Abstract
Plant beneficial microorganisms improve the health and growth of the associated plants. Application of beneficial microbes triggers an enhanced resistance state, also termed as induced systemic resistance (ISR), in the host, against a broad range of pathogens. Upon the activation of ISR, plants employ long-distance systemic signaling to provide protection for distal tissue, inducing rapid and strong immune responses against pathogens invasions. The transmission of ISR signaling was commonly regarded to be a jasmonic acid- and ethylene-dependent, but salicylic acid-independent, transmission. However, in the last decade, the involvement of both salicylic acid and jasmonic acid/ethylene signaling pathways and the regulatory roles of small RNA in ISR has been updated. In this review, the plant early recognition, responsive reactions, and the related signaling transduction during the process of the plant–beneficial microbe interaction was discussed, with reflection on the crucial regulatory role of small RNAs in the beneficial microbe-mediated ISR.
1. Introduction
With the rapid growth of the world’s population, people’s demand for agricultural products is increasing. Plants are sessile organisms, frequently exposed to a myriad of microorganisms, including pathogenic and beneficial ones. The pursuit of productivity has led to the abuse of fertilizers and pesticides, causing serious environmental pollution and ecological damage. During development, the main concerns in the agricultural industry have changed from yield to food quality and environmental impact. The use of environmentally friendly agricultural inputs has arisen since then. Biological control uses beneficial organisms to suppress harmful organisms and promote plant growth. Currently, many beneficial microorganisms, such as Bacillus, Pseudomonas, and Trichoderma, are used as biological control agents to control field plant diseases.
Plants possess an innate ability to sense and recognize potential invading microorganisms and to activate defense responses [1]. On the contrary, to perceive the beneficial microorganisms and form a symbiotic relationship with them, plants adopt similar, yet distinct, cell surface receptors [2]. Plants can recognize microbial- or pathogen-associated molecular patterns (MAMPs or PAMPs), such as bacterial flagellin and fungal chitin, through transmembrane pattern recognition receptors (PRRs), and this process triggers the first layer of immune defense, named pattern-triggered immunity (PTI) [3]. However, pathogens can overcome the first layer by suppressing PTI signaling or evading recognition of PRRs by secreting virulence effectors [4]. Effectors are a kind of virulence-associated molecule, delivered by pathogens via microbial secretion systems into plant cells or the apoplast to suppress host immunity [4]. In turn, the second layer of plant immunity, called effector-triggered immunity (ETI), evolved to recognize pathogen effectors through polymorphic NB-LRR proteins (possessing nucleotide-binding and leucine-rich repeat domains), resulting in hypersensitive reaction (HR) to limit the pathogen spread [5]. Interestingly, recent studies showed that PRRs are also required for ETI [6]. The complex and precise immune system built from host–pathogen competition allows beneficial microorganisms to induce plant immunity through targeting the key elements in the process of PTI and ETI by modulating host small RNAs.
Plant systemic resistance can be divided into induced systemic resistance (ISR) and systemic acquired resistance (SAR), induced by non-pathogenic microbes and pathogenic microbes, respectively [7,8]. Colonization by beneficial microbes induces a physiological state of plant host called “priming”. Upon the activation of “priming”, plants display stronger and faster defense responses against the following invasion of pathogens, demonstrated as a common feature of systemic resistance induced by beneficial microorganisms [9]. SAR was first discovered in 1961 and identified as a salicylic acid (SA)-dependent plant defense, featured by accumulation of SA and activation expression of pathogenic-related (PR) genes [10,11]. In 1991, three research groups independently and specifically evidenced that beneficial microbes enhanced plant immunity by ISR [12,13,14]. Among these three groups, Kloepper et al. found that plant growth-promoting rhizobacteria (PGPR) could induce cucumber systemic resistance to Fusarium-wilt, bacterial angular leaf spot, root-knot nematode, and cucumber mosaic cucumovirus [13,15,16,17,18]. In 1996, Pieterse et al. firstly reported that systemic resistance induced by PGPR was independent of SA and PR proteins in Arabidopsis thaliana, but depended on jasmonic acid (JA) and ethylene (ET) pathway [19,20], which was proposed to be the difference between ISR and SAR. Nevertheless, multiple following reports demonstrating activation of both SA and JA/ET signaling pathways in ISR triggered by beneficial microbes revealed the complexity and diversity of signal pathways involved in ISR [21,22,23,24].
Up to now, various beneficial microorganisms have shown the potential to induce systemic resistance. Beneficial bacteria, such as Bacillus spp. and Pseudomonas spp., can stimulate defense responses and help plants to obtain broad-spectrum disease resistance [14,25]. Beneficial fungi, such as Trichoderma spp. and arbuscular mycorrhizal fungi (AMFs), have been considered to be widespread potential biocontrol agents [26,27]. Root treatment with Trichoderma harzianum T39 induced ISR in bean against Botrytis cinerea [27]. AMFs, which form symbiotic associations with many plant root systems, have been proved to induce local and systemic resistance to Phytophthora parasitica in tomato roots [26].
In this review, we summarize the recognition of beneficial microorganisms and early events that occur during induced systemic resistance, highlighting reactive oxygen species burst, callose deposition that can inhibit the infection and expansion of pathogens, calcium signaling, and transcriptional factors, that play a significant role in regulating the expression of downstream defense-related genes and diseases control. The crosstalk of signaling transduction pathways and the function of secondary metabolites and stomatal regulation in ISR will be discussed. Finally, we will highlight recent advances about the role of small RNAs in rhizobacteria-induced ISR.
2. Recognition of Beneficial Microbes by Plants
Plants can sense the beneficial microbes by recognizing the common microbial compounds they produce, such as flagellin, lipopolysaccharides (LPS), exopolysaccharides, and chitin oligosaccharides, such as ligands [2,28,29,30]. Binding with ligands, the receptor proteins recruit co-receptors and form complexes to phosphorylate downstream substrates, leading a signal cascade involving oxidative burst, Ca2+ influx, MAPK activation, and hormone signaling activation [31].
The N-terminal part of flagellin, including the 22-amino acid epitope flg22, is highly conserved in a wide range of eubacteria [32]. The flagellin from beneficial microbes, such as Bacillus subtilis and Burkholderia phytofirmans, can be recognized by their host plants [32,33]. FLAGELLIN-SENSING 2 (FLS2) is the first reported receptor to recognize flagellin from PGPR [34]. The perception of flg22 results in the heterodimerization between FLS2 and its co-receptors, BRI1-associated kinase (BAK1) and BAK1-LIKE1 (BKK1), which phosphorylate their interacting receptor-like cytoplasmic kinase Botrytis-induced kinase1 (BIK1) to initiate the PTI signaling [35,36,37]. Arabidopsis thaliana bak1 mutations showed normal flagellin binding but abnormal PTI responses, indicating that BAK1 acted as a positive regulator in signaling [36]. BIK1 is phosphorylated upon flagellin perception and subsequently, transphosphorylates FLS2/BAK1 complex to transmit flagellin signaling and activate intracellular signaling cascades [37]. Similar to bak1 mutant, bik1 mutant is compromised in flagellin-mediated responses to the invasion of non-pathogenic microbes, indicating that BIK1 is an essential component in MAMP signal transduction and induced systemic resistance. In addition, rhizobia and AMF establish symbiosis with the host by means of chitin-derived oligosaccharides signals [2]. Nod factors, for instance, are acylated lipo-chitooligosaccharides, delivered by rhizobia and recognized by LysM receptor-like kinases to activate a common symbiotic pathway, which controls both the arbuscular mycorrhizal symbioses and the rhizobia-legume to form mycorrhization and nodulation [38,39].
Beneficial microbes produce a large number of MAMPs, such as flagellin and lipopolysaccharides (LPS), which can trigger host immunity [30]. Jacobs and associates demonstrated that PGPR could be recognized by the plant root immune system and triggered defense in a PTI-like manner at the early stage [40]. Leeman’s group [41] found that LPS, consisting of lipid A/innercore/O-antigen side chain, extracted from P. fluorescens WCS417 cell wall had the function of inducing systemic resistance against Fusarium wilt of radish. However, unlike pathogen-caused PTI, that often leads to severe cellular damage, beneficial microbes-induced immune responses were reported to be transient and relatively mild due to their host immune-manipulating mechanisms, performed in order to establish a mutually beneficial relationship with the host. It was shown that flg22 peptide, extracted from the beneficial Burkholderia phytofirmans, triggered only a small oxidative burst, which was enough to cause transient induction of defense genes without growth inhibition [32]. Furthermore, Millet and associates showed that the PGPR P. fluorescens WCS417 was able to suppress flagellin-triggered PTI responses in Arabidopsis roots via secretion of low molecular compounds [42]. Possibly, colonization of PGPR on the roots requires local suppression of PTI to protect the PGPR from MAMP-triggered antimicrobial compounds, which suggests a co-evolution leading to regulation of the host’s immune system after recognition of specific signals from beneficial microbes. Overall, these results demonstrated that beneficial microbes and their elicitors could induce plant defense responses, yet the mechanism of plant-specific recognition of beneficial microorganisms and immunity responses, that distinguish beneficial microorganisms from pathogens, is still unclear.
3. Early ISR Events Induced by Beneficial Microorganisms
Beneficial microorganisms are able to stimulate defense responses of host plants through different pathways, thereby endowing plants with resistance to multiple pathogens. Bacillus amyloliquefaciens, B. atrophaeus, B. cereus, Pseudomonas fluorescens, etc., were demonstrated to be effective against fungal, bacterial, and viral invasion through ISR (Table 1). Recent studies suggested that beneficial microbes induce early plant ISR events (Table 1), including, but not limited to, increased expression of pathogenesis-related PR genes, enhanced activities of defense-related substances, such as phenylalanine ammonia-lyase, polyphenol oxidase, peroxidase, β-1, 3 glucanase, and chitinase, and accumulating reactive oxygen species [43,44].

Table 1.
Resistant mechanism of beneficial microbes.
3.1. Reactive Oxygen Species
Under biotic or abiotic stress, plants produce a large number of reactive oxygen species (ROS), including superoxide anion (O2−), hydroxyl radical (OH), hydrogen peroxide (H2O2), and so on [71]. The induction of ROS is a significant signaling in control of various processes including immunity against pathogens, programmed cell death, and stomatal closure [72]. In Arabidopsis, the perception of MAMPs leads to a rapid, specific, and strong production of RBOHD-mediated ROS. RBOHD, a plant NADPH oxidase, is mainly controlled by Ca2+ via direct binding to EF hand motifs and phosphorylation by Ca2+-dependent protein kinases [73,74]. However, the accumulation of ROS also causes tissue cell damage [75]. Therefore, efficient scavenging of ROS by enzymatic and non-enzymatic reactions is necessary. Enzymatic ROS scavenging mechanisms in plants rely on peroxidase (POX), polyphenol oxidase (PPO), superoxide dismutase (SOD), ascorbate peroxidase (APX), glutathione peroxidase (GPX), and catalase (CAT), which are essential to the defense against ROS by reducing superoxide to H2O.
B. cereus AR156 activates plant defense response by inducing the accumulation of hydrogen peroxide and callose in plants and the activation of POD and SOD enzymes, mainly through SA and MAPK signaling pathways [76]. Pseudomonas aeruginosa 7NSK2 produced pyocyanin increases H2O2 in both local and distal leaves and induces resistance to blast disease (Magnaporthe grisea) but not sheath blight (Rhizoctonia solani). The opposite effect can be alleviated by co-application of pyocyanin and the antioxidant sodium ascorbate, suggesting that the reactive oxygen species can act as a double-edged sword in resistance against different diseases [56].
3.2. Callose Deposition
Callose is a β-1, 3 glucan polymer that accumulates in weak or compromised sections of plant cell walls under pathogen attack and plays an important role in plant sieve tube metabolism. Its synthesis and decomposition are directly related to the normal growth and metabolism of plants. Aniline blue staining was used to detect callose response to identify particular induced resistance-related genes involved in callose deposition. A study in 2009 illustrated the significance of PEN2 and PEN3 genes for pathogen resistance, required for callose deposition and consequently [77]. MAMPs released by PGPR generate ROS and increase the level of SA. High level of SA triggers callose deposition by regulating the PDLP5-dependent expression of callose synthase gene (CALS10) [78]. Endophytic bacterium Pseudomonas fluorescens strain 63–28 enhanced resistance to Fusarium oxysporum in tomato, through the rapid accumulation of callose and chitinases [79]. Trichoderma harzianum T-203 triggered plant systemic defense responses by increasing peroxidase and chitinase activities and forming barriers of callose [80].
3.3. Ca2+ Influx
Ion fluxes are immediately induced by elicitors, such as K+/H+ exchange, Cl− effluxes, and Ca2+ influx, which play an important role in cell development and signal transportation, as well as in plant immunity [81]. Among these ion fluxes, Ca2+ influx is regarded as one of the most significant events, because of its role of a second messenger for many diverse physiological changes and cellular processes [81]. Some reports show that elicitor-induced Ca2+ influx not only mediates subsequent events, but also further amplifies Ca2+ signaling through Ca2+-dependent production of H2O2, which is able to increase Ca2+ influx from extracellular sources [82,83]. Pretreatment on bean (Phaseolus vulgaris) with forskolin, dibutyryl cAMP or Ca2+ ionophore A23187 enhanced the production of ROS to antagonize Colletotrichum lindemuthianum. In contrast, the Ca2+ channel blocker decreased the oxidative burst [84], suggesting that Ca2+ influx is required for ROS.
Calmodulin is a ubiquitous Ca2+ sensor, which can be activated by Ca2+ binding. Ca2+ and activated calmodulin further activate Ca2+/calmodulin-dependent protein kinase and protein phosphatase, membrane-bound enzymes, or transcription factors [85]. A large kinase family, known as Ca2+-dependent protein kinases (CDPK), with essential roles in plant defense responses, is regulated by binding of Ca2+. The application and colonization of PGPR, Pseudomonas putida MTCC 5279, activated calcium-dependent signaling by upregulating the expression of calcium-dependent protein kinase (CPK32) [86]. The Ca2+ signal can be non-linearly amplified upon binding of Ca2+, Ca2+ sensor relay proteins, calmodulin-binding transcription activators, and regulated transcription in plants [87]. Besides the functions on ROS, protein kinase cascades further the transfer of lipid signaling messengers and amplification of the elicitor signals to downstream reactions; another significant effect of Ca2+ spiking is differential activation of transcription factors, which directly regulate extensive defense gene expression [87,88,89]. A regulatory mechanism linking Ca2+ signaling to salicylic acid level is EDS1, an established regulator of salicylic acid level modulated by Ca2+/calmodulin-binding transcription factors [90]. The beneficial root-colonizing fungus Mortierella hyalina activated a Ca2+-dependent signaling pathway to resist Alternaria brassicae [64]. Cell wall extract of Piriformospora indica, a growth-promoting root endosymbiont, transiently alleviated cytosolic Ca2+ in Arabidopsis and tobacco through activating an important Ca2+ channel encoded by CYCLIC NUCLEOTIDE GATED CHANNEL 19 (CNGC19) in the mutualistic interaction between beneficial microbe and plant [91,92].
3.4. Transcriptional Factors
Several crucial transcriptional factors are involved in the regulation network of ISR through JA or/and ET signaling pathway. WRKY transcription factors are implicated in the responses to plant–microbes interactions. The Arabidopsis thaliana WRKY genes are differentially expressed in a time-dependent manner during the plant interaction with beneficial fungus T. atroviride. The expression of positive regulators in JA-mediated pathway, such as AtWRKY8 and AtWRKY33, was more anticipated than the expression of the WRKY genes regulated by SA pathway [67]. WRKY11 and WRKY70 were involved in the regulation of B. cereus strain AR156-triggered ISR in Arabidopsis, through the JA and SA signaling pathways, respectively [93]. MYB family proteins function as transcriptional factors regulating plants development and responses to biotic and abiotic stress [94]. MYB72 was activated upon colonization of P. fluorescens WCS417r and was required in the early signaling steps of beneficial microbe-mediated ISR by acting upstream of ethylene in the signaling pathway [95]. The basic helix-loop-helix (bHLH) transcription factor MYC2 was required for beneficial microbe-triggered ISR, while its function was targeted by pathogens through effector-mediated suppression of host immunity [96]. Ethylene response factor1 (ERF1) encodes a transcription factor that regulates the expression of pathogen response genes that prevent disease progression. The expression of ERF1 can be activated rapidly and synergistically by both JA and ET [97]. There are two branches, the MYC branch and the ERF branch, in the JA signaling pathway responding to wounding stress and necrotrophic pathogen attack, regulated by MYC-type transcriptional regulator and APETALA2/ethylene response factor (AP2/ERF) family of transcriptional regulator, such as ERF1 and ORA59, respectively [98]. Future attempts to unravel more detailed regulatory mechanisms on transcription factors involved in beneficial microorganism-mediated ISR will improve our understanding of the formation and regulation of ISR.
3.5. Defense-Related Genes
Defense mechanisms of ISR depend on an accurate and context-specific regulation of gene expression. Interactions between genes and their products result in complex circuits and form a regulatory network. Timmermann et al. explored the regulatory mechanism of the ISR defense response triggered by the beneficial bacterium Paraburkholderia phytofirmans PsJN and drew a regulatory network according to gene expression and time series data [99]. The Plant Defensin 1.2 (encoded by PDF1.2; AT5G44420) has previously been proved to accumulate systemically via a SA-independent pathway in leaves of Arabidopsis upon challenge by fungal pathogens and play a role as a marker of the JA signaling pathway [100,101]. As previously mentioned, some SA-dependent PR genes express antimicrobial proteins. Notably, the activation of PR1, PR2, and PR5 depend on SA signaling, whereas PDF1.2, as well as PR3 and PR4 genes, are activated via an SA-independent and JA-dependent pathway [102]. Although it was proposed that PR genes were irrelevant with ISR after certain beneficial microbe treatment [19,103], pretreatment with non-pathogenic B. cereus AR156 triggered expression of PR1, PR2, PR5, and PDF1.2 of Arabidopsis thaliana, which indicated the activation of SA and JA/ET signaling pathways, respectively [21,104,105]. The loss-function mutant of NPR1, an important regulatory factor in the SA-dependent pathway [106,107], was able to express neither ISR nor SAR [103]. Based on the previous research results, NPR1 coordinates SA and JA signaling pathway and regulate downstream defense response genes [108].
3.6. Secondary Metabolites
Under natural conditions, plants produce a vast array of secondary metabolites, which are critical for plant adaptation to abiotic and/or biotic stresses. Plant secondary metabolites are able to interact with beneficial microbes and modulate plant growth and immune process, and inhibit growth or metabolism of pathogenic microorganisms. PGPR can be recruited by root exudates, which structure a special community of rhizosphere microorganisms and enhance biofilm formation of beneficial microbes [109]. Biochemical evidence showed that plant roots secreted L-malic acid (L-MA) to selectively recruit beneficial rhizobacteria, such as B. subtilis FB17 [110]. Metabolites derived from the tryptophan and phenylpropanoid pathways, such as flavonoids, play roles in plant interactions with beneficial and pathogenic microbes, and these pathways are regulated by nutrient availability [111]. The relative abundance of root-associated Acidobacteria, Gaiellales, Nocardioidaceae, and Thermomonosporaceae in the soil can be affected by the flavonoid (7,40-dihydroxyflavone) excreted from Medicago sativa [112]. Moreover, the flavonoids, such as luteolin, from the leguminous plants can act as growth regulators as well as signaling molecules for Rhizobium bacteria to initiate symbiosis [113]. Plants also release strigolactones that stimulate the branching of hyphae of arbuscular mycorrhizal fungi to establish beneficial symbiosis [114]. Camalexin and glucosinolates are required for the P. fluorescens SS101-induced SA signaling-dependent resistance against Pst [58].
In turn, the secondary metabolites secreted by beneficial microorganisms can directly antagonize pathogenic bacteria and act as immune elicitors to raise ISR [115]. Phenazines produced by beneficial Pseudomonas bacteria showed antifungal activity and were able to elicit ISR [116]. B. cereus AR156 extracellular polysaccharides (EPS) could induce systemic resistance to Pst DC3000 in Arabidopsis [76]. Lipopolysaccharides (LPS), as MAMP molecules, triggered the activation of signal transduction pathways involved in phytohormones SA and JA, and the associated methyl esters and sugar conjugates [117]. Harzianic acid produced by Trichoderma harzianum M10-induced, modulated signaling pathway and differentially expressed genes (DEGs) involving JA/ET- and SA-mediated signaling pathways, and increased reactive oxygen species (ROS) [69]. Microbial volatile compounds (MVCs) have been shown to promote plant growth via improved photosynthesis rates, enhanced immune system, and activated phytohormone signaling pathways [118]. Critical reviews have shown the effects of VOCs on ISR and their interactions with SA, JA/ET, and auxin signaling pathways [119,120,121]. Cyclic lipopeptides surfactin and VOC 2,3-butanediol, produced by Bacillus spp., have been identified as elicitors of ISR [46,122]. These results illustrate the network of interaction between plant and beneficial microorganisms, in which plants generate metabolites to recruit beneficial microbes and inhibit harmful microbes, and beneficial microbes secrete secondary metabolites to enhance resistance of host plants.
3.7. Stomatal Regulation
Stomata play an important role in plant photosynthesis, respiration, and transpiration. Although stomatal closure decreases gas exchange, resulting in the reduction in photosynthetic activity, this reaction is actually a part of a plant innate immune response to restrict bacterial invasion [123]. Abscisic acid (ABA) plays significant roles in the regulation of stomatal aperture. ABA is produced under stress. The cellular ABA receptors bind to ABA and interact with a group of type 2C protein phosphatases (PP2C) [124,125], inactivating the inhibitory regulatory function of PP2C, but activating SnRK2 protein kinase OST1 [126]. Activated OST1 binds directly to and phosphorylates the anion channel slow anion channel-associated1 (SLAC1), mediating anion release from the guard cells and promoting stomatal closure [127,128,129]. ROS play a key role in ABA-controlled, hyperpolarization-activated Ca2+ channels in the plasma membrane of guard cells [130]. The production of H2O2 can be catalyzed by OST1 [131,132]. Lipoxygenase encoding gene LOX1, also known as a JA-responsive gene, is expressed in guard cells in response to PAMPs and is required to trigger stomatal defense [133], indicating the JA signaling pathway participates in the regulation of stomatal defense. PGPR B. amyloliquefaciens FZB42 mediates ABA and JA pathways and produce acetoin and 2,3-butanediol to induce stomatal closure in response to biotic stress [45,134,135], which suggests multiple signaling components coordinate in stomatal regulation.
4. Induced Signaling Transduction Pathway
Systemic acquired resistance (SAR) is generally considered to be induced by pathogenic microbes, while induced systemic resistance (ISR) is caused by beneficial microbes. SAR often results in increasing level of SA and coordinating activation of pathogenesis-related (PR) genes, such as PR1, PR2, and PR5, and involves one or more long-distance signals that transduce an enhanced immune signal to undamaged plant parts [136]. ISR is commonly regarded as SA-independent and develops without accumulation of PR proteins [19]; however, there are a few exceptions where identified ISR occurs in an SA-dependent manner. For example, Pseudomonas aeruginosa 7NSK2 induce systemic resistance with higher innate SA accumulation and PAL activity by producing nanogram amounts of SA [137]. ISR is identified to be activated through the JA/ET-dependent signaling pathway, involving plant defensin 1.2 (PDF1.2) [101]. Our previous results showed that pretreatment with non-pathogenic B. cereus AR156 triggered expression of PR genes and PDF1.2 of Arabidopsis thaliana, which indicated that SAR and ISR were stimulated in SA and JA/ET signaling pathways, respectively [21]. It was demonstrated that simultaneous activation of SAR and ISR pathways resulted in an additive effect in an NPR1-dependent manner against Pseudomonas syringae pv. tomato (Pst) [138]. It is difficult to distinguish SAR and ISR, both of which activate the pathogenic related genes and increase the accumulation of reactive oxygen species (ROS) and callose. SAR stimulates a rapid response to pathogens, and this signal can be conferred in a short time. In contrast, plants activated by beneficial microbes are in a special ISR state called “priming”, ready to give faster and stronger defense responses.
Jasmonates (JAs) are fatty acid-derived signaling components involved in the regulation of development and defense response in plant [9]. It was reported that beneficial microbe-mediated ISR is JA/ET-dependent by enhancing sensitivity to hormones rather than enhancing the production level or expression of JA/ET-responsive genes [139]. In addition, activation of JA signaling by application of methyl jasmonate (MeJA) not only regulates the level of resistance, but also influences structure of rhizosphere microbial community, including the species known to suppress plant disease [140]. Salicylic acid (SA) has been shown to be a required signal molecule in SAR. SA level increased after microbes infection, and SA acts as an endogenous signal with rapid movement in phloem that triggers accumulation of PR proteins [141]. SA biosynthesis seems under direct control of SID2 and EDS5 genes, while the EDS1, EDS4, and PAD4 genes play regulation functions in the synthesis of SA [142,143]. It was reported that the Arabidopsis mutants enhanced disease susceptibility1 (eds1), eds4, eds5, phytoalexin deficient4 (pad4) and SA induction deficient2 (sid2) failed to accumulate SA and were more susceptible to P. syringae [144]. It is generally believed that salicylic acid (SA) signaling is linked with plant resistance against biotrophic and hemibiotrophic pathogens, while jasmonic acid (JA)/ethylene (ET) signaling provokes host resistance to necrotrophic pathogens [137,145]. Although the initial research disregarded the involvement of SA in beneficial microbe-induced systemic resistance, recent studies have shown that beneficial microorganisms can control plant disease through activating SA and JA/ET signaling pathways. Beneficial microbes, such as Bacillus and Trichoderma, showed the ability to increase the expression of SA and JA/ET marker genes PR1 and LOX2, respectively, and increased the content of SA and JA in plants [22,23,24].
Phytohormone crosstalk is crucial for plant defense against pathogens and insects. Crosstalk between SA- and JA-dependent pathways are generally considered to be antagonistic [146]. SA synthesis-deficient Arabidopsis plants produced 25-fold higher levels of JA and enhanced expression of the JA-responsive genes LOX2, PDF1.2 in response to infection by P. syringae DC3000 [106]. Mitogen-activated protein kinases (MAPKs) and their cascades were shown to transduce various extracellular stimuli into internal cellular responses. MPK3 and MPK6 are positive regulators of plant defense responses controlling JA and ET biosynthesis [138,147]. MAPKs are required in JA biosynthesis and ET production [148,149,150] and participate in the regulation of the ROS burst [151]—which, on the contrary, negatively regulates SA-induced defense responses [82]. However, recent studies also revealed the synergistic interactions of SA and JA/ET signaling pathways in beneficial microbe-induced systemic resistance. Simultaneous activation of SAR and ISR pathway resulted in an additive effect in a NPR1-dependent manner against P. syringae pv. tomato (Pst) [152]. B. cereus AR156 was able to activate SA- and JA/ET-dependent signaling pathways simultaneously [21], and rapidly activate MAPK signaling and FRK1/WRKY53 gene expression by leaf infiltration [104].
5. Regulatory Role of Small RNAs
Small RNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs), and piwi-interacting RNAs (piRNAs), are noncoding RNAs with approx. length of 20–30 nucleotides, with important roles in regulating biological processes, such as development, reproduction, and stress responses [153]—collectively termed RNA interference (RNAi). Small RNAs are generated by DICER or DICER-like (DCL) proteins and then loaded into RNAi effector proteins Argonautes (AGOs) for regulating the expression of target mRNA through transcription or translation inhibition [154]. Plant miRNA precursors, possessing imperfectly base-paired hairpin loop structures, are first transcribed by RNA polymerase II and then cut by DCL endonuclease to produce miRNA/miRNA* double-stranded RNA. The double strand consists of a guide strand (mature miRNA) and a passenger strand (miRNA*), one of which binds to AGOs to form an active RNA induced silencing complex (RISC) [155]. In contrast to miRNAs, siRNAs are derived from perfectly paired double-stranded RNA (dsRNA) precursors. These dsRNA precursors are derived either from antisense transcription or by the action of a cellular RNA-dependent RNA polymerase (RDR) [154].
It is evident that small RNAs play crucial roles in plant innate immunity against virus, bacteria, and fungi [135,156,157]. The first miRNA identified to involve in PTI is miR393, which is induced by flg22 to repress auxin signaling by silencing its receptors [158]. Emerging evidence indicates that plants microRNAs target conserved domains of NB-LRR-encoding genes and trigger ETI [159]. B. cereus AR156 pretreatment triggers ISR signaling and downregulates the miR825/miR825* pair, which targets toll-interleukin-like receptor NB-LRR (TNLs)-type resistance (R) genes [48,49]. In addition, B. cereus AR156 triggers ISR against P. syringae pv. tomato DC3000 by suppressing miR472 and activating coiled-coil NB-LRR-mediated basal immunity in Arabidopsis [160]. B. amyloliquefaciens FZB42 inoculation suppresses Arabidopsis specific miR846 expression to induce systemic resistance via a JA-dependent signaling pathway [135]. A total of 146 known miRNAs and 217 novel miRNAs were identified to be differentially expressed in maize in response to FZB42 and loss-of-function mutant FZB42 Δsfp Δalss (deficient in triggering ISR). Among those, four miRNAs (zma-miR169a-5p, zma-miR169c-5p, zma-miR169i-5p, and zma-miR395b-5p), specifically depressed in FZB42 treatment, were selected as candidates of ISR-associated miRNAs [161].
Small RNAs play a significant role in RNA silencing in universal eukaryotic gene expression regulation. Small RNA [162] and its function of mediating RNAi were first reported in Caenorhabditis elegans [163]. It has been shown that small RNAs can spread among different organisms and induce gene silencing of each other, which is also known as cross-kingdom RNAi [164]. Small RNAs from pathogens and pests move into the host plant to inhibit plant immunity; in turn, host-delivered RNA interference plays an important role in regulating host immunity against bacteria, fungi, oomycetes, viruses, and pests. Cross-kingdom, post-transcriptional gene silencing can also occur between the symbiotic organisms, such as AMF and the host plant, during the regulation of symbiosis [165,166]. Based on the naturally occurring cross-kingdom RNAi between the beneficial microorganisms/pathogens–plants, it is possible to achieve host-induced gene silencing (HIGS) by transgenic expression of genes encoding pathogenic double-stranded RNA (dsRNA) in the host to control plant diseases [167]. In addition, in vitro synthesized dsRNA can be directly sprayed to and absorbed by host plants or harvested fruits, circumventing the transgenic risk, and resulting in gene silencing of target pathogen/insect pests (called spray induced gene silencing, SIGS) [168,169]. The intrinsic advantages of HIGS and SIGS offer them the potential to develop new strategies for crop disease management.
6. Conclusions and Discussion
In this review, we discussed the recognition mechanisms of the plant to beneficial microbes. Beneficial microbes can be recognized as MAMPs by PRR and stimulate the host plant immune response. In order to build symbiosis relationship with the plant host, beneficial microbes evolved to be able to minimize stimulation of their host’s immune system. However, there is still an urgent need for detailed research about the mechanism on the balance between efficient recognition and strength of host immune response. The genes and transcriptional factors participating in defense response make up a complicated network through the signaling crosstalk. As mentioned, SA and JA can be activated by beneficial microorganisms at the same time in an NPR1-dependent pathway. In addition, SA, JA, ET, and MAPK cascades interact with each other, and coordinate in the downstream defense response. Moreover, non-coding RNAs, induced by beneficial microorganisms, play a vital role in regulating the host development and resistance to the pathogen (Figure 1). Therefore, genome-wide profiling of miRNA and the subsequent functional verification are two important projects to explore in the future, and RNA interference technology can be a sound method to control plant diseases and pests.
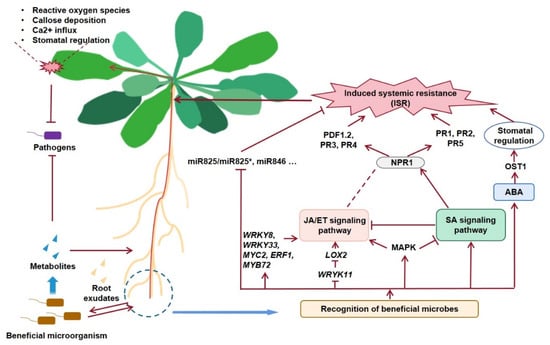
Figure 1.
Working model of beneficial microorganism-mediated ISRIn the next stage, there are still some problems to be solved. More microbial germplasm resources with biocontrol potential remains to be discovered; the formulation and shelf life of bacteria need to be improved; mining and identification of new antibacterial substances and analysis of their biosynthesis pathway, research on the genetic regulatory network of biosynthesis and microbial metabolites, and its application, based on genetic modification, are also interesting topics. Efficient and stable RNAi technology requires mastering the proper design and synthesis of dsRNA. The screening carriers of dsRNA are also indispensable to develop and improve the application of RNAi technology in plant disease control.
Author Contributions
D.N. conceived study; Y.Y., Y.G. and D.N. wrote the article; D.N., Y.Y., Z.L., C.J. and J.G. edited the manuscript; D.N., Y.Y. and Y.G. created and edited the figures. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of Jiangsu Province (BK20211524) and the National Natural Science Foundation of China (32072404).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pandey, P.; Irulappan, V.; Bagavathiannan, M.V.; Senthil-Kumar, M. Impact of Combined Abiotic and Biotic Stresses on Plant Growth and Avenues for Crop Improvement by Exploiting Physio-morphological Traits. Front. Plant Sci. 2017, 8, 537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zipfel, C.; Oldroyd, G.E.D. Plant signalling in symbiosis and immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling Mechanisms in Pattern-Triggered Immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Tian, F.; Wamboldt, Y.; Alfano, J.-R. The Majority of the Type III Effector Inventory of Pseudomonas syringae pv. tomato DC3000 Can Suppress Plant Immunity. Mol. Plant Microbe Interact. 2009, 22, 1069–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.-M.; He, S.Y.; Xin, X.-F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.F. Systemic acquired resistance induced by localized virus infections in plants. Virology 1961, 14, 340–358. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Tuzun, S.; Kuc, J.A. Proposed definitions related to induced disease resistance. Biocontrol Sci. Technol. 1992, 2, 349–351. [Google Scholar] [CrossRef]
- Conrath, U.; Beckers, G.J.M.; Flors, V.; García-Agustín, P.; Jakab, G.; Mauch, F.; Newman, M.-A.; Pieterse, C.M.J.; Poinssot, B.; Pozo, M.J.; et al. Priming: Getting ready for battle. Mol. Plant Microbe Interact. 2006, 19, 1062–1071. [Google Scholar] [CrossRef] [Green Version]
- Gaffney, T.; Friedrich, L.; Vernooij, B.; Negrotto, D.; Nye, G.; Uknes, S.; Ward, E.; Kessmann, H.; Ryals, J. Requirement of salicylic-acid for the induction of systemic acquired-resistance. Science 1993, 261, 754–756. [Google Scholar] [CrossRef]
- Vanloon, L.C. Pathogenesis-related proteins. Plant Mol. Biol. 1985, 4, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Alstrom, S. Induction of disease resistance in common bean susceptible to halo blight bacterial pathogen after seed bacterization with rhizosphere Pseudomonads. J. Gen. Appl. Microbiol. 1991, 37, 495–501. [Google Scholar] [CrossRef] [Green Version]
- Gang, W.; Kloepper, J.W.; Tuzun, S. Induction of systemic resistance of cucumber to Colletotrichum orbiculare by select strains of plant growth-promoting rhizobacteria. Phytopathology 1991, 81, 1508–1512. [Google Scholar]
- Vanpeer, R.G.; Niemann, J.; Schippers, B. Induced Resistance and Phytoalexin Accumulation in Biological Control of Fusarium Wilt of Carnation by Pseudomonas sp. Strain WCS417r. Phytopathology 1991, 81, 728–734. [Google Scholar]
- Liu, L.; Kloepper, J.W.; Tuzun, S. Induction of systemic resistance in cucumber against Fusarium-wilt by plant growth-promoting rhizobacteria. Phytopathology 1995, 85, 695–698. [Google Scholar] [CrossRef]
- Zehnder, G.W.; Yao, C.; Murphy, J.F.; Sikora, E.R.; Kloepper, J.W. Induction of resistance in tomato against cucumber mosaic cucumovirus by plant growth-promoting rhizobacteria. Biocontrol 2000, 45, 127–137. [Google Scholar] [CrossRef]
- Martinez-Ochoa, N.; Kloepper, J.W.; Rodriguez-Kabana, R. PGPR-mediated induced systemic resistance against root-knot nematode (Meloidogyne incognita) on cucumber. Phytopathology 1995, 85, 1154. [Google Scholar]
- Raupach, G.S.; Murphy, J.F.; Kloepper, J.W. Biological control of cucumber mosaic cucumovirus in Cucumis sativus L. by PGPR-mediated induced systemic resistance. Phytopathology 1995, 85, 1167. [Google Scholar]
- Pieterse, C.M.; Van Wees, S.C.; Hoffland, E.; Van Pelt, J.A.; Van Loon, L.C. Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 1996, 8, 1225–1237. [Google Scholar]
- Knoester, M.; Pieterse, C.M.; Bol, J.F.; Van Loon, L.C. Systemic resistance in Arabidopsis induced by rhizobacteria requires ethylene-dependent signaling at the site of application. Mol. Plant Microbe Interact. 1999, 12, 720–727. [Google Scholar] [CrossRef] [Green Version]
- Niu, D.D.; Liu, H.X.; Jiang, C.H.; Wang, Y.P.; Wang, Q.Y.; Jin, H.L.; Guo, J.H. The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylee-dependent signaling pathways. Mol. Plant Microbe Interact. 2011, 24, 533–542. [Google Scholar] [CrossRef] [Green Version]
- Samaras, A.; Roumeliotis, E.; Ntasiou, P.; Karaoglanidis, G. Bacillus subtilis MBI600 Promotes Growth of Tomato Plants and Induces Systemic Resistance Contributing to the Control of Soilborne Pathogens. Plants 2021, 10, 1113. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Huang, Y.; Ge, W.; Jia, Z.; Song, S.; Zhang, L.; Huang, Y. Involvement of jasmonic acid, ethylene and salicylic acid signaling pathways behind the systemic resistance induced by Trichoderma longibrachiatum H9 in cucumber. BMC Genom. 2019, 20, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barakat, I.; Chtaina, N.; Grappin, P.; El, G.M.; Ezzahiri, B.; Aligon, A.; Neveu, M.; Marchi, M. Induced Systemic Resistance (ISR) in Arabidopsis thaliana by Bacillus amyloliquefaciens and Trichoderma harzianum Used as Seed Treatments. Agriculture 2019, 9, 166. [Google Scholar]
- Ongena, M.; Duby, F.; Jourdan, E.; Beaudry, T.; Jadin, V.; Dommes, J.; Thonart, P. Bacillus subtilis M4 decreases plant susceptibility towards fungal pathogens by increasing host resistance associated with differential gene expression. Appl. Microbiol. Biot. 2005, 67, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Cordier, C.; Pozo, M.J.; Barea, J.M.; Gianinazzi, S.; Gianinazzi-Pearson, V. Cell defense responses associated with localized and systemic resistance to Phytophthora parasitica induced in tomato by an arbuscular mycorrhizal fungus. Mol. Plant Microbe Interact. 1998, 11, 1017–1028. [Google Scholar] [CrossRef] [Green Version]
- Bigirimana, J.; De Meyer, G.; Poppe, J.; Elad, Y.; Hofte, M. Induction of systemic resistance on bean (Phaseolus vulgaris) by Trichoderma harzianum. Meded. Fac. Landbouwkd. Toegep. Biol. Wet. Univ. Gent 1997, 62, 1001–1007. [Google Scholar]
- Zhang, J.; Zhou, J.-M. Plant Immunity Triggered by Microbial Molecular Signatures. Mol. Plant 2010, 3, 783–793. [Google Scholar] [CrossRef] [Green Version]
- Saijo, Y.; Loo, E.P.-I.; Yasuda, S. Pattern recognition receptors and signaling in plant-microbe interactions. Plant J. 2018, 93, 592–613. [Google Scholar] [CrossRef]
- Zamioudis, C.; Pieterse, C.M.J. Modulation of Host Immunity by Beneficial Microbes. Mol. Plant Microbe Interact. 2012, 25, 139–150. [Google Scholar] [CrossRef] [Green Version]
- Bazin, J.; Mariappan, K.; Jiang, Y.; Blein, T.; Voelz, R.; Crespi, M.; Hirt, H. Role of MPK4 in pathogen-associated molecular pattern-triggered alternative splicing in Arabidopsis. PLoS Pathog. 2020, 16, e1008401. [Google Scholar] [CrossRef] [PubMed]
- Felix, G.; Duran, J.D.; Volko, S.; Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999, 18, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Chen, H.; Li, C.; Xu, J.; Qi, Q.; Xu, Y.; Zhu, Y.; Zheng, J.; Peng, D.; Ruan, L.; et al. Endophyte Bacillus subtilis evade plant defense by producing lantibiotic subtilomycin to mask self-produced flagellin. Commun. Biol. 2019, 2, 368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trdá, L.; Fernandez, O.; Boutrot, F.; Héloir, M.C.; Kelloniemi, J.; Daire, X.; Adrian, M.; Clément, C.; Zipfel, C.; Dorey, S.; et al. The grapevine flagellin receptor VvFLS2 differentially recognizes flagellin-derived epitopes from the endophytic growth-promoting bacterium Burkholderia phytofirmans and plant pathogenic bacteria. New Phytol. 2014, 201, 1371–1384. [Google Scholar] [CrossRef]
- Segonzac, C.; Zipfel, C. Activation of plant pattern-recognition receptors by bacteria. Curr. Opin. Microbiol. 2011, 14, 54–61. [Google Scholar] [CrossRef]
- Chinchilla, D.; Zipfel, C.; Robatzek, S.; Kemmerling, B.; Nürnberger, T.; Jones, J.D.G.; Felix, G.; Boller, T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 2007, 448, 497–500. [Google Scholar] [CrossRef]
- Lu, D.; Wu, S.; Gao, X.; Zhang, Y.; Shan, L.; He, P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 496–501. [Google Scholar] [CrossRef] [Green Version]
- Madsen, E.B.; Madsen, L.H.; Radutoiu, S.; Olbryt, M.; Rakwalska, M.; Szczyglowski, K.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 2003, 425, 637–640. [Google Scholar] [CrossRef]
- Gough, C.; Cullimore, J. Lipo-chitooligosaccharide Signaling in Endosymbiotic Plant-Microbe Interactions. Mol. Plant Microbe Interact. 2011, 24, 867–878. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, S.; Zechmann, B.; Molitor, A.; Trujillo, M.; Petutschnig, E.; Lipka, V.; Kogel, K.-H.; Schäfer, P. Broad-Spectrum Suppression of Innate Immunity Is Required for Colonization of Arabidopsis Roots by the Fungus Piriformospora indica. Plant Physiol. 2011, 156, 726–740. [Google Scholar] [CrossRef] [Green Version]
- Leeman, M.; Van Pelt, J.A.; Den Ouden, F.M.; Heinsbroek, M.; Bakker, P.A.H.M.; Schippers, B. Induction of systemic resistance against Fusarium-wilt of radish by lipopolysaccharides of Pseudomonas-fluorescens. Phytopathology 1995, 85, 1021–1027. [Google Scholar] [CrossRef]
- Millet, Y.A.; Danna, C.H.; Clay, N.K.; Songnuan, W.; Simon, M.D.; Werck-Reichhart, D.; Ausubel, F.M. Innate Immune Responses Activated in Arabidopsis Roots by Microbe-Associated Molecular Patterns. Plant Cell 2010, 22, 973–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Q.; Li, Y.; Lou, Y.; Shi, M.; Jiang, Y.; Zhou, J.; Sun, Y.; Xue, Q.; Lai, H. Bacillus amyloliquefaciens Ba13 induces plant systemic resistance and improves rhizosphere microecology against tomato yellow leaf curl virus disease. Appl. Soil Ecol. 2019, 137, 154–166. [Google Scholar] [CrossRef]
- Wang, M.; Xue, J.; Ma, J.; Feng, X.; Ying, H.; Xu, H. Streptomyces lydicusM01 Regulates Soil Microbial Community and Alleviates Foliar Disease Caused by Alternaria alternataon Cucumbers. Front. Microbiol. 2020, 11, 942. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Huang, Z.; Li, X.; Ma, L.; Gu, Q.; Wu, H.; Liu, J.; Borriss, R.; Wu, Z.; Gao, X. Stomatal Closure and SA-, JA/ET-Signaling Pathways Are Essential for Bacillus amyloliquefaciens FZB42 to Restrict Leaf Disease Caused by Phytophthora nicotianae in Nicotiana benthamiana. Front. Microbiol. 2018, 9, 847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, S.P.; Uhl, J.; Grosch, R.; Alquéres, S.; Pittroff, S.; Dietel, K.; Schmitt-Kopplin, P.; Borriss, R.; Hartmann, A. Cyclic Lipopeptides of Bacillus amyloliquefaciens subsp plantarum Colonizing the Lettuce Rhizosphere Enhance Plant Defense Responses Toward the Bottom Rot Pathogen Rhizoctonia solani. Mol. Plant Microbe Interact. 2015, 28, 984–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayaz, M.; Ali, Q.; Farzand, A.; Khan, A.; Ling, H.; Gao, X. Nematicidal Volatiles from Bacillus atrophaeus GBSC56 Promote Growth and Stimulate Induced Systemic Resistance in Tomato against Meloidogyne incognita. Int. J. Mol. Sci. 2021, 22, 5049. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.; Xia, J.; Jiang, C.; Qi, B.; Ling, X.; Lin, S.; Zhang, W.; Guo, J.; Jin, H.; Zhao, H. Bacillus cereus AR156 primes induced systemic resistance by suppressing miR825/825* and activating defense-related genes in Arabidopsis. J. Integr. Plant Biol. 2016, 58, 426–439. [Google Scholar] [CrossRef] [Green Version]
- Nie, P.; Chen, C.; Yin, Q.; Jiang, C.; Guo, J.; Zhao, H.; Niu, D. Function of miR825 and miR825* as Negative Regulators in Bacillus cereus AR156-elicited Systemic Resistance to Botrytis cinerea in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 5032. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.J.; Tsay, J.F.; Chang, S.Y.; Yang, H.P.; Wu, W.S.; Chen, C.Y. Dimethyl disulfide is an induced systemic resistance elicitor produced by Bacillus cereus C1L. Pest Manag. Sci. 2012, 68, 1306–1310. [Google Scholar] [CrossRef]
- Chakraborty, U.; Chakraborty, B.; Basnet, M. Plant growth promotion and induction of resistance in Camellia sinensis by Bacillus megaterium. J. Basic Microb. 2006, 46, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, V.; Castaneda, R.; Rudrappa, T.; Bais, H.P. Root transcriptome analysis of Arabidopsis thaliana exposed to beneficial Bacillus subtilis FB17 rhizobacteria revealed genes for bacterial recruitment and plant defense independent of malate efflux. Planta 2013, 238, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Chowdappa, P.; Kumar, S.P.M.; Lakshmi, M.J.K.; Upreti, K. Growth stimulation and induction of systemic resistance in tomato against early and late blight by Bacillus subtilis OTPB1 or Trichoderma harzianum OTPB3. Biol. Control. 2013, 65, 109–117. [Google Scholar] [CrossRef]
- García-Gutiérrez, L.; Zeriouh, H.; Romero, D.; Cubero, J.; de Vicente, A.; Pérez-García, A. The antagonistic strain Bacillus subtilis UMAF6639 also confers protection to melon plants against cucurbit powdery mildew by activation of jasmonate-and salicylic acid-dependent defence responses. Microb. Biotechnol. 2013, 6, 264–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tjamos, S.E.; Flemetakis, E.; Paplomatas, E.J.; Katinakis, P. Induction of resistance to Verticillium dahliae in Arabidopsis thaliana by the biocontrol agent K-165 and pathogenesis-related proteins gene expression. Mol. Plant Microbe Interact. 2005, 18, 555–561. [Google Scholar] [CrossRef] [Green Version]
- De Vleesschauwer, D.; Hoefte, P.M. Redox-active pyocyanin secreted by Pseudomonas aeruginosa 7NSK2 triggers systemic resistance to Magnaporthe grisea but enhances Rhizoctonia solani susceptibility in rice. Mol. Plant Microbe Interact. 2006, 19, 1406–1419. [Google Scholar] [CrossRef] [Green Version]
- De Meyer, G.; Audenaert, K.; Hofte, M. Pseudomonas aeruginosa 7NSK2-induced systemic resistance in tobacco depends on in planta salicylic acid accumulation but is not associated with PR1a expression. Eur. J. Plant Pathol. 1999, 105, 513–517. [Google Scholar] [CrossRef]
- Van de Mortel, J.E.; de Vos, R.C.; Dekkers, E.; Pineda, A.; Guillod, L.; Bouwmeester, K.; van Loon, J.J.A.; Dicke, M.; Raaijmakers, J.M. Metabolic and Transcriptomic Changes Induced in Arabidopsis by the Rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol. 2012, 160, 2173–2188. [Google Scholar] [CrossRef] [Green Version]
- Lakkis, S.; Trotel-Aziz, P.; Rabenoelina, F.; Schwarzenberg, A.; Nguema-Ona, E.; Clément, C.; Aziz, A. Strengthening Grapevine Resistance by Pseudomonas fluorescens PTA-CT2 Relies on Distinct Defense Pathways in Susceptible and Partially Resistant Genotypes to Downy Mildew and Gray Mold Diseases. Front. Plant Sci. 2019, 10, 1112. [Google Scholar] [CrossRef]
- Desrut, A.; Moumen, B.; Thibault, F.; Le Hir, R.; Coutos-Thévenot, P.; Vriet, C. Beneficial rhizobacteria Pseudomonas simiae WCS417 induce major transcriptional changes in plant sugar transport. J. Exp. Bot. 2020, 71, 7301–7315. [Google Scholar] [CrossRef]
- Morcillo, R.J.; Singh, S.K.; He, D.; An, G.I.; Vílchez, J.; Tang, K.; Yuan, F.; Sun, Y.; Shao, C.; Zhang, S.; et al. Rhizobacterium-derived diacetyl modulates plant immunity in a phosphate-dependent manner. EMBO J. 2020, 39, e102602. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, Q.; Li, Y.; Sun, Y.; Xue, Q.; Lai, H. Streptomyces pactum Act12 controls tomato yellow leaf curl virus disease and alters rhizosphere microbial communities. Biol. Fert. Soils 2019, 55, 149–169. [Google Scholar] [CrossRef]
- Daroodi, Z.; Taheri, P.S. Direct antagonistic activity and tomato resistance induction of the endophytic fungus Acrophialophora jodhpurensis against Rhizoctonia solani. Biol. Control. 2021, 160, 104696. [Google Scholar] [CrossRef]
- Johnson, J.M.; Ludwig, A.; Furch, A.C.U.; Mithöfer, A.; Scholz, S.; Reichelt, M.; Oelmüller, R. The Beneficial Root-Colonizing Fungus Mortierella hyalina Promotes the Aerial Growth of Arabidopsis and Activates Calcium-Dependent Responses That Restrict Alternaria brassicae-Induced Disease Development in Roots. Mol. Plant Microbe Interact. 2019, 32, 351–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarkar, D.; Rovenich, H.; Jeena, G.; Nizam, S.; Tissier, A.; Balcke, G.U.; Mahdi, L.K.; Bonkowski, M.; Langen, G.; Zuccaro, A. The inconspicuous gatekeeper: Endophytic Serendipita vermifera acts as extended plant protection barrier in the rhizosphere. New Phytol. 2019, 224, 886–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-López, M.D.C.; Jijón-Moreno, S.; Dautt-Castro, M.; Ovando-Vázquez, C.; Ziv, T.; Horwitz, B.A.; Casas-Flores, S. Secretome Analysis of Arabidopsis-Trichoderma atroviride Interaction Unveils New Roles for the Plant Glutamate:Glyoxylate Aminotransferase GGAT1 in Plant Growth Induced by the Fungus and Resistance against Botrytis cinerea. Int. J. Mol. Sci. 2021, 22, 6804. [Google Scholar] [CrossRef] [PubMed]
- Saenz-Mata, J.; Berenice Salazar-Badillo, F.; Francisco Jimenez-Bremont, J. Transcriptional regulation of Arabidopsis thaliana WRKY genes under interaction with beneficial fungus Trichoderma atroviride. Acta Physiol. Plant 2014, 36, 1085–1093. [Google Scholar] [CrossRef]
- Singh, U.B.; Malviya, D.; Singh, S.; Kumar, M.; Sahu, P.K.; Singh, H.V.; Kumar, S.; Roy, M.; Imran, M.; Rai, J.P.; et al. Trichoderma harzianum-And Methyl Jasmonate-Induced Resistance to Bipolaris sorokiniana Through Enhanced Phenylpropanoid Activities in Bread Wheat (Triticum aestivum L.). Front. Microbiol. 2019, 10, 1697. [Google Scholar] [CrossRef] [Green Version]
- Manganiello, G.; Sacco, A.; Ercolano, M.R.; Vinale, F.; Lanzuise, S.; Pascale, A.; Napolitano, M.; Lombardi, N.; Lorito, M.; Woo, S.L. Modulation of Tomato Response to Rhizoctonia solani by Trichoderma harzianum and Its Secondary Metabolite Harzianic Acid. Front. Microbiol. 2018, 9, 1966. [Google Scholar] [CrossRef]
- Ruocco, M.; Lanzuise, S.; Lombardi, N.; Woo, S.L.; Vinale, F.; Marra, R.; Varlese, R.; Manganiello, G.; Pascale, A.; Scala, V.; et al. Multiple Roles and Effects of a Novel Trichoderma Hydrophobin. Mol. Plant Microbe Interact. 2015, 28, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.X.; Silva, H.; Klessig, D.F. Active oxygen species in the induction of plant systemic acquired-resistance by salicylic-acid. Science 1993, 262, 1883–1886. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Chen, D.D.; Zhang, X.P.; Liu, D.; Cheng, Y.Y.; Shen, F.F. Role of plant respiratory burst oxidase homologs in stress responses. Free Radic. Res. 2018, 52, 826–839. [Google Scholar] [CrossRef] [PubMed]
- Yasuhiro, K.; Ken, S.; Cyril, Z. Regulation of the NADPH Oxidase RBOHD During Plant Immunity. Plant Cell Physiol. 2015, 56, 1472–1480. [Google Scholar]
- Dat, J.; Vandenabeele, S.; Vranová, E.; Van Montagu, M.; Inzé, D.; Van Breusegem, F. Dual action of the active oxygen species during plant stress responses. Cell. Mol. Life Sci. 2000, 57, 779–795. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.H.; Fan, Z.H.; Xie, P.; Guo, J.H. Bacillus cereus AR156 Extracellular Polysaccharides Served as a Novel Micro-associated Molecular Pattern to Induced Systemic Immunity to Pst DC3000 in Arabidopsis. Front. Microbiol. 2016, 7, 664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clay, N.K.; Adi, A.M.; Denoux, C.; Jander, G.; Ausubel, F.M. Glucosinolate Metabolites Required for an Arabidopsis Innate Immune Response. Science 2009, 323, 95–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakthivel, A.; Balachandar, D. Rhizobacteria-mediated root architectural improvement: A hidden potential for agricultural sustainability. In Plant Growth Promoting Rhizobacteria for Agricultural Sustainability; Springer: Singapore, 2019; pp. 111–128. [Google Scholar]
- Mpiga, P.; Belanger, R.R.; Paulitz, T.C.; Benhamou, N. Increased resistance to Fusarium oxysporum f. sp. radicis-lycopersici in tomato plants treated with the endophytic bacterium Pseudomonas fluorescens strain 63-28. Physiol. Mol. Plant P 1997, 50, 301–320. [Google Scholar]
- Yedidia, I.; Benhamou, N.; Chet, I. Induction of defense responses in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum. Appl. Environ. Microbiol. 1999, 65, 1061–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trewavas, A.J.; Malho, R. Ca2+ signalling in plant cells: The big network! Curr. Opin. Plant Biol. 1998, 1, 428–433. [Google Scholar] [CrossRef]
- Price, A.H.; Taylor, A.; Ripley, S.J.; Griffiths, A.; Trewavas, A.J.; Knight, M.R. Oxidative signals in tobacco increase cytosolic calcium. Plant Cell. 1994, 6, 1301–1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecourieux, D.; Mazars, C.; Pauly, N.; Ranjeva, R.; Pugin, A. Analysis and effects of cytosolic free calcium increases in response to elicitors in Nicotiana plumbaginifolia cells. Plant Cell. 2002, 14, 2627–2641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bindschedler, L.; Minibayeva, F.; Gardner, S.L.; Gerrish, C.; Davies, D.R.; Bolwell, G.P. Early signalling events in the apoplastic oxidative burst in suspension cultured French bean cells involve cAMP and Ca2+. New Phytol. 2001, 151, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Davis, L.C.; Verpoorte, R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 2005, 23, 283–333. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Chaudhry, V.; Mishra, A.; Chauhan, P.S.; Rehman, A.; Yadav, A.; Tuteja, N.; Nautiyal, C.S. Gene expression profiling through microarray analysis in Arabidopsis thaliana colonized by Pseudomonas putida MTCC5279, a plant growth promoting rhizobacterium. Plant Signal. Behav. 2012, 7, 235–245. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, Z.; Shariq Iqbal, M.; Singh, S.P.; Buaboocha, T. Ca2+/Calmodulin Complex Triggers CAMTA Transcriptional Machinery Under Stress in Plants: Signaling Cascade and Molecular Regulation. Front. Plant Sci. 2020, 11, 598327. [Google Scholar] [CrossRef]
- Dolmetsch, R.E.; Lewis, R.S.; Goodnow, C.C.; Healy, J.I. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature 1997, 386, 855–858. [Google Scholar] [CrossRef]
- Yang, T.B.; Poovaiah, B.W. A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 2002, 277, 45049–45058. [Google Scholar] [CrossRef] [Green Version]
- Du, L.; Ali, G.S.; Simons, K.A.; Hou, J.; Yang, T.; Reddy, A.S.N.; Poovaiah, B.W. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 2009, 457, 1154–1158. [Google Scholar] [CrossRef]
- Vadassery, J.; Ranf, S.; Drzewiecki, C.; Mithöfer, A.; Mazars, C.; Scheel, D.; Lee, J.; Oelmüller, R. A cell wall extract from the endophytic fungus Piriformospora indica promotes growth of Arabidopsis seedlings and induces intracellular calcium elevation in roots. Plant J. 2009, 59, 193–206. [Google Scholar] [CrossRef]
- Jogawat, A.; Meena, M.K.; Kundu, A.; Varma, M.; Vadassery, J. Calcium channel CNGC19 mediates basal defense signaling to regulate colonization by Piriformospora indica in Arabidopsis roots. J. Exp. Bot. 2020, 71, 2752–2768. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.H.; Huang, Z.Y.; Xie, P.; Gu, C.; Li, K.; Wang, D.C.; Yu, Y.-Y.; Fan, Z.-H.; Wang, C.-J.; Wang, Y.-P.; et al. Transcription factors WRKY70 and WRKY11 served as regulators in rhizobacterium Bacillus cereus AR156-induced systemic resistance to Pseudomonas syringae pv. tomato DC3000 in Arabidopsis. J. Exp. Bot. 2016, 67, 157–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Van der Ent, S.; Verhagen, B.W.; Van Doorn, R.; Bakker, D.; Verlaan, M.G.; Pel, M.J.; Joosten, R.G.; Proveniers, M.C.G.; Van Loon, L.C.; Ton, J.; et al. MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in arabidopsis. Plant Physiol. 2008, 146, 1293–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazan, K.; Manners, J.M. MYC2: The Master in Action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenzo, O.; Piqueras, R.; Sanchez-Serrano, J.J.; Solano, R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 2003, 15, 165–178. [Google Scholar] [CrossRef] [Green Version]
- Lorenzo, O.; Chico, J.M.; Sanchez-Serrano, J.J.; Solano, R. Jasmonate-insensitive1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004, 16, 1938–1950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timmermann, T.; Gonzalez, B.; Ruz, G.A. Reconstruction of a gene regulatory network of the induced systemic resistance defense response in Arabidopsis using boolean networks. BMC Bioinform. 2020, 21, 142. [Google Scholar] [CrossRef] [Green Version]
- Penninckx, I.A.; Eggermont, K.; Terras, F.R.; Thomma, B.P.; De Samblanx, G.W.; Buchala, A.; Métraux, J.P.; Manners, J.M.; Broekaert, W.F. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 1996, 8, 2309–2323. [Google Scholar]
- Manners, J.M.; Penninckx, I.A.; Vermaere, K.; Kazan, K.; Brown, R.L.; Morgan, A.; Maclean, D.J.; Curtis, M.D.; Cammue, B.P.A.; Broekaert, W.F. The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol. Biol. 1998, 38, 1071–1080. [Google Scholar] [CrossRef]
- Thomma, B.P.H.J.; Eggermont, K.; Penninckx, I.A.M.A.; Mauch-Mani, B.; Vogelsang, R.; Cammue, B.P.A.; Broekaert, W.F. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 1998, 95, 15107–15111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pieterse, C.M.; Van Wees, S.C.; Van Pelt, J.A.; Knoester, M.; Laan, R.; Gerrits, H.; Weisbeek, P.J.; Van Loon, L.C. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 1998, 10, 1571–1580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, D.; Wang, X.; Wang, Y.; Song, X.; Wang, J.; Guo, J.; Zhao, H. Bacillus cereus AR156 activates PAMP-triggered immunity and induces a systemic acquired resistance through a NPR1-and SA-dependent signaling pathway. Biochem. Biophys. Res. Commun. 2016, 469, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Nie, P.; Li, X.; Wang, S.; Guo, J.; Zhao, H.; Niu, D. Induced Systemic Resistance against Botrytis cinerea by Bacillus cereus AR156 through a JA/ET- and NPR1-Dependent Signaling Pathway and Activates PAMP-Triggered Immunity in Arabidopsis. Front. Plant Sci. 2017, 8, 238. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Glazebrook, J.; Clarke, J.D.; Volko, S.; Dong, X.N. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 1997, 88, 57–63. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Bowling, S.A.; Gordon, A.S.; Dong, X. Characterization of an Arabidopsis Mutant That Is Nonresponsive to Inducers of Systemic Acquired Resistance. Plant Cell 1994, 6, 1583–1592. [Google Scholar] [CrossRef]
- Spoel, S.; Koornneef, A.; Claessens, S.M.C.; Korzelius, J.P.; Van Pelt, J.A.; Mueller, M.J.; Buchala, A.J.; Métraux, J.-P.; Brown, R.; Kazan, K.; et al. NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 2003, 15, 760–770. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Wang, D.; Liu, Y.; Li, S.; Shen, Q.; Zhang, R. Effects of different plant root exudates and their organic acid components on chemotaxis, biofilm formation and colonization by beneficial rhizosphere-associated bacterial strains. Plant Soil. 2014, 374, 689–700. [Google Scholar] [CrossRef]
- Rudrappa, T.; Czymmek, K.J.; Pare, P.W.; Bais, H.P. Root-Secreted Malic Acid Recruits Beneficial Soil Bacteria. Plant Physiol. 2008, 148, 1547–1556. [Google Scholar] [CrossRef] [Green Version]
- Hiruma, K. Roles of Plant-Derived Secondary Metabolites during Interactions with Pathogenic and Beneficial Microbes under Conditions of Environmental Stress. Microorganisms 2019, 7, 362. [Google Scholar] [CrossRef] [Green Version]
- Szoboszlay, M.; White-Monsant, A.; Moe, L.A. The effect of root exudate 7,4′-dihydroxyflavone and naringenin on soil bacterial community structure. PLoS ONE 2016, 11, e0146555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Lateif, K.; Bogusz, D.; Hocher, V. The role of flavonoids in the establishment of plant roots endosymbioses with arbuscular mycorrhiza fungi, rhizobia and Frankia bacteria. Plant Signal. Behav. 2012, 7, 636–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Babili, S.; Bouwmeester, H.J. Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 2015, 66, 161–186. [Google Scholar] [CrossRef] [PubMed]
- Prsic, J.; Ongena, M. Elicitors of Plant Immunity Triggered by Beneficial Bacteria. Front. Plant Sci. 2020, 11, 594530. [Google Scholar] [CrossRef] [PubMed]
- Chin-A-Woeng, T.F.C.; Bloemberg, G.V.; Lugtenberg, B.J.J. Phenazines and their role in biocontrol by Pseudomonas bacteria. New Phytol. 2003, 157, 503–523. [Google Scholar] [CrossRef] [Green Version]
- Finnegan, T.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. The Lipopolysaccharide-Induced Metabolome Signature in Arabidopsis thaliana Reveals Dynamic Reprogramming of Phytoalexin and Phytoanticipin Pathways. PLoS ONE 2016, 11, e0163572. [Google Scholar] [CrossRef]
- Kong, H.G.; Shin, T.S.; Kim, T.H.; Ryu, C.-M. Stereoisomers of the Bacterial Volatile Compound 2,3-Butanediol Differently Elicit Systemic Defense Responses of Pepper against Multiple Viruses in the Field. Front. Plant Sci. 2018, 9, 90. [Google Scholar] [CrossRef] [Green Version]
- Tyagi, S.; Mulla, S.I.; Lee, K.-J.; Chae, J.-C.; Shukla, P. VOCs-mediated hormonal signaling and crosstalk with plant growth promoting microbes. Crit. Rev. Biotechnol. 2018, 38, 1277–1296. [Google Scholar] [CrossRef]
- Cellini, A.; Spinelli, F.; Donati, I.; Ryu, C.M.; Kloepper, J.W. Bacterial volatile compound-based tools for crop management and quality. Trends Plant Sci. 2021, 26, 968–983. [Google Scholar] [CrossRef]
- Garbeva, P.; Weisskopf, L. Airborne medicine: Bacterial volatiles and their influence on plant health. New Phytol. 2020, 226, 32–43. [Google Scholar] [CrossRef] [Green Version]
- Ryu, C.-M.; Farag, M.A.; Hu, C.-H.; Reddy, M.S.; Kloepper, J.W.; Paré, P.W. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 2004, 134, 1017–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-F.F.; et al. Abscisic Acid Inhibits Type 2C Protein Phosphatases via the PYR/PYL Family of START Proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1266. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geiger, D.; Scherzer, S.; Mumm, P.; Stange, A.; Marten, I.; Bauer, H.; Ache, P.; Matschi, S.; Liese, A.; Al-Rasheid, K.A.S.; et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. USA 2009, 106, 21425–21430. [Google Scholar] [CrossRef] [Green Version]
- Brandt, B.; Brodsky, D.E.; Xue, S.; Negi, J.; Iba, K.; Kangasjärvi, J.; Ghassemian, M.; Stephan, A.B.; Hu, H.; Schroeder, J.I. Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. USA 2012, 109, 10593–10598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.C.; Lan, W.; Buchanan, B.B.; Luan, S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. USA 2009, 106, 21419–21424. [Google Scholar] [CrossRef] [Green Version]
- Pei, Z.M.; Murata, Y.; Benning, G.; Thomine, S.; Klüsener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 2000, 406, 731–734. [Google Scholar] [CrossRef]
- Sirichandra, C.; Gu, D.; Hu, H.-C.; Davanture, M.; Lee, S.; Djaoui, M.; Valot, B.; Zivy, M.; Leung, J.; Merlot, S.; et al. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 2009, 583, 2982–2986. [Google Scholar] [CrossRef] [Green Version]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Montillet, J.-L.; Leonhardt, N.; Mondy, S.; Tranchimand, S.; Rumeau, D.; Boudsocq, M.; Garcia, A.V.; Douki, T.; Bigeard, J.; Laurière, C.; et al. An Abscisic Acid-Independent Oxylipin Pathway Controls Stomatal Closure and Immune Defense in Arabidopsis. PLoS Biol. 2013, 11, e1001513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Li, X.; Ma, L.; Borriss, R.; Wu, Z.; Gao, X. Acetoin and 2,3-butanediol from Bacillus amyloliquefaciens induce stomatal closure in Arabidopsis thaliana and Nicotiana benthamiana. J. Exp. Bot. 2018, 69, 5625–5635. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Jiang, H.; Ding, T.; Xu, Q.; Chai, W.; Cheng, B. Bacillus amyloliquefaciens FZB42 represses plant miR846 to induce systemic resistance via a jasmonic acid-dependent signalling pathway. Mol. Plant Pathol. 2018, 19, 1612–1623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Z.Q.; Dong, X.N. Systemic Acquired Resistance: Turning Local Infection into Global Defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef] [Green Version]
- De Meyer, G.; Capieau, K.; Audenaert, K.; Buchala, A.; Métraux, J.P.; Höfte, M. Nanogram amounts of salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 activate the systemic acquired resistance pathway in bean. Mol. Plant Microbe Interact. 1999, 12, 450–458. [Google Scholar] [CrossRef] [Green Version]
- Tena, G.; Boudsocq, M.; Sheen, J. Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 2011, 14, 519–529. [Google Scholar] [CrossRef] [Green Version]
- Pieterse, C.M.; Van Pelt, J.A.; Ton, J.; Parchmannb, S.; Mueller, M.J.; Buchala, A.J.; Métrauxc, J.P.; Van Loon, L.C. Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis requires sensitivity to jasmonate and ethylene but is not accompanied by an increase in their production. Physiol. Mol. Plant P. 2000, 57, 123–134. [Google Scholar] [CrossRef] [Green Version]
- Carvalhais, L.C.; Dennis, P.G.; Badri, D.V.; Tyson, G.W.; Vivanco, J.M.; Schenk, P.M. Activation of the Jasmonic Acid Plant Defence Pathway Alters the Composition of Rhizosphere Bacterial Communities. PLoS ONE 2013, 8, e56457. [Google Scholar]
- Yalpani, N.; Silverman, P.; Wilson, T.M.A.; Kleier, D.A.; Raskin, I. Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. Plant Cell 1991, 3, 809–818. [Google Scholar]
- Glazebrook, J. Genes controlling expression of defense responses in Arabidopsis. Curr. Opin. Plant Biol. 2001, 4, 301–308. [Google Scholar] [CrossRef]
- Nawrath, C.J.; Metraux, P. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 1999, 11, 1393–1404. [Google Scholar] [PubMed] [Green Version]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhao, F.; Jiang, L.; Chen, C.; Wu, L.; Liu, Z. Different Pathogen Defense Strategies in Arabidopsis: More than Pathogen Recognition. Cells 2018, 7, 252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L.-J. Signaling Crosstalk between Salicylic Acid and Ethylene/Jasmonate in Plant Defense: Do We Understand What They Are Whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.Z.; Zhang, S.Q. MAPK Cascades in Plant Disease Resistance Signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Schweighofer, A.; Meskiene, I. Regulation of stress hormones jasmonates and ethylene by MAPK pathways in plants. Mol. Biosyst. 2008, 4, 799–803. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.R.; Zhang, S.Q.; Stacey, G. Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol. Plant Pathol. 2004, 5, 125–135. [Google Scholar] [CrossRef]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.-L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Asai, S.; Ohta, K.; Yoshioka, H. MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 2008, 20, 1390–1406. [Google Scholar] [CrossRef] [Green Version]
- Van Wees, S.C.; De Swart, E.A.; Van Pelt, J.A.; Van Loon, L.C.; Pieterse, C.M. Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2000, 97, 8711–8716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katiyar-Agarwal, S.; Jin, H.L. Role of Small RNAs in Host-Microbe Interactions. Annu. Rev. Phytopathol. 2010, 48, 225–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, K.; Chen, X. Biogenesis, Turnover, and Mode of Action of Plant MicroRNAs. Plant Cell. 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Yuan, Y.R.; Pei, Y.; Lin, S.S.; Tuschl, T.; Patel, D.J.; Chua, N.H. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006, 20, 3255–3268. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Meng, J.; Zhai, J.M.; Xu, P.S.; Luan, Y.S. MicroRNA396a-5p and-3p induce tomato disease susceptibility by suppressing target genes and upregulating salicylic acid. Plant Sci. 2017, 265, 177–187. [Google Scholar] [CrossRef]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D.G. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Park, J.H.; Shin, C. The role of plant small RNAs in NB-LRR regulation. Brief. Funct. Genom. 2015, 14, 268–274. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Fan, Z.; Li, Z.; Niu, D.; Li, Y.; Zheng, M.; Wang, Q.; Jin, H.; Guo, J. Bacillus cereus AR156 triggers induced systemic resistance against Pseudomonas syringae pv. tomato DC3000 by suppressing miR472 and activating CNLs-mediated basal immunity in Arabidopsis. Mol. Plant Pathol. 2020, 21, 854–870. [Google Scholar]
- Xie, S.; Yu, H.; Li, E.; Wang, Y.; Liu, J.; Jiang, H. Identification of miRNAs Involved in Bacillus velezensis FZB42-Activated Induced Systemic Resistance in Maize. Int. J. Mol. Sci. 2019, 20, 5057. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The, C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Weiberg, A.; Wang, M.; Lin, F.-M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.-D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silvestri, A.; Fiorilli, V.; Miozzi, L.; Accotto, G.P.; Turina, M.; Lanfranco, L. In silico analysis of fungal small RNA accumulation reveals putative plant mRNA targets in the symbiosis between an arbuscular mycorrhizal fungus and its host plant. BMC Genom. 2019, 20, 169. [Google Scholar] [CrossRef] [PubMed]
- Weiberg, A.; Bellinger, M.; Jin, H. Conversations between kingdoms: Small RNAs. Curr. Opin. Bio. 2015, 32, 207–215. [Google Scholar] [CrossRef] [Green Version]
- Koch, A.; Kumar, N.; Weber, L.; Keller, H.; Imani, J.; Kogel, K.-H. Host-induced gene silencing of cytochrome P450 lanosterol C14alpha-demethylase-encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. USA 2013, 110, 19324–19329. [Google Scholar] [CrossRef] [Green Version]
- Cai, Q.; He, B.; Kogel, K.H.; Jin, H. Cross-kingdom RNA trafficking and environmental RNAi-nature’s blueprint for modern crop protection strategies. Curr. Opin. Microbiol. 2018, 46, 58–64. [Google Scholar] [CrossRef]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-Based Control of Fusarium graminearum Infections through Spraying of Long dsRNAs Involves a Plant Passage and Is Controlled by the Fungal Silencing Machinery. PLoS Pathog. 2016, 12, e1005901. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).