Abstract
The target of rapamycin (TOR) protein phosphorylates its downstream effector p70kDa ribosomal protein S6 kinases (S6K1) for ribosome biogenesis and translation initiation in eukaryotes. However, the molecular mechanism of TOR-S6K1-ribosomal protein (RP) signaling is not well understood in plants. In the present study, we report the transcriptional upregulation of ribosomal protein large and small subunit (RPL and RPS) genes in the previously established TOR overexpressing transgenic lines of rice (in Oryza sativa ssp. indica, variety BPT-5204, TR-2.24 and TR-15.1) and of Arabidopsis thaliana (in Col 0 ecotype, ATR-1.4.27 and ATR-3.7.32). The mRNA levels of RP genes from this study were compared with those previously available in transcriptomic datasets on the expression of RPs in relation to TOR inhibitor and in the TOR-RNAi lines of Arabidopsis thaliana. We further analyzed TOR activity, i.e., S6K1 phosphorylation in SALK lines of Arabidopsis with mutation in rpl6, rpl18, rpl23, rpl24 and rps28C, where the rpl18 mutant showed inactivation of S6K1 phosphorylation. We also predicted similar putative Ser/Thr phosphorylation sites for ribosomal S6 kinases (RSKs) in the RPs of Oryza sativa ssp. indica and Arabidopsis thaliana. The findings of this study indicate that the TOR pathway is possibly interlinked in a cyclic manner via the phosphorylation of S6K1 as a modulatory step for the regulation of RP function to switch ‘on’/‘off’ the translational regulation for balanced plant growth.
1. Introduction
The TOR is a central regulator of ribosome biogenesis and protein synthesis in eukaryotes [1,2]. Several TOR substrates involved in the translational regulation have been identified previously, which include S6 kinases (S6Ks), eIF4E-binding proteins (4E-BPs), eIF4G initiation factors and La-related protein 1 (LARP1) [3,4,5,6,7]. S6Ks are members of the AGC family (which includes PKA, PKG and PKC) and are conserved substrates of TOR signaling [8,9]. Both animals and plants have two members of S6Ks, which are S6K1 and S6K2 [9,10]. TOR phosphorylates p70kDa ribosomal S6 kinase (S6K)-1 in Thr-389 residue and Thr-449 residue in animals and plants, respectively [11,12,13,14,15,16]. The activation of mammalian S6K requires phosphorylation at three conserved sites, namely the T-loop, the TM site and the HM site, which show high homology to plant S6Ks, except that the four proline residues directing Ser/Thr sites in the C-terminal domain are absent in plants. In contrast, plant S6Ks can be activated by phosphorylation at two of these three sites [10].
Multisite phosphorylation fully activates S6K1, which further phosphorylates several targets including ribosomal protein S6 in 40S ribosome. After the assembly of 40S with 60 S converts into an 80S ribosome, S6K is dephosphorylated and dissociated from the complex allowing the ribosome to proceed for the peptide elongation step [17]. Global phosphoproteomic study has predicted that LARP1 plays an important intermediate role in the translational regulation of the 5′-terminal oligopyrimidine tract (5′TOP) mRNA, when TOR is inactivated and the S6K-pT449/S6K ratios are noticeably lower [6,7]. Ribosomal proteins (RPs) are heterogeneous in nature and each RP is encoded by one of the multiple homologous partners for a given protein [18], where the function of which is determined and influenced by the tissue and the environmental conditions of the plant. The 80 S ribosomal large and small subunits consist of a heterogeneous incorporation of RPs and various rRNAs. Some cytosolic RP genes are also reported as 5′TOP mRNAs, which are regulated by either TOR-LARP1 or TOR-S6K1 signaling [7]. However, the molecular mechanisms of other S6K1 substrates are unknown and the interaction between RPs with S6K1 and their contribution to translation initiation is poorly understood in plants. The TOR also plays a crucial role in the transcription of nuclear-encoded mRNAs coding for plastidic RPs in Arabidopsis [19]. The inactivation of the TOR in two TOR-RNAi lines of Arabidopsis resulted in a coordinated decrease in transcription and translation of plastidic ribosomal protein genes, whereas the genes coding for cytosolic ribosomal proteins were interestingly upregulated [19]. In Saccharomyces cerevisiae, the mechanism of TOR-mediated regulated RP genes is well studied. The TOR controls the Forkhead transcription factor (FHL1) along with its co-activator, IFH1, and a repressor, CRF1, and triggers the transcription of RP genes under nutrient availability and nutrient-deprived conditions in yeast [20,21]. Despite having fundamental importance, the functions of TOR signaling in the regulation of genes encoding large and small subunit RPs are not well understood in plants. We, therefore, have explored the effects of the overexpression of TOR in the homologous and heterologous systems in Arabidopsis and rice, respectively, on the changes in the expression levels of RP genes.
We have also previously reported that rice and Arabidopsis plants ectopically overexpressing Arabidopsis thaliana TOR (AtTOR) exhibited enhanced growth, increased seed yield, leaf size and improved stress tolerance [2,22,23,24,25]. In the present study, we have therefore used these TOR overexpressing lines of Oryza sativa ssp. indica var. BPT-5204 and Arabidopsis thaliana for analyzing the mRNA levels of ribosomal proteins encoding genes. The lines represented with TR are TOR overexpressing lines of rice and the lines represented with ATR are TOR overexpressing lines of Arabidopsis thaliana [23,25]. In addition to the transcriptional regulation of RP genes, the results from a Western blot analysis in this study also show that TOR-mediated S6K1 phosphorylation is affected by a loss of RP function in Arabidopsis.
2. Results
2.1. Expression Analysis of RP Genes in the TOR-OE Lines of Rice and Arabidopsis
Although the involvement of TOR signaling in the modulation of RP genes transcription has been demonstrated previously in Arabidopsis [2,19,26,27], no such reports are available with regard to TOR-overexpressing crop-plants. We, therefore, performed an expression analysis of RPL and RPS genes in two high AtTOR-expressing rice lines, TR-2.24 and TR-15.1, and two Arabidopsis lines, ATR-1.4.27 and ATR-3.7.32 [23,25], to assess the effects of ectopic expression of AtTOR on the regulation of RP transcription in both of the species. In the present study, the paralogs of RPL and RPS genes were selected based on their expression levels in various developmental stages of shoots and roots of rice and Arabidopsis plants. The transcript levels of the RP genes were significantly upregulated in the rice transgenic lines. The genes RPL4, RPL14, RPL18A, RPL19.3, RPL36.2, RPL51, RPS3A, RPS6, RPS6A, RPS25A and RPS30 display upregulation in the rice transgenic plants that expressed AtTOR ectopically (Figure 1a and Figure 2a,b). The transcript levels of RPL18A, RPL19.3, RPL51, RPS25A and RPS30 were significantly increased up to ten-fold and RPL4, RPL14, RPL24B, RPL26.1, RPL30e, RPL38A, RPL44, RPS3A, RPS6, RPS6A, RPS27 and RPS27a were upregulated by more than five-fold (Figure 1a and Figure 2a,b). The transcript levels of RPL and RPS genes were significantly upregulated in the Arabidopsis TOR-OE lines, ATR-1.4.27 and ATR-3.7.32, except for the genes RPS2, RPS4, RPL12, RPL17, RPL26 and RPL39 that exhibited lower expression levels (Figure 1b and Figure 2c,d).
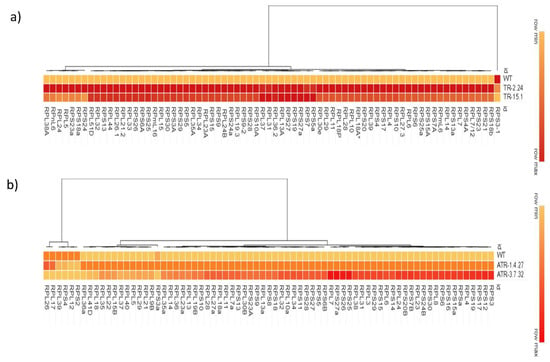
Figure 1.
Heatmap representation of expression of RP genes in the TOR-OE lines. Heatmap represents hierarchical clustering of the expression of RPL and RPS genes in TOR-overexpressing lines of rice and Arabidopsis. The RT-qPCR is used to determine the expression levels of (a) RPL and RPS genes in the TOR-OE lines TR-2.24 and TR-15.1 of rice and (b) in the lines ATR-1.4.27 and ATR-3.7.32 of Arabidopsis. Red and orange colors indicate increased RP gene expression, and the yellow color indicates decreased RP gene expression. The fold change was normalized using the 2−∆∆CT method relative to their corresponding WT control.
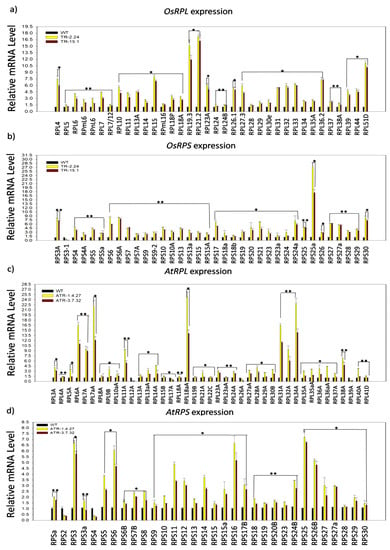
Figure 2.
Transcriptional regulation of ribosomal protein large subunit (RPL) and small subunit (RPS) genes in the TOR-OE lines of rice and Arabidopsis. The 7 DAG plants of two high AtTOR-expressing rice transgenic lines TR-2.24 and TR-15.1 were used to analyze the expression of RPL and RPS genes. The WT plants were used as controls. (a) Expression analysis of rice RPL genes in two high AtTOR overexpression lines of rice, TR-2.24 and TR-15.1. The RPL4, RPL14, RPL18A, RPL19.3, RPL36.2 and RPL51 genes were highly upregulated by 20-fold in both of the transgenic lines. (b) Expression analysis of rice RPS genes in two transgenic lines. The significant upregulation of RPS gene transcripts in two transgenic lines was observed, where the RPS3A, RPS6, RPS6A, RPS25A and RPS30 genes were highly upregulated by more than 7-fold in the transgenic plants. (c,d) The expression of RPL and RPS genes was also analyzed in two TOR-OE transgenic lines, ATR-1.4.27 and ATR-3.7.32, of Arabidopsis. The fold change was normalized using the ΔΔCT method relative to the WT plants. Rice Actin (Act1) and Arabidopsis Actin (Act2) were used as internal controls. Three biological and three technical replicates were included in this study. Vertical bars indicate the mean ± SE of three independent experiments and ANOVA analysis indicated the statistically significant differences, represented by asterisks (*) p < 0.05 and (**) p < 0.001.
The data of RP gene expression of AtTOR-OE rice and Arabidopsis lines obtained using RT-qPCR analysis were compared with the reports on the transcriptome analysis of RP genes by Dong et al. (2015) and Dobrenel et al. (2016) in TOR-RNAi lines of Arabidopsis (Figure 3a,b and Figure 4a,b) [19,27]. The results showed that the transcript levels of the cytoplasmic genes RPS5, RPS 8, RPS 14, RPS 20, RPS 23, RPS 29, RPS 30, RPL7, RPL13, RPL13a, RPL17, RPL26 and RPL29 were either slightly modulated or remained unchanged. Altogether, these data strongly suggest a positive correlation between the TOR expression levels and RP gene transcription in plants.
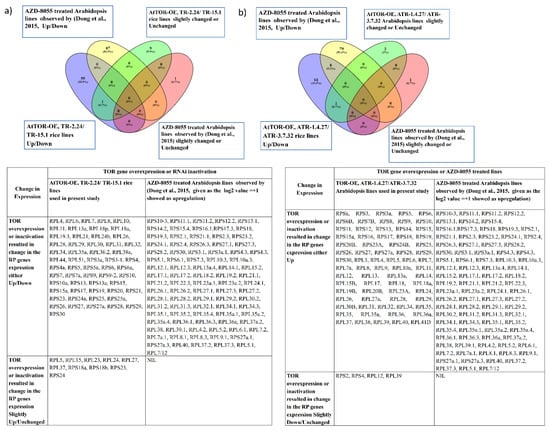
Figure 3.
Comparison of the overlapping expression patterns of RPL and RPS genes in TOR-OE lines of rice and Arabidopsis with the AZD-8055-treated Arabidopsis lines. The RP genes exhibiting a transcript level of ≥1-fold on log2 scale were considered as significantly upregulated and the transcript level below 1-fold was considered downregulated or unchanged in expression. Venn diagrams are used to show the overlaps between the RP gene expression (a) in the AZD-8055-treated lines of Arabidopsis (Dong et al., 2015 [27]) and TOR-OE lines of rice (TR-2.24 and TR-15.1) and (b) in the AZD-8055-treated lines of Arabidopsis and TOR-OE Arabidopsis lines, ATR-1.4.27 and ATR-3.7.32.

Figure 4.
Comparison of the overlapping expression patterns of RPL and RPS genes in TOR-OE lines of rice and Arabidopsis with the AtTOR-RNAi Arabidopsis lines. The RP genes exhibiting a transcript level of ≥1-fold on log2 scale were considered as significantly upregulated and the transcript level below 1-fold was considered downregulated or unchanged in expression. Venn diagrams are used to show the overlaps between the RP gene expression (a) in the TOR-RNAi lines of Arabidopsis (Dobrenel et al., 2016 [19]) and TOR-OE lines of rice, TR-2.24 and TR-15.1, (b) in the TOR-RNAi lines of Arabidopsis and TOR-OE Arabidopsis lines, ATR-1.4.27 and ATR-3.7.32, and (c) in the TOR-OE lines of rice, TR-2.24 and TR-15.1, and ATR-1.4.27 and ATR-3.7.32 Arabidopsis transgenic lines.
Among the analyzed RP genes in rice (71 RP genes) and in Arabidopsis (73 RP genes), AtTOR-OE lines TR-2.24, TR-15.1, ATR-1.4.27 and ATR-3.7.32, the transcript levels of the 28 overlapping RP genes were significantly upregulated by more than 2-fold (Figure 4c). The transcript levels of RP genes were significantly reduced in the AZD-8055-treated Arabidopsis lines [27], suggesting a positive influence of TOR expression on RP gene expression. However, contrary to this expectation, the TOR-RNAi lines of Arabidopsis showed significant upregulation of RP gene expression [19]. AZD-8055, an ATP competitive TOR inhibitor, was screened as the strongest active site TOR inhibitor (asTORi) in Arabidopsis compared with the other second-generation TOR inhibitors TORIN1 and KU63794 (KU) [27]. These inhibitors potentially target the ATP-binding pocket of the TOR kinase domain [28]. The differences between these two studies might be caused by the mode of silencing efficiencies between the TOR-RNAi lines and AZD-8055 treatment, or by selection of different growth conditions used in the study. Altogether, the results indicate that the TOR dynamically regulates the RP genes transcription and plays a crucial role in ribosome biogenesis.
2.2. Identification of Putative Phosphorylation Sites and Protein Kinase Binding Motifs in RPL and RPS Proteins
The ribosomal S6 protein kinases’ (RSKs) family proteins are serine/threonine kinases that regulate cell growth and proliferation. The two subfamilies of RSKs, p90 RSK and p70 S6K, phosphorylate RPS6 in Ser/Thr residues for the modulation of ribosome biogenesis and protein translation [7,12,28,29]. The RPS6 phosphorylation sites via S6K are identified in Ser237, Ser 238, Ser240 and Ser241 in plants [29,30]. Previously, the RPS6 was identified as the only phosphorylated protein among the ribosome small subunit proteins but the study on post-translational modifications in the ribosomal proteins of Arabidopsis identified the phosphorylation of the RPL13 protein in Ser137 residue [31]. The TOR inactivation in the RNAi lines of Arabidopsis showed inhibition of phosphorylation of RPS6A and RPS6B proteins in Ser240 and Ser237 residue at the C-terminal of peptide sequence pSRLpSSAPAKPVAA [19]. We observed a similar peptide sequence pSKLpSSAAKA at the C-terminal of rice RPS6A and RPS6B proteins in Ser 240 and Ser 241 residues, indicating the conserved nature of phosphorylation sites in plants. Similarly, the pSKLpSQGIK peptide was predicted in the RPL6 protein in Ser5 residue. Although validation through biochemical assays is further required for their phosphorylation, the predicted peptide sequences indicated the possibility of their phosphorylation by S6K (Table 1; Supplementary Figures S1–S6). The predicted peptide sequences with the Ser/Thr phosphorylation sites were then compared with the conserved RPS6 peptide sequences in Arabidopsis. The sequence alignment of rice (Oryza sativa ssp. japonica) RPS6A/B proteins from ssp. japonica shared the highest similarity with Arabidopsis thaliana RPS6A/B proteins in their Ser/Thr phosphorylation sites in similar amino acid positions (Supplementary Figures S1–S6). The OsRPL6 protein has twenty-five Ser/Thr phosphorylation sites for various AGC kinases (PKA, PKB, PKC, PKG and RSK) when compared with the AtRPL6A/B proteins having twenty phosphorylation sites, and it exhibited maximum variation in its phosphorylation sites and its amino acid sequence was adjacent to the Ser/Thr sites. A conserved Guanine residue is present in the peptide sequence of AtRPS6 in Thr 81 residue (RGTP) and in Thr 93(PGTV) of the AtRPL6 protein. Similarly, a conserved Leucine was also observed in Ser 119 (QLSL) of AtRPL6 and Ser 109 (DLSV) of AtRPS6. The presence of conserved amino acids adjacent to the phosphorylation sites in Ser/Thr residues in AtRPS6 and other RPs indicates their interaction with RSK (S6K1/2) proteins. The other ribosomal proteins consisted of similar conserved sites such as AtRPS6-Thr 91(RTGE)/AtRPL18-Thr 72(MTGK) and AtRPS6-Thr 129 (DTEK)/AtRPL18- Thr 105(FTER). The results of sequence alignment of RPs with AtRPS6 also showed the replacement of Ser or Thr to Thr or Ser phosphorylation sites with similar peptide sequences (AtRPS6-Ser105-VSPDL to AtRPL18-Thr86-ITDDL).

Table 1.
Identification of Ser/Thr phosphorylation sites in RPs mediated by the AGC kinase family (e.g., PKC, PKG, RSK, PKA, PKB).
2.3. Genotyping of Arabidopsis Mutant Lines
T-DNA insertions in the Arabidopsis mutants were detected by performing PCRs using the left border-specific primer (LB) and the corresponding gene specific right primer primer to identify homozygous or hemizygous lines and using the Left Primer + Right Primer combination for gene-specific amplification in each mutant following the protocol described at http://signal.salk.edu/tdna_FAQs.html accessed on 14 March 2019 (Supplementary Figure S7).
The position of the T-DNA insertion in the rpl23aA mutant was observed in the third exon and the rpl6 (emb2394) mutant had the T-DNA insertion in the first intron of the RPL6 gene (At1g05190). The homozygous lines of the rpl23aA knockouts were shown earlier to have embryo lethality [32,33]. As previously reported, the homozygous rpl6 (emb2394) mutant lines displayed yellow–green cotyledons [33]. The tor/tor homozygous allele showed the earliest insertion in the 47th exon of the kinase domain. The homozygous tor knockout lines also showed embryo lethality and the embryo growth was arrested at the 16–32 cell stage as previously reported [2]. Similarly, the T-DNA insertion in the rpl24 mutant was observed in the third intron of the RPL24 gene, the rpl18 mutant line had insertion in the third exon, the rps28 had insertion in the second exon and the s6k1 mutant line had insertion in the fourth exon of the gene (Supplementary Figure S8a–g).
A set of LB and each gene-specific right primers were used separately to amplify the site of insertion from the T-DNA left border to the 3′ end of the gene in the homozygous mutant lines of rpl6 (670bp), rpl18 (880bp), rpl23 (789bp), rpl24 (853bp), rps28 (894bp), s6k1 (752bp) and tor (730bp). The WT Arabidopsis genome showed no amplification with LB + Right gene specific Primers, showing the specificity by depicting the absence of T-DNA. In each case, the gene-specific Left Primers + Right Primers were used to amplify the RPL6 (1201bp), RPL18 (1194bp), RPL23 (1156bp), RPL24 (1200bp), RPS28 (1271bp), S6K1 (1105bp) and TOR (1054bp) genes without T-DNA insertion in the WT, whereas the primers had no amplification in the mutant lines (Supplementary Figure S9a–g; Supplementary Table S1). Furthermore, the transcript levels of S6K1, TOR, RPL6, RPL18, RPL23, RPL24 and RPS28 genes were analyzed in the total RNA extracted from 4DAG seeds of the SALK lines. The levels of the S6K1, TOR, RPL6, RPL18, RPL23, RPL24 and RPS28 transcripts displayed a more strongly reduced expression than the WT (Supplementary Figure S10a–g).
2.4. Ribosomal Protein Inhibition Modulates Feedback Regulation of S6K1 Phosphorylation
The S6K1 phosphorylation in Threonine 449 was strongly inhibited in the TOR-deficient RNAi silenced line (35-7) of Arabidopsis when compared to the WT (Col0) and was increased in the TOR-overexpressing line G548, suggesting that TOR kinase expression levels are major effectors of regulating the S6K1 phosphorylation in Arabidopsis [13,22]. To gain more insights into the involvement of RPs in the phosphorylation of the S6K protein, we performed a Western blot analysis of S6K1 in Arabidopsis insertional mutants for some of the important ribosomal proteins and observed that the mutation of RPL and RPS genes in Arabidopsis corroborated the differential regulation of S6K1 phosphorylation in Arabidopsis. Simultaneously, the mutation in RPs also resulted in the loss of total S6K1 stability besides its phosphorylation. The TOR and S6K1 mutants were used as negative controls and both mutants showed inhibited S6K1 phosphorylation and reduced stability of total S6K1 protein. However, the TOR mutant showed slight phosphorylation of the S6K1 protein in comparison with the S6K1 mutant, in which the phosphorylation was completely inhibited. The rpl6 mutants had an equally phosphorylated S6K1 protein, whereas the phosphorylation of S6K1 in rpl23a, rpl24, rpl24a and rps28a mutants was reduced to some extent. The stability of total S6K1 protein in rpl6, rpl18a and rpl23a was almost similar to that in the WT sample, whereas the rpl6, rpl18a and rpl23a mutants had increased stability of total S6K1 protein. The mutation of rpl18 had completely inhibited S6K1 phosphorylation and the rpl24a mutant had moderate inhibition, whereas the mutation in rpl23 and rps28 had no effect on S6K1 phosphorylation (Figure 5a–e). The WT (Col0) protein was used as a positive control for phosphorylation study and GAPDH was used as an endogenous loading control (Figure 5c). Our Western blot results clearly suggest that ribosomal proteins are interlinked and involved in the regulation of S6K1 phosphorylation in plants, as with the animal systems.
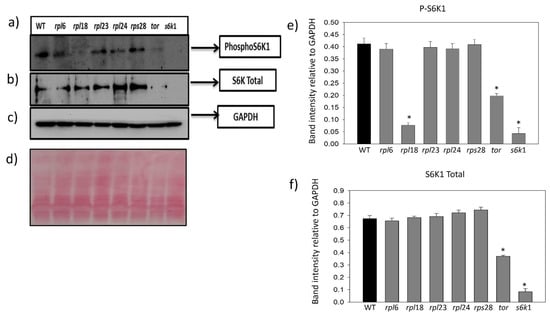
Figure 5.
S6K1 phosphorylation assay in Arabidopsis T- DNA insertional mutants. Phosphorylation of p70kDa S6K1 in Thr389 residue was detected in Arabidopsis T-DNA insertional mutants of tor and s6k1 protein along with mutants of ribosomal proteins rpl6, rpl18, rpl23a, rpl24 and rps28, and total protein isolated from WT (Col 0) Arabidopsis was taken as the control. (a) Phospho S6K1 detection in all of the mutants; (b) Western blot analysis of total S6K protein; (c) GAPDH protein used as loading control; (d) ponceau stain of the blot (e) the relative band intensity of S6K1 phosphorylation and (f) total S6K1 to band intensity of GAPDH was analyzed in the Arabidopsis T-DNA insertion mutants using Image J software. One-way ANOVA analysis was performed using the mean values (n = 3) with ±SE. Significantly reduced S6K1 phosphorylation compared with Col0 (WT, control) at p < 0.001 are marked with asterisks (*).
2.5. Identification of Putative Networking Partners of TOR-S6K1-RP Signaling
The protein–protein interactions (PPI) network were predicted between TOR, S6K1, S6K2, RPL6, RPL18, RPL23, RPL24 and RPS28 using the STRING v11 database search tool (Supplementary Figure S11a–h). STRING v11 identifies the PPI network based on the curated literature, experimentally determined/predicted interactions, co-expression or co-occurrence of proteins and the protein homology. The PPI networks of RPL6, RPL18, RPL23, RPL24 and RPS28 were also predicted separately (Supplementary Figure S11a–g; Supplementary Table S6). The PPI between TOR, S6K1, S6K2, RPS6A, RPS6B, RPL6, RPL18, RPL23.1, RPL23.2, RPL24A, RPL24B and RPS28 proteins showed interactions between 115 nodes with 5707 edges with an average 99.3 node degree, average local clustering coefficient of 0.971 and <1.0 × 10−16 p-value of the PPI enrichment (Supplementary Figure S11h, Supplementary Table S7). The 50S RPL6 (emb2394, AT1G05190.1) protein showed interaction with a confidence level of ≥0.90 with the RPL19e (emb2386, AT1G02780.1) family protein, a probable ribosome biogenesis protein RLP24 (AT2G44860.1, which is involved in the biogenesis of the 60S ribosomal subunit) and EMB2207 (AT1G43170.8) which encodes a universal cytoplasmic ribosomal protein uL3 family. RPL18 (AT1G29965) also interacts with RPL24 (AT2G36620) and RPS28C (AT5G64140). The RPL23 (AT2G39460) protein interacts with proteins essential for ribosome assembly, such as RPL24 (AT2G36620), RPS17 (AT3G10610), ATARCA (AT1G18080.1, a Transducin/WD40 repeat-like super family protein, which is also a major component of the RACK1 regulatory proteins that play a role in multiple signal transduction pathways and are involved in the MAPK cascade scaffolding in the protease IV), RACK1B_AT (AT1G48630.1, a receptor for activated C kinase 1B, whose function is to shuttle activated protein kinase C to different subcellular sites and may also function as a scaffold through physical interactions with other proteins), and the RACK1 subfamily translation elongation factor EF1B/ribosomal protein S6 family protein, which binds together with S18 to 16S ribosomal RNA (AT1G64510), and also binds to a S6K1 homolog (AT3G08850.1). Similarly, RPL24 and RPS28C (AT5G64140) interact with ribosomal constituent 40S RPSa-1, which is required for the 40S ribosome subunit assembly and processing of the 20S rRNA- precursor to mature 18S rRNA, RPuS2 family protein P40 (AT1G72370.1), LOS1 (RPS5/elongation factor G/III/V family protein, which catalyzes the GTP-dependent ribosomal translocation step during translation elongation), EIF3G1 (AT3G11400.2, an eukaryotic translation initiation factor 3 subunit G and RNA-binding component of the eukaryotic translation initiation factor 3 (eIF-3) complex, which is involved in the protein synthesis of a specialized repertoire of mRNAs and, together with other initiation factors, stimulates the binding of mRNA and methionyl-tRNAi to the 40S ribosome), ATARCA (AT1G18080.1), RPL10 (AT3G11250.1), zinc-binding ribosomal protein family protein (AT3G10950), RPS17 (AT3G10610), RPS26e (AT2G40590), RPS5 (AT2G41840), RPL16A (AT2G42740.1) and RPL18e/L15 (AT2G47570). The S6K1 and S6K2 proteins also interact with the LOS1 protein with a confidence level of 0.499. The LOS1 appears to be a downstream effector of the TOR signaling pathway via interaction with the S6K1 or S6K2 protein (Supplementary Tables S6 and S7). The S6K1 also shows interaction with a chloroplastic homolog of RPL23 (At4g18520) containing a pentatricopeptide repeat (PPR) protein. The binding of the S6K1 protein with the RPL23 having the ≥0.90 confidence level in the predicted PPI showed a possible feedback regulation of S6K1 via direct binding of the RPs.
3. Discussion
Arabidopsis RP genes are multigene families of more than two members [34]. The expression of multiple RP genes of a family is required for high translational demand during plant development. We performed a comparative analysis of transcriptional regulation of RPs via the TOR pathway in rice and Arabidopsis. Our expression analysis of RP genes clearly suggested that TOR overexpression significantly upregulates the transcription of RP genes in two diverse plant species (Figure 3a,b). Dong et al. (2015) observed that 114 RP genes have been shown in the differentially expressed genes (DEGs) and that these genes were downregulated in the Arabidopsis seedlings treated with active site TOR inhibitors (asTORis) and AZD-8055, except for one RP gene which showed upregulation [27]. The TOR inactivation in TOR-RNAi Arabidopsis lines resulted in a coordinated downregulation of the transcription and translation of nuclear-encoded mRNAs coding for plastidic RPs and a lower phosphorylation of the conserved Ser240 residue in the C-terminal region of the 40S ribosomal protein S6 (RPS6) [19]. These results suggest that TOR acts differently in the regulation of RP genes’ transcription when overexpressed or inhibited.
Our western blot analysis showed unchanged TOR activity in rpl6, rpl23 and rpl24 mutant lines, except for in the rpl18 mutant line which showed inactivation of S6K1 phosphorylation in Thr-449. Previous studies in plants and animals suggest the importance of extra-ribosomal association of RPs with RNAs and other proteins and provide a link for the possible interaction with the S6K1 for translational control [17,18,27,29,35,36,37]. Bacillus subtilis, a bacterium, reportedly requires the essential binding of RPL6 with a GTPase (RbgA) for the assembly of a ribosome large subunit [38]. The studies on S6K-RP interactions have been well documented in the animal systems [38,39]. In animal cells, RPS6 is associated with mRNAs of 5’-TOP tract such as RPL11 and RPS16 and negatively regulates their translation [39]. The association of ribosomal proteins such as RPL6, RPL18 and RPL24 along with other RPs has been reported as an essential step in translational transactivation in plants and animals upon viral infection [40,41,42,43]. Although there is no supporting evidence of an interaction between the S6K1 and 60S large subunit of ribosome in plants, several reports have demonstrated that S6K1 can also interact with other RPs. A co-immunoprecipitation assay predicted RPS3, RPS6, RPS7, RPS10, RPS11 and RPS17 and RPL13A, RPL18, RPL18A, RPL19 and RPL23 as S6K-interacting proteins with conserved phosphorylation sites, RXRXXT/S [44]. Additionally, a study showed that mutation or loss of RPS19 and other RPs induces S6K phosphorylation with an increase in ROS (reactive oxygen species) levels in zebrafish [45]. Similarly, the silencing of RPS27L led to autophagy in mouse fibroblasts and human breast cancer cells via the inhibition of S6K1 phosphorylation and mTOR Complex1 activity [46]. However, these studies were conducted using animal systems and analogous data are not available in plant systems. In addition, the binding of bacterial RPL18 to L5 and 5S rRNA is an essential step for the final phase of ribosome assembly [47,48,49]. Additionally, the TOR inactivation in Arabidopsis TOR-RNAi showed decreased cytoplasmic RPL18a- transcript levels [19]. This suggests that RPL18 plays an important role in ribosome biogenesis and translation. However, S6K1 phosphorylation might also be affected by several other protein kinases (PKA or PKB), its regulation is variable in plant systems and the exact mechanism of this is still not known, but repeated events of activation of S6Ks via multisite phosphorylation and its inactivation by dephosphorylation can switch the regulation of catalytic activity on or off and can influence target substrate selection and specificity [17,50]. Taking a cue from these results, the first possibility of unchanged S6K1 phosphorylation in other rpl mutants could be the occurrence of the dose compensation effect via the function of other paralogs of these genes. The second possibility could be the mutation in the rpl18 protein function which might be reflected in the feedback regulation of S6K1 phosphorylation in Thr-449 residue, also suggesting a possible interaction between the other 60S large ribosomal subunit RPs and the S6K1 protein. Additionally, a third possibility of impaired ribosome biogenesis and improper association in the rpl18 mutant line might lead to the dephosphorylation of S6K1. However, further investigation about plant RPs’ structures and binding partners will uncover their association with various pathways (Figure 6).
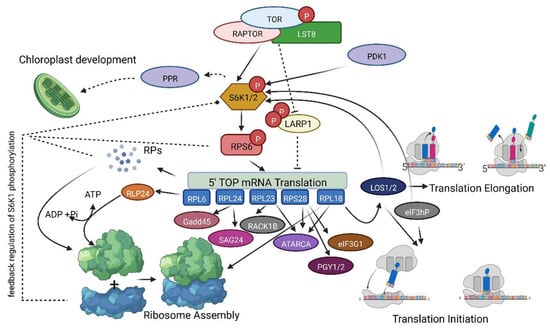
Figure 6.
Possible feedback regulation of S6K1 phosphorylation via the TOR pathway. An illustration of TOR Complex 1-mediated regulation of S6K1 phosphorylation and translational initiation by further phosphorylation and activation of RPS6 protein and other RPL and RPS proteins. Dashed lines represent signaling pathways or intermediates that are not fully revealed. The observations from the predicted PPI networks and the inhibition of RP genes suggest that the S6K1 phosphorylation is differentially regulated. Possibly, the TOR and RPs are interlinked for the regulation of S6K1 phosphorylation, where RPs also have an independent role in differentially regulating the S6K phosphorylation and modulating protein translation in the plant cell. The figure represents a model for the regulation of S6K1 phosphorylation by loss of RPs in plants, which is possibly mediated via the association of RPs with the S6K1 protein or the other regulatory proteins in the TOR pathway. The illustration was created using www.biorender.com (accessed on 21 December 2022) and exported using a free trial subscription.
4. Materials and Methods
4.1. Generation of TOR-OE Lines of Rice and Arabidopsis
The full-length TOR cDNA (7.4 kb) was amplified from Arabidopsis thaliana (Col 0) and cloned into pEarleyGate-203 [2]. The transgenic lines overexpressing the full-length Arabidopsis thaliana TOR (AtTOR) gene in rice (Oryza sativa, BPT-5204 variety of ssp. indica) and Arabidopsis thaliana (Col 0 ecotype) were developed using an in-planta transformation and floral dip method, respectively [23,25]. The high TOR-expressing transgenic lines of rice (TR-15.1 and TR-2.24) and Arabidopsis (ATR-1.4.27 and ATR-3.7.32) were used in the expression analysis of RP genes [23,25].
4.2. Growth Conditions of Rice and Arabidopsis Lines
The seeds of rice and Arabidopsis TOR-OE lines, TR-2.24, TR-15.1 and ATR-1.4.27, ATR-3.7.32, respectively, and their corresponding wild type controls (WTs) were surface sterilized with 4% sodium hypochlorite for 20 min followed by three washes with sterile double-distilled water. The rice seeds were germinated on solid Murashige and Skoog (MS) medium and were grown at 28 ± 2 °C in 16 h light/8 h dark photoperiods. The seeds of Arabidopsis thaliana SALK-T-DNA mutant lines, TOR-OE lines (ATR-1.4.27 and ATR-3.7.32) and WT plants were surface sterilized and grown on solid ½ strength of MS medium at 22 ± 2 °C of temperature and 100–150 μmol m−2 s−1 of light intensity following 16/8 h of light/dark photoperiods. Seven-day-(7 d)-old rice and Arabidopsis TOR-OE transgenic lines and their corresponding WT control counterparts were used for the expression analysis of RP genes. The heterozygous T-DNA mutant lines were grown in a growth chamber and seeds were harvested.
4.3. Nucleotide Sequence Retrieval of RPS and RPL Genes of Rice and Arabidopsis
The sequences of ribosomal protein large and small subunit genes of rice (RPL and RPS) were retrieved from RGAP-DB. The sequences were validated using RAP-DB, NCBI and various other databases to ensure that the sequences were gene specific as described by Moin et al. (2016 and 2017) and Saha et al. (2017) [35,36,37]. The Arabidopsis thaliana RPL and RPS gene sequences were retrieved from Ensemble and the NCBI database and validated using the TAIR database.
4.4. Realtime-qPCR (RT-qPCR) Analyses
Total RNA was extracted from the 7 d-grown plants of TOR-OE rice transgenic lines TR-2.24 and TR-15.1 [23], Arabidopsis TOR-OE transgenic lines ATR-1.4.27 and ATR-3.7.32 [25] and their corresponding WT plants using Tri-Reagent (Takara Bio, London, UK) following the manufacturer’s protocol. The quality and quantity of extracted RNA were checked on 1.2% agarose gel prepared in TBE (Tris-borate-EDTA) buffer and quantified using a Nano-Drop Spectrophotometer 2000 (Thermo Scientific, Waltham, MA, USA). Total RNA (2 μg) was used to synthesize the first strand cDNA using SMARTTM MMLV Reverse Transcriptase (Takara Bio, London, UK). The Actin1 and β-tubulin genes were used as endogenous reference genes in RT-qPCR analysis of RP genes in the rice transgenic lines, whereas the Actin2 and α-tubulin genes were used as endogenous reference genes for normalization of RT-qPCR analysis of RP genes in TOR-OE Arabidopsis lines. Specific primers were designed for studying the expression of RP genes in Arabidopsis and rice lines using primer3 (v.0.4.0) online tool (Supplementary Tables S1–S3). The RT-qPCR data were analyzed using three biological and three technical replicates according to the 2−ΔΔCT method [51].
4.5. In Silico Prediction of Putative Ser/Thr Phosphorylation Sites in the ribosomal protein Genes of Rice and Arabidopsis
To identify the similarity between the Ser/Thr phosphorylation sites in rice and Arabidopsis RPs, the RPS6, RPL6, RPL18, RPL23, RPL24 and RPS28 protein sequences were retrieved from the databases NCBI, UNIPROT and Ensemble Plants and the retrieved sequences were validated using the TAIR and RAPDB databases. The obtained sequences were then analyzed for the presence of Ser/Thr phosphorylation sites for PKA, PKB, PKC, PKG and RSK protein kinases of the AGC kinase family using NetPhos 3.1 Server (Table 1; http://www.cbs.dtu.dk/services/NetPhos/ accessed on 22 November 2019). The multiple sequence alignment was performed using Clustal Omega (Supplementary Figures S1–S6; https://www.ebi.ac.uk/Tools/msa/clustalo/ accessed on 17 November 2019) with the selected RPL and RPS proteins to check the similarity between various serine/threonine phosphorylation sites (Table 1; Supplementary Figures S1–S6).
4.6. Genotyping of Arabidopsis Mutants
The RP gene mutants rpl6 (CS16176), rpl18 (SALK_134424), rpl23A (SALK_091329), rpl24a (SALK_064513), rps28A (SALK_094189), s6k1 (SALK_113295) and tor (SALK_138622) in the Col 0 background were obtained in a heterozygous condition from Arabidopsis Biological Resource Center (Ohio State University, USA). The T-DNA insertions in the mutant lines were confirmed using PCR. These lines were grown by successive inbreeding (the T1-generation plants were self-pollinated, the T2-seeds were collected, and the process was repeated until collection of T3-generation seeds) up to T3-generation until they became homozygous insertion lines. As these lines are reported to be embryo lethal when they achieve homozygosity [2,32,33], this shows that the functional nature of the gene amongst their paralogs is essential for embryo development. We, therefore, used the mature seeds from the heterozygous mutant lines in T2 generation (their homozygosity was confirmed through genotyping) to check the mRNA levels of the mutated genes in these lines and for protein extraction to detect the S6K1 phosphorylation. The transcript accumulation of genes in the SALK lines was analyzed using RT-qPCR. The mature seeds obtained in T3 generation of each SALK line were used to extract total RNA [52]. The cDNAs were synthesized and used to analyze the transcript levels of genes in the homozygous s6k1−/−, tor−/−, rpl6−/−, rpl18−/−, rpl23−/−, rpl24−/− and rps28−/− (−/− represents that both alleles were absent in the homozygous mutant lines) SALK lines. The Actin2 and α-tubulin genes were used as endogenous reference genes for normalization and WT was used as the control. The primers used in genotyping and RT-qPCR are detailed in the Supplementary Tables S4 and S5.
4.7. Western Blot Analysis
Total protein was extracted from the mature seeds of Arabidopsis WT plants and homozygous T-DNA insertion mutants of Arabidopsis RP genes: rpl6 (Emb2394, CS16176), rpl18 (SALK_134424C), rpl23A (SALK_091329), rpl24a (SALK_064513), rps28A (SALK_094189), Ats6k1 (SALK_113295) and tor (SALK_138622), in T3-generation using a standard phenol extraction method. As reported earlier, the homozygous tor−/− and rpl23aA−/− knockout mutants had defective embryo development [2,32]. To avoid phenotypic aberrations in the homozygous mutants, we used seeds harvested from T2 plants for S6K1 phosphorylation assay. The protein sample from the WT-Col 0 was used as a positive control, whereas protein samples from tor and s6k1 mutants were used as negative controls in the phosphorylation assay. The protein precipitate was dissolved in rehydration buffer (7 M Urea, 2 M Thiourea, 4% CHAPS and 30 mM DTT) and quantified using the Bradford method with BSA standards. About 30 μg of total protein was loaded in SDS-PAGE and Western blot analysis. The phosphorylation site of human and Arabidopsis p70kDa-S6K1 proteins is homologous and conserved for Thr-389 and Thr-449 residues [28]. We further performed S6K1 phosphorylation in homozygous T-DNA insertion mutants of the genes encoding for cytosolic ribosomal proteins rpl6−/−, rpl18−/−, rpl23A−/−, rpl24a−/−, rps28A−/−, s6k1−/− and tor−/− to determine the cross-link between TOR and RPs. The anti-human S6K1 antibody raised in mice (anti phospho70kDa-S6K1-Thr(P)-389) (Cell Signaling Technologies, Danvers, MA, USA; cat# 9206) was used for detecting S6K1 phosphorylation in the mutants, and WT control, the anti-70kDa S6K1 (CST, Danvers, MA, USA; cat# 9202) and anti-GAPDH (Santa Cruz, Dallas, Texas, USA; FL-335#SC25778) were used as loading controls. The membrane was incubated with secondary HRP-conjugated antibodies and signals were detected using a chemi-luminescence method (ChemiDoc XRS, Bio-Rad, USA).
4.8. Identification of Interaction between TOR and Ribosomal Proteins
A protein–protein interactions (PPI) between the TOR protein and RPs were identified through the bioinformatic approach using STRING v11 [53]. The STRING v11 (https://string-db.org/ accessed on 24 July 2021) identifies the interaction based on the evidence from Arabidopsis homologs in other species. The search tool of STRING v11 was used for the retrieval of interacting genes/proteins associated with TOR and ribosomal proteins and to construct the PPI network. The TOR and RP genes used in the study were listed as inputs in the STRING dB and were searched for their neighbor interactors at a high level of confidence (sources: experiments, databases; score ≥ 0.90).
4.9. Bioinformatic Analysis
The results from expression analyses of RP genes were depicted as heatmaps using the MORPHEUS program (https://software.broadinstitute.org/morpheus/ accessed on 24 November 2020). Venn diagrams were depicted using VENNY 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/index.html accessed on 17 November 2020). Quantification of phosphorylation of S6K1 protein in the Western blot analysis was performed using Image J software (https://imagej.nih.gov/ij/index.html accessed on 17 November 2020). The putative Ser/Thr phosphopeptide sequences were identified using NetPhos 3.1 Server (http://www.cbs.dtu.dk/services/NetPhos/ accessed on 22 November 2019) and STRING v11 (https://string-db.org/ accessed on 24 July 2021) was used to predict protein–protein interactions.
4.10. Statistical Analysis
Statistical analysis was performed with the mean values using one-way ANOVA in the SigmaPlotv11 software. Mean values are represented with standard error (±SE) with the significance level p < 0.001 represented as asterisks ‘**’ and p < 0.05 represented as an asterisk ‘*’.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants12010176/s1, Figure S1: Multiple sequence alignment of RPL and RPS proteins identifies conserved Ser/ Thr phosphorylation sites; Figure S2: Sequence alignment of RPS6A/B and RPL6 proteins of Arabidopsis thaliana and Oryza sativa ssp. japonica; Figure S3: Alignment of RPS6A/B and RPL18 proteins of Arabidopsis thaliana and Oryza sativa ssp. japonica; Figure S4: Alignment of RPS6A/B and RPL23 proteins of Arabidopsis thaliana and Oryza sativa ssp. japonica; Figure S5: Alignment of RPS6A/B and RPL24 proteins of Arabidopsis thaliana and Oryza sativa ssp. japonica; Figure S6: Alignment of RPS6A/B and RPS28 proteins of Arabidopsis thaliana and Oryza sativa ssp. japonica; Figure S7: Phenotype of heterozygous T-DNA Arabidopsis mutants of ribosomal protein genes in T1 generation; Figure S8: Positions of T-DNA insertion site in the SALK Arabidopsis mutant lines; Figure S9: Genotyping of mutants for confirmation of T-DNA insertion; Figure S10: Measurement of transcript level of the mutant allele in the SALK lines; Figure S11: Identification of interacting proteins of TOR-S6K-RPs signaling; Table S1: List of primers used in RT-qPCR analysis of RPS genes in TOR-OE lines of rice; Table S2: List of primers used in RT-qPCR analysis of RPL genes in TOR-OE lines of rice; Table S3: List of primers used in RT-qPCR analysis of RPL/RPS genes in the TOR-OE lines of Arabidopsis thaliana; Table S4: List of primers used in genotyping of T-DNA mutants; Table S5: List of primers used in RT-qPCR of the T-DNA insertion in the SALK lines; Table S6: Identification of the predicted functionally interacting partners of TOR-S6K-RP signaling for ribosome biogenesis. Table S7: The TOR, RPS6A, RPS6B, RPL6, RPL18, RPL23.1, RPL23.2, RPL24A, RPL24B, RPS28, S6K1, and S6K2 protein enrichment analysis.
Author Contributions
A.B., M.S.M., R.D. and P.B.K. designed the work; A.B. performed all the experiments and analyses; M.M. helped with the RT-qPCR analysis; A.B.M.R. and M.B.G. helped with S6K1 phosphorylation; A.B., M.S.M., R.D. and P.B.K. prepared the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Department of Biotechnology, Government of India under Research Associate program in Biotechnology and Life Sciences (2-29/RA/Bio/2018/550) and the APC was funded by Global Institute for Food Security.
Data Availability Statement
Data availability is reported in the text. All the data in the study is available in the main text and in the supplementary material.
Acknowledgments
A.B. acknowledges financial support from the Department of Biotechnology Research Associate program in Biotechnology and Life Sciences, Government of India (2-29/RA/Bio/2018/550), and A.B. and R.D. acknowledge Global Institute for Food Security, Saskatoon, Canada. A.B., M.M. and M.S.M. also acknowledge the Department of Biotechnology, ICAR-Indian Institute of Rice Research (IIRR), Hyderabad, India. The authors acknowledge Gassmann Walter of Christopher S. Bond Life Sciences Center, University of Missouri, USA for providing the Arabidopsis SALK mutant lines of RPs and TOR. A.B.M.R. and M.G.B. acknowledge the Department of Animal Biology, University of Hyderabad, India. P.B.K. acknowledges the National Academy of Sciences, India, for the grant for the NASI-Platinum Jubilee Senior Scientist position.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
TOR, target of rapamycin; TOR-OE, TOR overexpressing; RPs, ribosomal proteins; RPL, ribosomal protein large subunit; RPS, ribosomal protein small subunit; WT, wild type; DAG, days after germination; S6K1, ribosomal protein small subunit 6 kinase 1.
References
- Urban, J.; Soulard, A.; Huber, A.; Lippman, S.; Mukhopadhyay, D.; Deloche, O.; Wanke, V.; Anrather, D.; Ammerer, G.; Riezman, H.; et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 2007, 26, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Qiu, S.; Venglat, P.; Xiang, D.; Li, F.; Datla, R. Target of Rapamycin Regulates Development and Ribosomal RNA Expression through Kinase Domain in Arabidopsis. Plant Physiol. 2011, 155, 1367–1382. [Google Scholar] [CrossRef] [PubMed]
- Thoreen, C.C.; Chantranupong, L.; Keys, H.R.; Wang, T.; Gray, N.S.; Sabatini, D.M. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012, 485, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Meyuhas, O.; Kahan, T. The race to decipher the top secrets of TOP mRNAs. Biochim. Biophys. Acta 2015, 1849, 801–811. [Google Scholar] [CrossRef]
- Berman, A.J.; Thoreen, C.C.; Dedeic, Z.; Chettle, J.; Roux, P.P.; Sarah, B.P. Controversies around the function of LARP1. RNA Biol. 2020, 11, 1733787. [Google Scholar] [CrossRef]
- Philippe, L.; van den Elzen, A.M.G.; Watson, M.J.; Thoreen, C.C. Global analysis of LARP1 translation targets reveals tunable and dynamic features of 5′ TOP motifs. Proc. Natl. Acad. Sci. USA 2020, 117, 5319–5328. [Google Scholar] [CrossRef]
- Scarpin, M.R.; Leiboff, S.; Brunkard, J.O. Parallel global profiling of plant TOR dynamics reveals a conserved role for LARP1 in translation. eLife 2020, 9, e58795. [Google Scholar] [CrossRef]
- Henriques, R.; Magyar, Z.; Monardes, A.; Khan, S.; Zalejski, C.; Orellana, J.; Szabados, L.; de la Torre, C.; Koncz, C.; Bogre, L. Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1-E2F pathway activity. Embo J. 2010, 29, 2979–2993. [Google Scholar] [CrossRef]
- Magnuson, B.; Ekim, B.; Fingar, D.C. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem. J. 2012, 441, 1–21. [Google Scholar] [CrossRef]
- Yaguchi, M.; Ikeya, S.; Kozaki, A. The activation mechanism of plant S6 kinase (S6K), a substrate of TOR kinase, is different from that of mammalian S6K. FEBS Lett. 2020, 594, 776–787. [Google Scholar] [CrossRef]
- Xiong, Y.; Sheen, J. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J. Biol. Chem. 2012, 287, 2836–2842. [Google Scholar] [CrossRef] [PubMed]
- Mahfouz, M.M.; Kim, S.; Delauney, A.J.; Verma, D.P. Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 2006, 18, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Schepetilnikov, M.; Dimitrova, M.; Mancera-Martínez, E.; Geldreich, A.; Keller, M.; Ryabova, L.A. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 2013, 32, 1087–1102. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, B.D.; Smith, E.M.; Yelle, N.; Alain, T.; Bushell, M.; Pause, A. The ever-evolving role of mTOR in translation. Semin. Cell Dev. Biol. 2014, 36, 102–112. [Google Scholar]
- Ahn, C.S.; Ahn, H.K.; Pai, H.S. Overexpression of the PP2A regulatory subunit Tap46 leads to enhanced plant growth through stimulation of the TOR signalling pathway. J. Exp. Bot. 2015, 66, 827–840. [Google Scholar] [CrossRef]
- Xiong, F.; Zhang, R.; Meng, Z.; Deng, K.; Que, Y.; Zhuo, F.; Feng, L.; Guo, S.; Datla, R.; Ren, M. Brassinosteriod insensitive 2 (BIN2) acts as a downstream effector of the target of rapamycin (TOR) signaling pathway to regulate photoautotrophic growth in Arabidopsis. New Phytol. 2017, 213, 233–249. [Google Scholar] [CrossRef]
- Bohlen, J.; Roiuk, M.; Teleman, A.A. Phosphorylation of ribosomal protein S6 differentially affects mRNA translation based on ORF length. Nucleic Acids Res. 2021, 49, 13062–13074. [Google Scholar] [CrossRef]
- Wool, I.G. The structure and function of eukaryotic ribosomes. Annu. Rev. Biochem. 1979, 48, 719–754. [Google Scholar] [CrossRef]
- Dobrenel, T.; Mancera-Martínez, E.; Forzani, C.; Azzopardi, M.; Davanture, M.; Moreau, M.; Schepetilnikov, M.; Chicher, J.; Langella, O.; Zivy, M.; et al. The Arabidopsis TOR Kinase Specifically Regulates the Expression of Nuclear Genes Coding for Plastidic Ribosomal Proteins and the Phosphorylation of the Cytosolic Ribosomal Protein S6. Front. Plant Sci. 2016, 7, 1611. [Google Scholar] [CrossRef]
- Martin, D.E.; Soulard, A.; Hall, M.N. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 2004, 119, 969–979. [Google Scholar] [CrossRef]
- Wade, J.T.; Hall, D.B.; Struhl, K. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 2004, 432, 1054–1058. [Google Scholar] [CrossRef] [PubMed]
- Deprost, D.; Yao, L.; Sormani, R.; Moreau, M.; Leterreux, G.; Nicolaı, M.; Bedu, M.; Robaglia, C.; Meyer, C. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007, 8, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, A.; Moin, M.; Kumar, M.U.; Reddy, A.B.; Ren, M.; Datla, R.; Siddiq, E.A.; Kirti, P.B. Ectopic expression of Arabidopsis Target of Rapamycin (AtTOR) improves water-use efficiency and yield potential in rice. Sci. Rep. 2017, 23, 42835. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, A.; Moin, M.; Madhav, M.S.; Kirti, P.B. Target of Rapamycin (TOR), a master regulator of multiple signaling pathways and a potential candidate gene for crop improvement. Plant Biol. 2019, 21, 190–205. [Google Scholar] [PubMed]
- Bakshi, A.; Moin, M.; Madhav, M.S.; Datla, R.; Kirti, P.B. Target of rapamycin (tor) negatively regulates chlorophyll degradation and lipid peroxidation and controls responses under abiotic stress in Arabidopsis thaliana. Plant Stress 2021, 2, 100020. [Google Scholar] [CrossRef]
- Kojima, H.; Suzuki, T.; Kato, T.; Enomoto, K.-I.; Sato, S.; Kato, T.; Tabata, S.; Sa’ez-Vasquez, J.; Echeverría, M.; Nakagawa, T.; et al. Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 2007, 49, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Xiong, F.; Que, Y.; Wang, K.; Yu, L.; Li, Z.; Ren, M. Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis. Front. Plant Sci. 2015, 6, 677. [Google Scholar] [CrossRef]
- Xiong, Y.; McCormack, M.; Li, L.; Hall, Q.; Xiang, C.; Sheen, J. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 2013, 496, 181–186. [Google Scholar] [CrossRef]
- Williams, A.J.; Werner-Fraczek, J.; Chang, I.F.; and Bailey-Serres, J. Regulated phosphorylation of 40S ribosomal protein S6 in root tips of maize. Plant Physiol. 2003, 132, 2086–2097. [Google Scholar] [CrossRef]
- Chang, I.F.; Szick-Miranda, K.; Pan, S.; Bailey-Serres, J. Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiol. 2005, 137, 848–862. [Google Scholar] [CrossRef]
- Carroll, A.J.; Heazlewood, J.L.; Ito, J.; Millar, A.H. Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol. Cell Proteom. 2008, 7, 347–369. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, R.F.; Bonham-Smith, P.C. Transcript profiling demonstrates absence of dosage compensation in Arabidopsis following loss of a single RPL23a paralog. Planta 2008, 228, 627–640. [Google Scholar] [CrossRef] [PubMed]
- Aryamanesh, N.; Ruwe, H.; Sanglard, L.V.; Eshraghi, L.; Bussell, J.D.; Howell, K.A.; Small, I.; des Francs-Small, C.C. The Pentatricopeptide Repeat Protein EMB2654 Is Essential for Trans-Splicing of a Chloroplast Small Ribosomal Subunit Transcript. Plant Physiol. 2017, 173, 1164–1176. [Google Scholar] [CrossRef] [PubMed]
- Barakat, A.; Szick-Miranda, K.; Chang, I.F.; Guyot, R.; Blanc, G.; Cooke, R.; Delseny, M.; Bailey-Serres, J. The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 2001, 127, 398–415. [Google Scholar] [CrossRef] [PubMed]
- Moin, M.; Bakshi, A.; Saha, A.; Dutta, M.; Kirti, P.B. Rice Ribosomal Protein Large Subunit Genes & their Spatio-temporal & stress regulation. Front. Plant Sci. 2016, 7, 1284. [Google Scholar] [PubMed]
- Moin, M.; Bakshi, A.; Madhav, M.S.; Kirti, P.B. Expression Profiling of Ribosomal Protein Gene Family in Dehydration Stress Responses and Characterization of Transgenic Rice Plants Overexpressing RPL23A for Water-Use Efficiency and Tolerance to Drought and Salt Stresses. Front. Chem. 2017, 5, 97. [Google Scholar] [CrossRef]
- Saha, A.; Das, S.; Moin, M.; Dutta, M.; Bakshi, A.; Madhav, M.S.; Kirti, P.B. Genome-wide identification and comprehensive expression profiling of Ribosomal protein small subunit (RPS) genes and their comparative analysis with the large subunit (RPL) genes in rice. Front. Plant Sci. 2017, 8, 1553. [Google Scholar] [CrossRef]
- Gulati, M.; Jain, N.; Davis, J.H.; Williamson, J.R.; Britton, R.A. Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly. PLoS Genet. 2014, 10, e1004694. [Google Scholar] [CrossRef][Green Version]
- Hagner, P.R.; Mazan-Mamczarz, K.; Dai, B.; Balzer, E.M.; Corl, S.; Martin, S.S.; Zhao, X.F.; Gartenhaus, R.B. Ribosomal protein S6 is highly expressed in non-Hodgkin lymphoma and associates with mRNA containing a 5 terminal oligopyrimidine tract. Oncogene 2011, 30, 1531–1541. [Google Scholar] [CrossRef]
- Leh, V.; Yot, P.; Keller, M. The cauliflower mosaic virus translational transactivator interacts with the 60S ribosomal subunit protein L18 of Arabidopsis thaliana. Virology 2000, 266, 1–7. [Google Scholar] [CrossRef]
- Wang, J.; Yang, X.; Zhou, P.; Han, H. Cloning of mouse genomic ribosomal protein L6 gene and analysis of its promoter. Biochim. Biophys. Acta 2002, 1576, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Martínez, F.; Daròs, J.A. Tobacco etch virus protein P1 traffics to the nucleolus and associates with the host 60S ribosomal subunits during infection. J. Virol. 2014, 88, 10725–10737. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, X.; Zhou, Y. Ribosomal protein L18 is an essential factor that promote rice stripe virus accumulation in small brown planthopper. Virus Res. 2018, 247, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Pavan, I.C.B.; Yokoo, S.; Granato, D.C.; Meneguello, L.; Carnielli, C.M.; Tavares, M.R.; do Amara, C.L.; de Freitas, L.B.; PaesLeme, A.F.; Luchessi, A.D.; et al. Different interactomes for p70-S6K1 and p54-S6K2 revealed by proteomic analysis. Proteomics 2016, 16, 2650–2666. [Google Scholar] [CrossRef] [PubMed]
- Heijnen, H.F.; van Wijk, R.; Pereboom, T.C.; Goos, Y.J.; Seinen, C.W.; van Oirschot, B.A.; van Dooren, R.; Gastou, M.; Giles, R.H.; van Solinge, W.; et al. Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin Pathway. PLoS Genet. 2014, 10, e1004371. [Google Scholar] [CrossRef]
- Xiong, X.; Liu, X.; Li, H.; He, H.; Sun, Y.; Zhao, Y. Ribosomal protein S27-like regulates autophagy via the β-TrCP-DEPTORmTORC1 axis. Cell Death Dis. 2018, 9, 1131. [Google Scholar] [CrossRef]
- Steitz, J.A.; Berg, C.; Hendrick, J.P.; La Branche-Chabot, H.; Metspalu, A.; Rinke, J.; Yario, T. A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J. Cell Biol. 1988, 106, 545–556. [Google Scholar] [CrossRef]
- Deshmukh, M.; Stark, J.; Yeh, L.C.C.; Lee, J.C.; Woolford, J.L. Multiple regions of yeast ribosomal protein L1 are important for its interaction with 5 S rRNA and assembly into ribosomes. J. Biol. Chem. 1995, 270, 30148–30156. [Google Scholar] [CrossRef]
- Almutairi, Z.M. Molecular characterization and expression analysis of ribosomal L18/L5e gene in Pennisetum glaucum (L.) R. Br. Saudi J. Biol. Sci. 2021, 28, 3585–3593. [Google Scholar] [CrossRef]
- Arif, A.; Jia, J.; Willard, B.; Li, X.; Fox, L.P. Multisite Phosphorylation of S6K1 Directs a Kinase Phospho-code that Determines Substrate Selection. Mol. Cell 2019, 73, 446–457. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Oñate-Sánchez, L.; Vicente-Carbajosa, J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes 2008, 1, 93. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, 607–613. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).