Abstract
Fluctuations in oxygen (O2) availability occur as a result of flooding, which is periodically encountered by terrestrial plants. Plant respiration and mitochondrial energy generation rely on O2 availability. Therefore, decreased O2 concentrations severely affect mitochondrial function. Low O2 concentrations (hypoxia) induce cellular stress due to decreased ATP production, depletion of energy reserves and accumulation of metabolic intermediates. In addition, the transition from low to high O2 in combination with light changes—as experienced during re-oxygenation—leads to the excess formation of reactive oxygen species (ROS). In this review, we will update our current knowledge about the mechanisms enabling plants to adapt to low-O2 environments, and how to survive re-oxygenation. New insights into the role of mitochondrial retrograde signaling, chromatin modification, as well as moonlighting proteins and mitochondrial alternative electron transport pathways (and their contribution to low O2 tolerance and survival of re-oxygenation), are presented.
1. Introduction
The increasing frequency of floods resulting from severe rainfall, a rise in the seawater level, and the glacial melt caused by climate change, has devastating effects on plant growth and productivity [1,2,3,4]. As a result of restricted gas diffusion underwater, flooded plants experience dramatic variations in molecular oxygen (O2) availability, ranging from partial O2 deficiency (hypoxia; usually between 1% and 5% O2) to total O2 depletion (anoxia, 0% O2). Two types of hypoxia can be distinguished: environmental hypoxia can be caused by specific environmental cues, while developmental hypoxia can also occur in tissues and organs under normal O2 availability. On the one hand, environmental hypoxia results from soil waterlogging, which occurs when only the root zone is flooded and cannot respire due to excessive water in the soil [5]. Under these conditions, nutrient uptake via the root zone and stomata conductance, as well as CO2 assimilation in aerial parts of the plant, are reduced. In contrast to terrestrial plants, wetland plants tolerate longer and stronger hypoxic conditions through a combination of escape (increased metabolism and growth) and quiescence (slowing growth and saving metabolic resources) strategies [6]. On the other hand, environmental hypoxia is generated by submergence. In this case, the entire plant (roots and aerial parts of the plant) is under water [5], which may, similarly to wetland plants, trigger quiescence or escape strategies as well as ethylene entrapment [7,8]. However, hypoxic and anoxic conditions even occur during normal plant growth and development. Hence, O2 concentrations differ in plant tissues and plant species. Since the anatomy of specific tissues or organs, such as seeds, fruits and roots, may limit O2 diffusion due to dense tissue, hypoxic conditions can occur [9,10,11]. For instance, the O2 concentrations in the centers of growing potato tubers can decrease to around 5% [12], while O2 is undetected in seeds of barley (Hordeum vulgare L.) and pea (Pisum sativum L.) [9,10,13,14]. Most plant organs possess a relatively high surface-to-volume ratio, allowing for efficient O2 diffusion, thereby preventing the occurrence of hypoxic conditions [9]. However, due to high metabolic activity, even tissues in which O2 diffusion is unlimited may become hypoxic. For example, high local rates of O2 consumption have been shown to occur in the phloem of castor bean (Ricinus communis L.), which reaches O2 levels of about 7% [9], and in the shoot apical meristem of Arabidopsis and Solanum lycopersicum, which shows O2 levels below 5% [15]. Furthermore, an O2 gradient is formed within developing monocot anthers as a result of rapid cell proliferation, which has been shown using Zea mays mutants (Zmmsca1, male sterile converted anther1), characterized by an early developmental defect after anther primordial formation [7,16].
During evolution, plants have developed several morphological and physiological adaptations that allow for plant survival during environmentally low O2 conditions [17,18,19]. The anatomical alterations of plants grown under these conditions include the formation of lateral roots (e.g., Arabidopsis thaliana), root aerenchyma or adventitious roots (e.g., Zea mays, Oryza sativa), and the elongation of the coleoptile (e.g., Oryza sativa, Echinochloa spp.), internode (e.g., deepwater rice) or petiole (e.g., Rumex palustris) to re-establish gas exchange [20]. Furthermore, plants can limit their growth to save carbohydrate and energy reserves until O2 becomes available again in post-hypoxic or anoxic conditions [21]. On the physiological level, limitations in O2 availability lead to restrictions of aerobic metabolism. Therefore, plants have to adapt their metabolism in order to guarantee sufficient energy supply for survival, but at the expense of growth. Under low O2 conditions, ATP generation via oxidative phosphorylation (OXPHOS) in plant mitochondria, as well as the regeneration of NAD+ via the mitochondrial electron transport chain (mtETC), are reduced. Under these conditions, the anaerobic metabolism is upregulated instead, in which ATP and NAD+ supplies solely depend on glycolytic fermentation. While aerobic metabolism generates 36–38 molecules ATP per molecule glucose, anaerobic metabolism generates 2 to 3 molecules ATP only. Hence, the cellular ATP content is extremely low under O2 deficiency [19]. The re-routing of plant energy metabolism under limited O2 conditions ultimately leads to cell acidification and the accumulation of reducing equivalents, causing excess generation of reactive oxygen species (ROS) and reactive nitrogen species (RNS) that potentially lead to cell damage [22,23].
To prevent cell damage under low-O2 conditions, plant cells execute various stress responses, including major metabolic re-routing, hormonal regulation, changes in mitochondrial biogenesis, as well as the reprograming of gene expression [24,25,26,27,28,29,30,31,32,33,34,35]. The latter includes the upregulation of transcripts that are required to meet the energy demands of plant cells via fermentative glycolysis, such as alcohol dehydrogenase (ADH), pyruvate decarboxylase (PDC) and alanine aminotransferase (ALAT), while downregulation of gene transcription associated with ATP-consuming pathways, such as lipid metabolism, secondary metabolism, transport, signaling, and redox regulation, is initiated [15,30,31,36,37]. During fermentative glycolysis, ethanol and lactate accumulation (catalyzed by ADH, PDC and lactate dehydrogenase (LDH)) can reach toxic levels in plant cells. However, comparative analyses of flood-tolerant and flood-sensitive plant species have demonstrated that tolerance to flooding is related to ethanol tolerance in plants [38].
While a lot of information has been published concerning physiological and molecular events occurring during hypoxia, there is still a big gap in our knowledge about re-oxygenation in post-hypoxic conditions. Post-hypoxic and -anoxic stresses as experienced during re-oxygenation and simultaneous re-illumination cause photoinhibition that is accompanied by an increased generation of excess ROS and RNS, which leads to cell damage. To counteract this, the aerobic metabolism is suppressed during recovery from O2 deficiency, leading to an inhibition of metabolic functions [39,40,41,42].
The present review aims to update current knowledge about mitochondrial mechanisms enabling plants to adapt to low-O2 stress and re-oxygenation post hypoxia. We will summarize timely relevant research that has been carried out to reveal plant mitochondrial strategies to survive low O2 concentrations as well as re-oxygenation. In this context, we will focus on the metabolic, transcriptional as well as post-translational levels.
2. Re-Routing of Plant Energy Metabolism as a Strategy to Survive O2 Deprivation
The energetic requirements of plants are fulfilled by photosynthesis and respiration, the latter supplying 100% of the ATP required in darkness, non-green or heterotrophic tissues. Respiration comprises glycolysis, the tricarboxylic acid (TCA) cycle, and the mtETC (Figure 1). It provides metabolites and plays a role in the maintenance of photosynthesis. Even in green tissues, respiration is essential. Respiration is responsive to abiotic and biotic stresses, demonstrating its crucial role in plant stress acclimation to adverse growth conditions [43]. It can vary dramatically depending on plant species, tissue and growth conditions [44,45]. Therefore, plant respiration represents an important hub, linking energy generation for growth and stress tolerance. Plant respiration is highly flexible, to meet the varying demands of plant growth and defense, especially under low O2 conditions.
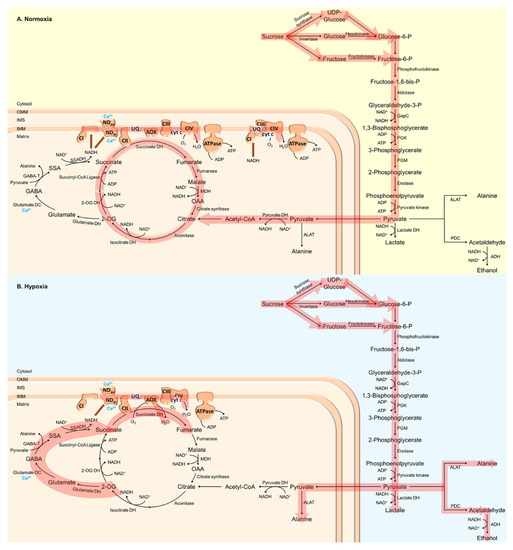
Figure 1.
Schematic visualization of plant respiratory metabolism under (A) normoxic and (B) hypoxic conditions. Under low O2 conditions, ATP generation via oxidative phosphorylation (OXPHOS) in plant mitochondria, as well as regeneration of NAD+ via the mitochondrial electron transport chain (mtETC), are reduced. Under these conditions, the anaerobic metabolism is upregulated instead, in which ATP and NAD+ supply solely depend on glycolytic fermentation. 2-OG, 2-oxoglutarate; 2-OG DH, 2-oxoglutarate dehydrogenase; ADH, alcohol dehydrogenase; ALAT, alanine aminotransferase; AOX, alternative oxidase; CI, complex I (NADH dehydrogenase); CII, complex II (succinate dehydrogenase); CIII, complex III (cytochrome bc1 complex); CIV, complex IV (cytochrome c oxidase, COX); cyt c, cytochrome c; GABA, γ-aminobutyric acid; GABA-T, GABA-transaminase; GapC, cytosolic glyceraldehyde-3-phosphate dehydrogenase; glutamate DC, glutamate decarboxylase; glutamate DH, glutamate dehydrogenase; IMM, inner mitochondrial membrane; IMS, intermembrane space; isocitrate DH, isocitrate dehydrogenase; lactate DH, lactate dehydrogenase; MDH, malate dehydrogenase; NDin/ex, internal/external alternative NAD(P)H dehydrogenase; OAA, oxaloacetate; OMM, outer mitochondrial membrane; PDC, pyruvate decarboxylase; PGK, phosphoglycerate kinase; PGM, phosphoglycerate mutase; pyruvate DH, pyruvate dehydrogenase; SSA, succinic semialdehyde; SSADH, succinic semialdehyde dehydrogenase; succinate DH, succinate dehydrogenase; UQ, ubiquinone.
A major factor affecting plant respiration is O2 depletion, since O2 is the final electron acceptor in the mtETC [46]. Low O2 conditions have been shown to inactivate OXPHOS and inhibit mtETC terminal oxidase (cytochrome c oxidase, COX) (Figure 1) [47,48,49,50,51]. To circumvent an energy crisis and the excess generation of ROS at complexes I and III, plants switch from aerobic to anaerobic metabolism (fermentation) to generate ATP through glycolysis and to regenerate NAD+ through ethanolic fermentation (Figure 1) [19]. In the cytosol, pyruvate is reduced to lactate by LDH, thus maintaining the redox balance without the loss of carbon that is associated with alcoholic fermentation. However, the accumulation of lactate in turn leads to a decrease in pH in the cytosol that deactivates LDH and activates PDC [52]. PDC catalyzes the decarboxylation of pyruvate, thereby generating acetaldehyde that is finally reduced to ethanol by ADH (Figure 1) [53]. Under low O2 conditions, the increased expression and activity of ADH as well as ethanol accumulation could be observed (Figure 1) [28,54,55]. The overexpression of PDC and ADH in Arabidopsis has been shown to promote tolerance against O2 deficiency [56]. In contrast, adh and pdc loss-of-function mutants of maize, rice and Arabidopsis are more sensitive towards O2 deprivation, indicating the importance of ethanolic fermentation for low-O2 stress tolerance [37,57,58,59]. In addition to fermentation, the Pasteur effect contributes to plant survival under low O2 conditions by inhibiting OXPHOS. Thereby, carbohydrate metabolism is accelerated and energy can be maintained [60,61,62].
An even higher flexibility of plant metabolism can be reached by the action of the TCA cycle. For instance, ALAT directly links glycolysis and the TCA cycle. The interconversion of 2-oxoglutarate (2-OG) to succinate catalyzed by ALAT generates NADH, which is in turn used by malate dehydrogenase (MDH) to reduce oxaloacetate (OAA) to malate (Figure 1). Together with succinate-coenzyme A (CoA) ligase, these enzymes contribute to the generation of ATP under hypoxic conditions [19,63]. As mentioned above, various enzymes involved in the TCA cycle increase plant survival under low O2 conditions, leading to an increased concentration of fumarate, aconitate, citrate and succinate in wheat and rice [55,64,65]. Furthermore, hypoxia has been demonstrated to promote malate, γ-aminobutyric acid (GABA) and alanine biosynthesis (Figure 1B) [66]. O2 deprivation leads to a rapid accumulation of GABA, as has been shown in various plant species [67,68,69,70,71,72]. GABA synthesis is stimulated by low O2 conditions due to increased cytosolic calcium (Ca2+) concentrations and the activation of glutamate decarboxylase by the calmodulin (CaM)-dependent pathway (Figure 1B) [73]. However, GABA can also be generated via polyamine degradation or the decarboxylation of proline [74,75]. In the mitochondrial matrix, GABA can be catabolized by GABA transaminase, leading to the generation of succinic semi-aldehyde and alanine (Figure 1) [74,76]. To date, the physiological role of hypoxia-induced GABA production is unknown. However, the GABA shunt has been suggested as an important adaptive mechanism to store carbon and nitrogen that would otherwise be lost under O2 deprivation [35,77,78]. Furthermore, the GABA shunt could be responsible for the maintenance of osmotic potential, the prevention of ROS accumulation and the inhibition of cell death [79,80]. Interestingly, GABA accumulation has also been hypothesized to be involved in the regulation of cytosolic pH and increased energy generation via the activation of the GABA shunt and the TCA cycle [73,81]. Recently, Wu et al. [82] demonstrated that a hypoxia-induced increase in GABA levels is essential for the restoration of the membrane potential, and to prevent ROS-induced disturbances to ion homeostasis.
About 50 years ago, the first reports were published describing mitochondrial degeneration and developmental failure during O2 deprivation that could be reversed upon re-oxygenation [83,84,85,86,87]. Interestingly, the dimerization and subsequent oligomerization of the ATP synthase has been demonstrated to contribute to cristae formation, thereby directly linking mitochondrial morphology and supercomplex formation in the mtETC in plants [88]. The function of supercomplexes for respiratory electron transport is still puzzling, but they have been hypothesized to (i) increase the stability of individual complexes, (ii) affect membrane structure and increase the dense packing of proteins within the membranes, (iii) channel electrons between the reactive sites of the mtETC complexes within supercomplexes, and (iv) affect the direction of electron flow from and towards the various components of alternative mtETC (alternative NAD(P)H dehydrogenases (NDs) and alternative oxidase (AOX)), which would enable the fine-tuning of plant energy metabolism and ATP yield under adverse growth conditions such as hypoxia [89,90,91,92,93,94]. However, Ramírez-Aguilar et al. [95] demonstrated that, under low O2 conditions, complex I dissociates from the respirasomes to support electron transfer from cytosolic NAD(P)H to the mtETC via NDs (Figure 1). Cell acidification and the accumulation of organic acids, as found during hypoxia, inhibit complex I activity and activate NDs. Thereby, the oxidation of cytosolic NADH shifts from complex I to the NDs (Figure 1) to keep glycolysis running when OXPHOS is reduced during O2 deprivation [96,97,98].
While extensive research has been carried out using roots or whole seedlings kept in darkness under hypoxic or anoxic conditions, knowledge about the effect of O2 deprivation on photosynthetic tissues or organs such as leaves and shoots is scarce. However, distinct responses of roots and shoots to limited O2 availability have been identified [35,99,100,101,102,103,104,105]. While roots exhibit a typical hypoxic response, such as the accumulation of GABA and alanine as well as a strong decrease in raffinose, sucrose, hexoses, and pentoses, leaves exhibit a strong increase in starch, sugars, sugar derivatives, and phenolics [105]. In addition, decreased expressions of nitrite reductase and declines in several amino acids could be observed in the leaves, but not the roots, of waterlogged plants [106,107,108]. Together, these data indicate that limited O2 availability leads to the inhibition of sugar export and N assimilation. Furthermore, low O2 conditions induce a decrease in stomatal conductance, a reduced leaf area, leaf senescence and lower chlorophyll content, which altogether lead to a rapid decrease in the photosynthetic rate [109,110,111,112,113]. However, shoots with access to partial photosynthesis or dissolved O2 are less perturbed, but still express hypoxia-responsive genes and perform anaerobic metabolism [35].
3. Signaling Events Initiating Plant Hypoxic Responses
Second messengers, such as ROS and RNS, as well as Ca2+, have been widely discussed to play an important role under adverse growth conditions [114,115]. Since most cellular functions are compromised under hypoxic conditions that lead to cytosolic acidification and the elevation of cytosolic Ca2+ concentrations, as well as the accumulation of ROS and RNS [22,23], these factors may be considered as signal transducers when O2 availability is limited.
3.1. The Cytosolic pH Decreases during Hypoxia and Promotes Anaerobic Metabolism
Low O2 conditions lead to a decrease in cytosolic pH, which drops from a pH value of 7.5 under normoxia to pH 6.0 during hypoxia [116]. While cytosolic acidification provides optimal conditions for anaerobic metabolism, aerobic metabolism is suppressed [117,118]. The origin of cytosolic acidification remains controversial. However, different mechanisms have been suggested to contribute to cytosolic acidification: (i) the switch to ethanol production and (ii) accumulation of lactate generated via anaerobic carbohydrate catabolism, (iii) ATP depletion, which reduces the activity of the plasma membrane and tonoplast proton pumps, (iv) induced membrane depolarization leading to potassium (K+) loss via Kout+ channels, and (v) the inhibition of K+ uptake into root tip cells [119,120,121,122].
3.2. The Rapid Increase in Cytosolic Ca2+ Concentrations Enables Adaptation to Low O2 Conditions
Ca2+ is one of the most important ubiquitous intracellular second messengers that are involved in several signal transduction pathways in plants. Free cytosolic Ca2+ concentrations increase in response to various physiological stimuli of an abiotic and biotic nature [123,124,125]. A rapid increase in cytosolic Ca2+ concentrations in response to low O2 conditions has been observed in several plant species [116,126,127,128,129]. Ca2+ waves that are generated and spread during hypoxia result in the activation of several enzymes and genes, enabling adaptive responses at the cellular, tissue and plant levels [130,131,132]. For instance, Ca2+ is involved in the activation of glutamate decarboxylase (initiation of the GABA shunt), as well as internal and external NDs in plant mitochondria (Figure 1). Interestingly, cytosolic acidification has been demonstrated to be an important activator of external NADPH oxidation, by decreasing the Ca2+ requirements for external NDs [133]. Glutamate decarboxylase catalyzes the interconversion of glutamate to GABA (Figure 1). Its Ca2+-dependent activation therefore leads to GABA accumulation, causing a shift from lactate to alanine formation, which in turn leads to the stabilization of the cytosolic pH under hypoxic conditions [80,134]. On the transcript level, Ca2+ initiates transcription of ADH and PDC, which are both involved in hypoxic fermentation [126].
3.3. Reactive Oxygen and Nitrogen Species Accumulate during Hypoxia
Low O2 stress affects respiration, and has a severe impact on mitochondrial metabolism, the ATP/ADP ratio, the NAD(P)H redox state, the proton motive force, the antioxidant pool, as well as ROS/RNS homeostasis, which initiates mitochondrial retrograde responses [135,136,137,138]. Under hypoxic conditions, ROS accumulation occurs in mitochondria due to the lack of electron acceptors in the mtETC and the apoplast via respiratory burst oxidase homologs (RBOHs) that belong to the group of NADPH oxidases [139]. In mitochondria, limited O2 conditions activate the so-called ROS burst, as well as mitogen-activated protein kinases 3 and 6 (MPK3 and MPK6) [104]. Furthermore, mitochondria-generated hydrogen peroxide (H2O2) can cross the mitochondrial membrane and can be released into the cytosol [140]. In the apoplast, NADPH oxidases catalyze the electron flow from NADPH to O2. Thereby, superoxide anions are generated that are rapidly converted to H2O2 [141]. Torres et al. [142] showed that superoxide production as well as H2O2 accumulation is reduced in rbohd knockout mutants. In addition, these plants have been shown to be more sensitive to anoxia compared to the wildtype [143,144,145]. Interestingly, RBOHD has been shown to interact with the hypoxia-responsive universal stress protein 1 (HRU1), indicating that HRU1 might function as a hub connecting low O2 sensing and ROS signaling [146]. Another important fact of ROS signaling during O2 deprivation is that ROS are able to activate the Ca2+ channels located in the plasma membrane, slow down anion channels, and inhibit K+ influx channels via Ca2+, pH as well as nitric oxide (NO) signaling [147,148,149].
A process known as the hemoglobin (Hb)/NO cycle describes an alternative type of respiration via the mtETC under low O2 stress [150,151]. Both molecules increase under hypoxic conditions and maintain the ATP gradient by consuming NADH, although the detailed mechanism is unknown so far. However, Hb and NO are involved in the regulation of the energy level in plant cells under low O2 conditions. Hence, Hb has been hypothesized to trigger a signaling cascade through ethylene and NO [150]. Increased levels of ethylene as well as NO both promote aerenchyma formation, thereby maintaining the energy and redox status [152]. Furthermore, AOX was shown to prevent NO and superoxide production under normoxia, while superoxide generation was prevented and NO production was increased by AOX under hypoxic conditions [153].
4. Hypoxia-Induced Transcriptional Reprograming as a Strategy to Prevent Oxidative Stress
Transcription factors (TFs) play key roles in the cellular adaptation to low O2 stress, since many tolerance-mediating genes, including those related to metabolic adjustment, are strongly upregulated upon O2 deficiency [154]. The TF families participating in transcriptional reprogramming under hypoxia include group-VII ethylene-response factors (ERF-VIIs) [155,156], no apical meristem (NAM), Arabidopsis transcription activation factor (ATAF), and cup-shaped cotyledon (CUC) (NAC domain proteins) [157,158], WRKY [157,159], HEAT-SHOCK FACTOR (HSF) [160] and MYB [161] factors. ERF-VII factors are currently of the utmost importance and the subject of extensive research.
4.1. Hypoxia-Related Transcription Factors Initiate Gene Expression in Response to Low O2 Conditions
ERF-VII family members are found to be conserved within the plant kingdom, and share sequential/structural similarities and a common function under low O2 stress in rice [29], barley [162], wheat [163], maize [164], Arabidopsis [155,156] and other species. The conserved N-termini of ERF-VII factors possess a pentultimate cysteine (Cys) residue, rendering some of these TFs substrates of the N-degron pathway. This multi-step enzymatic pathway O2-dependently oxidizes the Cys residue and, following other modification steps, such as arginylation and ubiquitination, supplies the protein for proteasomal degradation [156,165,166]. Hence, under aerobic conditions, the ERF-VII protein is degraded constantly, while under hypoxic conditions, where Cys oxidation is hampered, protein stabilization is achieved. This protein degradation mechanism enables the integration of the internal cellular O2 concentration into the ERF-VII signaling pathways. However, one has to distinguish between (i) high metabolic activity resulting in low O2 levels within plant cells or tissues and (ii) environmental hypoxia, e.g., due to flooding stress, leading to an actual energy crisis for which metabolic adjustment is required. For discrimination, other intracellular signals next to O2 availability have to be integrated in order to provoke a cellular reaction tailored to the current (stress) situation [167,168]. Indeed, many cytosolic parameters, such as ATP level and Ca2+ level, pH and redox status, change rapidly during hypoxia and have the capacity to participate in signaling processes [116]. In addition, the ethylene formed under hypoxia promotes ERF-VII protein stabilization [169]. The drop in ATP levels under low O2 induces the release of plasma membrane-stored RAP2.12, an Arabidopsis ERF-VII factor, and its accumulation in the nucleus (Figure 2). Here, the energy deficiency signal is transmitted via the fatty acid precursor C18:1-CoA, which binds as a specific ligand to the ACBP:RAP2.12 complex at the plasma membrane, resulting in TF dissociation (Figure 2). Thereby, the integration of the actual O2 concentration and the energy status within the ERF-VII signaling pathway is achieved [170].
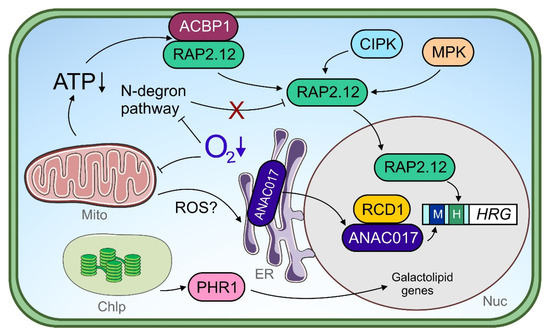
Figure 2.
Transcriptional regulators under hypoxia. Under O2 limitation, the mitochondrial function is impaired leading to lower ATP production. An energy crisis induces dissociation of the ACBP1-RAP2.12 complex at the plasma membrane. Released RAP2.12 translocates to the nucleus and free RAP2.12 protein is stabilized upon a dysfunctional N-degron pathway, which uses O2 as a co-substrate. The post-translational modification of RAP2.12 by mitogen-activated protein kinases (MPKs) and calcineurin-b-like interacting kinases (CIPKs) may regulate transcription factor (TF) function. In the nucleus, RAP2.12 activates hypoxia-responsive genes (HRG) and binds to the hypoxia-responsive promoter element (HRPE, shown as “H”) within target promoters. Endoplasmic reticulum-(ER-)localized ANAC017 is mobilized upon a mitochondrial dysfunction signal under hypoxia, potentially by reactive oxygen species (ROS). Nuclear ANAC017 is regulated in its function via physical interaction with radical-induced cell death 1 (RCD1) and binds to target gene promoters harboring the mitochondrial dysfunction motif (MDM, shown as “M”). Chloroplast signals under hypoxia are suggested to be transmitted to the nucleus by phosphate starvation response 1 (PHR1) and lead to the activation of galactolipid genes. Chlp, chloroplast; ER, endoplasmic reticulum; Mito, mitochondrion; Nuc, nucleus.
Strikingly, various studies in Arabidopsis have demonstrated that ERF-VII factor stabilization and translocation alone are insufficient to activate downstream hypoxia-responsive genes fully [169,170]. Instead, ERF-VII factors are presumably regulated in their activity under hypoxia by additional mechanisms that exceed our current state of knowledge. As one post-translational modification strategy, the phosphorylation of ERF-VII proteins in a non-hypoxic context has been reported, namely, the phosphorylation of RAP2.2 by CIPK11 and CIPK14 and RAP2.3 by MPK3 and MPK6 in Arabidopsis (Figure 2) [171,172]. Remarkably, the MPK-assisted phosphorylation of RAP2.3 at the serine residue at position 151 (Ser-151) results in enhanced TF activity. The ERF-VII factor SUB1A in rice represents another potential target of MPK-dependent modification, as its flooding tolerance-conferring SUB1A1 allele produces a protein harboring a Ser-186 in a variable region C-terminal to the ERF domain, which constitutes a putative MPK phosphorylation site. Interestingly, this site is not present in the SUB1A2-specific TF protein correlating with only moderate flooding tolerance [27]. Moreover, phosphorylation of the SUB1A1- but not SUB1A2-derived protein by MPK3 was demonstrated [173]. Since CIPK11 and CIPK14 belong to Ca2+-regulated kinases, typically activated by intracellular Ca2+ influx [174], and MPK3 and MPK6, at least in Arabidopsis, are redox-sensitive proteins directly activated by H2O2 and induced under O2 deficiency upon a mitochondrial ROS signal (Figure 2) [104,175], the action of these different kinase families on ERF-VII factors may represent entry points for multiple Ca2+ and ROS signals into the ERF-VII transduction pathway under low O2 stress. In turn, ERF-VII factors themselves partially act on kinase gene expression under low O2 stress, as shown with RAP2.12 inducing CIPK25 in Arabidopsis and the SUB1A1 allele-specific protein inducing MPK3 expression in rice [173,176].
ERF-VII factors interact with other ERF-VII factors, but also other TF families, to orchestrate hypoxia-responsive gene expression. For instance, SUB1A from rice is capable of inducing two downstream ERF-VII genes, ERF66 and ERF67, through direct DNA interaction, thereby forming a transcriptional triad [177]. While SUB1A itself is not a target of the N-degron pathway [165], the protein stability of ERF66 and ERF67 is O2-dependently regulated. Thereby, both downstream ERF-VII factors strengthen the transcriptional response to submergence orchestrated by SUB1A [177].
LOB domain containing protein 41 (LBD41) is a transcriptional repressor in Arabidopsis [178], whose transcript level is dramatically induced by hypoxia [154]. As LBD41 shows upregulation in N-degron pathway-deficient mutants under aerobic conditions [165,179], its expression level under hypoxia is clearly ERF-VII-dependent. Furthermore, RAP2.2 and RAP2.12 activate the LBD1 promoter via the hypoxia-responsive promoter element (HRPE), since a lack of the cis-element prevents ERF-VII-mediated LBD41 responses to hypoxia [180].
A more recent study in Arabidopsis unveiled that WRKY33 and WRKY12 act upstream of ERF-VII-dependent gene expression under hypoxia [159]. Both WRKY factors synergistically activate RAP2.2 expression under hypoxia by binding directly to the W-box within its promoter, and thereby contribute to submergence tolerance. Interestingly, while three other Arabidopsis ERF-VII genes (RAP2.12, HRE1 and HRE2) contain such a W-box in their gene-regulatory regions as well, only RAP2.2 induction by dark submergence appears to be WRKY33- and WRKY12-dependent. RAP2.2 overexpression in turn could complement the submergence-sensitive phenotypes of wrky33 or wrky12 knock-out mutants. That the functions of WRKY and ERF-VII factors under hypoxia are intertwined at multiple levels is, in addition, reflected by the finding that RAP2.2 activates the WRKY33 promoter directly via an ERF-VII-specific HRPE copy, while RAP2.12 does so in an indirect manner. Thereby, both ERF-VII factors establish a negative feedback loop [159].
RAP2.12 activity is fine-tuned by repressing TF HRA1, whose transcript level is induced upon hypoxia by RAP2.12; thereby, a negative feedback loop is established [181,182]. HRA1 itself is capable of directly binding to hypoxia-responsive target promoters of RAP2.12, including its own endogenous promoter. The authors propose that the physical interaction of HRA1 with target DNA may dampen RAP2.12 activity on precisely that [181]. The action of HRA1 is, interestingly, RAP2.12-specific, and does not cover other ERF-VII factors. It remains open through which molecular mechanism HRA1 is repressing RAP2.12 function and, in case other repressors exist, whether their expression levels depend on ERF-VII factors as well.
4.2. Mitochondrial Retrograde Signals Induce Nuclear Gene Expression in Response to Hypoxia
The mitochondrion, as the primary organelle being affected in its function by O2 limitation, is capable of inducing nuclear gene expression through retrograde signaling [157,158]. Many hypoxia-induced genes respond to pharmacologically induced mitochondrial dysfunction, such as antimycin-A treatment, including the Arabidopsis ERF-VII gene HRE2 [138], which transcriptionally links mitochondrial retrograde signaling with ERF-VII transduction pathways. Endoplasmic reticulum (ER)-located NAC transcription factors, such as ANAC017 and ANAC013 in particular, facilitate mitochondrial retrograde signals in order to induce nuclear gene expression [183,184]. Under submergence, ANAC017 improves tolerance by assisting in retrograde signaling-coordinated chloroplast function [157]. Notably, in anac017 knock-out mutants, photosynthesis rate and chlorophyll content were lower and ROS production higher than in the wildtype under submergence followed by reaeration. ANAC017 likely conducts transcriptional reprogramming under low O2 stress by inducing other TF genes, including WRKY40 and WRKY45 [157]. While the role of both WRKYs in signaling in general and retrograde signaling in particular is largely unexplored under hypoxia, WRKY40 or WRKY45 overexpression improved, and their knockout lowered submergence tolerance in Arabidopsis, respectively [157]. Another recent study reports the regulation of mitochondrion-related, ROS-induced genes by ANAC017 under submergence (Figure 2) [158]. Here, the action of ANAC017 is, interestingly, more apparent at younger than at later plant developmental stages, and positively correlates with a higher submergence tolerance in the juvenile phase.
ER-bound NAC factors are presumably cleaved at their transmembrane domain by intramembrane proteases, and the protease activity might be regulated in a mitochondrion-dependent manner [183]. Under non-stress conditions, the ectopic expression of a truncated ANAC017 lacking its transmembrane domain, i.e., the TF is soluble and accumulates in the nucleus even in the absence of a mitochondrial stress signal, leads to elevated transcript levels of multiple anaerobic genes, including HRE2, PDC1 and PGB1 [185]. This suggests not only an involvement of ANAC017 in hypoxia-responsive gene expression in general, but also underlines the importance of a tightly controlled release of the TF from the ER under stress. The cleavage of ANAC017 under submergence has indeed been observed, but after a 24 h treatment [158], suggesting a role of ANAC017 in rather slow transcriptional responses. Moreover, as antimycin-A treatment promoted ANAC017 cleavage [158] (mitochondrial), ROS signals represent promising candidates for low O2 stress-induced ANAC017 release from the ER. Exciting questions still to be answered in terms of mitochondrial retrograde signaling under low O2 stress are: (i) which mitochondrial protein(s) initiate(s) retrograde signaling, (ii) which signal(s) mediate(s) the mitochondrion-to-ER transfer, (iii) how is the release of ANAC017 from the ER mechanistically initiated and, finally, what is the role of the ER itself in mitochondrial retrograde signaling?
ANAC017 is linked to the cellular ROS system by activating ROS-related genes, even under submergence [158], and interacts with ROS-associated proteins [186]. One of these interactors is radical-induced cell death 1 (RCD1), an (ADP-ribosyl) transferase containing the typical poly (ADP-ribose) polymerase (PARP) domain, whose enzymatic function is inactive [186,187]. RCD1 interacts with a variety of TFs [187]; however, it is not understood at all through which molecular mechanism RCD1 interferes with TF function. Remarkably, RCD1 contains seven Cys residues within its interdomain linkers, rendering the protein redox-sensitive. Upon oxidative stress, the oxidation of RCD1 results in its protein decay [186]. It may be speculated that, via the RCD1-ANAC107 module, redox signals generated under low O2 stress are integrated into retrograde signaling and affect downstream gene expression (Figure 2). In addition, TFs of the HSF family potentially contribute to redox-dependent hypoxia signaling. HSFA1a, HSFA1b and HSFA2 are well-known positive regulators of Arabidopsis’ low O2 stress tolerance [160], of which HSFA1a is activated upon thiol oxidation [188]. Still, redox-dependent signaling under hypoxia is an emerging field and awaits further research effort (reviewed in [189]).
While mitochondrial retrograde signaling is becoming more and more the subject of interest under low O2 stress, retrograde signaling events derived from the chloroplast are much less explored. Notably, ANAC017 and its repressor RCD1 together establish a communication link between the chloroplast and the mitochondrion (Figure 2) [186,190], which strengthens the already suggested role of ANAC017 in chloroplast function coordination [157]. The loss of RCD1 increases the expression of mitochondrial dysfunction stimulon (MDS) genes typically regulated by ANAC017 [186,191]. It would be worthwhile to test whether the ANAC017-RCD1 module also participates in plastid-to-mitochondrion communication pathways during low O2 stress. Under hypoxia, the rcd1 mutant shows an impressive tolerance towards methyl viologen (MV), which leads to ROS formation in the chloroplast in the light [190]. This enhanced phenotype is interestingly not the consequence of altered plastidial ROS scavenging abilities, but can be traced back to a disrupted electron transfer from photosystem I (PSI) to MV found in the wildtype. Likewise, a correlation between improved MV tolerance under aerobic conditions and enhanced MDS expression is observed for ANAC017 overexpression plants [191]. It is worth mentioning in the context of illuminated hypoxia-specific chloroplast signaling the MYB factor phosphate starvation 1 (PHR1), which mediates both transcriptional responses to O2 deficiency and phosphate starvation, and is potentially post-translationally activated by a retrograde signal upon lowered photosynthesis (Figure 2). Interestingly, PHR1, which mainly regulates galactolipid-related genes (present in chloroplast membranes) but not anaerobic core genes, acts completely independently of ERF-VII factors or the N-degron pathway under hypoxia [161].
4.3. Gene Regulatory Mechanisms and Chromatin Modifications during O2 Deprivation
The upstream regulation of ERF-VII factors and other TFs under low O2 stress is the subject of extensive ongoing research, but less light is shed on the downstream mechanisms of hypoxia gene induction. The process of transcription initiation typically involves recruitment of the general transcriptional machinery, i.e., general TFs and RNA polymerase II (RNAPII) [192] via multiple co-factors. Remarkably, ANAC017 has been linked mechanistically to regulator of alternative oxidase 1 (RAO1), also known as cyclin-dependent kinase e1 (CDKE1), which is a kinase unit of the Mediator complex [193]. The Mediator complex is conserved among eukaryotes and functions as a bridge between (stress-related) TFs and the general transcriptional machinery of the cell [192]. The flooding-sensitive phenotype of the rao1 knock-out mutant suggests a regulatory role of the Mediator complex under low O2 stress [157]. Moreover, as CDKE1 interacts with sucrose non-fermenting related kinase 1 (SnRK1/KIN10), it integrates information on the cellular energy status into mitochondrial retrograde signaling, at least under non-stress conditions [193]. Whether the employment of the Mediator complex in gene regulation under low O2 stress covers only mitochondrial retrograde signaling-related TFs such as ANAC017, or is extended to other important TF families, has yet to be explored. This assumption is, however, likely, since RAP2.2 as an ERF-VII representative interacts with the Mediator subunit MED25 in a non-hypoxia context [194].
Interestingly, a proportion of anaerobic core genes in Arabidopsis, including LBD41, PGB1 and PCO2, rapidly respond to hypoxia within minutes of exposure. Thereby, genes respond earlier than ERF-VII factors appear in the nucleus (after 3 hours of hypoxia) [195]. While these findings are solely based on a single ERF-VII factor—RAP2.12—and need further exploration, the possibility exists that those rapidly responding transcripts, at least in the first three hours of stress, are not induced by ERF-VII factors, but by early-acting, so far unidentified TFs.
Hypoxia not only stimulates the transcription and translation of anaerobic core genes, but also translationally blocks unrelated transcripts that are either subjected to degradation or kept in stress granules for further storage [196]. Preferentially non-uracil-rich mRNA is bound by oligouridylate binding protein 1 (UBP1) under hypoxia, and its aggregation results in granule formation. By the discrimination between hypoxia-related and -unrelated transcripts, an energy-saving adaptation strategy is followed under O2 constraints, and UBP1 knockout seedlings are consistently impaired in low O2 tolerance. Notably, while transient hypoxia promotes (UBP1-dependent) stress granule formation, re-oxygenation is accompanied by a concomitant decay of UBP1-stress granules [196]. The production but also autophagy of stress granules specific to hypoxia is furthermore regulated in a Ca2+-dependent manner, and involves the Ca2+ sensor CALMODULIN-LIKE 38 (CML38) [197]. CML38 was found to co-localize with the mRNP stress granule marker RNA Binding Protein 47 (RBP47), and furthermore interacts with proteins associated with messenger RNA ribonucleoprotein (mRNP) complexes. The stress granule disorganization in cml38 mutants points to the multiple functions of CML38 in granule formation, maintenance and degradation under long-term hypoxia, and, in addition, indicates that CML38 promotes increased autophagy during re-oxygenation [198].
The accessibility of regulatory target DNA to stress-related TFs, as well as those acting under hypoxia, is mandatory in order to induce gene expression upon stress stimulus. Chromatin modifications and DNA methylation are important aspects of gene regulation under stress. Under submergence, the occurrence of repressing histone modifications, and thus a lower chromatin accessibility, in ANAC017-regulated promoters correlates with decreased submergence tolerance in older plant stages [158], indicating that epigenetic effects may represent an important aspect of low O2 adaptation in plants. Indeed, transcription-activating histone 3 (H3) Lysine 4 trimethylation (H3K4me3) has been reported under submergence for the two hypoxia-responsive promoters PDC and ADH in rice [199]. A recent study in Arabidopsis on chromatin modification patterns under hypoxia revealed that the gene-regulatory regions of strongly hypoxia-responding genes are exempted from the transcription-repressing Histone 2A variant H2A.Z and, in addition, show Histone 3 modifications, such as H3 Lysine 9 acetylation (H3K9ac), but also H3K4me3, both promoting transcription [200]. Interestingly, the removal of H2A.Z is partially ERF-VII-dependently regulated, as multiple ERF-VII factors physically interact with BRAHMA, an H2A.Z-related chromatin remodeling ATPase, and thereby oppose its function [201,202].
To date, chromatin-modifying components largely await identification under hypoxia. In this context, it may be helpful to consider potential similarities between hypoxia and other stresses. For instance, temperature stress, similar to hypoxia, promotes the reduced association of the repressing histone variant H2A.Z prior to the transcriptional activation of heat-responsive genes [203]. The O2-sensing mechanism provided by the N-degron pathway directly accesses epigenetic gene repression by regulating the stability of polycomb repressor complex 2 (PRC2) subunit vernalisation 2 (VRN2) [204]. VRN2 protein abundance accumulates under both hypoxia and cold stress, and, likewise, anaerobic core genes show overlapping transcriptional responses to both stresses, hinting at similar epigenetic regulatory mechanisms under low O2 and low temperatures. VRN2 may have an exciting role in stress memory under hypoxia, as it does under cold stress, where a continuously stabilized VRN2 protein keeps the PRC2 complex active under extended cold periods and induces flowering when warmer temperatures arise [204]. It would be highly interesting to explore in detail the phenomenon of stress memory under O2 constraints, and to investigate to what extent VRN2, but also other epigenetic regulators, participate in it.
5. Post-Translational Modifications Enable a Rapid and Fine-Tuned Response to Hypoxia
Under hypoxic conditions, the COX pathway is inhibited, while glycolysis, fermentation and the GABA shunt are stimulated (Figure 1B). This leads to the accumulation of various metabolic intermediates such as succinate, citrate and malate [205,206]. On the transcriptional level, citrate has been shown to induce Arabidopsis AOX1A expression [206]. In addition, several studies have shown that the AOX amount can change in response to hypoxia, anoxia, or re-oxygenation after low O2 treatment [207,208,209,210,211,212]. Jayawardhane et al. [153] also demonstrated that AOX was particularly important in preventing nitro-oxidative stress during re-oxygenation.
While the post-translational regulation of AOX activity relies on redox mechanisms and the formation of a thiohemiacetal at CysI with 2-oxo acids in most plant species, in some instances, AOX isoforms can be stimulated by succinate, possibly through the formation of an ester bond [213,214,215,216,217,218,219]. Succinate activation of AOX isoforms relies on the presence of a Ser residue at the position of CysI, and leads to insensitivity to 2-oxo acid activation. However, Oryza sativa (rice), Nelumbo nucifera (lotus), Zea mays (maize), Solanum lycopersicum (tomato) and Chlamydomonas reinhardtii naturally possess AOX isoforms containing a Ser residue at the position of CysI (for alignment see [218]). The succinate-dependent post-translational activation of AOX isoforms possessing the Ser residue at the position of CysI may allow for an increased capacity of the AOX pathway, and may enable plants to survive during hypoxic conditions. This mechanism might even play an important role in adaptation to wet or flooded habitats, as well as recovery following reaeration. Furthermore, the tight cuticle of tomato fruits and maize kernels leads to hypoxic metabolism in these organs, additionally pinpointing the hypothesized role of succinate activation of AOX isoforms under hypoxic conditions. In addition to the potential role of succinate in AOX activation, the succinylation of TCA cycle and glycolytic enzymes has been suggested as an important regulatory mechanism to control metabolic processes under low O2 conditions [138,220]. Therefore, succinate accumulation might play a key role under limited O2 and post-hypoxic conditions to guarantee plant survival. Similar to other post-translational modifications, such as arginylation and ubiquitination (ERF-VII factors), phosphorylation and protein acetylation critically rely on substrates that can be derived from mitochondrial energy metabolism and are therefore involved in the regulation of plant metabolism under limiting O2 conditions.
There is a big gap in our knowledge about post-translational modifications in other cell compartments that might contribute to the plant stress response to O2 deprivation. However, using a redox sensor such as the redox-sensitive green fluorescent protein 2 (roGFP2), Wagner et al. [116] demonstrated that low O2 stress leads to a strong transition from reducing to oxidizing conditions in the cytosol. There is increasing evidence demonstrating that cytosolic enzymes of the central metabolism, such as glyceraldehyde 3-phosphate dehydrogenase, MDH, enolase, LDH and isocitrate dehydrogenase, are able to change their properties in the oxidized and post-translationally modified form, translocate to other subcellular compartments and take over new tasks (for review see [221]). These so-called moonlighting functions directly link energy metabolism to adaptive stress responses that are required for the maintenance of redox homeostasis and rapid responses that might also be of high importance during low O2 conditions and re-oxygenation. However, this hypothesis needs experimental proof and should be assessed in the future.
6. Re-Oxygenation—A Challenge of Plant Survival Post-Hypoxic Conditions
Since flooding is a biphasic stress, with an initial reduction of molecular O2 during submergence followed by re-oxygenation post-flooding, re-exposure of plants to normoxic conditions imposes more challenges on the plant, which may be as severe as or even more severe than the O2 deficiency itself [42]. During submergence, plant tissues adapt to low O2 and low light in turbid water, while re-exposure to atmospheric O2 after the low O2 phase results in post-anoxic injuries, specifically due to the excess generation of ROS [157,222,223]. Prolonged hypoxia or anoxia during submergence leads to a saturated mtETC, low adenylate energy charge, and high levels of reducing equivalents (Figure 3), which can have a direct effect on intracellular ROS production once O2 becomes available during re-oxygenation [224]. Re-oxygenation restores aerobic metabolism in plant cells to meet the high energy demand. However, the accelerated activity of mitochondrial respiration additionally stimulates the generation of ROS [225,226,227,228].
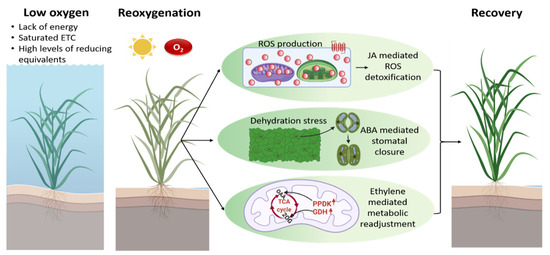
Figure 3.
Under water, plants experience a shortage in O2 that ultimately affects the mtETC. Conditions such as a lack of energy, saturated mtETC and high levels of reducing equivalent prevail during hypoxia. A shift to normal environmental conditions with intense light and higher O2 after flood water retreats results in excess ROS generation in chloroplasts and mitochondria, respectively. On top of that, plants also encounter dehydration stress and an energy crisis immediately post-submergence. The re-oxygenation response is mediated via JA, ABA and ethylene signaling to counteract excess ROS generation, to overcome dehydration stress and to generate energy through metabolic readjustment. 2-OG, 2-oxoglutarate; GDH, glutamate dehydrogenase; OAA, oxaloacetate; PPDK, pyruvate phosphate dikinase. The figure was created using BioRender.com, accessed on 10 January 2022.
7. Plants Have to Cope with Several Injuries Occurring Post-Hypoxic Conditions
7.1. Oxidative Stress Is Increased during Reaeration
Post-hypoxic injuries are mainly associated with the excess generation of ROS leading to the oxidation of lipid membranes, proteins, nucleic acids, and carbohydrates [40,229]. Besides peroxisomes, chloroplasts and mitochondria are the primary sites of ROS production in photosynthetically active tissues, predominantly due to their high metabolic activities and increased rate of electron transfer. The PSI and PSII reaction centers are major sites of ROS production in the chloroplast [230,231], while complex I and III of the mtETC are major sites for mitochondrial superoxide (O2−) production. Roughly 2–5% of the total O2 consumed by mitochondria is converted to O2− [232]. In Arabidopsis, reaeration induces RBOHD transcripts, which results in the generation of O2− in the apoplastic space [223]. In the wetland species Alternanthera philoxeroides, increased abundance of ROS-generating acetaldehyde was observed after desubmergence [233]. In addition, increased generation of ROS radicals was observed in Arabidopsis leaves within 3 h of exposure to post-hypoxic conditions [223]. The roots of lupine (Lupinus luteus) exposed to a hypoxia–re-oxygenation regime caused an increase in the level of free radicals and H2O2, which confirms an increased ROS production during re-oxygenation. Re-oxygenation also triggered a higher activity of superoxide dismutase (SOD) and catalase (CAT) in roots of lupine [227]. Furthermore, increased malondialdehyde (MDA) content as a consequence of ROS-mediated lipid peroxidation has been observed post-hypoxic conditions in Arabidopsis [223,234], rice seedlings [41,235], and soybean [236]. Excess accumulation of ROS during reaeration is a combinatorial effect of increased ROS generation due to an increased electron pressure in the chloroplast and mitochondrial electron transport chains, as well as a result of reduced scavenging capacity [237,238].
7.2. Photoinhibition Results from Increased Light Intensities Experienced Post-Submergence
Submergence results in the severe loss of carbon reserves required to meet the physiological energy demand and limited light conditions. Furthermore, limited CO2 diffusion reduces photosynthesis and the restoration of carbon reserves. However, reaeration leads to a sudden increase in O2 concentrations and a simultaneous increase in light intensity due to the outflow of water. In particular, the shift from low to high light conditions experienced by plants during re-oxygenation can lead to phototoxic damage at PSII, resulting in the inactivation of the reaction centers [239]. In Hemarthria altissima, A. philoxeroides and rice, a considerable decline in the maximum quantum yield of PSII (Fv/Fm) and nonphotochemical energy quenching (NPQ) could be observed during desubmergence [235,240,241]. Furthermore, a significant reduction in Fv/Fm was observed after 5 h post-hypoxia in Arabidopsis accessions [223]. In the grass species Agrostis stolonifera, Cynodon dactylon, and Zoysia japonica, reduced Fv/Fm values could be observed after 1 day of re-oxygenation [242]. Therefore, recovery from PSII photodamage is essential to restore submergence-depleted energy reserves, which in turn prevent leaf senescence and cell death [243].
7.3. Fine-Tuning of Stomatal Conductance Is Essential during Re-Oxygenation
A decrease in water conductivity and desiccation stress on desubmergence was observed despite the soil being soaked with water [41]. Submergence and reaeration affect the optimal function of roots due to decreased root hydraulic conductance, which reflects the ability of roots to absorb water [244,245]. Dehydration leads to a decreased relative water content (RWC), loss of leaf turgidity, leaf rolling, and wilting [41,223,235,246]. Therefore, ABA-mediated stomatal closure plays an important role to prevent transpirational water loss in leaves during recovery (Figure 3) [223,235]. In the intolerant Arabidopsis accession Bay-0 and rice cultivar M202, severe dehydration symptoms have been observed due to decreased stomatal conductance and the inability to close the stomata [41,223,234]. However, plants must keep a fine balance between the gradual opening and closing of the stomata, since closed stomata also result in limited CO2 uptake, which in turn affects photosynthesis and recovery.
7.4. Plants Accumulate Various Toxic Metabolites during Reaeration
In addition to factors such as excess ROS generation caused by re-oxygenation, re-illumination, and water loss, plants also accumulate toxic compounds and show lipid peroxidation during submergence and desubmergence [225]. The roots of wheat and rice seedlings showed increased lipid peroxidation (LPO) and accumulation of end-LPO products during re-oxygenation after prolonged anaerobiosis [225,247]. Moreover, the accumulation of diene and triene conjugates of fatty acids (primary intermediates of LPO) and thiobarbituric acid (TBA)-reactive end products of LPO was observed in roots of sensitive Iris germanica, Triticum aestivum and Avena sativa [248]. Similarly, intensive lipid hydrolysis was observed, along with the accumulation of LPO products, during re-oxygenation in potato cells and potato plants [226,249]. The tubers of sensitive potato plants accumulated 5–7-fold increased acetaldehyde during re-oxygenation [46].
7.5. Senescence Is Induced under Post-Hypoxic Conditions
Energy reduction, cytoplasmic acidosis, electron leakage and ROS production, moisture loss, and the accumulation of toxic products of anaerobic metabolism together significantly impair plant survival, contributing to post-anoxic damage and leading to programmed cell death or necrosis [236,250]. Senescence usually happens to be the last phase of leaf development and is typically characterized by leaf yellowing through chlorophyll degradation [251,252,253]. In the intolerant Arabidopsis accession Bay-0, desubmergence induced the expression of NAC domain-containing protein6/Oresara1 (ORE1) [223], a positive regulator of leaf senescence [254,255,256]. In rice, the presence of the SUB1A loci significantly reduces chlorophyll breakdown during submergence [26]. The retention of chlorophyll was observed in the forage grass Melilotus siculus simultaneously with senescence avoidance in submergence recovery [257].
8. Re-Adjustment of Plant Metabolism Is Initiated during Re-Oxygenation
A significant decrease in ATP production via mtETC [138,258] is probably one of the most influential metabolic processes for successive re-oxygenation survival. Decreased ATP is linked to increased cytoplasmic acidity, potentially hindering recovery upon re-oxygenation [53]. Various metabolic processes such as glycolysis, fermentation and TCA cycle replenishment were fine-tuned during re-oxygenation. While hypoxia results in the accumulation of alanine, re-oxygenation leads to the production of pyruvate and NADH that can be directed to the TCA cycle. During re-oxygenation, ethylene signaling mediated by ethylene-insensitive 3 (EIN3) enhances the expression of glutamate dehydrogenase (GDH) that catalyzes the interconversion of glutamate and 2-OG (Figure 3) [258]. Via the transaminase reaction, 2-OG reacts with alanine to allow the refueling of pyruvate in the TCA cycle. Interestingly, it has been reported that the absence of GDH results in the disturbance of carbohydrate metabolism, phytosterol biosynthesis, and energy regeneration. α-Tocopherol and phytosterols are vital components that are needed for the stability of cell membranes [259,260,261]. Phytosterols are known to maintain membrane integrity and fluidity in response to large temperature variations [262]. Hence, it has been suggested that re-oxygenation results in the activation of a membrane repair system that incorporates newly synthesized α-tocopherol and phytosterols to strengthen damaged membranes [258,263]. In addition, the enzyme pyruvate phosphate dikinase (PPDK) is regulated by ethylene during re-oxygenation (Figure 3). The PPDK enzyme catalyzes the conversion of phosphoenolpyruvate to pyruvate, which is further converted to OAA. Therefore, ethylene overall supports TCA cycle replenishment and carbohydrate metabolism during re-oxygenation [258,263].
Ethanol that has been produced during low O2 conditions is oxidized to acetaldehyde during re-oxygenation. Acetaldehyde is then further metabolized by aldehyde dehydrogenase to aid plant recovery [264]. The accumulation of metabolites such as arabinose and trehalose during re-oxygenation has also been reported [55,265]. Furthermore, plant recovery after hypoxia or anoxia also involves several other mitochondrial metabolic processes, such as polyamine production based on basic amino acid metabolism, respiratory chain function, and alternative respiration. In addition, changes in the morphology of plant mitochondria under hypoxia or anoxia and re-oxygenation are well documented by Shingaki-Wells et al. [250]. There is also evidence of mitochondria disintegration in the absence of O2 in anoxia-intolerant wheat [85]. However, anoxia-tolerant rice and Echinochloa phyllopogon were able to maintain the ultrastructure and shape of their mitochondria [266]. Besides changes in ultrastructure, mitochondrial ROS/RNS generation is a key element in anoxia and re-oxygenation stress signaling. The involvement of respiratory chain components in NO and ROS signaling under low O2 has also been reported [225,267]. Thus, apart from playing a major role in energy production, the mtETC could play a signaling role in anoxic and post-anoxic survival. Mitochondria—and especially its alternative pathways—have been shown to be involved in NO homeostasis during hypoxia [268]. As part of the alternative electron transport pathway, AOX aids nitrite-dependent NO production in mitochondria during hypoxia. However, under normoxia, AOX minimizes NO synthesis, ROS generation and peroxynitrite formation [269]. Therefore, AOX might function as a crucial switch in the hypoxia–re-oxygenation transition by regulating ROS/RNS-dependent protein modifications during re-oxygenation. Recently, the role of AOX in preventing nitro-oxidative stress during the re-oxygenation period has been reported, which allows for the recovery of the energy status following hypoxia [153].
The alternative electron transport pathway components, AOX and NDs, provide high flexibility to plant mitochondria [270]. Alternative mtETC components enhance the electron flow from reducing equivalents generated via the TCA cycle and photosynthesis to O2, specifically when the COX pathway is inhibited by abiotic or biotic stresses. Thus, the alternative mtETC components can counteract excess ROS production during re-oxygenation by the fast removal of excess reductive power build-up during hypoxia. At the same time, alternative mtETC components enable the avoidance of oxidative stress, since these components function as a safety valve preventing the excess generation of ROS [271,272]. It has been reported that AOX and NDs are upregulated at the transcriptional, protein, and activity levels when the basal mtETC is compromised [273,274,275]. Hence, alternative mtETC components are likely to play a vital role during re-oxygenation and adaptation. However, further research is needed to prove their function during hypoxic stress and re-oxygenation.
9. Survival Strategies of Plants during Re-Oxygenation
One of the primary survival strategies of plants during recovery from anoxia or hypoxia is the upregulation of antioxidative defense mechanisms to minimize the occurrence of oxidative stress. There have been several reports of different plant species upregulating their antioxidative defense system during re-oxygenation [234,236,263]. In Arabidopsis, a high level of ascorbate and glutathione levels was observed in genotypes exhibiting better recovery during re-oxygenation [223,234]. In soybean roots and the hypocotyl, the enzymatic activities of peroxidases were suppressed during flooding, but significantly increased during recovery [276,277]. Alternanthera philoxeroides is a submergence-resistant wetland species that uses the escape strategy during submergence via rapid stem elongation. Upon re-oxygenation, A. philoxeroides leaves exhibit increased ascorbic acid, CAT, and SOD activities that are indispensable for ROS scavenging [278]. The M202 (Sub1) rice cultivar containing the SUB1A submergence-tolerance gene displayed reduced ROS accumulation and less lipid peroxidation upon re-oxygenation [41]. Chickpea seedlings treated with ascorbic acid showed improved plant survival during anoxia and re-oxygenation [279]. Furthermore, a gradual increase in SOD activity was observed in the rhizomes of Iris pseudacorus during re-oxygenation compared to the normoxic control. Moreover, the tolerant I. pseudacorus showed a higher activity of SOD in comparison to sensitive I. germanica [280].
Jasmonic acid (JA) signaling is involved in maintaining the oxidative stress response during re-oxygenation (Figure 3). Re-oxygenation results in the induction of JA biosynthesis genes and the accumulation of JA in Arabidopsis rosettes within a few hours. Mutants deficient in JA signaling or biosynthesis exhibited a weak recovery phenotype. Moreover, the pre-treatment of JA prior to submergence results in better survival after re-oxygenation. The JA-driven re-oxygenation response is under the control of MYC2, a basic helix–loop–helix leucine-zipper TF that promotes the expression of genes involved in ascorbate and glutathione biosynthesis (Figure 3) [234]. Soybean roots treated with JA showed better ROS detoxification and recovery by promoting enzymes involved in nucleotide metabolism [254]. Post-submergence recovery requires a fine-tuned balance between ROS detoxification and alleviation. Since ROS detoxification is important during recovery, complete ROS elimination is harmful for plants, which points towards a signaling role for ROS during reaeration. In this context, it is worth mentioning that the recovery of Arabidopsis plants was hindered when ROS production was significantly restrained upon desubmergence [223]. SUB1A transcripts were also substantially induced in response to MV treatment [41]. Hence, the initial ROS burst post-submergence could act as signal for the transition to re-oxygenation and allow plants to trigger adaptive stress responses.
A better antioxidant defense or enhanced ROS scavenging upon re-oxygenation might also be useful for photosynthesis recovery. Efficient ROS scavenging reduces the damage of PS II, which in turn limits photoinhibition and allows for the faster refueling of energy reserves through photosynthetic carbon fixation during recovery. In comparison to the submergence-sensitive Arabidopsis accession Bay-0 and submergence-sensitive M202 in rice, the tolerant accession Lp2-6 and M202 (Sub1) exhibited an optimal Fv/Fm ratio after re-oxygenation [223,235]. Therefore, the replenishment of energy reserves through reactivated photosynthesis during desubmergence is vital for recovery. A correlation is shown between post-submergence carbohydrate status, restoration of photosynthetic ability and flooding tolerance [3,281]. Interestingly, resistant species were able to maintain the functionality of PSII complexes even after long periods of O2 deficiency or darkness [26,233,251].
Besides ROS detoxification, the dehydration of areal tissues is another key issue in post-hypoxic stress recovery. Plants induce dehydration-responsive genes to maintain osmotic homeostasis and water loss and enhance the survival of shoots. Shoot ABA levels and ABA-and drought-responsive transcripts increased during desubmergence in Arabidopsis and SUB1A rice [41,223,234]. The submergence-sensitive accession Bay-0 in Arabidopsis showed rapid water loss and leaf dehydration even though shoot ABA levels were high. Bay-0 shoots also showed higher ethylene release upon desubmergence, which countered ABA-mediated stomatal closure through the induction of senescence-associated gene 113 (SAG113), resulting in a higher water loss (Figure 3) [223]. Ethylene also enhanced ORE1 during desubmergence, which directs chlorophyll breakdown. Correspondingly, the knockout mutants sag113 and ore1 showed reduced water loss and increased chlorophyll levels, respectively, in comparison to wildtype plants during desubmergence. Similar results were obtained by blocking ethylene perception during recovery [223]. Hence, ethylene induction post-submergence induces senescence and reduces survival in the sensitive Arabidopsis Bay-0 accession [223].
There are still several open questions about the role of ethylene during re-oxygenation. Ethylene signaling during re-oxygenation is also required for replenishing TCA cycle intermediates [263]. Interestingly, metabolome studies show that SUB1A was induced by ethylene in rice, which alters the TCA cycle flux by stimulating free amino acid synthesis during low O2 conditions, which is then quickly overturned on desubmergence [282,283]. It might be possible that the post-submergence response is mediated by hormonal crosstalk between ethylene and JA (Figure 3). MYC2 overexpressors are known to have enhanced JA signaling and show reduced leaf withering during reaeration, indicating an important role of JA in dehydration regulation. Moreover, DNA-binding activity and downstream ethylene signaling by the TFs EIN3 and EIN3-Like1 (EIL1) are inhibited by MYC2. In Rumex palustris and Rumex maritimus, JA could alter EIN3 and EIL1 activity to regulate ethylene-mediated re-oxygenation responses [284]. Hence, the re-oxygenation response in Arabidopsis could be maintained by fine-tuning ethylene and JA, and consequently by balancing oxidative stress and energy metabolism [42].
Apart from submergence, recovery post-submergence is also an important aspect of flooding tolerance. Thus, careful investigation of the post-hypoxic recovery phase at the transcriptomic, proteomic and metabolic levels will help us to understand signals and downstream responses to reveal effective recovery strategies. Post-submergence injuries are characterized by excess ROS generation in mitochondria due to the intensive activation of the mtETC in the presence of O2 and in chloroplasts due to photoinhibition. Besides this, reduced carbohydrate replenishment, dehydration stress, and senescence also contribute to post-submergence injury. Plants employ strategies such as upregulating ROS-scavengers, antioxidative enzymes and drought-responsive genes to overcome oxidative and dehydration stress. Effective recovery additionally requires an adequate balance in crosstalk between the plant hormones JA and ethylene. However, only a few studies on adaptation and acclimation to the post-hypoxic phase have been made available so far, especially in comparison to studies on the hypoxic or anoxic phase. Although the major signals and factors involved in the regulation of re-oxygenation have been identified, knowledge about the molecular mechanisms directing efficient post-submergence recovery is still very minimal. In order to strengthen our understanding of these processes, along with comparative studies of species with varying degrees of resistance to re-oxygenation damage, the involvement of phytohormones, TFs, and other regulatory responses is necessary.
Author Contributions
Conceptualization, J.S.; writing—review and editing, J.J., R.R.S.; M.S., J.S.; funding acquisition, J.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the University of Bielefeld (RRS) and the University of Kiel (JS) (Tenure-Track Program, BMBF).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| 2-OG | 2-oxoglutarate |
| 2-OG DH | 2-oxoglutarate dehydrogenase |
| ADH | alcohol dehydrogenase |
| ALAT | alanine aminotransferase |
| AOX | alternative oxidase |
| ATAF | Arabidopsis transcription activation factor |
| Ca2+ | calcium |
| CaM | calmodulin |
| CAT | catalase |
| CDKE | cyclin-dependent kinase e |
| CIPK | calcineurin-b-like interacting kinase |
| CML | calmodulin-like |
| CoA | coenzyme A |
| COX | cytochrome c oxidase |
| CUC | cup-shaped cotyledon |
| Cys | cysteine |
| cyt c | cytochrome c |
| EIN | ethylene insensitive |
| ER | endoplasmic reticulum |
| ERF-VII | group-VII ethylene-response factor |
| Fv/Fm | maximum quantum yield of PSII |
| GABA | γ-aminobutyric acid |
| GABA-T | GABA-transaminase |
| GapC | cytosolic glyceraldehyde-3-phosphate dehydrogenase |
| GDH | glutamate dehydrogenase |
| glutamate DC | glutamate decarboxylase |
| H3 | histone 3 |
| H2O2 | hydrogen peroxide |
| Hb | hemoglobin |
| HRG | hypoxia-responsive genes |
| HRPE | hypoxia-responsive promoter element |
| HRU | hypoxia-responsive universal stress protein |
| HSF | heat-shock factor |
| IMM | inner mitochondrial membrane |
| IMS | intermembrane space |
| isocitrate DH | isocitrate dehydrogenase |
| JA | jasmonic acid |
| LBD | LOB domain containing protein |
| LDH | lactate dehydrogenase |
| LPO | lipid peroxidation |
| MDA | malondialdehyde |
| MDH | malate dehydrogenase |
| MDM | mitochondrial dysfunction motif |
| MDS | mitochondrial dysfunction stimulon |
| MPK | mitogen-activated protein kinase |
| mRNP | messenger RNA ribonucleoprotein |
| mtETC | mitochondrial electron transport chain |
| MV | methyl viologen |
| NAM | no apical meristem |
| NDin/ex | internal/external alternative NAD(P)H dehydrogenase |
| NO | nitric oxide |
| NPQ | nonphotochemical energy quenching |
| O2 | oxygen |
| O2− | superoxide |
| OAA | oxaloacetate |
| OMM | outer mitochondrial membrane |
| ORE | oresara |
| OXPHOS | oxidative phosphorylation |
| PARP | poly (ADP-ribose) polymerase |
| PDC | pyruvate decarboxylase |
| PGK | phosphoglycerate kinase |
| PGM | phosphoglycerate mutase |
| PHR | phosphate starvation response |
| PPDK | pyruvate phosphate dikinase |
| PRC | polycomb repressor complex |
| PSI/PSII | photosystem I/II |
| pyruvate DH | pyruvate dehydrogenase |
| RAO | regulator of alternative oxidase |
| RBOH | respiratory burst oxidase homolog |
| RBP | RNA binding protein |
| RCD | radical-induced cell death |
| RNAP | RNA polymerase |
| RNS | reactive nitrogen species |
| roGFP | redox-sensitive green fluorescent protein |
| ROS | reactive oxygen species |
| RWC | relative water content |
| SAG | senescence associated gene |
| Ser | serine |
| SnRK | sucrose non-fermenting related kinase |
| SOD | superoxide dismutase |
| SSA | succinic semialdehyde |
| SSADH | succinic semialdehyde dehydrogenase |
| succinate DH | succinate dehydrogenase |
| TBA | thiobarbituric acid |
| TCA | tricarboxylic acid |
| TF | transcription factor |
| UBP | oligouridylate binding protein |
| UQ | ubiquinone |
| VRN | vernalization |
References
- Bailey-Serres, J.; Lee, S.C.; Brinton, E. Waterproofing crops: Effective flooding survival strategies. Plant Physiol. 2012, 160, 1698–1709. [Google Scholar] [CrossRef]
- Hirabayashi, Y.; Mahendran, R.; Koirala, S.; Konoshima, L.; Yamazaki, D.; Watanabe, S.; Kim, H.; Kanae, S. Global flood risk under climate change. Nat. Clim. Change 2013, 3, 816–821. [Google Scholar] [CrossRef]
- Voesenek, L.A.C.J.; Bailey-Serres, J. Flood adaptive traits and processes: An overview. New Phytol. 2015, 206, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of climate change on crops adaptation and strategies to tackle its outcome: A review. Plants 2019, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Bailey-Serres, J.; Ashikari, M.; Atwell, B.J.; Colmer, T.D.; Fagerstedt, K.; Fukao, T.; Geigenberger, P.; Hebelstrup, K.H.; Hill, R.D.; et al. Community recommendations on terminology and procedures used in flooding and low oxygen stress research. New Phytol. 2017, 214, 1403–1407. [Google Scholar] [CrossRef]
- Nakamura, M.; Noguchi, K. Tolerant mechanisms to O2 deficiency under submergence conditions in plants. J. Plant. Res. 2020, 133, 343–371. [Google Scholar] [CrossRef] [PubMed]
- Loreti, E.; Perata, P. The many facets of hypoxia in plants. Plants 2020, 9, 745. [Google Scholar] [CrossRef]
- Weits, D.A.; van Dongen, J.T.; Licausi, F. Molecular oxygen as a signaling component in plant development. New Phytol. 2021, 229, 24–35. [Google Scholar] [CrossRef] [PubMed]
- van Dongen, J.T.; Schurr, U.; Pfister, M.; Geigenberger, P. Phloem metabolism and function have to cope with low internal oxygen. Plant Physiol. 2003, 131, 1529–1543. [Google Scholar] [CrossRef]
- Borisjuk, L.; Rolletschek, H. The oxygen status of the developing seed. New Phytol. 2009, 182, 17–30. [Google Scholar] [CrossRef]
- Ho, Q.T.; Verboven, P.; Verlinden, B.E.; Schenk, A.; Delele, M.A.; Rolletschek, H.; Vercammen, J.; Nicolaï, B.M. Genotype effects on internal gas gradients in apple fruit. J. Exp. Bot. 2010, 61, 2745–2755. [Google Scholar] [CrossRef]
- Geigenberger, P.; Fernie, A.R.; Gibon, Y.; Christ, M.; Stitt, M. Metabolic activity decreases as an adaptive response to low internal oxygen in growing potato tubers. Biol. Chem. 2000, 381, 723–740. [Google Scholar] [CrossRef]
- Porterfield, D.M.; Kuang, A.; Smith, P.J.; Crispi, M.L.; Musgrave, M.E. Oxygen-depleted zones inside reproductive structures of Brassicaceae: Implications for oxygen control of seed development. Can. J. Bot. 1999, 77, 1439–1446. [Google Scholar] [CrossRef]
- Rolletschek, H.; Borisjuk, L.; Koschorreck, M.; Wobus, U.; Weber, H. Legume embryos develop in a hypoxic environment. J. Exp. Bot. 2002, 53, 1099–1107. [Google Scholar] [CrossRef]
- Weits, D.A.; Kunkowska, A.B.; Kamps, N.C.W.; Portz, K.M.S.; Packbier, N.K.; Nemec Venza, Z.; Gaillochet, C.; Lohmann, J.U.; Pedersen, O.; van Dongen, J.T.; et al. An apical hypoxic niche sets the pace of shoot meristem activity. Nature 2019, 569, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Dukowic-Schulze, S.; van der Linde, K. Oxygen, secreted proteins and small RNAs: Mobile elements that govern anther development. Plant Reprod 2021, 34, 1–19. [Google Scholar] [CrossRef]
- Kende, H.; van der Knaap, E.; Cho, H.T. Deepwater rice: A model plant to study stem elongation. Plant Physiol. 1998, 118, 1105–1110. [Google Scholar] [CrossRef] [PubMed]
- Drew, M.C.; He, C.J.; Morgan, P.W. Programmed cell death and aerenchyma formation in roots. Trends Plant Sci. 2000, 5, 123–127. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Voesenek, L.A. Flooding stress: Acclimations and genetic diversity. Annu. Rev. Plant Biol. 2008, 59, 313–339. [Google Scholar] [CrossRef]
- Fukao, T.; Bailey-Serres, J. Plant responses to hypoxia—Is survival a balancing act? Trends Plant Sci. 2004, 9, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Hinz, M.; Wilson, I.W.; Yang, J.; Buerstenbinder, K.; Llewellyn, D.; Dennis, E.S.; Sauter, M.; Dolferus, R. Arabidopsis RAP2.2: An ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 2010, 153, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Hebelstrup, K.H.; Møller, I.M. Mitochondrial signaling in plants under hypoxia: Use of reactive oxygen species (ROS) and reactive nitrogen species (RNS). In Reactive Oxygen and Nitrogen Species Signaling and Communication in Plants; Gupta, K.J., Igamberdiev, A.U., Eds.; Springer: Cham, Switzerland, 2015; pp. 63–77. [Google Scholar] [CrossRef]
- Turkan, I. ROS and RNS: Key signalling molecules in plants. J. Exp. Bot. 2018, 69, 3313–3315. [Google Scholar] [CrossRef] [PubMed]
- Branco-Price, C.; Kawaguchi, R.; Ferreira, R.B.; Bailey-Serres, J. Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann. Bot. 2005, 96, 647–660. [Google Scholar] [CrossRef]
- Loreti, E.; Poggi, A.; Novi, G.; Alpi, A.; Perata, P. A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol. 2005, 137, 1130–1138. [Google Scholar] [CrossRef]
- Fukao, T.; Xu, K.; Ronald, P.C.; Bailey-Serres, J. A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 2006, 18, 2021–2034. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef]
- Branco-Price, C.; Kaiser, K.A.; Jang, C.J.; Larive, C.K.; Bailey-Serres, J. Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J. 2008, 56, 743–755. [Google Scholar] [CrossRef]
- Fukao, T.; Bailey-Serres, J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 16814–16819. [Google Scholar] [CrossRef]
- van Dongen, J.T.; Frohlich, A.; Ramirez-Aguilar, S.J.; Schauer, N.; Fernie, A.R.; Erban, A.; Kopka, J.; Clark, J.; Langer, A.; Geigenberger, P. Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Ann. Bot. 2009, 103, 269–280. [Google Scholar] [CrossRef]
- Rocha, M.; Licausi, F.; Araujo, W.L.; Nunes-Nesi, A.; Sodek, L.; Fernie, A.R.; van Dongen, J.T. Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol. 2010, 152, 1501–1513. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Beard, K.F.; Nunes-Nesi, A.; Fernie, A.R.; Ratcliffe, R.G. Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 2010, 15, 462–470. [Google Scholar] [CrossRef]
- Mustroph, A.; Lee, S.C.; Oosumi, T.; Zanetti, M.E.; Yang, H.; Ma, K.; Yaghoubi-Masihi, A.; Fukao, T.; Bailey-Serres, J. Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 2010, 152, 1484–1500. [Google Scholar] [CrossRef]
- Licausi, F.; Weits, D.A.; Pant, B.D.; Scheible, W.R.; Geigenberger, P.; van Dongen, J.T. Hypoxia responsive gene expression is mediated by various subsets of transcription factors and miRNAs that are determined by the actual oxygen availability. New Phytol. 2011, 190, 442–456. [Google Scholar] [CrossRef] [PubMed]
- Mustroph, A.; Barding, G.A., Jr.; Kaiser, K.A.; Larive, C.K.; Bailey-Serres, J. Characterization of distinct root and shoot responses to low-oxygen stress in Arabidopsis with a focus on primary C- and N-metabolism. Plant Cell Environ. 2014, 37, 2366–2380. [Google Scholar] [CrossRef]
- Klok, E.J.; Wilson, I.W.; Wilson, D.; Chapman, S.C.; Ewing, R.M.; Somerville, S.C.; Peacock, W.J.; Dolferus, R.; Dennis, E.S. Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell 2002, 14, 2481–2494. [Google Scholar] [CrossRef] [PubMed]
- Ismond, K.P.; Dolferus, R.; de Pauw, M.; Dennis, E.S.; Good, A.G. Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiol. 2003, 132, 1292–1302. [Google Scholar] [CrossRef] [PubMed]
- Maricle, B.R.; White, S.J.; Meraz, A.; Maforo, N.G.; Biggs, T.N.; Martin, N.M.; Caudle, K.L. Effect of ethanol toxicity on enzyme activity in anaerobic respiration in plants. Trans. KAS 2014, 117, 237–244. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Chang, R. Sensing and signalling in response to oxygen deprivation in plants and other organisms. Ann. Bot. 2005, 96, 507–518. [Google Scholar] [CrossRef]
- Santosa, I.; Ram, P.; Boamfa, E.; Laarhoven, L.; Reuss, J.; Jackson, M.; Harren, F. Patterns of peroxidative ethane emission from submerged rice seedlings indicate that damage from reactive oxygen species takes place during submergence and is not necessarily a post-anoxic phenomenon. Planta 2007, 226, 193–202. [Google Scholar] [CrossRef]
- Fukao, T.; Yeung, E.; Bailey-Serres, J. The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 2011, 23, 412–427. [Google Scholar] [CrossRef]
- Yeung, E.; Bailey-Serres, J.; Sasidharan, R. After the deluge: Plant revival post-flooding. Trends Plant Sci. 2019, 24, 443–454. [Google Scholar] [CrossRef]
- Rizhsky, L.; Liang, H.; Mittler, R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 2002, 130, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Atkin, O.K.; Tjoelker, M.G. Thermal acclimation and the dynamic response of plant respiration to temperature. Trends Plant Sci. 2003, 8, 343–351. [Google Scholar] [CrossRef]
- Hauben, M.; Haesendonckx, B.; Standaert, E.; van der Kelen, K.; Azmi, A.; Akpo, H.; van Breusegem, F.; Guisez, Y.; Bots, M.; Lambert, B.; et al. Energy use efficiency is characterized by an epigenetic component that can be directed through artificial selection to increase yield. Proc. Natl. Acad. Sci. USA 2009, 106, 20109–20114. [Google Scholar] [CrossRef]
- Gibbs, J.; Greenway, H. Review: Mechanisms of anoxia tolerance in plants. I. Growth, survival and anaerobic catabolism. Funct. Plant Biol. 2003, 30, 353. [Google Scholar] [CrossRef] [PubMed]
- Moller, I.M. Plant mitochondria and oxidative stress: Electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 561–591. [Google Scholar] [CrossRef]
- Liao, C.T.; Lin, C.H. Physiological adaptation of crop plants to flooding stress. Proc. Natl. Sci. Counc. Repub. China B 2001, 25, 148–157. [Google Scholar]
- Greenway, H.; Gibbs, J. Review: Mechanisms of anoxia tolerance in plants. II. Energy requirements for maintenance and energy distribution to essential processes. Funct. Plant. Biol. 2003, 30, 999–1036. [Google Scholar] [CrossRef]
- Mancuso, S.; Marras, A.M. Adaptative response of Vitis root to anoxia. Plant Cell Physiol. 2006, 47, 401–409. [Google Scholar] [CrossRef]
- Mailloux, R.J.; Bériault, R.; Lemire, J.; Singh, R.; Chénier, D.R.; Hamel, R.D.; Appanna, V.D. The tricarboxylic acid cycle, an ancient metabolic network with a novel twist. PLoS ONE 2007, 2, e690. [Google Scholar] [CrossRef]
- Roberts, J.K.; Callis, J.; Jardetzky, O.; Walbot, V.; Freeling, M. Cytoplasmic acidosis as a determinant of flooding intolerance in plants. Proc. Natl. Acad. Sci. USA 1984, 81, 6029–6033. [Google Scholar] [CrossRef] [PubMed]
- Felle, H.H. pH regulation in anoxic plants. Ann. Bot. 2005, 96, 519–532. [Google Scholar] [CrossRef]
- Narsai, R.; Rocha, M.; Geigenberger, P.; Whelan, J.; van Dongen, J.T. Comparative analysis between plant species of transcriptional and metabolic responses to hypoxia. New Phytol. 2011, 190, 472–487. [Google Scholar] [CrossRef] [PubMed]
- Shingaki-Wells, R.N.; Huang, S.; Taylor, N.L.; Carroll, A.J.; Zhou, W.; Millar, A.H. Differential molecular responses of rice and wheat coleoptiles to anoxia reveal novel metabolic adaptations in amino acid metabolism for tissue tolerance. Plant Physiol. 2011, 156, 1706–1724. [Google Scholar] [CrossRef]
- Kato-Noguchi, H.; Morokuma, M. Ethanolic fermentation and anoxia tolerance in four rice cultivars. J. Plant Physiol. 2007, 164, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, R.A.; Rumpho, M.E.; Fox, T.C. Anaerobic metabolism in plants. Plant Physiol. 1992, 100, 1–6. [Google Scholar] [CrossRef]
- Rahman, M.; Grover, A.; Peacock, W.J.; Dennis, E.S.; Ellis, M.H. Effects of manipulation of pyruvate decarboxylase and alcohol dehydrogenase levels on the submergence tolerance of rice. Funct. Plant Biol. 2001, 28, 1231–1241. [Google Scholar] [CrossRef]
- Kürsteiner, O.; Dupuis, I.; Kuhlemeier, C. The pyruvate decarboxylase1 gene of Arabidopsis is required during anoxia but not other environmental stresses. Plant Physiol. 2003, 132, 968–978. [Google Scholar] [CrossRef]
- Huang, S.; Colmer, T.D.; Millar, A.H. Does anoxia tolerance involve altering the energy currency towards PPi? Trends Plant Sci 2008, 13, 221–227. [Google Scholar] [CrossRef]
- Camacho-Pereira, J.; Meyer, L.E.; Machado, L.B.; Oliveira, M.F.; Galina, A. Reactive oxygen species production by potato tuber mitochondria is modulated by mitochondrially bound hexokinase activity. Plant Physiol. 2009, 149, 1099–1110. [Google Scholar] [CrossRef]
- Wegner, L.H. Oxygen transport in waterlogged plants. In Waterlogging Signalling and Tolerance in Plants; Mancuso, S., Shabala, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 3–22. [Google Scholar] [CrossRef]
- Geigenberger, P. Response of plant metabolism to too little oxygen. Curr. Opin. Plant Biol. 2003, 6, 247–256. [Google Scholar] [CrossRef]
- Howell, K.A.; Millar, A.H.; Whelan, J. Ordered assembly of mitochondria during rice germination begins with pro-mitochondrial structures rich in components of the protein import apparatus. Plant Mol. Biol. 2006, 60, 201–223. [Google Scholar] [CrossRef]
- Nakamura, T.; Yamamoto, R.; Hiraga, S.; Nakayama, N.; Okazaki, K.; Takahashi, H.; Uchimiya, H.; Komatsu, S. Evaluation of metabolite alteration under flooding stress in soybeans. JARQ 2012, 46, 237–248. [Google Scholar] [CrossRef]
- Fan, T.W.; Higashi, R.M.; Lane, A.N. An in vivo 1H and 31P NMR investigation of the effect of nitrate on hypoxic metabolism in maize roots. Arch. Biochem. Biophys. 1988, 266, 592–606. [Google Scholar] [CrossRef]
- Aurisano, N.; Bertani, A.; Reggiani, R. Anaerobic accumulation of 4-aminobutyrate in rice seedlings; Causes and significance. Phytochemistry 1995, 38, 1147–1150. [Google Scholar] [CrossRef]
- Bai, Q.; Chai, M.; Gu, Z.; Cao, X.; Li, Y.; Liu, K. Effects of components in culture medium on glutamate decarboxylase activity and γ-aminobutyric acid accumulation in foxtail millet (Setaria italica L.) during germination. Food Chem. 2009, 116, 152–157. [Google Scholar] [CrossRef]
- Yang, R.; Guo, Q.; Gu, Z. GABA shunt and polyamine degradation pathway on γ-aminobutyric acid accumulation in germinating fava bean (Vicia faba L.) under hypoxia. Food Chem. 2013, 136, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Salvatierra, A.; Pimentel, P.; Almada, R.; Hinrichsen, P. Exogenous GABA application transiently improves the tolerance to root hypoxia on a sensitive genotype of Prunus rootstock. Environ. Exp. Bot. 2016, 125, 52–66. [Google Scholar] [CrossRef]
- Lü, G.; Liang, Y.; Wu, X.; Li, J.; Ma, W.; Zhang, Y.; Gao, H. Molecular cloning and functional characterization of mitochondrial malate dehydrogenase (mMDH) is involved in exogenous GABA increasing root hypoxia tolerance in muskmelon plants. Sci. Hort. 2019, 258, 108741. [Google Scholar] [CrossRef]
- Shelp, B.J.; Aghdam, M.S.; Flaherty, E.J. γ-aminobutyrate (GABA) regulated plant defense: Mechanisms and opportunities. Plants 2021, 10, 1939. [Google Scholar] [CrossRef]
- Bouché, N.; Fromm, H. GABA in plants: Just a metabolite? Trends Plant Sci. 2004, 9, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Shelp, B.J.; Mullen, R.T.; Waller, J.C. Compartmentation of GABA metabolism raises intriguing questions. Trends Plant Sci. 2012, 17, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Signorelli, S.; Dans, P.D.; Coitiño, E.L.; Borsani, O.; Monza, J. Connecting proline and γ-aminobutyric acid in stressed plants through non-enzymatic reactions. PLoS ONE 2015, 10, e0115349. [Google Scholar] [CrossRef]
- Palanivelu, R.; Brass, L.; Edlund, A.F.; Preuss, D. Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 2003, 114, 47–59. [Google Scholar] [CrossRef]
- Shelp, B.J.; Walton, C.S.; Snedden, W.A.; Tuin, L.G.; Oresnik, I.J.; Layzell, D.B. GABA shunt in developing soybean seeds is associated with hypoxia. Physiol. Plant 1995, 94, 219–228. [Google Scholar] [CrossRef]
- Ricoult, C.; Echeverria, L.O.; Cliquet, J.B.; Limami, A.M. Characterization of alanine aminotransferase (AlaAT) multigene family and hypoxic response in young seedlings of the model legume Medicago truncatula. J. Exp. Bot. 2006, 57, 3079–3089. [Google Scholar] [CrossRef]
- Fait, A.; Yellin, A.; Fromm, H. GABA shunt deficiencies and accumulation of reactive oxygen intermediates: Insight from Arabidopsis mutants. FEBS Lett. 2005, 579, 415–420. [Google Scholar] [CrossRef]
- Miyashita, Y.; Good, A.G. Contribution of the GABA shunt to hypoxia-induced alanine accumulation in roots of Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 92–102. [Google Scholar] [CrossRef]
- Carillo, P. GABA shunt in durum wheat. Front. Plant Sci. 2018, 9, 100. [Google Scholar] [CrossRef]
- Wu, Q.; Su, N.; Huang, X.; Cui, J.; Shabala, L.; Zhou, M.; Yu, M.; Shabala, S. Hypoxia-induced increase in GABA content is essential for restoration of membrane potential and preventing ROS-induced disturbance to ion homeostasis. Plant Commun. 2021, 2, 100188. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Tsuji, H. Ultrastructural changes of organelles in coleoptile cells during anaerobic germination of rice seeds. Protoplasma 1971, 73, 203–215. [Google Scholar] [CrossRef]
- Vartapetian, B.B.; Andreeva, I.N.; Kozlova, G.I. The resistance to anoxia and the mitochondrial fine structure of rice seedlings. Protoplasma 1976, 88, 215–224. [Google Scholar] [CrossRef]
- Oliveira, L. Changes in the ultrastructure of mitochondria of roots of Triticale subjected to anaerobiosis. Protoplasma 1977, 91, 267–280. [Google Scholar] [CrossRef]
- Vartapetian, B.B.; Zakhmylova, N.A.; Generozova, I.P. Ultrastructure of wheat coleoptile mitochondria at short-term anoxia and post-anoxia. Plant Cell Environ. 1985, 8, 65–67. [Google Scholar] [CrossRef]
- Fox, T.C.; Kennedy, R.A. Mitochondrial enzymes in aerobically and anaerobically germinated seedlings of Echinochloa and rice. Planta 1991, 184, 510–514. [Google Scholar] [CrossRef]
- Dudkina, N.V.; Heinemeyer, J.; Sunderhaus, S.; Boekema, E.J.; Braun, H.P. Respiratory chain supercomplexes in the plant mitochondrial membrane. Trends Plant Sci. 2006, 11, 232–240. [Google Scholar] [CrossRef]
- Schägger, H. Respiratory chain supercomplexes. IUBMB Life 2001, 52, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Eubel, H.; Heinemeyer, J.; Sunderhaus, S.; Braun, H.P. Respiratory chain supercomplexes in plant mitochondria. Plant Physiol. Biochem. 2004, 42, 937–942. [Google Scholar] [CrossRef]
- Diaz, F.; Fukui, H.; Garcia, S.; Moraes, C.T. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol. Cell. Biol. 2006, 26, 4872–4881. [Google Scholar] [CrossRef] [PubMed]
- Boekema, E.J.; Braun, H.P. Supramolecular structure of the mitochondrial oxidative phosphorylation system. J. Biol. Chem. 2007, 282, 1–4. [Google Scholar] [CrossRef]
- Acín-Pérez, R.; Fernández-Silva, P.; Peleato, M.L.; Pérez-Martos, A.; Enriquez, J.A. Respiratory active mitochondrial supercomplexes. Mol. Cell 2008, 32, 529–539. [Google Scholar] [CrossRef]
- Gupta, K.J.; Zabalza, A.; van Dongen, J.T. Regulation of respiration when the oxygen availability changes. Physiol. Plant 2009, 137, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Aguilar, S.J.; Keuthe, M.; Rocha, M.; Fedyaev, V.V.; Kramp, K.; Gupta, K.J.; Rasmusson, A.G.; Schulze, W.X.; van Dongen, J.T. The composition of plant mitochondrial supercomplexes changes with oxygen availability. J. Biol. Chem. 2011, 286, 43045–43053. [Google Scholar] [CrossRef] [PubMed]
- Abad, M.F.; Di Benedetto, G.; Magalhães, P.J.; Filippin, L.; Pozzan, T. Mitochondrial pH monitored by a new engineered green fluorescent protein mutant. J. Biol. Chem. 2004, 279, 11521–11529. [Google Scholar] [CrossRef]
- Rosca, M.G.; Vazquez, E.J.; Kerner, J.; Parland, W.; Chandler, M.P.; Stanley, W.; Sabbah, H.N.; Hoppel, C.L. Cardiac mitochondria in heart failure: Decrease in respirasomes and oxidative phosphorylation. Cardiovasc. Res. 2008, 80, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Orij, R.; Postmus, J.; Ter Beek, A.; Brul, S.; Smits, G.J. In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology 2009, 155, 268–278. [Google Scholar] [CrossRef]
- Ellis, M.H.; Dennis, E.S.; Peacock, W.J. Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol. 1999, 119, 57–64. [Google Scholar] [CrossRef]
- Mustroph, A.; Boamfa, E.I.; Laarhoven, L.J.; Harren, F.J.; Albrecht, G.; Grimm, B. Organ-specific analysis of the anaerobic primary metabolism in rice and wheat seedlings. I: Dark ethanol production is dominated by the shoots. Planta 2006, 225, 103–114. [Google Scholar] [CrossRef]
- Mustroph, A.; Boamfa, E.I.; Laarhoven, L.J.; Harren, F.J.; Pörs, Y.; Grimm, B. Organ specific analysis of the anaerobic primary metabolism in rice and wheat seedlings II: Light exposure reduces needs for fermentation and extends survival during anaerobiosis. Planta 2006, 225, 139–152. [Google Scholar] [CrossRef]
- Hsu, F.C.; Chou, M.Y.; Peng, H.P.; Chou, S.J.; Shih, M.C. Insights into hypoxic systemic responses based on analyses of transcriptional regulation in Arabidopsis. PLoS ONE 2011, 6, e28888. [Google Scholar] [CrossRef]
- Lee, S.C.; Mustroph, A.; Sasidharan, R.; Vashisht, D.; Pedersen, O.; Oosumi, T.; Voesenek, L.A.; Bailey-Serres, J. Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol. 2011, 190, 457–471. [Google Scholar] [CrossRef]
- Chang, R.; Jang, C.J.H.; Branco-Price, C.; Nghiem, P.; Bailey-Serres, J. Transient MPK6 activation in response to oxygen deprivation and reoxygenation is mediated by mitochondria and aids seedling survival in Arabidopsis. Plant Mol. Biol. 2012, 78, 109–122. [Google Scholar] [CrossRef]
- Lothier, J.; Diab, H.; Cukier, C.; Limami, A.M.; Tcherkez, G. Metabolic responses to waterlogging differ between roots and shoots and reflect phloem transport alteration in Medicago truncatula. Plants 2020, 9, 1373. [Google Scholar] [CrossRef]
- Cui, J.; Abadie, C.; Carroll, A.; Lamade, E.; Tcherkez, G. Responses to K deficiency and waterlogging interact via respiratory and nitrogen metabolism. Plant Cell Environ. 2019, 42, 647–658. [Google Scholar] [CrossRef]
- Cui, J.; Lamade, E.; Fourel, F.; Tcherkez, G. δ15 N values in plants are determined by both nitrate assimilation and circulation. New Phytol. 2020, 226, 1696–1707. [Google Scholar] [CrossRef]
- Christianson, J.A.; Llewellyn, D.J.; Dennis, E.S.; Wilson, I.W. Global gene expression responses to waterlogging in roots and leaves of cotton (Gossypium hirsutum L.). Plant Cell Physiol. 2010, 51, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Araki, H.; Hossain, M.A.; Takahashi, T. Waterlogging and hypoxia have permanent effects on wheat root growth and respiration. J. Agron. Crop. Sci. 2012, 198, 264–275. [Google Scholar] [CrossRef]
- Gomes, A.R.; Kozlowski, T.T. Growth responses and adaptations of Fraxinus pennsylvanica seedlings to flooding. Plant Physiol. 1980, 66, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Ismail, M.R.; Noor, K.M. (1996). Growth and physiological processes of young starfruit (Averrhoa carambola L.) plants under soil flooding. Sci. Hortic. 1996, 65, 229–238. [Google Scholar] [CrossRef]
- Malik, A.I.; Colmer, T.D.; Lambers, H.; Schortemeyer, M. Changes in physiological and morphological traits of roots and shoots of wheat in response to different depths of waterlogging. Func. Plant Biol. 2001, 28, 1121–1131. [Google Scholar] [CrossRef]
- Lal, B.; Gautam, P.; Rath, L.; Haldar, D.; Panda, B.B.; Raja, R.; Shahid, M.; Tripathi, R.; Bhattacharyya, P.; Mohanty, S.; et al. Effect of nutrient application on growth, metabolic and enzymatic activities of rice seedlings during flooding stress and subsequent re-aeration. J. Agron. Crop Sci. 2015, 201, 138–151. [Google Scholar] [CrossRef]
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Nagar, P.; Goel, P.; Singh, A.K.; Kumari, S.; Mustafiz, A. Second messengers: Central regulators in plant abiotic stress response. In Abiotic Stress-Mediated Sensing and Signaling in Plants: An Omics Perspective; Zargar, S., Zargar, M., Eds.; Springer: Singapore, 2018; pp. 47–94. [Google Scholar] [CrossRef]
- Wagner, S.; Steinbeck, J.; Fuchs, P.; Lichtenauer, S.; Elsässer, M.; Schippers, J.H.M.; Nietzel, T.; Ruberti, C.; van Aken, O.; Meyer, A.J.; et al. Multiparametric real-time sensing of cytosolic physiology links hypoxia responses to mitochondrial electron transport. New Phytol. 2019, 224, 1668–1684. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.D.; Grego, S.; Kenworthy, P. The control of the production of lactate and ethanol by higher plants. Planta 1974, 118, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.K.; Callis, J.; Wemmer, D.; Walbot, V.; Jardetzky, O. Mechanisms of cytoplasmic pH regulation in hypoxic maize root tips and its role in survival under hypoxia. Proc. Natl. Acad. Sci. USA 1984, 81, 3379–3383. [Google Scholar] [CrossRef]
- Gout, E.; Boisson, A.; Aubert, S.; Douce, R.; Bligny, R. Origin of the cytoplasmic pH changes during anaerobic stress in higher plant cells. Carbon-13 and phosphorous-31 nuclear magnetic resonance studies. Plant Physiol. 2001, 125, 912–925. [Google Scholar] [CrossRef] [PubMed]
- Felle, H.H. pH: Signal and messenger in plant cells. Plant Biol. 2001, 3, 577–591. [Google Scholar] [CrossRef]
- Becker, D.; Geiger, D.; Dunkel, M.; Roller, A.; Bertl, A.; Latz, A.; Carpaneto, A.; Dietrich, P.; Roelfsema, M.R.; Voelker, C.; et al. AtTPK4, an Arabidopsis tandem-pore K+ channel, poised to control the pollen membrane voltage in a pH- and Ca2+-dependent manner. Proc. Natl. Acad. Sci. USA 2004, 101, 15621–15626. [Google Scholar] [CrossRef]
- Zeng, F.; Konnerup, D.; Shabala, L.; Zhou, M.; Colmer, T.D.; Zhang, G.; Shabala, S. Linking oxygen availability with membrane potential maintenance and K+ retention of barley roots: Implications for waterlogging stress tolerance. Plant Cell Environ. 2014, 37, 2325–2338. [Google Scholar] [CrossRef]
- Reddy, A.S.N.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses: Roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef]
- Steinhorst, L.; Kudla, J. Signaling in cells and organisms—Calcium holds the line. Curr. Opin. Plant Biol. 2014, 22, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.Y.; Dunand, C.; Snedden, W.; Galaud, J.P. CaM and CML emergence in the green lineage. Trends Plant Sci. 2015, 20, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Subbaiah, C.C.; Zhang, J.K.; Sachs, M.M. Involvement of intracellular calcium in anaerobic gene-expression and survival of maize seedlings. Plant Physiol. 1994, 105, 369–376. [Google Scholar] [CrossRef]
- Yemelyanov, V.V.; Shishova, M.F.; Chirkova, T.V.; Lindberg, S.M. Anoxia-induced elevation of cytosolic Ca2+ concentration depends on different Ca2+ sources in rice and wheat protoplasts. Planta 2011, 234, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, S.; Kader, M.A.; Yemelyanov, V. Calcium signalling in plant cells under environmental stress. In Environmental Adaptations and Stress Tolerance of Plants in the Era of Climate Change; Ahmad, P., Prasad, M.M.V., Eds.; Springer Science & Business Media: New York, NY, USA, 2012; pp. 325–360. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Hill, R.D. Elevation of cytosolic Ca2+ in response to energy deficiency in plants: The general mechanism of adaptation to low oxygen stress. Biochem. J. 2018, 475, 1411–1425. [Google Scholar] [CrossRef]
- Subbaiah, C.C.; Sachs, M.M. Molecular and cellular adaptations of maize to flooding stress. Ann. Bot. 2003, 91, 119–127. [Google Scholar] [CrossRef]
- Shabala, S.; Shabala, L.; Barcelo, J.; Poschenrieder, C. Membrane transporters mediating root signalling and adaptive responses to oxygen deprivation and soil flooding. Plant Cell Environ. 2014, 37, 2216–2233. [Google Scholar] [CrossRef]
- Wang, F.; Chen, Z.H.; Liu, X.; Colmer, T.D.; Zhou, M.; Shabala, S. Tissue-specific root ion profiling reveals essential roles for the CAX and ACA calcium transport systems for hypoxia response in Arabidopsis. J. Exp. Bot. 2016, 67, 3747–3762. [Google Scholar] [CrossRef]
- Hao, M.S.; Jensen, A.M.; Boquist, A.S.; Liu, Y.J.; Rasmusson, A.G. The Ca2+-regulation of the mitochondrial external NADPH dehydrogenase in plants is controlled by cytosolic pH. PLoS ONE 2015, 10, e0139224. [Google Scholar] [CrossRef][Green Version]
- António, C.; Päpke, C.; Rocha, M.; Diab, H.; Limami, A.M.; Obata, T.; Fernie, A.R.; van Dongen, J.T. Regulation of primary metabolism in response to low oxygen availability as revealed by carbon and nitrogen isotope redistribution. Plant Physiol. 2016, 170, 43–56. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Mittler, R. The roles of reactive oxygen species in plant cells. Plant Physiol. 2006, 141, 311. [Google Scholar] [CrossRef] [PubMed]
- Schwarzländer, M.; Finkemeier, I. Mitochondrial energy and redox signaling in plants. Antioxid. Redox. Signal. 2013, 18, 2122–2144. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Watanabe, K.; Fukazawa, A.; Mori, H.; Abe, F.; Kawaguchi, K.; Oyanagi, A.; Nakazono, M. Ethylene and reactive oxygen species are involved in root aerenchyma formation and adaptation of wheat seedlings to oxygen-deficient conditions. J. Exp. Bot. 2014, 65, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; van Aken, O.; Elsässer, M.; Schwarzländer, M. Mitochondrial energy signaling and its role in the low-oxygen stress response of plants. Plant Physiol. 2018, 176, 1156–1170. [Google Scholar] [CrossRef]
- Baxter-Burrell, A.; Yang, Z.B.; Springer, P.S.; Bailey-Serres, J. RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science 2002, 296, 2026–2028. [Google Scholar] [CrossRef] [PubMed]
- Hamanaka, R.B.; Chandel, N.S. Mitochondrial reactive oxygen species regulate hypoxic signaling. Curr. Opin. Cell Biol. 2009, 21, 894–899. [Google Scholar] [CrossRef]
- Sagi, M.; Fluhr, R. Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 2006, 141, 336–340. [Google Scholar] [CrossRef]
- Torres, M.A.; Dangl, J.L.; Jones, J.D. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 2002, 99, 517–522. [Google Scholar] [CrossRef]
- Pucciariello, C.; Parlanti, S.; Banti, V.; Novi, G.; Perata, P. Reactive oxygen species-driven transcription in Arabidopsis under oxygen deprivation. Plant Physiol. 2012, 159, 184–196. [Google Scholar] [CrossRef]
- Chen, L.; Liao, B.; Qi, H.; Xie, L.J.; Huang, L.; Tan, W.J.; Zhai, N.; Yuan, L.B.; Zhou, Y.; Yu, L.J.; et al. Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 2015, 11, 2233–2246. [Google Scholar] [CrossRef]
- Wang, F.; Chen, Z.H.; Liu, X.; Colmer, T.D.; Shabala, L.; Salih, A.; Zhou, M.; Shabala, S. Revealing the roles of GORK channels and NADPH oxidase in acclimation to hypoxia in Arabidopsis. J. Exp. Bot. 2017, 68, 3191–3204. [Google Scholar] [CrossRef]
- Gonzali, S.; Loreti, E.; Cardarelli, F.; Novi, G.; Parlanti, S.; Pucciariello, C.; Bassolino, L.; Banti, V.; Licausi, F.; Perata, P. Universal stress protein HRU1 mediates ROS homeostasis under anoxia. Nat. Plants 2015, 1, 15151. [Google Scholar] [CrossRef]
- Kim, T.H.; Böhmer, M.; Hu, H.; Nishimura, N.; Schroeder, J.I. Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 2010, 61, 561–591. [Google Scholar] [CrossRef]
- Hedrich, R. Ion channels in plants. Physiol. Rev. 2012, 92, 1777–1811. [Google Scholar] [CrossRef]
- Chen, Z.H.; Chen, G.; Dai, F.; Wang, Y.; Hills, A.; Ruan, Y.L.; Zhang, G.; Franks, P.J.; Nevo, E.; Blatt, M.R. Molecular evolution of grass stomata. Trends Plant Sci. 2017, 22, 124–139. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Hill, R.D. Nitrate, NO and haemoglobin in plant adaptation to hypoxia: An alternative to classic fermentation pathways. J. Exp. Bot. 2004, 55, 2473–2482. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Baron, K.; Manac’H-Little, N.; Stoimenova, M.; Hill, R.D. The haemoglobin/nitric oxide cycle: Involvement in flooding stress and effects on hormone signalling. Ann. Bot. 2005, 96, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.D. Non-symbiotic haemoglobins—What’s happening beyond nitric oxide scavenging? AoB Plants 2012, 2012, pls004. [Google Scholar] [CrossRef] [PubMed]
- Jayawardhane, J.; Cochrane, D.W.; Vyas, P.; Bykova, N.V.; Vanlerberghe, G.C.; Igamberdiev, A.U. Roles for plant mitochondrial alternative oxidase under normoxia, hypoxia, and reoxygenation conditions. Front. Plant Sci. 2020, 11, 566. [Google Scholar] [CrossRef] [PubMed]
- Mustroph, A.; Zanetti, M.E.; Jang, C.J.; Holtan, H.E.; Repetti, P.P.; Galbraith, D.W.; Girke, T.; Bailey-Serres, J. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 18843–18848. [Google Scholar] [CrossRef]
- Licausi, F.; van Dongen, J.T.; Giuntoli, B.; Novi, G.; Santaniello, A.; Geigenberger, P.; Perata, P. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 2010, 62, 302–315. [Google Scholar] [CrossRef]
- Licausi, F.; Kosmacz, M.; Weits, D.A.; Giuntoli, B.; Giorgi, F.M.; Voesenek, L.A.; Perata, P.; van Dongen, J.T. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 2011, 479, 419–422. [Google Scholar] [CrossRef]
- Meng, X.; Li, L.; Narsai, R.; De Clercq, I.; Whelan, J.; Berkowitz, O. Mitochondrial signalling is critical for acclimation and adaptation to flooding in Arabidopsis thaliana. Plant J. 2020, 103, 227–247. [Google Scholar] [CrossRef]
- Bui, L.T.; Shukla, V.; Giorgi, F.M.; Trivellini, A.; Perata, P.; Licausi, F.; Giuntoli, B. Differential submergence tolerance between juvenile and adult Arabidopsis plants involves the ANAC017 transcription factor. Plant J. 2020, 104, 979–994. [Google Scholar] [CrossRef]
- Tang, H.; Bi, H.; Liu, B.; Lou, S.; Song, Y.; Tong, S.; Chen, N.; Jiang, Y.; Liu, J.; Liu, H. WRKY33 interacts with WRKY12 protein to up-regulate RAP2.2 during submergence induced hypoxia response in Arabidopsis thaliana. New Phytol. 2021, 229, 106–125. [Google Scholar] [CrossRef] [PubMed]
- Banti, V.; Mafessoni, F.; Loreti, E.; Alpi, A.; Perata, P. The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol. 2010, 152, 1471–1483. [Google Scholar] [CrossRef]
- Klecker, M.; Gasch, P.; Peisker, H.; Dörmann, P.; Schlicke, H.; Grimm, B.; Mustroph, A. A shoot-specific hypoxic response of Arabidopsis sheds light on the role of the phosphate-responsive transcription factor PHOSPHATE STARVATION RESPONSE1. Plant Physiol. 2014, 165, 774–790. [Google Scholar] [CrossRef]
- Mendiondo, G.M.; Gibbs, D.J.; Szurman-Zubrzycka, M.; Korn, A.; Marquez, J.; Szarejko, I.; Maluszynski, M.; King, J.; Axcell, B.; Smart, K.; et al. Enhanced waterlogging tolerance in barley by manipulation of expression of the N-end rule pathway E3 ligase PROTEOLYSIS6. Plant Biotechnol. J. 2016, 14, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Xu, H.; Rong, W.; Ye, X.; Zhang, Z. Constitutive expression of a stabilized transcription factor group VII ethylene response factor enhances waterlogging tolerance in wheat without penalizing grain yield. Plant Cell Environ. 2019, 42, 1471–1485. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Liang, K.; Fang, T.; Zhao, H.; Han, X.; Cai, M.; Qiu, F. A group VII ethylene response factor gene, ZmEREB180, coordinates waterlogging tolerance in maize seedlings. Plant Biotechnol. J. 2019, 17, 2286–2298. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Lee, S.C.; Isa, N.M.; Gramuglia, S.; Fukao, T.; Bassel, G.W.; Correia, C.S.; Corbineau, F.; Theodoulou, F.L.; Bailey-Serres, J.; et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 2011, 479, 415–418. [Google Scholar] [CrossRef]
- Weits, D.A.; Giuntoli, B.; Kosmacz, M.; Parlanti, S.; Hubberten, H.M.; Riegler, H.; Hoefgen, R.; Perata, P.; van Dongen, J.T.; Licausi, F. Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 2014, 5, 3425. [Google Scholar] [CrossRef]
- Schmidt, R.R.; Weits, D.A.; Feulner, C.F.J.; van Dongen, J.T. Oxygen sensing and integrative stress signaling in plants. Plant Physiol. 2018, 176, 1131–1142. [Google Scholar] [CrossRef]
- Schmidt, R.R.; van Dongen, J.T. The ACBP1-RAP2.12 signalling hub: A new perspective on integrative signalling during hypoxia in plants. Plant Signal Behav. 2019, 14, e1651184. [Google Scholar] [CrossRef] [PubMed]
- Hartman, S.; Liu, Z.; van Veen, H.; Vicente, J.; Reinen, E.; Martopawiro, S.; Zhang, H.; van Dongen, N.; Bosman, F.; Bassel, G.W.; et al. Ethylene-mediated nitric oxide depletion pre-adapts plants to hypoxia stress. Nat. Commun. 2019, 10, 4020. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.R.; Fulda, M.; Paul, M.V.; Anders, M.; Plum, F.; Weits, D.A.; Kosmacz, M.; Larson, T.R.; Graham, I.A.; Beemster, G.T.S.; et al. Low-oxygen response is triggered by an ATP-dependent shift in oleoyl-CoA in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E12101–E12110. [Google Scholar] [CrossRef]
- Lumba, S.; Toh, S.; Handfield, L.F.; Swan, M.; Liu, R.; Youn, J.Y.; Cutler, S.R.; Subramaniam, R.; Provart, N.; Moses, A.; et al. A mesoscale abscisic acid hormone interactome reveals a dynamic signaling landscape in Arabidopsis. Dev Cell 2014, 29, 360–372. [Google Scholar] [CrossRef]
- Li, Y.; Liu, K.; Tong, G.; Xi, C.; Liu, J.; Zhao, H.; Wang, Y.; Ren, D.; Han, S. MPK3/MPK6-mediated ERF72 phosphorylation positively regulates resistance to Botrytis cinerea through directly and indirectly activating the transcription of camalexin-biosynthesis enzymes. J. Exp. Bot. 2021, 73, 413–428. [Google Scholar] [CrossRef]
- Singh, P.; Sinha, A.K. A positive feedback loop governed by SUB1A1 interaction with MITOGEN-ACTIVATED PROTEIN KINASE3 impacts submergence tolerance in rice. Plant Cell 2016, 28, 1127–1143. [Google Scholar] [CrossRef]
- Tang, R.J.; Wang, C.; Li, K.; Luan, S. The CBL–CIPK calcium signaling network: Unified paradigm from 20 years of discoveries. Trends Plant Sci. 2020, 25, 604–617. [Google Scholar] [CrossRef] [PubMed]
- Dóczi, R.; Brader, G.; Pettkó-Szandtner, A.; Rajh, I.; Djamei, A.; Pitzschke, A.; Teige, M.; Hirt, H. The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell 2007, 19, 3266–3279. [Google Scholar] [CrossRef] [PubMed]
- Tagliani, A.; Tran, A.N.; Novi, G.; Di Mambro, R.; Pesenti, M.; Sacchi, G.A.; Perata, P.; Pucciariello, C. The calcineurin β-like interacting protein kinase CIPK25 regulates potassium homeostasis under low oxygen in Arabidopsis. J. Exp. Bot. 2020, 71, 2678–2689. [Google Scholar] [CrossRef]
- Lin, C.C.; Chao, Y.T.; Chen, W.C.; Ho, H.Y.; Chou, M.Y.; Li, Y.R.; Wu, Y.L.; Yang, H.A.; Hsieh, H.; Lin, C.S.; et al. Regulatory cascade involving transcriptional and N-end rule pathways in rice under submergence. Proc. Natl. Acad. Sci. USA 2019, 116, 3300–3309. [Google Scholar] [CrossRef]
- Mitsuda, N.; Ohme-Takagi, M. Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 2009, 50, 1232–1248. [Google Scholar] [CrossRef]
- Choy, M.K.; Sullivan, J.A.; Theobald, J.C.; Davies, W.J.; Gray, J.C. An Arabidopsis mutant able to green after extended dark periods shows decreased transcripts of seed protein genes and altered sensitivity to abscisic acid. J. Exp. Bot. 2008, 59, 3869–3884. [Google Scholar] [CrossRef]
- Gasch, P.; Fundinger, M.; Müller, J.T.; Lee, T.; Bailey-Serres, J.; Mustroph, A. Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 2016, 28, 160–180. [Google Scholar] [CrossRef] [PubMed]
- Giuntoli, B.; Lee, S.C.; Licausi, F.; Kosmacz, M.; Oosumi, T.; van Dongen, J.T.; Bailey-Serres, J.; Perata, P. A trihelix DNA binding protein counterbalances hypoxia-responsive transcriptional activation in Arabidopsis. PLoS Biol. 2014, 12, e1001950. [Google Scholar] [CrossRef]
- Giuntoli, B.; Licausi, F.; van Veen, H.; Perata, P. Functional balancing of the hypoxia regulators RAP2.12 and HRA1 takes place in vivo in Arabidopsis thaliana plants. Front Plant Sci. 2017, 8, 591. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; Ivanova, A.; Duncan, O.; Law, S.R.; van Aken, O.; De Clercq, I.; Wang, Y.; Carrie, C.; Xu, L.; Kmiec, B.; et al. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 2013, 25, 3450–3471. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, I.; Vermeirssen, V.; van Aken, O.; Vandepoele, K.; Murcha, M.W.; Law, S.R.; Inzé, A.; Ng, S.; Ivanova, A.; Rombaut, D.; et al. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 2013, 25, 3472–3490. [Google Scholar] [CrossRef]
- Meng, X.; Li, L.; De Clercq, I.; Narsai, R.; Xu, Y.; Hartmann, A.; Claros, D.L.; Custovic, E.; Lewsey, M.G.; Whelan, J.; et al. ANAC017 coordinates organellar functions and stress responses by reprogramming retrograde signaling. Plant Physiol. 2019, 180, 634–653. [Google Scholar] [CrossRef]
- Shapiguzov, A.; Vainonen, J.P.; Hunter, K.; Tossavainen, H.; Tiwari, A.; Järvi, S.; Hellman, M.; Aarabi, F.; Alseekh, S.; Wybouw, B.; et al. Arabidopsis RCD1 coordinates chloroplast and mitochondrial functions through interaction with ANAC transcription factors. eLife 2019, 15, e43284. [Google Scholar] [CrossRef]
- Jaspers, P.; Blomster, T.; Brosché, M.; Salojärvi, J.; Ahlfors, R.; Vainonen, J.P.; Reddy, R.A.; Immink, R.; Angenent, G.; Turck, F.; et al. Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J. 2009, 60, 268–279. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, C.; Chen, J.; Guo, L.; Li, X.; Li, W.; Yu, Z.; Deng, J.; Zhang, P.; Zhang, K.; et al. Arabidopsis heat shock factor HsfA1a directly senses heat stress, pH changes, and hydrogen peroxide via the engagement of redox state. Plant Physiol. Biochem. 2013, 64, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Schippers, J.H.M.; Schmidt, R.R. Redox and low-oxygen stress: Signal integration and interplay. Plant Physiol. 2021, 186, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Shapiguzov, A.; Nikkanen, L.; Fitzpatrick, D.; Vainonen, J.P.; Gossens, R.; Alseekh, S.; Aarabi, F.; Tiwari, A.; Blokhina, O.; Panzarová, K.; et al. Dissecting the interaction of photosynthetic electron transfer with mitochondrial signaling and hypoxic response in the Arabidopsis rcd1 mutant. Phil. Trans. R Soc. B 2020, 375, 20190413. [Google Scholar] [CrossRef]
- van Aken, O.; De Clercq, I.; Ivanova, A.; Law, S.R.; van Breusegem, F.; Millar, A.H.; Whelan, J. Mitochondrial and chloroplast stress responses are modulated in distinct touch and chemical inhibition phases. Plant Physiol. 2016, 171, 2150–2165. [Google Scholar] [CrossRef]
- Kornberg, R.D. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 2005, 30, 235–239. [Google Scholar] [CrossRef]
- Ng, S.; Giraud, E.; Duncan, O.; Law, S.R.; Wang, Y.; Xu, L.; Narsai, R.; Carrie, C.; Walker, H.; Day, D.A.; et al. Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J. Biol. Chem. 2013, 288, 3449–3459. [Google Scholar] [CrossRef] [PubMed]
- Ou, B.; Yin, K.Q.; Liu, S.N.; Yang, Y.; Gu, T.; Wing Hui, J.M.; Zhang, L.; Miao, J.; Kondou, Y.; Matsui, M.; et al. A high-throughput screening system for Arabidopsis transcription factors and its application to MED25-dependent transcriptional regulation. Mol. Plant 2011, 4, 546–555. [Google Scholar] [CrossRef]
- Kosmacz, M.; Parlanti, S.; Schwarzländer, M.; Kragler, F.; Licausi, F.; van Dongen, J.T. The stability and nuclear localization of the transcription factor RAP2.12 are dynamically regulated by oxygen concentration. Plant Cell Environ. 2015, 38, 1094–1103. [Google Scholar] [CrossRef]
- Sorenson, R.; Bailey-Serres, J. Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 2373–2378. [Google Scholar] [CrossRef]
- Lokdarshi, A.; Conner, W.C.; McClintock, C.; Li, T.; Roberts, D.M. Arabidopsis CML38, a calcium sensor that localizes to ribonucleoprotein complexes under hypoxia stress. Plant Physiol. 2016, 170, 1046–1059. [Google Scholar] [CrossRef] [PubMed]
- Field, S.; Gulledge, W.; Roberts, D.M. The Arabidopsis calcium sensor calmodulin-like 38 regulates stress granule autophagy and dynamics during low oxygen stress and re-aeration recovery. bioRxiv 2021. [Google Scholar] [CrossRef]
- Tsuji, H.; Saika, H.; Tsutsumi, N.; Hirai, A.; Nakazono, M. Dynamic and reversible changes in histone H3-Lys4 methylation and H3 acetylation occurring at submergence-inducible genes in rice. Plant Cell Physiol. 2006, 47, 995–1003. [Google Scholar] [CrossRef]
- Lee, T.A.; Bailey-Serres, J. Integrative analysis from the epigenome to translatome uncovers patterns of dominant nuclear regulation during transient stress. Plant Cell 2019, 31, 2573–2595. [Google Scholar] [CrossRef]
- Vicente, J.; Mendiondo, G.M.; Movahedi, M.; Peirats-Llobet, M.; Juan, Y.T.; Shen, Y.Y.; Dambire, C.; Smart, K.; Rodriguez, P.L.; Charng, Y.Y.; et al. The Cys-Arg/N-end rule pathway is a general sensor of abiotic stress in flowering plants. Curr. Biol. 2017, 27, 3183–3190. [Google Scholar] [CrossRef]
- Torres, E.S.; Deal, R.B. The histone variant H2A.Z and chromatin remodeler BRAHMA act coordinately and antagonistically to regulate transcription and nucleosome dynamics in Arabidopsis. Plant J. 2019, 99, 144–162. [Google Scholar] [CrossRef] [PubMed]
- Sura, W.; Kabza, M.; Karlowski, W.M.; Bieluszewski, T.; Kus-Slowinska, M.; Pawełoszek, Ł.; Sadowski, J.; Ziolkowski, P.A. Dual role of the histone variant H2A.Z in transcriptional regulation of stress-response genes. Plant Cell 2017, 29, 791–807. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Tedds, H.M.; Labandera, A.M.; Bailey, M.; White, M.D.; Hartman, S.; Sprigg, C.; Mogg, S.L.; Osborne, R.; Dambire, C.; et al. Oxygen-dependent proteolysis regulates the stability of angiosperm polycomb repressive complex 2 subunit VERNALIZATION 2. Nat. Commun. 2018, 9, 5438. [Google Scholar] [CrossRef]
- Perata, P.; Armstrong, W.; Voesenek, L.A. Plants and flooding stress. New Phytol. 2011, 190, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.J.; Shah, J.K.; Brotman, Y.; Jahnke, K.; Willmitzer, L.; Kaiser, W.M.; Bauwe, H.; Igamberdiev, A.U. Inhibition of aconitase by nitric oxide leads to induction of the alternative oxidase and to a shift of metabolism towards biosynthesis of amino acids. J. Exp. Bot. 2012, 63, 1773–1784. [Google Scholar] [CrossRef] [PubMed]
- Amora, Y.; Chevionb, M.; Levinea, A. Anoxia pretreatment protects soybean cells against H2O2-induced cell death: Possible involvement of peroxidases and of alternative oxidase. FEBS Lett. 2000, 477, 175–180. [Google Scholar] [CrossRef]
- Tsuji, H.; Nakazono, M.; Saisho, D.; Tsutsumi, N.; Hirai, A. Transcript levels of the nuclear-encoded respiratory genes in rice decrease by oxygen deprivation: Evidence for involvement of calcium in expression of the alternative oxidase 1a gene. FEBS Lett. 2000, 471, 201–204. [Google Scholar] [CrossRef]
- Szal, B.; Jolivet, Y.; Hasenfratz-Sauder, M.P.; Dizengremel, P.; Rychter, A.M. Oxygen concentration regulates alternative oxidase expression in barley roots during hypoxia and post-hypoxia. Physiol. Plant 2003, 119, 494–502. [Google Scholar] [CrossRef]
- Millar, A.H.; Trend, A.E.; Heazlewood, J.L. Changes in the mitochondrial proteome during the anoxia to air transition in rice focus around cytochrome-containing respiratory complexes. J. Biol. Chem. 2004, 279, 39471–39478. [Google Scholar] [CrossRef]
- Vishwakarma, A.; Kumari, A.; Mur, L.A.J.; Gupta, K.J. A discrete role for alternative oxidase under hypoxia to increase nitric oxide and drive energy production. Free Radic. Biol. Med. 2018, 122, 40–51. [Google Scholar] [CrossRef]
- Panozzo, A.; Dal Cortivo, C.; Ferrari, M.; Vicelli, B.; Varotto, S.; Vamerali, T. Morphological changes and expressions of AOX1A, CYP81D8, and putative PFP genes in a large set of commercial maize hybrids under extreme waterlogging. Front Plant Sci. 2019, 10, 62. [Google Scholar] [CrossRef]
- Ito, Y.; Saisho, D.; Nakazono, M.; Tsutsumi, N.; Hirai, A. Transcript levels of tandem-arranged alternative oxidase genes in rice are increased by low temperature. Gene 1997, 203, 121–129. [Google Scholar] [CrossRef]
- Djajanegara, I.; Holtzapffel, R.; Finnegan, P.M.; Hoefnagel, M.H.; Berthold, D.A.; Wiskich, J.T.; Day, D.A. A single amino acid change in the plant alternative oxidase alters the specificity of organic acid activation. FEBS Lett. 1999, 454, 220–224. [Google Scholar] [CrossRef]
- Karpova, O.V.; Kuzmin, E.V.; Elthon, T.E.; Newton, K.J. Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 2002, 14, 3271–3284. [Google Scholar] [CrossRef]
- Holtzapffel, R.C.; Castelli, J.; Finnegan, P.M.; Millar, A.H.; Whelan, J.; Day, D.A. A tomato alternative oxidase protein with altered regulatory properties. Biochim. Biophys. Acta 2003, 1606, 153–162. [Google Scholar] [CrossRef]
- Grant, N.; Onda, Y.; Kakizaki, Y.; Ito, K.; Watling, J.; Robinson, S. Two cys or not two cys? That is the question; Alternative oxidase in the thermogenic plant sacred Lotus. Plant Physiol. 2009, 150, 987–995. [Google Scholar] [CrossRef]
- Selinski, J.; Hartmann, A.; Kordes, A.; Deckers-Hebestreit, G.; Whelan, J.; Scheibe, R. Analysis of posttranslational activation of alternative oxidase isoforms. Plant Physiol. 2017, 174, 2113–2127. [Google Scholar] [CrossRef] [PubMed]
- Selinski, J.; Scheibe, R.; Day, D.A.; Whelan, J. Alternative oxidase is positive for plant performance. Trends Plant Sci. 2018, 23, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.; O’Neill, L.A. Succinate: A metabolic signal in inflammation. Trends Cell Biol. 2014, 24, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Selinski, J.; Scheibe, R. Central metabolism in mammals and plants as a hub for controlling cell fate. Antioxid. Redox Signal. 2021, 34, 1025–1047. [Google Scholar] [CrossRef]
- Rasmusson, A.G.; Møller, I.M. Mitochondrial electron transport and plant stress. In Plant Mitochondria; Kempken, F., Ed.; Springer: New York, NY, USA, 2011; pp. 357–381. [Google Scholar] [CrossRef]
- Yeung, E.; van Veen, H.; Vashisht, D.; Sobral Paiva, A.L.; Hummel, M.; Rankenberg, T.; Steffens, B.; Steffen-Heins, A.; Sauter, M.; de Vries, M.; et al. A stress recovery signaling network for enhanced flooding tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2018, 115, E6085–E6094. [Google Scholar] [CrossRef]
- Rasmusson, A.G.; Geisler, D.A.; Møller, I.M. The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion 2008, 8, 47–60. [Google Scholar] [CrossRef]
- Blokhina, O.; Virolainen, E.; Fagerstedt, K.V. Antioxidants, oxidative damage and oxygen deprivation stress: A review. Ann. Bot. 2003, 91, 179–194. [Google Scholar] [CrossRef]
- Pavelic, D.; Arpagaus, S.; Rawyler, A.; Brändle, R. Impact of post-anoxia stress on membrane lipids of anoxia-pretreated potato cells. A re-appraisal. Plant Physiol. 2000, 124, 1285–1292. [Google Scholar] [CrossRef]
- Garnczarska, M.; Bednarski, W.; Morkunas, I. Re-aeration–induced oxidative stress and antioxidative defenses in hypoxically pretreated lupine roots. J Plant Physiol. 2004, 161, 415–422. [Google Scholar] [CrossRef]
- Huang, S.; van Aken, O.; Schwarzländer, M.; Belt, K.; Millar, A.H. The roles of mitochondrial reactive oxygen species in cellular signaling and stress response in plants. Plant Physiol. 2016, 171, 1551–1559. [Google Scholar] [CrossRef] [PubMed]
- Blokhina, O.B.; Virolainen, E.; Fagerstedt, K.V.; Hoikkala, A.; Wähälä, K.; Chirkova, T.V. Antioxidant status of anoxia-tolerant and-intolerant plant species under anoxia and reaeration. Physiol. Plant 2001, 109, 396–403. [Google Scholar] [CrossRef]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef]
- Dietz, K.J.; Turkan, I.; Krieger-Liszkay, A. Redox-and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiol. 2016, 171, 1541–1550. [Google Scholar] [CrossRef]
- Taylor, N.L.; Tan, Y.F.; Jacoby, R.P.; Millar, A.H. Abiotic environmental stress induced changes in the Arabidopsis thaliana chloroplast, mitochondria and peroxisome proteomes. J. Proteom. 2009, 72, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.L.; Thiele, B.; Janzik, I.; Zeng, B.; Schurr, U.; Matsubara, S. De-submergence responses of antioxidative defense systems in two wetland plants having escape and quiescence strategies. J. Plant Physiol. 2012, 169, 1680–1689. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.B.; Dai, Y.S.; Xie, L.J.; Yu, L.J.; Zhou, Y.; Lai, Y.X.; Yang, Y.C.; Xu, L.; Chen, Q.F.; Xiao, S. Jasmonate regulates plant responses to postsubmergence reoxygenation through transcriptional activation of antioxidant synthesis. Plant Physiol. 2017, 173, 1864–1880. [Google Scholar] [CrossRef]
- Alpuerto, J.B.; Hussain, R.M.F.; Fukao, T. The key regulator of submergence tolerance, SUB1A, promotes photosynthetic and metabolic recovery from submergence damage in rice leaves. Plant Cell Environ. 2016, 39, 672–684. [Google Scholar] [CrossRef]
- Tamang, B.G.; Magliozzi, J.O.; Maroof, M.A.S.; Fukao, T. Physiological and transcriptomic characterization of submergence and reoxygenation responses in soybean seedlings. Plant Cell Environ. 2014, 37, 2350–2365. [Google Scholar] [CrossRef] [PubMed]
- Blokhina, O.; Fagerstedt, K.V. Oxidative metabolism, ROS and NO under oxygen deprivation. Plant Physiol. Biochem. 2010, 48, 359–373. [Google Scholar] [CrossRef] [PubMed]
- Shapiguzov, A.; Vainonen, J.P.; Wrzaczek, M.; Kangasjärvi, J. ROS-talk—How the apoplast, the chloroplast, and the nucleus get the message through. Front. Plant Sci. 2012, 3, 292. [Google Scholar] [CrossRef] [PubMed]
- Pospíšil, P. Production of reactive oxygen species by photosystem II. Biochim. Biophys. Acta 2009, 1787, 1151–1160. [Google Scholar] [CrossRef]
- Sone, C.; Sakagami, J.I. Physiological mechanism of chlorophyll breakdown for leaves under complete submergence in rice. Crop Sci. 2017, 57, 2729–2738. [Google Scholar] [CrossRef]
- Luo, F.L.; Matsubara, S.; Chen, Y.; Wei, G.W.; Dong, B.C.; Zhang, M.X.; Yu, F.H. Consecutive submergence and de-submergence both impede growth of a riparian plant during water level fluctuations with different frequencies. Environ. Exp. Bot. 2018, 155, 641–649. [Google Scholar] [CrossRef]
- Pompeiano, A.; Huarancca Reyes, T.; Moles, T.M.; Villani, M.; Volterrani, M.; Guglielminetti, L.; Scartazza, A. Inter- and intraspecific variability in physiological traits and post-anoxia recovery of photosynthetic efficiency in grasses under oxygen deprivation. Physiol. Plant 2017, 161, 385–399. [Google Scholar] [CrossRef]
- Kato, Y.; Collard, B.C.Y.; Septiningsih, E.M.; Ismail, A.M. Physiological analyses of traits associated with tolerance of long-term partial submergence in rice. AoB Plants 2014, 6, plu058. [Google Scholar] [CrossRef]
- Vadez, V. Root hydraulics: The forgotten side of roots in drought adaptation. F Crop Res. 2014, 165, 15–24. [Google Scholar] [CrossRef]
- Shahzad, Z.; Canut, M.; Tournaire-Roux, C.; Martinière, A.; Boursiac, Y.; Loudet, O.; Maurel, C. A potassium-dependent oxygen sensing pathway regulates plant root hydraulics. Cell 2016, 167, 87–98.e14. [Google Scholar] [CrossRef]
- Striker, G.G.; Casas, C.; Kuang, X.; Grimoldi, A.A. No escape? Costs and benefits of leaf de-submergence in the pasture grass Chloris gayana under different flooding regimes. Funct. Plant Biol. 2017, 44, 899–906. [Google Scholar] [CrossRef]
- Chirkova, T.V.; Novitskaya, L.; Blokhina, O. Lipid peroxidation and antioxidant systems under anoxia in plants differing in their tolerance to oxygen deficiency. Russ. J. Plant Physiol. 1998, 45, 55–62. [Google Scholar]
- Blokhina, O.B.; Fagerstedt, K.V.; Chirkova, T.V. Relationships between lipid peroxidation and anoxia tolerance in a range of species during post-anoxic reaeration. Physiol. Plant 1999, 105, 625–632. [Google Scholar] [CrossRef]
- Sieber, M.; Brändle, R. Energy metabolism in rhizomes of Acorus calamus (L.) and in tubers of Solanum tuberosum (L.) with regard to their anoxia tolerance. Bot. Acta 1991, 104, 279–282. [Google Scholar] [CrossRef]
- Shingaki-Wells, R.; Millar, A.H.; Whelan, J.; Narsai, R. What happens to plant mitochondria under low oxygen? An omics review of the responses to low oxygen and reoxygenation. Plant Cell Environ. 2014, 37, 2260–2277. [Google Scholar] [CrossRef]
- Gautam, P.; Lal, B.; Tripathi, R.; Shahid, M.; Baig, M.J.; Raja, R.; Maharana, S.; Nayak, A.K. Role of silica and nitrogen interaction in submergence tolerance of rice. Environ. Exp. Bot. 2016, 125, 98–109. [Google Scholar] [CrossRef]
- Wang, M.; Shen, Y.; Tao, F.; Yang, S.; Li, W. Submergence induced changes of molecular species in membrane lipids in Arabidopsis thaliana. Plant Divers. 2016, 38, 156–162. [Google Scholar] [CrossRef]
- Ye, X.Q.; Meng, J.L.; Zeng, B.; Wu, M.; Zhang, Y.Y.; Zhang, X.P. Submergence causes similar carbohydrate starvation but faster post-stress recovery than darkness in Alternanthera philoxeroides plants. PLoS ONE 2016, 11, e0165193. [Google Scholar] [CrossRef]
- He, X.J.; Mu, R.L.; Cao, W.H.; Zhang, Z.G.; Zhang, J.S.; Chen, S.Y. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005, 44, 903–916. [Google Scholar] [CrossRef]
- Balazadeh, S.; Wu, A.; Mueller-Roeber, B. Salt-triggered expression of the ANAC092-dependent senescence regulon in Arabidopsis thaliana. Plant Signal. Behav. 2010, 5, 733–735. [Google Scholar] [CrossRef]
- Qiu, K.; Li, Z.; Yang, Z.; Chen, J.; Wu, S.; Zhu, X.; Gao, S.; Gao, J.; Ren, G.; Kuai, B.; et al. EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet. 2015, 11, e1005399. [Google Scholar] [CrossRef]
- Striker, G.G.; Kotula, L.; Colmer, T.D. Tolerance to partial and complete submergence in the forage legume Melilotus siculus: An evaluation of 15 accessions for petiole hyponastic response and gas-filled spaces, leaf hydrophobicity and gas films, and root phellem. Ann. Bot. 2019, 123, 169–180. [Google Scholar] [CrossRef]
- Tsai, K.J.; Lin, C.Y.; Ting, C.Y.; Shih, M.C. Ethylene-regulated glutamate dehydrogenase fine-tunes metabolism during anoxia-reoxygenation. Plant Physiol. 2016, 172, 1548–1562. [Google Scholar] [CrossRef] [PubMed]
- Hincha, D.K. Effects of α-tocopherol (vitamin E) on the stability and lipid dynamics of model membranes mimicking the lipid composition of plant chloroplast membranes. FEBS Lett. 2008, 582, 3687–3692. [Google Scholar] [CrossRef] [PubMed]
- Roche, Y.; Gerbeau-Pissot, P.; Buhot, B.; Thomas, D.; Bonneau, L.; Gresti, J.; Mongrand, S.; Perrier-Cornet, J.; Simon-Plas, F. Depletion of phytosterols from the plant plasma membrane provides evidence for disruption of lipid rafts. FASEB J. 2008, 22, 3980–3991. [Google Scholar] [CrossRef] [PubMed]
- Mlayeh, L.; Chatkaew, S.; Léonetti, M.; Homblé, F. Modulation of plant mitochondrial VDAC by phytosterols. Biophys J. 2010, 99, 2097–2106. [Google Scholar] [CrossRef] [PubMed]
- Dufourc, E.J. The role of phytosterols in plant adaptation to temperature. Plant Signal Behav. 2008, 3, 133–134. [Google Scholar] [CrossRef]
- Tsai, K.J.; Chou, S.J.; Shih, M.C. Ethylene plays an essential role in the recovery of Arabidopsis during post-anaerobiosis reoxygenation. Plant Cell Environ. 2014, 37, 2391–2405. [Google Scholar] [CrossRef]
- Tsuji, H.; Meguro, N.; Suzuki, Y.; Tsutsumi, N.; Hirai, A.; Nakazono, M. Induction of mitochondrial aldehyde dehydrogenase by submergence facilitates oxidation of acetaldehyde during re-aeration in rice. FEBS Lett. 2003, 546, 369–373. [Google Scholar] [CrossRef]
- Narsai, R.; Howell, K.A.; Carroll, A.; Ivanova, A.; Millar, A.H.; Whelan, J. Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol. 2009, 151, 306–322. [Google Scholar] [CrossRef]
- Kennedy, R.A.; Barrett, S.C.H.; Zee, D.V.; Rumpho, M.E. Germination and seedling growth under anaerobic conditions in Echinochloa crus-galli (barnyard grass). Plant Cell Environ. 1980, 3, 243–248. [Google Scholar] [CrossRef]
- Gupta, K.J.; Igamberdiev, A.U.; Manjunatha, G.; Segu, S.; Moran, J.F.; Neelawarne, B.; Bauwe, H.; Kaiser, W.M. The emerging roles of nitric oxide (NO) in plant mitochondria. Plant Sci. 2011, 181, 520–526. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Ratcliffe, R.G.; Gupta, K.J. Plant mitochondria: Source and target for nitric oxide. Mitochondrion 2014, 19, 329–333. [Google Scholar] [CrossRef]
- Kumari, A.; Pathak, P.K.; Bulle, M.; Igamberdiev, A.U.; Gupta, K.J. Alternative oxidase is an important player in the regulation of nitric oxide levels under normoxic and hypoxic conditions in plants. J. Exp. Bot. 2019, 70, 4345–4354. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.H.; Whelan, J.; Soole, K.L.; Day, D.A. Organization and regulation of mitochondrial respiration in plants. Annu. Rev. Plant Biol. 2011, 62, 79–104. [Google Scholar] [CrossRef] [PubMed]
- Cvetkovska, M.; Vanlerberghe, G.C. Alternative oxidase impacts the plant response to biotic stress by influencing the mitochondrial generation of reactive oxygen species. Plant Cell Environ. 2013, 36, 721–732. [Google Scholar] [CrossRef]
- Cvetkovska, M.; Dahal, K.; Alber, N.A.; Jin, C.; Cheung, M.; Vanlerberghe, G.C. Knockdown of mitochondrial alternative oxidase induces the ‘stress state’ of signaling molecule pools in Nicotiana tabacum, with implications for stomatal function. New Phytol. 2014, 203, 449–461. [Google Scholar] [CrossRef]
- Maxwell, D.P.; Wang, Y.; McIntosh, L. The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. USA 1999, 96, 8271–8276. [Google Scholar] [CrossRef] [PubMed]
- Rasmusson, A.G.; Escobar, M.A. Light and diurnal regulation of plant respiratory gene expression. Physiol. Plant 2006, 129, 57–67. [Google Scholar] [CrossRef]
- Vanlerberghe, G.C. Alternative oxidase: A mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int. J. Mol. Sci. 2013, 14, 6805–6847. [Google Scholar] [CrossRef]
- Khan, M.N.; Sakata, K.; Hiraga, S.; Komatsu, S. Quantitative proteomics reveals that peroxidases play key roles in post-flooding recovery in soybean roots. J. Proteome Res. 2014, 13, 5812–5828. [Google Scholar] [CrossRef]
- Khan, M.N.; Komatsu, S. Characterization of post-flooding recovery-responsive enzymes in soybean root and hypocotyl. J. Plant Biol. 2016, 59, 478–487. [Google Scholar] [CrossRef]
- Luo, F.L.; Nagel, K.A.; Scharr, H.; Zeng, B.; Schurr, U.; Matsubara, S. Recovery dynamics of growth, photosynthesis and carbohydrate accumulation after de-submergence: A comparison between two wetland plants showing escape and quiescence strategies. Ann. Bot. 2010, 107, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Crawford, R.M.M.; Wollenweber-Ratzer, B. Influence of L-ascorbic acid on post-anoxic growth and survival of chickpea seedlings (Cicer arietinum L.). J. Exp. Bot. 1992, 43, 703–708. [Google Scholar] [CrossRef]
- Monk, L.S.; Fagerstedt, K.V.; Crawford, R.M.M. Superoxide dismutase as an anaerobic polypeptide: A key factor in recovery from oxygen deprivation in Iris pseudacorus? Plant Physiol. 1987, 85, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Das, K.K.; Sarkar, R.K.; Ismail, A.M. Elongation ability and non-structural carbohydrate levels in relation to submergence tolerance in rice. Plant Sci. 2005, 168, 131–136. [Google Scholar] [CrossRef]
- Barding, G.A.J.; Béni, S.; Fukao, T.; Bailey-Serres, J.; Larive, C.K. Comparison of GC-MS and NMR for metabolite profiling of rice subjected to submergence stress. J. Proteome Res. 2013, 12, 898–909. [Google Scholar] [CrossRef]
- Locke, A.M.; Barding, G.A.J.; Sathnur, S.; Larive, C.K.; Bailey-Serres, J. Rice SUB1A constrains remodelling of the transcriptome and metabolome during submergence to facilitate post-submergence recovery. Plant Cell Environ. 2018, 41, 721–736. [Google Scholar] [CrossRef]
- Song, S.; Huang, H.; Gao, H.; Wang, J.; Wu, D.; Liu, X.; Yang, S.; Zhai, Q.; Li, C.; Qi, T.; et al. Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 2014, 26, 263–279. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).