; Kidder, B.L., Ed.; Springer: New York, NY, USA, 2014; pp. 97–111. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).