High-Throughput Phenotyping Accelerates the Dissection of the Phenotypic Variation and Genetic Architecture of Shank Vascular Bundles in Maize (Zea mays L.)
Abstract
:1. Introduction
2. Methods
2.1. Material and Sample Collection
2.2. CT Image Acquisition and Phenotypic Analysis of Maize Shank
2.3. Phenotypic Data Analysis
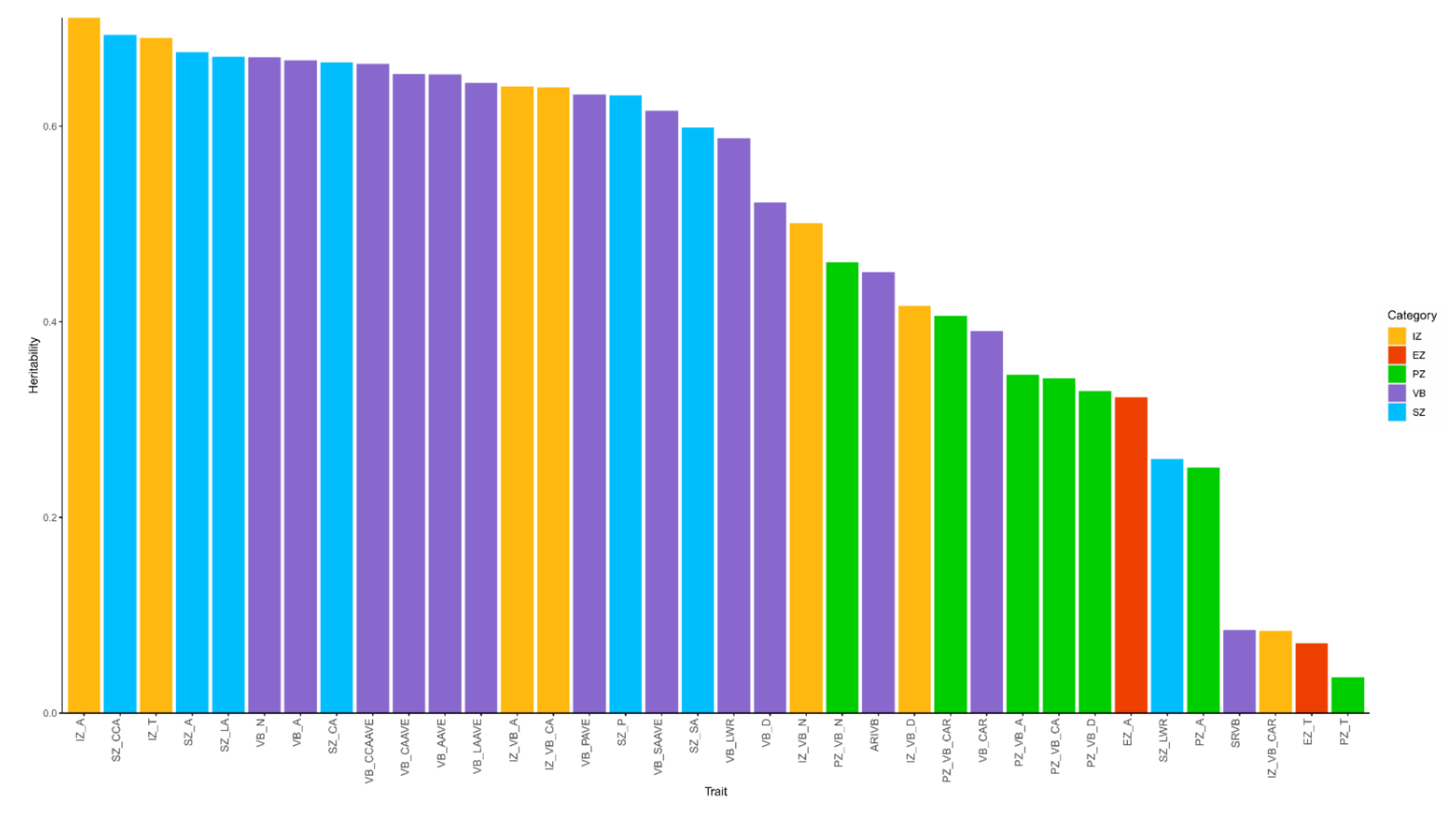
2.4. Heritability Analysis
2.5. Genome-Wide Association Study
3. Results
3.1. Phenotypic Analysis of Shank and Vascular Bundle
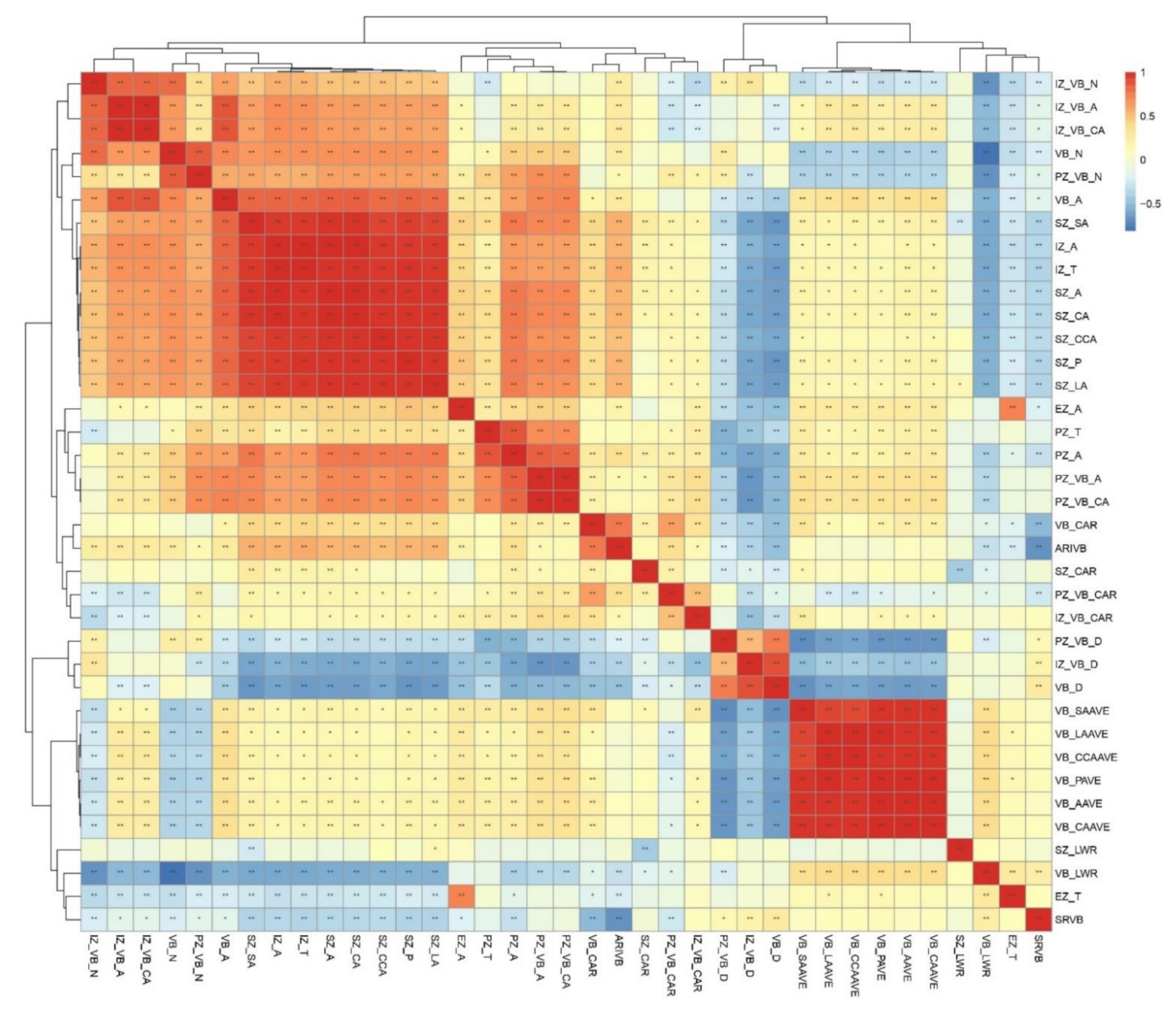
3.2. Phenotypic Variations of Shank Vascular Bundles among a Group of 202 Inbred Lines
3.3. Heritability Analysis
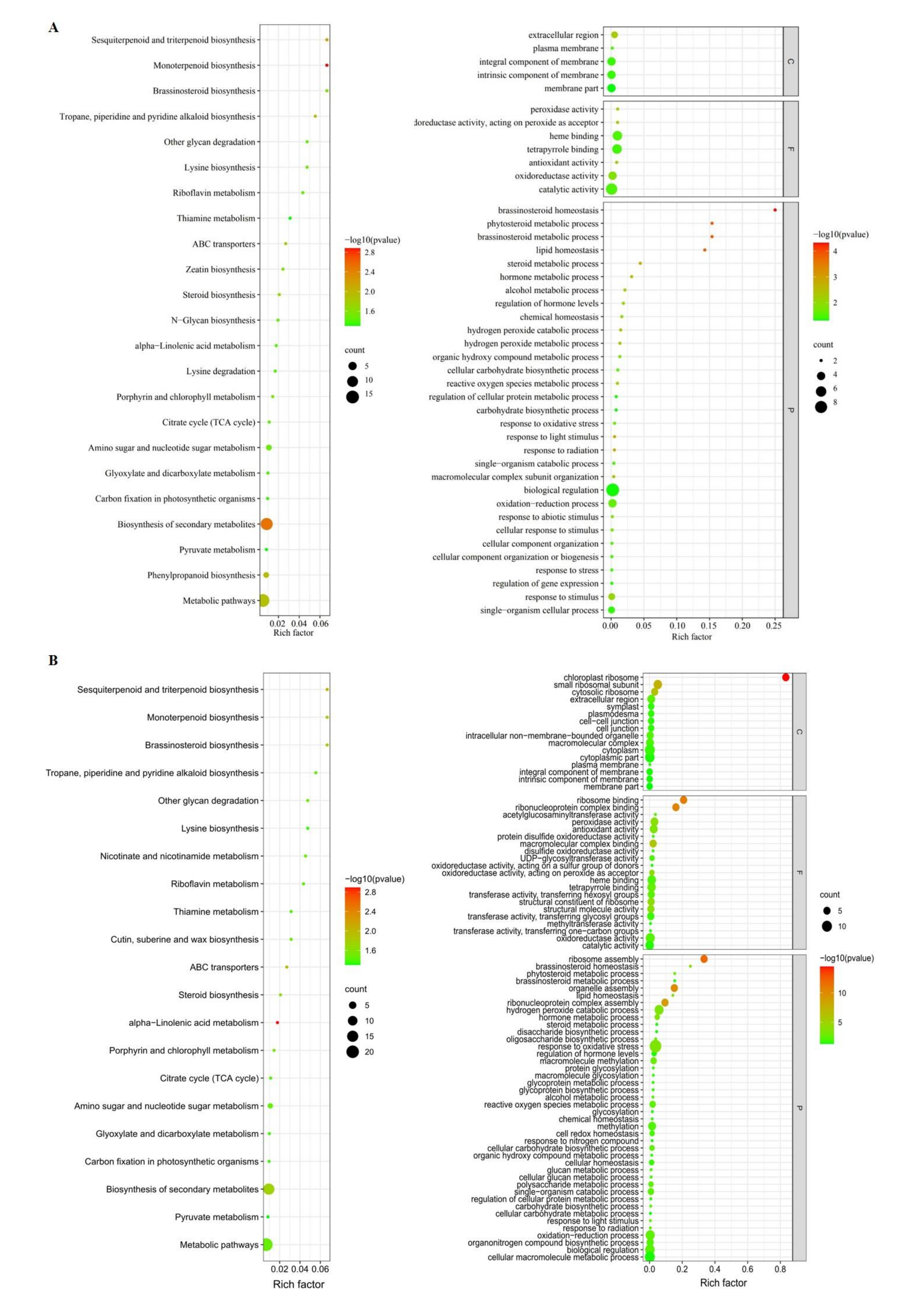
3.4. Genome-Wide Association Analysis of Shank Vascular Bundles
3.4.1. SNP Loci Localization and Candidate Gene Identifying
3.4.2. Pathways Enriched by Functional Enrichment Analysis
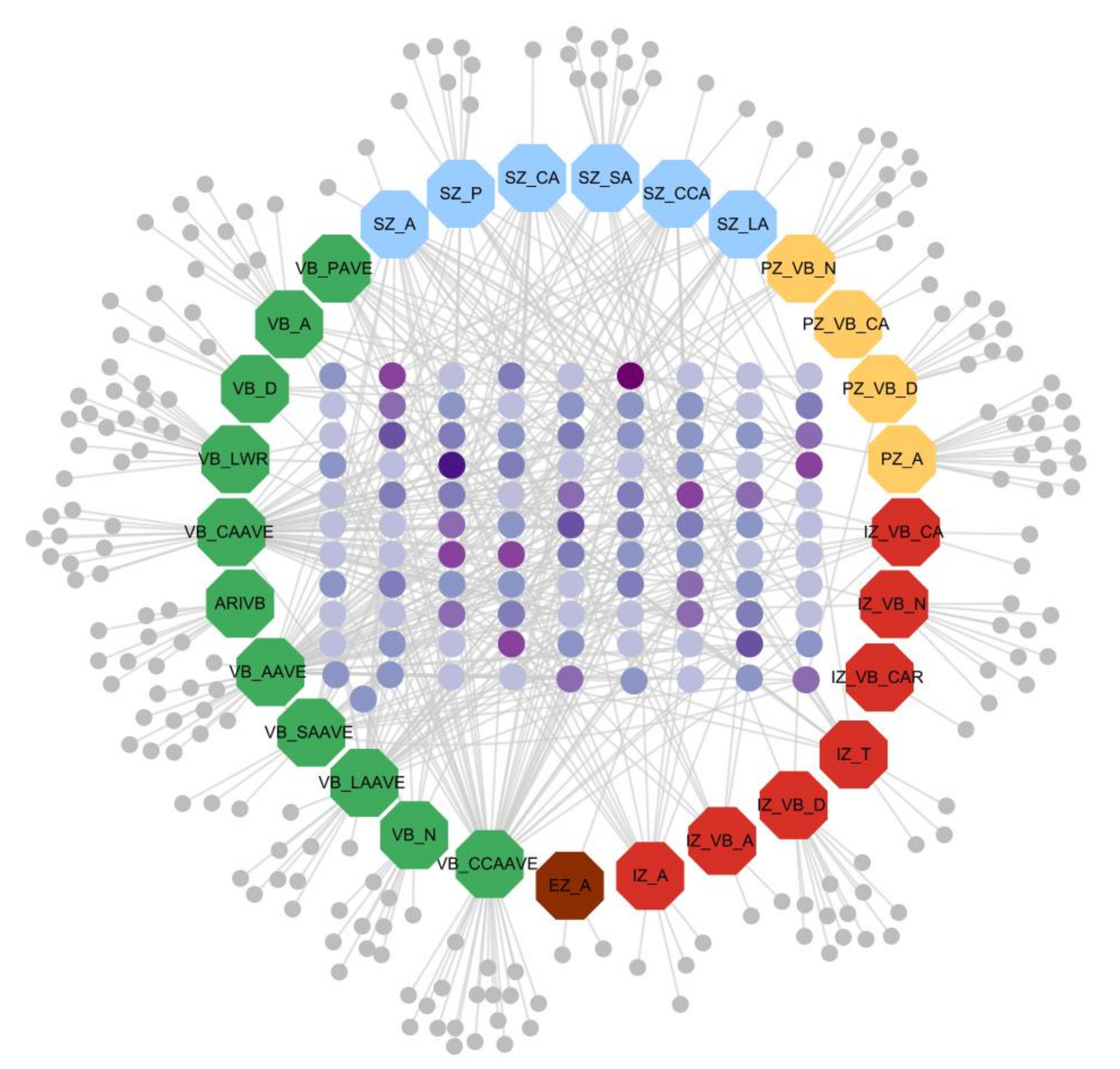
3.4.3. Trait-Gene Network Visualization
4. Discussion
4.1. Advances in Detecting Phenotypic Traits of Shank Vascular Bundle
4.2. Phenotypic Variation of Shank and Vascular Bundles
4.3. Candidate Genes Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, H.; Wang, X.; Warburton, M.L.; Wen, W.; Yan, J. Genomic, transcriptomic, and phenomic variation reveals the complex adaptation of modern maize breeding. Mol. Plant 2015, 8, 871–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.Y. Morphology and Anatomy of Gramineae Crops; Shanghai Science and Technology Press: Shanghai, China, 1979. [Google Scholar]
- Wang, L.M.; Guo, Q.F.; Wang, Q.C. Maize Cultivation in China; Shanghai Science and Technology Press: Shanghai, China, 1986. [Google Scholar]
- Housley, T.L.; Peterson, D.M. Oat stem vascular size in relation to kernel number and weight. I. Controlled environment. Crop Sci. 1982, 22, 2. [Google Scholar] [CrossRef]
- Peterson, D.M.; Housley, T.L.; Luk, T.M. Oat stem vascular size in relation to kernel number and weight. II. Field environment1. Crop Sci. 1982, 22, 274–278. [Google Scholar] [CrossRef]
- Nátrová, Z. Anatomical characteristics of the uppermost internode of winter wheat genoypes differing in stem length. Biol. Plant. 1991, 33, 491. [Google Scholar] [CrossRef]
- Cui, K.; Peng, S.; Xing, Y.; Yu, S.; Xu, C.; Zhang, Q. Molecular dissection of the genetic relationships of source, sink and transport tissue with yield traits in rice. Theor. Appl. Genet. 2003, 106, 649. [Google Scholar] [CrossRef]
- He, Q.P.; Dong, S.T.; Gao, R.Q. Relationship between development of spike vascular bundle and sink capacity of ear and kernel in maize (Zea mays L.). Acta Agron. Sin. 2005, 8, 995–1000. [Google Scholar]
- Suns, G.Y. Genome-Wide Association Analysis of Handle Vascular and Plant-Type Traits in Maize. Master Thesis, Henan Agricultural University, Zhengzhou, China, 2017. [Google Scholar]
- Liu, M.; He, W.; Zhang, A.; Zhang, L.; Ruan, Y. Genetic analysis of maize shank length by QTL mapping in three recombinant inbred line populations. Plant Sci. 2021, 303, 110767. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, J.L.; Du, J.J.; Zhao, Y.X.; Lu, X.J.; Wen, W.; Gu, S.H.; Fan, J.; Wang, C.; Wu, S.; et al. Dissecting the phenotypic components and genetic architecture of maize stem vascular bundles using high-throughput phenotypic analysis. Plant Biotechnol. J. 2021, 19, 35–50. [Google Scholar] [CrossRef]
- Zhang, Y.; Legay, S.; Barrière, Y.; Méchin, V.; Legland, D. Color quantification of stained maize stem section describes lignin spatial distribution within the whole stem. Sci. Food Agric. 2013, 61, 3186–3192. [Google Scholar] [CrossRef]
- Legland, D.; Devaux, M.F.; Guillon, F. Statistical mapping of maize bundle intensity at the stem scale using spatial normalisation of replicated images. PLoS ONE 2014, 9, e90673. [Google Scholar] [CrossRef]
- Heckwolf, S.; Heckwolf, M.; Kaeppler, S.M.; De Leon, N.; Spalding, E.P. Image analysis of anatomical traits in stem transections of maize and other grasses. Plant Methods 2015, 11, 26. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Zhang, Y.; Guo, X.; Ma, L.; Shao, M.; Pan, X.; Zhao, C. Micronscale phenotyping quantification and three-dimensional microstructure reconstruction of vascular bundles within maize stems based on micro-CT scanning. Funct. Plant Biol. 2016, 44, 10–22. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, H.; Wu, L.; Warburton, M.; Yan, J. Genome-wide association studies in maize: Praise and stargaze. Mol. Plant 2017, 10, 374. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Zhang, F.; Zhao, C.; Lu, G.; Chen, F. Genome-wide association study of six quality traits reveals the association of the tarpp13l1 gene with flour colour in Chinese bread wheat. Plant Biotechnol. J. 2019, 17, 2106–2122. [Google Scholar] [CrossRef] [Green Version]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The b73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss YBuckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Riedelsheimer, C.; Lisec, J.; Czedik-Eysenberg, A.; Sulpice, R.; Flis, A.; Grieder, C.; Altmann, T.; Stitt, M.; Willmitzer, L.; Melchinger, A.E. Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc. Natl. Acad. Sci. USA 2012, 109, 8872–8877. [Google Scholar] [CrossRef] [Green Version]
- Peiffer, J.A.; Romay, M.C.; Gore, M.A.; Flint-Garcia, S.A.; Zhang, Z.; Millard, M.J.; Gardner, C.A.C.; McMullen, M.D.; Holland, J.B.; Bradbury, P.J.; et al. The genetic architecture of maize height. Genetics 2014, 196, 1337–1356. [Google Scholar] [CrossRef] [Green Version]
- Dell’Acqua, M.; Gatti, D.M.; Pea, G.; Cattonaro, F.; Coppens, F.; Magris, G.; Hlaing, A.L.; Aung, H.H.; Nelissen, H.; Baute, J.; et al. Genetic properties of the MAGIC maize population: A new platform for high defifinition QTL mapping in Zea mays. Genome Biol. 2015, 16, 167. [Google Scholar] [CrossRef] [Green Version]
- Hung, H.Y.; Shannon, L.M.; Tian, F.; Bradbury, P.J.; Chen, C.; Flint-Garcia, S.A.; McMullen, M.D.; Ware, D.; Buckler, E.S.; Doebley, J.F.; et al. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 2012, 109, E1913–E1921. [Google Scholar] [CrossRef] [Green Version]
- Yang, N.; Lu, Y.; Yang, X.; Huang, J.; Zhou, Y.; Ali, F.; Wen, W.; Liu, J.; Li, J.; Yan, J. Genome wide association studies using a new nonparametric model reveal the genetic architecture of 17 agronomic traits in an enlarged maize association panel. PLoS Genet. 2014, 10, e1004573. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Guo, Z.; Huang, C.; Duan, L.; Chen, G.; Jiang, N.; Fang, W.; Feng, H.; Xie, W.; Lian, X.; et al. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat. Commun. 2014, 5, 5087. [Google Scholar] [CrossRef]
- Huang, C.; Chen, Q.; Xu, G.; Xu, D.; Tian, J.; Tian, F. Identification and fine mapping of quantitative trait loci for the number of vascular bundle in maize stem. Integr. Plant Biol. 2016, 58, 81–90. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Gao, S.; Xu, S.; Zhang, Z.; Prasanna, B.M.; Li, L.; Li, J.; Yan, J. Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol. Breed 2011, 28, 511–526. [Google Scholar] [CrossRef]
- David, B. Asreml—Fit the Linear Mixed Model, R Package Version 3.0; VSN International Ltd.: Hemel Hempstead, UK, 2009; Available online: http://www.vsni.co.uk (accessed on 29 March 2022).
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1633–1664. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.W.; Li, P.; Ren, W.L.; Ni, Y.L.; Zhang, Y.M. MrMLM: Multi-Locus Random-SNP-Effect Mixed Linear Model Tools for Genome-Wide Association Study, R Package Version 4.0; Huazhong Agricultural University: Wuhan, China, 2019; Available online: https://CRAN.R-project.org/package=mrMLM.GUI (accessed on 28 December 2021).
- He, Q.P.; Dong, S.T.; Gao, R.Q. Comparison of ear vascular bundles in different maize cultivars. Crops 2007, 7, 1187–1196. [Google Scholar]
- Cochard, H.; Delzon, S.; Badel, E. X-ray microtomography (micro-CT): A reference technology for high-resolution quantification of xylem embolism in trees. Plant Cell Environ. 2015, 38, 201–206. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, Y.; Du, J.; Guo, X.; Wen, W.; Gu, S.; Wang, J.; Fan, J. Crop phenomics: Current status and perspectives. Front. Plant Sci. 2019, 10, 714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, M.; Zhang, Y.; Guo, X.Y. Research progress on phenotype of transport tissue of ear and kernel in maize. J. Agric. Sci. Technol. 2016, 18, 38–45. [Google Scholar]
- Campbell, M.T.; Knecht, A.C.; Berger, B.; Brien, C.J.; Wang, D.; Walia, H. Integrating image-based phenomics and association analysis to dissect the genetic architecture of temporal salinity responses in rice. Plant Physiol. 2015, 168, 1476–1489. [Google Scholar] [CrossRef] [Green Version]
- Campbell, M.T.; Du, Q.; Liub, K.; Brien, C.J.; Berger, B.; Zhang, C.; Walia, H. A comprehensive image-based phenomic analysis reveals the complex genetic architecture of shoot growth dynamics in rice (Oryza sativa). Plant Genome 2017, 10, 1–14. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, C.; Li, L.; Meng, L.; Singh, J.; Jiang, N.; Deng, X.; He, Z.-H.; Lemaux, P.G. Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 2005, 139, 1107–1124. [Google Scholar] [CrossRef] [Green Version]
- Guillet-Claude, C.; Birolleau-Touchard, C.; Manicacci, D.; Rogowsky, P.M.; Rigau, J.; Murigneux, A.; Martinant, J.; Barrière, Y. Nucleotide diversity of the zmpox3 maize peroxidase gene: Relationships between a mite insertion in exon 2 and variation in forage maize digestibility. BMC Genet. 2004, 51, 19. [Google Scholar]
- Jia, X.L.; Wang, G.L.; Xong, F.; Yu, X.R.; Xu, Z.S.; Wang, F.; Xiong, A.S. De novo assembly, transcriptome characterization, lignin accumulation and anatomic characteristics novel insights into lignin biosynthesis during celery leaf development. Sci. Rep. 2015, 5, 8259. [Google Scholar] [CrossRef] [Green Version]
- De Obeso, M.; Caparros-Ruiz, D.; Vignols, F.; Puigdomenech, P.; Rigau, J. Characterisation of maize peroxidases having differential patterns of mRNA accumulation in relation to lignifying tissues. Gene 2003, 309, 23–33. [Google Scholar] [CrossRef]
- Wagner, T.A.; Kohorn, B.D. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 2001, 13, 303–318. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, R.K.; Aguirre, J.A.; Singh, J. Genome-wide analysis of wall associated kinase (WAK) gene family in barley. Genomics 2021, 113, 523–530. [Google Scholar] [CrossRef]
- Holland, N.; Holland, D.; Helentjaris, T.; Dhugga, K.S.; Delmer, D.P. A comparative analysis of the plant cellulose synthase (cesa) gene family. Plant Physiol. 2000, 123, 1313–1324. [Google Scholar] [CrossRef] [Green Version]
- Arooli, T.; Peng, L.; Betzner, A.S.; Burn, J.; Wittke, W.; Herth, W.; Camilleri, C.; Höfte, H.; Plazinski, J.; Birch, R.; et al. Molecular analysis of Cellulose biosynthesis in Arabidopsis. Science 1998, 79, 717–720. [Google Scholar] [CrossRef] [Green Version]
- Delmer, D.P. Cellulose biosynthesis: Exciting times for a difficult field of study. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 245–276. [Google Scholar] [CrossRef]
- Liebsch, D.; Sunaryo, W.; Holmlund, M.; Norberg, M.; Zhang, J.; Hall, H.C.; Helizon, H.; Jin, X.; Helariutta, Y.; Nilsson, O.; et al. Class I KNOX transcription factors promote differentiation of cambial derivatives into xylem fibers in the Arabidopsis hypocotyl. Development 2014, 141, 4311–4319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, Y.; Sentoku, N.; Miura, Y.; Hirochika, H.; Kitano, H.; Matsuoka, M. Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J. 1999, 18, 992–1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, R.; Hanak, T.; Persson, S.; Voigt, C.A. Cellulose and callose synthesis and organization in focus, what’s new? Curr. Opin. Plant Biol. 2016, 34, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Hyun, T.K.; Zhang, M.; Kumar, R.; Koh, E.J.; Kang, B.H.; Lucas, W.J.; Kim, J.-Y. Auxin-callose-mediated plasmodesmal gating is essential for tropic auxin gradient formation and signaling. Dev. Cell 2014, 28, 132–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, W.; Lee, J.Y. Arabidopsis callose synthases CalS1/8 regulate plasmodesmal permeability during stress. Nat. Plants 2016, 2, 16034. [Google Scholar] [CrossRef] [PubMed]
- Barratt, D.H.; Kölling, K.; Graf, A.; Pike, M.; Calder, G.; Findlay, K.; Zeeman, S.C.; Smith, A.M. Callose synthase GSL7 is necessary for normal phloem transport and inflorescence growth in Arabidopsis. Plant Physiol. 2011, 155, 328–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Marco, F.; Pagliari, L.; Degola, F.; Buxa, S.V.; Loschi, A.; Dinant, S.; Le Hir, R.; Morin, H.; Santi, S.; Musetti, R. Combined microscopy and molecular analyses show phloem occlusions and cell wall modifications in tomato leaves in response to ‘Candidatus Phytoplasma solani’. J. Microsc. 2016, 263, 212–225. [Google Scholar] [CrossRef]
- Amsbury, S.; Kirk, P.; Benitez-Alfonso, Y. Emerging models on the regulation of intercellular transport by plasmodesmata-associated callose. J. Exp. Bot. 2017, 69, 105–115. [Google Scholar] [CrossRef] [Green Version]
- Scharf, K.D.; Rose, S.; Zott, W.; Schǒffl, F.; Nover, L. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J. 1990, 9, 4495–4501. [Google Scholar] [CrossRef]
- Huhel, A.; Schoffl, F. Arabidopsis heat shock factor: Isolation and characterization of the gene and the recombinant protein. Plant Mol. Biol. 1994, 26, 353–362. [Google Scholar]
- Yamanouchi, U.; Yano, M.; Lin, H.; Ashikari, M.; Yamada, K. A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc. Natl. Acad. Sci. USA 2002, 99, 7530–7535. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.X.; Jiang, H.Y.; Chu, Z.X.; Tang, X.L.; Zhu, S.W.; Cheng, B.J. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genom. 2011, 12, 76. [Google Scholar] [CrossRef] [Green Version]
- Tang, R.M.; Zhu, W.J.; Song, X.Y.; Lin, X.Z.; Cai, J.H.; Wang, M.; Yang, Q. Genome-wide identification and function analyses of heat shock transcription factors in potato. Front. Plant Sci. 2016, 7, 490. [Google Scholar] [CrossRef] [Green Version]
- Feng, L.; Hu, H.T. Heat shock transcription factors on defense responses in plant. J. Anhui Agric. Sci. 2011, 4, 1931–1934. [Google Scholar]
- Shen, C.; Yuan, J. Genome-wide characterization and expression analysis of the heat shock transcription factor family in pumpkin (Cucurbita moschata). BMC Plant Biol. 2020, 20, 471. [Google Scholar] [CrossRef]
- Almoguera, C.; Rojas, A.; Díaz-Martín, J.; Prieto-Dapena, P.; Carranco, R.; Jordano, J. A seed-specific heat-shock transcription factor involved in developmental regulation during embryogenesis in sunflower. J. Biol. Chem. 2002, 277, 43866–43872. [Google Scholar] [CrossRef] [Green Version]
- Peng, J. Gibberellin and jasmonate crosstalk during stamen development. J. Integr. Plant Biol. 2009, 51, 1064–1070. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Ma, X.; Guo, H.; Sukiran, N.L.; Guo, B.; Assmann, S.M.; Ma, H. Flower Development under Drought Stress: Morphological and Transcriptomic Analyses Reveal Acute Responses and Long-Term Acclimation in Arabidopsis. Plant Cell 2013, 25, 3785–3807. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Foster, R.; Ma, S.; Liao, S.-J.; Bliss, S.; Kartika, D.; Wang, L.; Wu, L.; Eamens, A.L.; Ruan, Y. Identification of transcription factors controlling cell wall invertase gene expression for reproductive development via bioinformatic and transgenic analyses. Plant J. 2021, 106, 1058–1074. [Google Scholar] [CrossRef]
- Kranz, H.D.; Denekamp, M.; Greco, R.; Jin, H.; Leyva, A.; Meissner, R.C.; Petroni, K.; Urzainqui, A.; Bevan, M.; Martin, C.; et al. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 1998, 16, 263–276. [Google Scholar] [CrossRef] [PubMed]
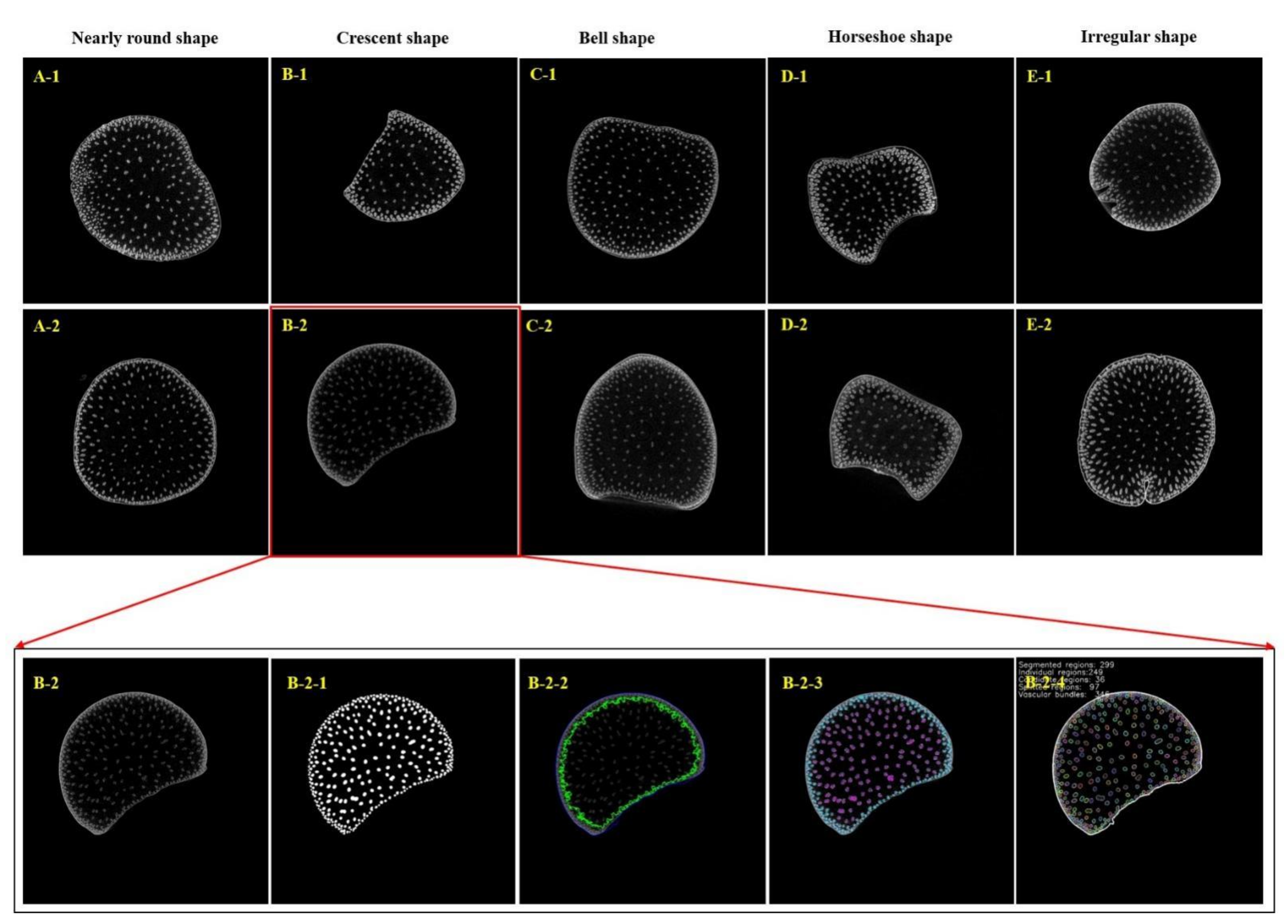
| Group | Phenotypes | Index | Unit | Median | Minimum | Maximum |
|---|---|---|---|---|---|---|
| Cross-section | The short axis length of the slice zone | SZ_SA | mm | 12.267 | 7.537 | 18.678 |
| The perimeter of the slice zone | SZ_P | mm | 44.903 | 34.325 | 128.45 | |
| The length-width ratio of the slice zone | SZ_LWR | - | 1.098 | 1.008 | 1.547 | |
| The long axis length of the slice zone | SZ_LA | mm | 13.557 | 8.649 | 21.574 | |
| The circumcircle area of the slice zone | SZ_CCA | mm2 | 132.604 | 55.483 | 318.124 | |
| The convex area of the slice zone | SZ_CA | mm2 | 137.63 | 59.087 | 232.311 | |
| The area of the slice zone | SZ_A | mm2 | 134.354 | 55.833 | 319.316 | |
| Epidermis zone | The thickness of the epidermis | EZ_T | mm | 0.076 | 0.052 | 0.144 |
| The area of epidermis | EZ_A | mm2 | 3.344 | 1.5 | 5.876 | |
| Periphery zone | The number of vascular bundles in the periphery zone | PZ_VB_N | - | 193 | 91 | 424 |
| The density of vascular bundles in the periphery zone | PZ_VB_D | number/mm2 | 7.861 | 3.181 | 15.198 | |
| The average convex area ratio of the vascular bundles in the periphery zone | PZ_VB_CAR | - | 0.944 | 0.482 | 0.962 | |
| The convex area of the vascular bundles in the periphery zone | PZ_VB_CA | mm2 | 12.494 | 5.581 | 25.588 | |
| The total area of the vascular bundles in the periphery zone | PZ_VB_A | mm2 | 11.731 | 5.311 | 24.114 | |
| The thickness of the periphery zone | PZ_T | mm | 0.687 | 0.291 | 1.209 | |
| The area of the periphery zone | PZ_A | mm2 | 25.675 | 9.536 | 47.102 | |
| Inner zone | The number of vascular bundles in the inner zone | IZ_VB_N | - | 154 | 67 | 338 |
| The density of the vascular bundles in the inner zone | IZ_VB_D | number/mm2 | 1.529 | 0.571 | 3.481 | |
| The average convex area ratio of the vascular bundles in the inner zone | IZ_VB_CAR | - | 0.976 | 0.964 | 0.98 | |
| The convex area of the vascular bundles in the inner zone | IZ_VB_CA | mm2 | 15.281 | 6.725 | 35.693 | |
| The total area of the vascular bundles in the inner zone | IZ_VB_A | mm2 | 14.912 | 6.561 | 34.76 | |
| The thickness of the inner zone | IZ_T | mm | 5.709 | 3.498 | 9.299 | |
| The area of the inner zone | IZ_A | mm2 | 108.220 | 64.688 | 214.114 | |
| Vascular bundles | The average short axis length of the vascular bundles | VB_SAave | mm | 0.274 | 0.222 | 0.395 |
| The average perimeter of the vascular bundles | VB_Pave | mm | 1.036 | 0.82 | 1.466 | |
| The total number of vascular bundles | VB_N | - | 357 | 196 | 722.5 | |
| The length-width ratio of vascular bundle | VB_LWR | - | 0.004 | 0.002 | 0.008 | |
| The average long axis length of the vascular bundles | VB_LAave | mm | 0.33 | 0.257 | 0.47 | |
| The density of the vascular bundles | VB_D | number/mm2 | 2.718 | 1.035 | 5.605 | |
| The average circumcircle area of the vascular bundles | VB_CCAave | mm2 | 0.107 | 0.064 | 0.213 | |
| The average convex area ratio of the vascular bundles | VB_CAR | - | 0.966 | 0.953 | 0.975 | |
| The average convex area of the vascular bundles | VB_CAave | mm2 | 0.08 | 0.051 | 0.158 | |
| The average area of the vascular bundles | VB_Aave | mm2 | 0.077 | 0.049 | 0.154 | |
| The total area of the vascular bundles | VB_A | mm2 | 27.16 | 13.587 | 56.158 | |
| The separation ratio of the vascular bundles | SRVB | - | 2.726 | 1.831 | 9.613 | |
| The area ratio of individual vascular bundles | ARIVB | - | 0.725 | 0.424 | 0.899 |
| Phenotypic Index | p-Value | Mixed | NSS | SS | TST |
|---|---|---|---|---|---|
| SZ_SA (mm2) | 0.00016 *** | 12.631 b | 13.094 a | 12.378 b | 12.266 b |
| SZ_P (mm) | 0.000841 *** | 45.812 b | 47.537 a | 45.229 b | 44.71 b |
| SZ_LA (mm) | 0.000143 *** | 13.918 b | 14.577 a | 13.806 b | 13.524 b |
| SZ_CCA (mm2) | 0.000274 *** | 175.382 b | 191.394 a | 170.184 b | 164 b |
| SZ_CA (mm2) | 8.19 × 10−5 *** | 145.432 b | 159.054 a | 140.723 b | 137.129 b |
| SZ_A (mm2) | 4.59× 10−5 *** | 143.297 b | 157.433 a | 138.59 b | 134.456 b |
| PZ_VB_D (number/mm2) | 3.82× 10−5 *** | 6.859 b | 6.661 c | 7.071 a | 6.997 ab |
| PZ_VB_CA (mm2) | 0.000825 *** | 13.594 ab | 13.86 a | 13.173 b | 13.16 b |
| PZ_VB_A (mm2) | 0.00085 *** | 12.72 ab | 12.976 a | 12.324 b | 12.308 b |
| PZ_T (mm) | 0.000152 *** | 0.779 ab | 0.781 a | 0.776 c | 0.777 bc |
| PZ_A (mm2) | 4.06× 10−6 *** | 30.92 b | 32.089 a | 29.936 b | 30.04 b |
| IZ_VB_D (number/mm2) | 0.00201 ** | 1.467 a | 1.393 b | 1.506 a | 1.5 a |
| IZ_T (mm) | 0.000684 *** | 5.784 b | 6.057 a | 5.733 b | 5.596 b |
| IZ_A (mm2) | 0.000395 *** | 108.716 b | 120.298 a | 106.58 b | 101.691 b |
| VB_SAave (mm) | 0.018 * | 0.29 ab | 0.292 a | 0.283 b | 0.285 b |
| VB_Pave (mm) | 0.0268 * | 1.095 ab | 1.103 a | 1.07 b | 1.075 b |
| VB_LAave (mm) | 0.0331 * | 0.352 ab | 0.354 a | 0.343 b | 0.345 b |
| VB_D (number/mm2) | 0.000197 *** | 2.549 a | 2.409 b | 2.621 a | 2.62 a |
| VB_CCAave (mm2) | 0.0267 * | 0.122 ab | 0.124 a | 0.116 b | 0.116 b |
| VB_CAave (mm2) | 0.0171 * | 0.089 ab | 0.091 a | 0.085 b | 0.086 b |
| VB_Aave (mm2) | 0.0156 * | 0.086 ab | 0.088 a | 0.082 b | 0.083 b |
| VB_A (mm2) | 0.000822 *** | 28.436 ab | 29.686 a | 27.568 b | 26.689 b |
| Trait | No. of Significant SNPs | No. of Annotated Genes | No. of Significant SNPs Validated by Multiple Methods | No. of Unique Annotated Genes Validated by Multiple Methods | No. of Genes Only Related to Specific Trait | No. of SNPs Shared between More than 2 Phenotypes | No. of Shared Associated Genes between More than 2 Phenotypes |
|---|---|---|---|---|---|---|---|
| SZ_A | 32 | 61 | 7 | 14 | 2 | 19 | 21 |
| SZ_P | 29 | 54 | 9 | 15 | 7 | 11 | 11 |
| SZ_LA | 35 | 67 | 5 | 10 | 2 | 10 | 12 |
| SZ_SA | 36 | 67 | 8 | 16 | 10 | 9 | 9 |
| SZ_CA | 23 | 43 | 9 | 17 | 1 | 15 | 18 |
| SZ_CCA | 36 | 67 | 7 | 14 | 2 | 17 | 19 |
| EZ_A | 18 | 32 | 1 | 2 | 2 | 1 | 1 |
| VB_N | 34 | 54 | 9 | 18 | 10 | 6 | 6 |
| VB_A | 35 | 60 | 7 | 12 | 10 | 8 | 8 |
| VB_AAVE | 109 | 186 | 22 | 42 | 13 | 40 | 47 |
| VB_PAVE | 28 | 52 | 6 | 11 | 0 | 10 | 13 |
| VB_LAAVE | 53 | 92 | 18 | 33 | 9 | 19 | 26 |
| VB_SAAVE | 55 | 96 | 6 | 10 | 4 | 14 | 16 |
| VB_CAAVE | 96 | 163 | 21 | 37 | 10 | 36 | 42 |
| VB_CCAAVE | 127 | 211 | 21 | 40 | 18 | 32 | 40 |
| VB_CAR | 1 | 2 | 0 | 0 | 0 | 0 | 0 |
| VB_LWR | 50 | 70 | 12 | 21 | 16 | 3 | 3 |
| PZ_VB_N | 43 | 73 | 7 | 12 | 10 | 3 | 3 |
| PZ_A | 37 | 66 | 12 | 23 | 15 | 1 | 1 |
| IZ_VB_N | 33 | 55 | 6 | 11 | 9 | 2 | 2 |
| IZ_T | 26 | 40 | 9 | 13 | 3 | 10 | 10 |
| IZ_VB_A | 26 | 44 | 6 | 12 | 2 | 7 | 7 |
| IZ_A | 15 | 24 | 5 | 10 | 4 | 11 | 12 |
| ARIVB | 30 | 56 | 7 | 13 | 9 | 0 | 0 |
| PZ_VB_D | 27 | 45 | 6 | 9 | 8 | 1 | 1 |
| PZ_VB_CA | 32 | 50 | 6 | 12 | 2 | 0 | 0 |
| IZ_VB_D | 18 | 34 | 8 | 16 | 12 | 2 | 2 |
| IZ_VB_CA | 19 | 33 | 7 | 12 | 2 | 7 | 7 |
| IZ_VB_CAR | 4 | 7 | 1 | 1 | 1 | 0 | 0 |
| VB_D | 27 | 47 | 6 | 12 | 6 | 4 | 4 |
| Summery | 806 | 1245 | 186 | 320 | 199 | 88 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Zhou, G.; Wang, J.; Lu, X.; Zhao, H.; Zhang, M.; Guo, X.; Zhang, Y. High-Throughput Phenotyping Accelerates the Dissection of the Phenotypic Variation and Genetic Architecture of Shank Vascular Bundles in Maize (Zea mays L.). Plants 2022, 11, 1339. https://doi.org/10.3390/plants11101339
Guo S, Zhou G, Wang J, Lu X, Zhao H, Zhang M, Guo X, Zhang Y. High-Throughput Phenotyping Accelerates the Dissection of the Phenotypic Variation and Genetic Architecture of Shank Vascular Bundles in Maize (Zea mays L.). Plants. 2022; 11(10):1339. https://doi.org/10.3390/plants11101339
Chicago/Turabian StyleGuo, Shangjing, Guoliang Zhou, Jinglu Wang, Xianju Lu, Huan Zhao, Minggang Zhang, Xinyu Guo, and Ying Zhang. 2022. "High-Throughput Phenotyping Accelerates the Dissection of the Phenotypic Variation and Genetic Architecture of Shank Vascular Bundles in Maize (Zea mays L.)" Plants 11, no. 10: 1339. https://doi.org/10.3390/plants11101339
APA StyleGuo, S., Zhou, G., Wang, J., Lu, X., Zhao, H., Zhang, M., Guo, X., & Zhang, Y. (2022). High-Throughput Phenotyping Accelerates the Dissection of the Phenotypic Variation and Genetic Architecture of Shank Vascular Bundles in Maize (Zea mays L.). Plants, 11(10), 1339. https://doi.org/10.3390/plants11101339






