Abstract
The sweet chestnut tree (Castanea sativa Mill.) is one of the most significant Mediterranean tree species, being an important natural resource for the wood and fruit industries. It is a monoecious species, presenting unisexual male catkins and bisexual catkins, with the latter having distinct male and female flowers. Despite the importance of the sweet chestnut tree, little is known regarding the molecular mechanisms involved in the determination of sexual organ identity. Thus, the study of how the different flowers of C. sativa develop is fundamental to understand the reproductive success of this species and the impact of flower phenology on its productivity. In this study, a C. sativa de novo transcriptome was assembled and the homologous genes to those of the ABCDE model for floral organ identity were identified. Expression analysis showed that the C. sativa B- and C-class genes are differentially expressed in the male flowers and female flowers. Yeast two-hybrid analysis also suggested that changes in the canonical ABCDE protein–protein interactions may underlie the mechanisms necessary to the development of separate male and female flowers, as reported for the monoecious Fagaceae Quercus suber. The results here depicted constitute a step towards the understanding of the molecular mechanisms involved in unisexual flower development in C. sativa, also suggesting that the ABCDE model for flower organ identity may be molecularly conserved in the predominantly monoecious Fagaceae family.
1. Introduction
Development of unisexual male and female flowers has been regarded as a successful mechanism to decrease inbreeding by promoting cross-pollination and gene fluidity [1]. The development of unisexual flowers has evolved, independently, multiple times, in some families being the predominant reproductive strategy. Such is the case of the Fagaceae, a large plant family comprising more than 900 species that constitute an important economic resource and are pivotal to the dynamics of the forest ecosystems [2].
In the Fagaceae, the development of unisexual male and female flowers has been studied in Quercus suber (cork oak) [3,4,5]. Sobral and Costa [4] have shown that changes in the dynamics of the ABCDE model of flower organ identity may occur during the development of the male and female flowers of Q. suber. The canonical ABCDE model postulates that the development of the flower reproductive organs (stamens or carpels) is controlled by distinct classes of genes: stamen identity is determined by the interaction of B-, C-, and E-class transcription factors, whereas C-, D-, and E-class gene activity specify carpel identity [6,7,8,9,10,11]. The ABCDE genes belong, predominantly, to the highly conserved MADS-box family. These genes encode for transcription factors with four domains: MADS (M), intervening (I), keratin-like (K), and C-terminal (C) [12]. The MADS domain, which is the DNA-binding domain, is the most conserved, conferring these proteins the ability to bind to specific DNA motifs, the CArG-boxes [12,13]. The I and K domains are involved in the mediation and establishment of multimeric protein–protein complexes [14]. Although the C-terminal domain of the MADS-box proteins is much less conserved, it contains lineage specific motifs that are functionally important. In the specific case of B-class proteins, their stability and ability to bind CArG-boxes is dependent on an obligatory heterodimerisation [15,16,17,18]. In cork oak, the expression of one of the B-class genes, QsPISTILLATA (QsPI), determines the development of the male flower by establishing a complex with other B-class and C-class proteins. In female flowers, complete lack of QsPI expression disables the obligatory heterodimerisation of the B-class protein complex, thus promoting carpel development [4].
The male and female unisexual flowers of the Fagaceae species are typically unisexual by inception and develop at different periods during the growing season [4,19,20,21]. Castanea and Lithocarpus species present two distinct inflorescences, unisexual male catkins and bisexual catkins, the latter bearing functionally distinct male and female flowers [22,23] suggesting different mechanisms controlling the development of unisexual flowers within the Fagaceae. Castanea sativa Mill. is a deciduous Fagaceae species, distributed throughout the Mediterranean region, spreading from the Caucasus to Portugal and from Southern England to the southern part of the Iberian Peninsula [24]. It is monoecious, bearing two types of inflorescences: unisexual male catkins that develop in the leaf axis of the current season growth, and bisexual catkins that develop near the terminal end of the shoot. Each unisexual male catkin is composed by staminate flowers gathered in glomerules of 3 to 7 flowers each, in an average number of 40 glomerules per catkin [25]. Each individual male flower has four to six tepals and 8 to 12 stamens [26]. Female inflorescences appear singly or in two or three clusters at the base of the bisexual catkins. Each inflorescence usually has three flowers [26,27]. Each female flower typically presents six to eight styles, emerging from enclosed scales. The ovaries have five to seven loci with two viable ovules each, although usually only one of the ovules becomes a mature seed [23,27,28]. The stigmatic region, which is exposed during the receptivity period, is covered with secretory cells, which facilitates pollen adhesion and germination [26,28]. C. sativa presents a protandrous habit, in which unisexual catkins mature before the bisexual ones, and pollen from the unisexual catkin is released before the female flowers are fully receptive: this delay between pollen shedding and female flower maturation takes more than two weeks [26,29]. Cross-pollination is required to ensure fertilisation and fruit development due to gametophytic self-incompatibility and morphological male sterility of some cultivars [27,30].
Most genetic resources related to the Castanea genus are derived from studies performed in its close relative Castanea mollissima (Chinese chestnut), whose transcriptome and whole-genome have been recently released [31,32]. However, there is still no available transcriptomic data of C. sativa, despite its economic and ecological importance. Thus, the molecular mechanisms controlling the development of the C. sativa unisexual and bisexual catkins are still unclear, and it is not known to what extent these regulatory mechanisms are comparable to what has been described for other Fagaceae species. In the present study, we obtained a comprehensive transcriptome of C. sativa and studied the dynamics of BCE-class gene expression and BCE-class protein complexes during the establishment of the male and female flowers of C. sativa.
2. Results
2.1. Castanea sativa ‘Judia’ Cultivar Flowering Phenology
Castanea sativa is a monoecious species with an accentuated protandrous habit, as unisexual male catkins and bisexual catkins develop with a significant delay. In this study, a detailed phenological characterisation of the C. sativa ‘Judia’ cultivar was performed. The ‘Judia’ cultivar was chosen because it is a predominant cultivar in Portugal due to its high-quality fruits and productivity [30]. Unisexual male catkins emerge during spring in the axils of the new leaves that developed within the bursting axillary buds (Figure 1A,B). At this stage of development, the glomerules are still covered by bracts. As spring progresses, both the shoot and the unisexual male catkins elongate exposing the glomerules (Figure 1C,D). At the same time, bisexual catkins start developing at the terminal end of the new shoot, with no clear observable separation between female and male flowers (Figure 1E,F). The development of the female and male organs in the bisexual catkin is asynchronous as the male flower is the first to be identifiable. Towards the beginning of summer, the glomerules in the unisexual male catkins are fully exposed as the bracts start to recede, the anthers are visible, and pollen grains start to develop (Figure 1G, white arrow). During the same period, the separation between the male and female flowers becomes clearer in the bisexual catkin (Figure 1G, blue arrow, and detailed in Figure 1H (male flower); Figure 1G, pink arrow, and detailed in Figure 1I (female flower)). The styles of the female flowers are not receptive (Figure 1J, pink arrow) when the pollen from the unisexual male catkins is shed (Figure 1K). Thus, there is a temporal separation between pollen shedding of the unisexual male catkin and the period in which female flower is receptive (Figure 1L). The male flower in the bisexual catkin is not mature (Figure 1M) during female flower receptivity. During mid-summer and after pollen shedding, the unisexual male catkins start to acquire a yellow colour and fall. At this stage, pollen shedding occurs in the male flower of the bisexual catkin, while the female flowers have already been fertilised and start to swell (Figure 1N).

Figure 1.
Flowering phenology of Castanea sativa. (A,B) In spring, following bud burst, the unisexual male catkins emerge in the axils of new leaves (white arrows). (C,D) As spring progresses, bracts start to open and expose the glomerules. (E,F) Bisexual catkin, with no clear distinction between the female and male flowers. (G) Shoot from June, in which the unisexual catkins reach their final stage of development (white arrows) and the differentiation between male (blue arrow, detailed in (H)) and female flowers (pink arrow, detailed in (I)) of the bisexual catkin is visible. (H) Detail of the male flower of the bisexual catkin. (I) Detail of the female flower of the bisexual catkin. (J) Developed bisexual catkin, with individualised male and female flowers (blue and pink arrows, respectively). (K) Glomerule of the unisexual male catkin, with anthers exposed. (L) Receptive female flower with exposed stigma. (M) Developed glomerule of the male flower of the bisexual catkin. (N) Shoot from July, in which the unisexual catkin is already turning yellow and dehiscent, having shed its pollen, and the female flower is swelling in order to develop the bur.
2.2. Castanea sativa De Novo Transcriptome Analysis
Despite the C. sativa economic and ecological importance, it is still not clear which molecular mechanisms are employed to control the flower phenology in this species as no genomic/transcriptomic resources were available. Therefore, the assembly of a de novo C. sativa transcriptome was vital to lay the foundations for molecular studies in this species. To obtain the transcriptome of C. sativa, we isolated total RNA from male flowers from the unisexual catkin and male and female flowers from the bisexual catkin, leaves, and buds at different developmental stages during the plant life cycle. The RNA samples were pooled together and sequenced using Illumina technology to generate 2 × 150 bp paired-end reads. After trimming, 134,286,842 sequences were obtained and used for de novo transcriptome assembly. A pipeline for de novo transcriptome assembly and processing proposed by Chabikwa et al. [33] was implemented, resulting in 164,926 contigs (Table S1). To evaluate the reliability and quality of this assembly, we mapped the reads used to generate the de novo transcriptome back to the transcriptome using bowtie2 [34]. The alignment rate of the input RNA-Seq reads was 97.49%, whereas alignment with the C. mollissima genome yielded an overall alignment rate of 62.93%.
After redundancy removal, 32,871 transcripts were obtained, with an average length of 906 bp, an N50 value of 1230 bp, and a GC percentage of 44% (Table S1). BUSCO is a tool that quantitatively assesses the quality and coverage of a transcriptome on the basis of evolutionarily informed expectations of gene content from eukaryotic genomes [35]. In the C. sativa transcriptome, 94.1% of the BUSCOS had a complete representation, indication a high-quality assembly, with 86.67% single copy genes (Table S2). The 32,871 transcript sequences were uploaded to Blast2GO [36], and blastx was performed against the NCBI nr database with an E value cut off of 1e-3; 29,607 sequences had blast hits, and 3264 had no hits. GO annotation categorised the transcripts in different functional groups according to cellular components, biological process, and molecular function (Figure S1).
2.3. ABCDE MADS-Box Transcription Factors Were Conserved in Castanea sativa
In cork oak, a closely related Fagaceae, the identity of the female and male sexual organs is dependent on the spatial activity of the ABCDE MADS-box genes [4]. It is not clear, however, as to the way in which in other Fagaceae, such as C. sativa, the ABCDE model is involved in the determination of flower unisexuality.
To check if sexual organ identity development in sweet chestnut is controlled by MADS-box genes, we screened the de novo transcriptome for homologs of Arabidopsis thaliana or Solanum lycopersicum MADS-box proteins (SlTM6 was used as a query because there is no TM6 homolog in A. thaliana). The blast queries included homologs for genes belonging to A-class (AtAPETALA1), B-class (AtPI, SlTM6, and AtAPETALA3), C- and D-classes (AtAGAMOUS, AtSHATTERPROOF1-2, and AtSEEDSTICK), and E-class (AtSEPALATTA1-4). The phylogeny of the C. sativa ABCDE MADS-box proteins was inferred using homologs of other angiosperms, and, whenever possible, homologs from an ancestral angiosperm (Amborella trichopoda) and a gymnosperm (Pinus radiata). On the basis of multiple alignments, we were able to conclude that all the retrieved sequences were complete and displayed the four conserved domains (M-, I-, K-, and C-terminals) (Figure S2).
In the A-class (Figure 2A), CsaAPETALA1 (CsaAP1) clusters with AP1 proteins from other perennials, in the same clade that includes AP1-like proteins from C. mollissima, Q. suber, Betula pendula, and Corylus avellana, species belonging to the order Fagales. The B-class phylogroup (Figure 2B) can be divided into three lineages: PI, euAP3, and paleoAP3 [37]. The PI and paleoAP3 lineages arose from a duplication event that took place before the emergence of angiosperms, whereas a later duplication event that took place at the base of higher dicots originated the euAP3 lineage [37] (reviewed in [38]). One sweet chestnut homolog was identified in each lineage (CsaPI, CsaAP3, and CsaTM6) clustering with homologs of C. mollissima, Quercus robur, Q. suber, and Quercus rubra, all of them monoecious Fagaceae. The PI lineage is characterised by a PI motif that is essential for protein function [37]. A comparative analysis of the PI motif between AtPI and CsaPI revealed that CsaPI has a complete PI motif in its C-terminal domain that seems to be conserved in the Fagaceae (Figure S3A). The core difference between the paleoAP3 and euAP3 lineages lies in the motifs contained in the C-terminal domain (Figure S2B) [37]. Comparison of the euAP3 motif of A. thaliana and C. sativa homologs suggests a partial conservation of the euAP3 motif in CsaAP3, similar to what happens in other Fagaceae (Figure S3). Similarly, there is a partial conservation of the paleoAP3 motif in CsaTM6. This motif is totally conserved in C. sativa and C. mollissima, but there is one amino acid difference between proteins of the Castanea and Quercus genus (QsTM6) (Figure S3C).
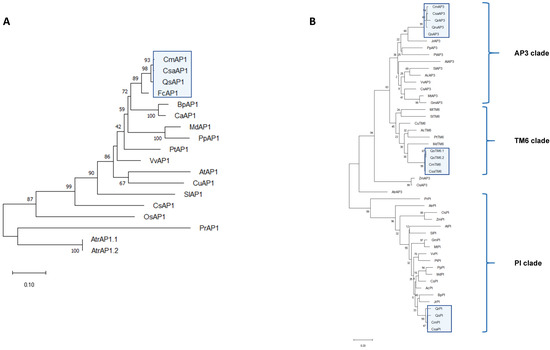
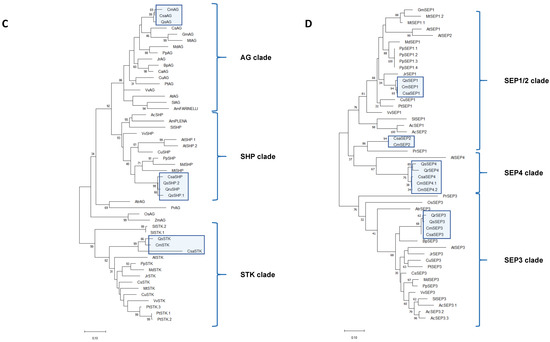
Figure 2.
Phylogenetic profiling of the Castanea sativa ABCDE genes. (A) A-class lineage; (B) B-class lineage; (C) C- and D-class lineages; (D) E-class lineage. Phylogenies were inferred using the maximum likelihood method (1000 replicates). The percentage of replicate trees in which the associated taxa clustered together is shown next to the branch nodes. The JTT method was used to compute evolutionary distances (number of aminoacids per site). The accession numbers for the proteins used in this analysis are presented in Table S3. Fagaceae species are highlighted in the blue box. The protein sequences used in this analysis were retrieved from the following species: Castanea mollissima (Cm), Quercus suber (Qs), Quercus robur (Qr), Quercus rubra (Qru), Fagus crenata (Fc), Betula pendula (Bp), Corylus avellana (Ca), Juglans regia (Jr), Malus domestica (Md), Prunus persica (Pp), Actinidia chinensis (Ac), Populus trichocarpa (Pt), Vitis vinifera (Vv), Arabidopsis thaliana (At), Citrus unshiu (Cu), Solanum lycopersicum (Sl), Cucumis sativus (Cs), Anthirrinum majus (Am), Medicago truncatula (Mt), Glycine max (Gm), Oryza sativa (Os), Zea mays (Zm), Pinus radiata (Pr), Amborella trichopoda (Atr).
The AG subfamily also underwent several duplication events throughout evolution: one duplication event originated the C and D lineages before the radiation of extant angiosperms [13]. The phylogenetic analysis for the C- and D-class proteins (Figure 2C) placed CsaAG in the same clade as C. mollissima and Q. suber AGAMOUS homologs. CsaSHP was also placed in the same clade as the homologs of the closely related Fagaceae Q. suber and Q. rubra (Figure 2C). There are two characteristic motifs (I and II) in the C-terminal domain of AG proteins [39]. CsaAG seems to have a partial conservation of these motifs, compared to its A. thaliana counterpart, and a complete amino acid conservation within the Fagaceae (Figure S2D). There is also partial conservation between CsaSHP and AtSHP AG motifs I and II, and a full conservation between the Fagaceae in motif I (Figure S2E). However, the same pattern is not followed regarding the AG motif II, where there is a complete deletion in QsSHP, which might suggest functional divergence.
The E-class comprises the SEPALLATA genes, which act as co-factors in the protein complexes that specify the identity of the different floral organs [40]. In the C. sativa transcriptome, four SEP homologs were identified (Figure 2D). Regarding the SEP1/2 lineage, both the CsaSEP1 and CsaSEP2 are closely related to the proteins of C. mollissima and Q. suber. CsaSEP3 also clusters with other Fagales; however, it is related to a closer degree with AtSEP3. CsaSEP4 is located in the same clade as SEP4 homologs of C. mollissima, Q. suber, and Q. robur.
2.4. C. sativa BCE-Like Gene Expression in Unisexual and Bisexual Catkins
During floral developmental, the coordinated spatial and temporal regulation of the MADS-box gene expression is pivotal for proper floral organ development (reviewed in [40]). The canonical ABCDE model postulates that stamen identity is determined by the expression of B-, C-, and E-class genes in the third whorl, whereas carpel identity is determined by expression of C- and E-class genes in the centre floral whorl (reviewed in [13]).
An expression analysis, by qRT-PCR, of the sweet chestnut BCE-like gene homologs was conducted using samples that included male unisexual catkins, as well as male and female flowers from the bisexual catkins, at different stages of development. The developmental stages were collected after the emergence of the unisexual male catkin primordia until their full development (MF S1–S4, represented in Figure 1A–D, respectively); after the emergence of bisexual catkins (BP, Figure 1E–F), in which there is no observable separation between the male and female flowers, and after complete separation and developed male (MB 1, represented in Figure 1H, and MB 2, represented in Figure 1M) and female flowers (FB 1, represented in Figure 1I, and FB 2, represented in Figure 1L).
The B-class genes, CsaAP3, CsaPI and CsaTM6 (Figure 3A–C), present similar expression patterns during the unisexual male flower development, increasing steadily from stages MF-S1 to MF-S4 (Figure 3A). The expression of CsaAP3, CsaPI and CsaTM6 is low during bisexual catkin primordia development (BP) and in early stages of its male and female flowers development (MB-S1 and FB-S1), but the expression peaks in the male portion of the bisexual catkin in stage MB-S2 (Figure 3A–C). There is expression of CsaPI in the female structure of the bisexual catkin in stage FB-S2, although it is not detected in stage FB-S1 (Figure 3B). The expression of the three B-class genes in female flowers led to a more thorough morphologic and histological analysis of these flowers, which revealed the presence of stamen-like structures fused at the base of the styles, concealed by the perianth (Figure 4A–C). Histological analysis of the male flowers revealed that no carpelloid structures can be found in these flowers (Figure 4D).
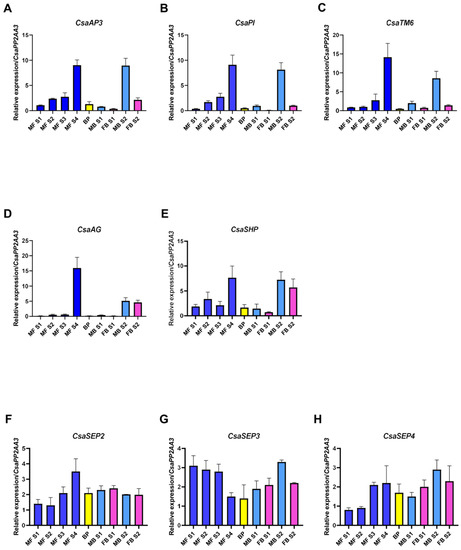
Figure 3.
Expression analysis of C. sativa BCE homolog genes in Castanea sativa flowers. (A) CsaAP3; (B) CsaPI; (C) CsaTM6; (D) CsaAG; (E) CsaSHP; (F) CsaSEP2; (G) CsaSEP3; (H) CsaSEP4. Error bars indicate standard deviation (SD) of three biological and technical replicates. CsaPP2AA3 was used as a reference gene. MF S1—unisexual male catkin primordia appearing in the axils of leaves of bursting buds (Figure 1A); MF S2—immature unisexual male catkins, starting to arise from bursting buds (Figure 1B); MF S3—mature unisexual male catkins (Figure 1C); MF S4—unisexual male catkin with developed glomerules, at a later stage of development (Figure 1D); BP—bisexual catkin primordia, with no definition between male and female flowers (Figure 1E,F); MB S1—male flowers of the bisexual catkin, early stage (Figure 1H); FB S1—female flower of the bisexual catkin, early stage (Figure 1I); MB S2—male flower of the bisexual catkin, fully developed (Figure 1M); FB S2—female flower of the bisexual catkin, fully developed (Figure 1L).
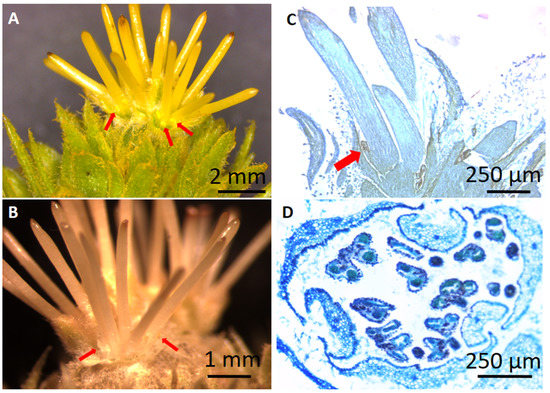
Figure 4.
Female flowers from the ‘Judia’ cultivar present stamen-like structures. (A,B) Detail of the stamen-like structures at the base of the styles of female flowers—the perianth was removed to expose these organs (red arrows). (C) Longitudinal section of a female flower, where it is possible to see the stamen-like organ fused with the style (red arrow). (D) Transversal section of a male flower, with no evidence of stylar-like organs.
CsaSHP expression follows a similar pattern to that of B-class genes, presenting low expression in early stages of male flower development and peaking at MF-S4 (Figure 3E). CsaSHP expression is low in bisexual primordia and in early stages after the separation of male and female structures, but increases in stages S2 for both tissues (MB-S2 and FB-S2), being higher in the male flowers than in the female flowers. The fact that CsaSHP is expressed also in the male flowers suggests that this gene might have retained a C-function after the duplication event that originated the C and D lineages. CsaAG is significantly more expressed in unisexual male flowers, mainly in the later stage (MF-S4), and in the bisexual catkins its expression follows a pattern similar to that of CsaSHP, with low expression levels in the early developmental stages, and higher in later stages of development (Figure 3D). CsaSTK expression was not detected in the flowers at the chosen developmental stages.
E-class genes CsaSEP2 (Figure 3F) and CsaSEP4 (Figure 3H) present low expression levels in early stages of unisexual male flower development, peaking in MF-S4. CsaSEP3 (Figure 3G) shows an inverse expression pattern, being significantly higher during early development (MF S1) and decreasing in later stages. In the bisexual catkin primordia (BP) all the SEP-like genes are expressed, and upon early male and female flower development, their expression is higher in female flowers (FB S1) than in male flowers (MB S1). CsaSEP1 was not detected in any of the flower tissues.
Overall, CsaAG, CsaSHP, CsaAP3, CsaPI, CsaTM6 and CsaSEPs showed higher expression in the unisexual male catkin flowers and the male flower of the bisexual catkin than in female flowers, particularly at later stages of development.
2.5. Dimerisation of C. sativa BCE-Like Proteins
According to the canonical ABCDE model, stamen development depends on the interaction of B-, C-, and E-class proteins, whereas carpel development depends on the interaction of C- and E-class proteins [41,42]. Different dynamics of the ABCDE model in the Fagaceae Q. suber, namely, the interaction of B-class QsAP3 and QsPI with C-class QsSHP, and not QsAG (C-class), may be one of the factors underlying unisexual flower development in this species [4]. It is not clear if redeployment of the canonical ABCDE model interactions could be the mechanism behind the development of all the unisexual flowers in C. sativa.
Dimerisation of C. sativa BCE-like proteins was assessed in a yeast-two-hybrid experiment, in which the coding regions of the C. sativa B-, C-, and E-like genes were fused to the activation or binding domain of the GAL4 transcription factor. Interactions between C. sativa B-, C-, and E-like proteins are compiled in Figure 5.
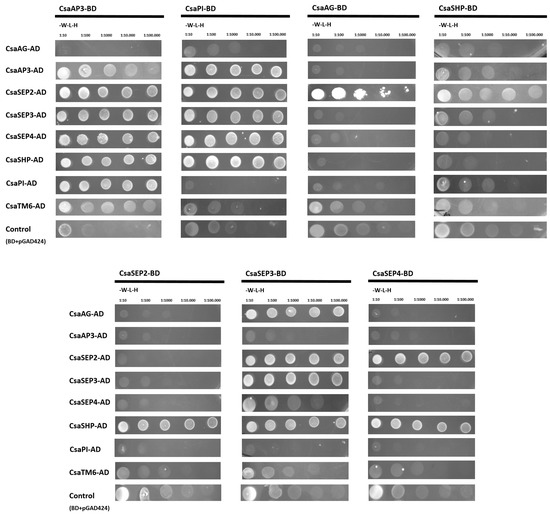
Figure 5.
Interaction between Castanea sativa BCE-like proteins. Double yeast transformations of C. sativa BCE-like proteins fused to GAD4-activation domain (AD) and GAL4-binding domain (BD) were performed and tested for plasmid presence by growing cells in synthetic defined (SD) medium without tryptophan and lecucine (-W-L). Interacting capacity was tested in SD medium without aminoacids supplemented with 3-aminotriazole (tryptophan, leucine, histidine). Interaction strength was evaluated through sequential dilutions (1:10, 1:100, 1:1000, 1:10,000, 1:100,000). Each cropped image corresponds to a matching single plate in which the double transformants were selected.
An important trademark of the ABCDE model is the functional obligatory heterodimerisation between the B-class proteins. In C. sativa, the obligatory heterodimerisation of AP3 and PI-like proteins is likely to be conserved, as CsaPI and CsaAP3 proteins were able to dimerise and activate the reporter gene HISTIDINE3. CsaAP3 also interacted with its closest relative CsaTM6, but there was no interaction detected between CsaPI and CsaTM6. Interaction between B- and E-class proteins seems to be conserved in C. sativa, as all B-class proteins were able to interact with CsaSEP2, CsaSEP3, or CsaSEP4. CsaSEP1 protein was not used in this assay because CsaSEP1 expression was not detected in flower tissues, as mentioned previously. The B-class proteins CsaPI and CsaAP3 were also able to interact with the C-class protein CsaSHP, but not with CsaAG (Figure 5).
The other C-class protein, CsaAG, was only able to interact with the E-class proteins CsaSEP2 and CsaSEP3, but the activation of the gene reporter was not sufficient to allow yeast growth in higher order dilutions, suggesting a weaker interaction between these proteins. The expression of CsSHP in male and female flowers combined with the non-canonical dimerisation of CsaSHP with B-class proteins suggests that CsaSHP might have retained a C-class function in the determination of floral reproductive organs.
3. Discussion
Changes to the ABCDE model dynamics is proposed as one of the factors controlling the development of unisexual flowers by inception in several families, including the predominantly monoecious Fagaceae [4,33,34,35]. In cork oak, several ABCDE gene homologs are differentially expressed in male and female flowers, suggesting that floral organ identity in this species is controlled by rearrangements in the expression dynamics of these genes. In the monoecious C. sativa, a similar scenario could also explain the development of its unisexual flowers. Therefore, it is of particular importance to characterise B- and C-function genes that specify male and female organ identity, in order to clarify the mechanism of sexual organ determination in C. sativa. In the present work, several genes homologous to the ABCDE model genes were identified and their role in the establishment of C. sativa unisexual flowers was assessed.
A de novo transcriptome of C. sativa was assembled and surveyed for ABCDE model genes homologs. According to this model, female flower identity is established by the coordinated activity of C- and E-class genes, whereas male flower development depends of B-, C-, and E-class genes [13]. In the present study, three B-class genes were identified and classified according to their phylogenetic clustering (CsaAP3, CsaPI and CsaTM6). B-class genes are expressed in the second and third whorl of the flower meristem, being involved in petal and stamen identity, respectively [13]. In C. sativa, the temporal expression profile for all B-class genes is similar, being higher in male flowers from the unisexual catkin and in the male flower of the bisexual catkin, and significantly lower in the female flower.
The expression pattern presented by the B-class genes CsaTM6 and CsaAP3 in C. sativa is similar to its close relative Q. suber: QsTM6.1/2 and QsAP3 present high expression levels in male flowers, being expressed in female flowers to a lesser extent [4]. However, this pattern is not mimicked by CsaPI, as there is expression of this gene in female flowers, in late developmental stages, which does not occur in Q. suber. Reports of hermaphroditism in C. sativa female flowers [26,36,37,38,39,40] suggest that the rudimentary stamens are sterile [27,36]. A pollen germination assay might be required to assess the fertility of these rudimentary stamens in the presently studied ‘Judia’ cultivar. The presence of these stamen-like organs in female flowers is not exclusive of C. sativa, having been also detected in other species of the Castanea genus. Morphologic observations in C. mollissima revealed the presence of 12 underdeveloped staminodes with short filaments and visible anthers capable of producing pollen, which did not dehisce [41,42]. In the Chinese chinquapin (Castanea henryi), female flowers experience a period of hermaphroditism, as 12 stamen primordia also develop during carpel development, arresting their elongation as the stigma enlarges [43]. It is thus likely that female flowers in the Castanea genus are hermaphroditic at inception and become unisexual by abortion or arrested growth of male organs. This is contrary to what was reported for other Fagaceae, in which the flowers are unisexual by inception as no aborted organs of the opposite sex are detected [4,19,20,21]. The expression of CsaPI and the other B- and C-class genes in the female flowers is likely associated to the development of the stamen-like structures in the female flowers of C. sativa. In the present study, CsaPI was found to be able to interact with CsaAP3 and CsaTM6, which is indicative that the obligatory heterodimerisation of B-class proteins is conserved also in this species. In the other Fagaceae Q. suber, this interaction was also shown [4]. Homodimerisation of AP3 homologs is also conserved in these two Fagaceae. However, the interaction between QsAP3 and QsTM6 was not reported for Q. suber, while in C. sativa, CsaAP3 and CsaTM6 were able to interact. Thus, it is likely that different interactions between Fagaceae B-class proteins might be species-specific. In A. thaliana, E-class genes have ubiquitous and redundant functions in flower meristem identity and flower organ development in the four whorls, as triple and quadruple mutants for these genes have floral organs converted into sepals and leaf-like organs [10,44]. In C. sativa, E-class proteins might have their function in flower organ identity conserved, as they are able to interact with B- and C-class proteins. Thus, similarly to what was determined in the close relative Q. suber, the heterodimerisation of B-class proteins and their interaction with C- and E-class proteins might be needed for male flower development in C. sativa (Figure 6).
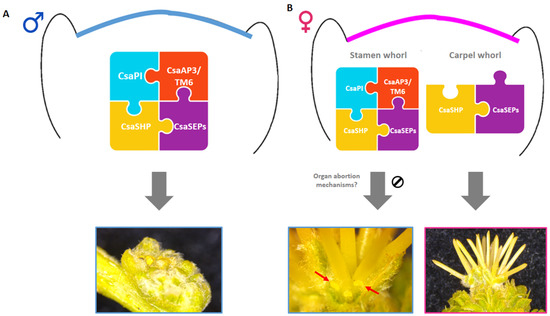
Figure 6.
Representative model of the potential role of MADS-box transcription factors during the development of Castanea sativa unisexual flowers. (A) Combinatorial action of B-class CsaPI/TM6/AP3 with C-class CsaSHP and E-class CsaSEPs confers male identity to the unisexual male flowers. (B) Female identity is conferred by a combination of CsaSHP and CsaSEPs. Unknown mechanisms, downstream of the MADS-box genes, may contribute to the development of staminodes in female flowers (blue box, red arrows).
In the C. sativa transcriptome, one AG protein (CsaAG) was inferred, which clustered in the euAG clade from the Fagaceae. In the PLENA lineage, one SHP-like protein was identified (CsaSHP). Expression analysis of these genes in C. sativa flowers revealed that CsaAG is mainly expressed in later developmental stages of male flowers, a pattern that has also been observed in other species with unisexual flowers, such as Populus trichocarpa [45], Q. suber [4], and B. pendula [46] with a strong association with pollen development. Unlike CsaAG, CsaSHP is expressed during unisexual male flower development, in the bisexual catkin primordia, and in the female and male flowers of the bisexual catkin. Similarly to CsaSHP, QsSHP was also detected in both the male and female flowers [4], suggesting that the Fagaceae SHP homologs, and not the AG homologs, could be acting as functional C-like genes.
Regarding the C-class proteins interactions, CsaAG only dimerises with CsaSEP3 and CsaSEP2. Similar results were found in Q. suber [4] and Prunus mume [47], in which AG-like proteins only interacted with SEP-like proteins. CsaSHP displayed interaction with all SEP-like proteins, which was also observed in species such as Arabidopsis, cork oak, and cucumber [4,48,49], indicative that the interactions between these proteins are relatively conserved among species. On the other hand, C. sativa B-class proteins CsaAP3 and CsaPI, but not CsaTM6, were also able to heterodimerise with C-class CsaSHP, as was previously reported for Q. suber, suggesting a conservation of this non-canonical interaction in the Fagaceae. The similar temporal expression patterns of CsaAG and CsaSHP, coupled with the fact that there was no observed interaction between B-class proteins and CsaAG, might indicate that CsaSHP may be required to act simultaneously with CsaAG in order to complete the C-function, as was observed in other species [4,50].
The temporal expression of the ABCDE model homologous genes in C. sativa and the ABCDE model interactome suggests that it is likely that the homeotic genes are involved in the determination of the male flower identity, both in the unisexual and bisexual catkins. However, the determination of unisexuality in the female flowers requires a different explanation. It is possible that the spatial expression of CsaPI in the female flower could be restricted to the non-reproductive and stamen whorls, allowing the development of stamens in the third whorl and the carpel in the centre whorl (Figure 6). Analysis of the spatial expression of CsaPI during the bisexual catkin primordia formation might help unveil the role of this gene during female flower development. Furthermore, the mechanisms that could be promoting the probable stamen abortion in this cultivar are still illusive. Thus, RNA-Seq studies concerning C. sativa inflorescences will be pivotal to clarify this hypothesis, as well as to identify genes exclusively expressed in female inflorescences that might be involved in the genetic basis of their development. It is possible that some of these genes are related to the developmental arrest of the stamens, as was reported in transcriptomic studies in other species in which sex determination is a late event [51,52,53,54].
Clarification of the mechanisms that lead to flower unisexuality are of particular interest in species of agronomical and ecological relevance, as it has biological implications, affecting fructification, seed production, and species perpetuation. C. sativa constitutes a particular case of variation of monoecy within the Fagaceae family, presenting unisexual male catkins and bisexual catkins. Furthermore, unlike most Fagaceae, C. sativa female flowers are not unisexual by inception. The likely temporary hermaphroditism in this species could be considered a relaxation that is counterbalanced by an additional layer to prevent inbreeding, which is the C. sativa gametophytic self-incompatibility. Thus, it is of utmost importance to study the genetic mechanisms for unisexual flower development in this species, which may differ significantly within the Fagaceae. The results here presented show that, even though phylogenetically close, ABCDE model genes may have different dynamics within distinct Fagaceae species.
4. Materials and Methods
4.1. De Novo Transcriptome Assembly
4.1.1. Plant Material
Tissue samples (leaves, buds, and flowers in several developmental stages) from Castanea sativa M. were harvested at the germplasm bank located at UTAD (Vila Real district, Portugal, 41.2863826, −7.7448634) from three adult trees from the ‘Judia’ cultivar located in Lagoa (Vila Pouca de Aguiar district, Portugal, 41.6292217, −7.2960124). The flower developmental stages were selected on the basis of the phenological stages described in [55]. The collected tissues were frozen in liquid nitrogen and kept at −80 °C until RNA extraction.
4.1.2. RNA Extraction
For de novo transcriptome assembly, total RNA was extracted from frozen tissue samples of young and mature leaves, dormant and active buds, and male and bisexual inflorescences in different stages of development. Samples were ground to powder using liquid nitrogen, and the RNA was extracted using the CTAB/LiCl method [56], with some modifications [57,58]. DNAse treatment was conducted using TURBO™ DNase (ThermoFischer, Waltham, MA, USA), following the manufacturer’s instructions, and integrity was analysed using the Experion™ RNA StdSens Analysis Kit (BioRad, Hercules, CA, USA), prior to library preparation.
4.1.3. Library Preparation and Sequencing
The individual RNA samples were pooled, and the cDNA libraries were prepared using Illumina TruSeq stranded mRNA kit and sequenced at Fasteris S.A. (Plain-les-Ouates, Switzerland), using Illumina technology to generate 2 × 150 bp paired-end reads.
4.1.4. De Novo Assembly of Castanea sativa Transcriptome
FastQC v0.11.8 [59] was applied to check the quality of the sequencing data. Low-quality reads were filtered and Illumina adapters removed using bbduk from the package bbtools [60]. All sequences with a quality score below 20 were removed and a forced trimming of the first 15 bp was performed. Overrepresented polyG sequences were removed using the option “literal”. Furthermore, in order to eliminate small reads that were interfering with the sequence length distribution, we filtered all reads with a length below 30 bp.
De novo transcriptome assembly was performed using Trinity (v. 2.9.1) [61,62] in a Galaxy server (https://usegalaxy.org/, accessed on 26 July 2021), with default settings. Assembly metrics were investigated using TransRate (v. 1.0.3) [63], a software that reports basic statistics such as assembly score, number of contigs, and N50. To examine the RNA-Seq read representation of the assembly, we mapped the reads used to generate the de novo assembly against the assembly using bowtie2 (v. 2.3.4.3) [64]. TransDecoder (Galaxy Version 3.0.1), with default settings, was used to predict coding regions within the transcriptome and remove redundancy. The single best open reading frame (ORF) per transcript longer than 100 peptides was used. CD-HIT-EST (v. 4.7) [65] was used with 95% similarity to reduce transcript redundancy and produce unique genes (henceforth “transcripts”). Benchmarking Universal Single-Copy Orthologs (BUSCO) [66], part of OmicsBox (v. 1.2.3, https://www.biobam.com/omicsbox, accessed on 26 July 2021), was used to validate the transcriptome assembly. Functional annotation of the whole de novo transcriptome was performed using Blast2GO (v. 5.2.5) [67]. The sequences were blasted against NCBI NR protein database (March, 2020) and were scanned with InterPro and mapped against the PSD, UniProt, Swiss-Prot, TrEMBL, RefSeq, GenPept, and PDB databases. The reads that originated the transcriptome were deposited in NCBI Sequence Read Archive under the accession number PRJNA744401.
4.2. Phylogenetic Analysis
Castanea sativa MADS-box transcript sequences were obtained using the blast+ software [68] by performing a BLAST in the C. sativa de novo transcriptome using A. thaliana sequences as query. Homologous protein sequences from other selected species were obtained by performing a BLASTx at the NCBI database (https://www.ncbi.nlm.nih.gov/, accessed on 26 July 2021), Hardwood Genomics (https://www.hardwoodgenomics.org/, accessed on 26 July 2021) and URGI (https://urgi.versailles.inra.fr/, accessed on 26 July 2021). Protein sequences were aligned using Clustal Omega [69], and distances were estimated using the Jones–Taylor–Thornton (JTT) model of evolution for a maximum likelihood tree with the MEGA X software [70]. A phylogenetic tree was generated from 1000 bootstrap datasets in order to provide statistical support for each node.
4.3. qRT-PCR Analysis
cDNA was synthesised using the Invitrogen cDNA synthesis kit SuperScript IV, according to manufacturer’s instructions. cDNA amplification was carried out using SsoFast EvaGreen Supermix (BioRad), 250 nM of each gene-specific primer, and 1 μL of 1/25 diluted cDNA. Quantitative real-time PCR (qRT-PCR) reactions were performed on the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Gene expression analysis was based on three biological and three technical replicates, and was normalised with the reference gene CsaPP2AA3 [71]. Following an initial period of 3 min/95 °C, each cycle (40 in total) consisted of a denaturation step of 10 s at 95 °C and annealing step of 10 s at specific primer temperature. Following the 40 cycles, a melting curve was obtained: the amplification products were heated in a gradient ranging from 65 to 95 °C in 5 s intervals. The primers used in the expression analysis are listed in Table S4.
4.4. Histological Analysis
For histological analysis, the flowers were collected and immediately fixed in 4% (w/v) paraformaldehyde in 1× PBS, under vacuum infiltration, followed by overnight incubation in the fixative solution at 4 °C. Samples were dehydrated, cleared, and embedded according to the protocol described by Coen et al. [72]. Tissue sections with 8 μm thickness were obtained using a microtome (SLEE) and mounted in pre-coated poly-L-lysine slides (VWR). The sections were deparaffined and stained according to Viejo et al. [73], with modifications.
4.5. Yeast-2-Hybrid Analysis
Protein–protein interactions were analysed using a GAL4-based yeast hybrid system (Matchmaker two-hybrid system; Clontech, Kusatsu, Shiga, Japan). Competent cells from Saccharomyces cerevisiae strain AH-109 were transformed with pGBT9 (bait vector; Clontech) and pGAD424 (prey plasmid; Clontech, Kusatsu, Shiga, Japan) derivatives, using the LiAc/DNA/PEG transformation method. Self-activation assays and selection of positive interactors were performed according to Causier and Davies [74]. To check for interaction, double transformations were selected first in SD medium without leucine and tryptophan and then in SD medium without leucine, tryptophan, and histidine supplemented with 3-aminotriazole. The primers used to construct the clones are listed in Table S4.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/plants10081538/s1, Figure S1: GO functional annotation of the Castanea sativa de novo transcriptome. Figure S2: Alignment of ABCDE-like amino acid sequences. Figure S3: Motif similarity in B- and C- MADS-box proteins between C. sativa and A. thaliana, Q. suber and C. mollissima (or S. lycopersicum in the case of TM6). Table S1: De novo assembly statistics of Castanea sativa transcriptome before and after redundancy removal. Table S2: Castanea sativa transcriptome completeness as determined by Benchmarking Universal Single-Copy Orthologous (BUSCO). Table S3: List of gene accessions. Table S4: List of primers.
Author Contributions
A.T.A. and R.S. designed and performed most of the experiments. M.R. and M.J.N.R. contributed to the transcriptome assembly and bioinformatics analysis. S.A., L.M.-C. and J.G.-L. contributed to the phenology characterisation. M.M.R.C. obtained funding, and designed and supervised all the experiments. A.T.A. and R.S. wrote the manuscript, with contributions from all the authors. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by FCT/COMPETE/FEDER with the project grant POCI-01-0145-FEDER-027980/PTDC/ASP-SIL/27980/2017—“FlowerCAST—Characterisation of genetic and environmental determinants involved in reproductive development of Castanea sativa”. A.T.A. and S.A. were supported by FCT with PhD grants (ref. SFRH/BD/136834/2018 and SFRH/BD/146660/2019, respectively).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The generated de novo reads used in this study are openly available in NCBI Sequence Read Archive under the accession number PRJNA744401.
Acknowledgments
The authors would like to acknowledge the Galaxy Project for the freely available resources that made the C. sativa transcriptome assembly possible. The authors are also grateful to Isaura Castanheiro for allowing access to the chestnut orchard.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Charlesworth, B.; Charlesworth, D. A Model for the Evolution of Dioecy and Gynodioecy. Am. Nat. 1978, 112, 975–997. [Google Scholar] [CrossRef]
- Kremer, A.; Abbott, A.G.; Carlson, J.E.; Manos, P.S.; Plomion, C.; Sisco, P.; Staton, M.E.; Ueno, S.; Vendramin, G.G. Genomics of Fagaceae. Tree Genet. Genomes 2012, 8, 583–610. [Google Scholar] [CrossRef] [Green Version]
- Rocheta, M.; Sobral, R.; Magalhães, J.; Amorim, M.I.; Ribeiro, T.; Pinheiro, M.; Egas, C.; Morais-Cecílio, L.; Costa, M.M.R. Comparative transcriptomic analysis of male and female flowers of monoecious Quercus suber. Front. Plant Sci. 2014, 5, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sobral, R.; Costa, M.M.R. Role of floral organ identity genes in the development of unisexual flowers of Quercus suber L. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Sobral, R.; Silva, H.G.; Laranjeira, S.; Magalhães, J.; Andrade, L.; Alhinho, A.T.; Costa, M.M.R. Unisexual flower initiation in the monoecious Quercus suber L.: A molecular approach. Tree Physiol. 2020, 40, 1260–1276. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genes directing flower development in Arabidopsis. Plant Cell 1989, 1, 37–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyerowitz, E.M.; Bowman, J.L.; Brockman, L.L.; Drews, G.N.; Jack, T.; Sieburth, L.E.; Weigel, D. A genetic and molecular model for flower development in Arabidopsis thaliana. Development 1991, 112, 157–167. [Google Scholar] [CrossRef]
- Bowman, J.L.; Alvarez, J.; Weigel, D.; Meyerowitz, E.M.; Smyth, D.R. Control of flower development in Arabidopsis thaliana by APETALA 1 and interacting genes. Development 1993, 119, 721–743. [Google Scholar] [CrossRef]
- Flanagan, C.A.; Hu, Y.; Ma, H. Specific expression of the AGL1 MADS-box gene suggests regulatory functions in Arabidopsis gynoecium and ovule development. Plant J. 1996, 10, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATTA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Liljegren, S.J.; Ditta, G.S.; Eshed, Y.; Savidge, B.; Bowmant, J.L.; Yanofsky, M.F. SHATTERPROOF MADS-box genes control dispersal in Arabidopsis. Nature 2000, 404, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Theißen, G.; Gramzow, L. Structure and Evolution of Plant MADS Domain Transcription Factors; Academic Press: Cambridge, MA, USA, 2016; ISBN 9780128011270. [Google Scholar]
- Theißen, G.; Melzer, R.; Ruümpler, F. MADS-domain transcription factors and the floral quartet model of flower development: Linking plant development and evolution. Development 2016, 143, 3259–3271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaufmann, K.; Melzer, R.; Theißen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Trobner, W.; Ramirez, L.; Motte, P.; Hue, I.; Huijser, P.; Lonnig, W.E.; Saedler, H.; Sommer, H.; Schwarz-Sommer, Z. GLOBOSA: A homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 1992, 11, 4693–4704. [Google Scholar] [CrossRef] [PubMed]
- McGonigle, B.; Bouhidel, K.; Irish, V.F. Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev. 1996, 10, 1812–1821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riechmann, J.L.; Krizek, B.A.; Meyerowitz, E.M. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 1996, 93, 4793–4798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenser, T.; Theißen, G.; Dittrich, P. Developmental robustness by obligate interaction of class B floral homeotic genes and proteins. PLoS Comput. Biol. 2009, 5. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, M. New Interpretation of the Inflorescence of Fagus Drawn From the Developmental Study of Fagus Crenata, With Description of an Extremely Monstrous Cupule. Am. J. Bot. 1989, 76, 14–22. [Google Scholar] [CrossRef]
- Ducousso, A.; Michaud, H.; Lumaret, R. Reproduction and gene flow in the genus Quercus L. Ann. Des. Sci. For. 1993, 50, 91s–106s. [Google Scholar] [CrossRef] [Green Version]
- Varela, M.C.; Valdiviesso, T. Phenological phases of Quercus suber L. flowering. For. Genet. 1996, 3, 93–102. [Google Scholar]
- Langdon, L.M. Ontogenetic and Anatomical Studies of the Flower and Fruit of the Fagaceae and Juglandaceae. Bot. Gaz. 1939, 101, 301–327. [Google Scholar] [CrossRef]
- Kubitzki, K.; Fagaceae, B.T. Flowering Plants Dicotyledons: Magnoliid, Hamamelid and Caryophyllid Families; Kubitzki, K., Rohwer, J.G., Bittrich, V., Eds.; Springer: Berlin/Heidelberg, Germany, 1993; ISBN 978-3-662-02899-5. [Google Scholar]
- Mellano, M.G.; Beccaro, G.L.; Donno, D.; Marinoni, D.T.; Boccacci, P.; Canterino, S.; Cerutti, A.K.; Bounous, G. Castanea spp. biodiversity conservation: Collection and characterization of the genetic diversity of an endangered species. Genet. Resour. Crop Evol. 2012, 59, 1727–1741. [Google Scholar] [CrossRef]
- Bounous, G.; Marinoni, D.T. Chestnut: Botany, Horticulture, and Utilization. Hortic. Rev. (Am. Soc. Hortic. Sci). 2004, 291–347. [Google Scholar]
- Valdiviesso, T. Estudo Sobre a Reprodução Sexuada e Caracterização de Cultivares de Castanea sativa Mill.; Instituto Nacional de Investigação Agrária: Oeiras, Portugal, 1999. [Google Scholar] [CrossRef]
- Botta, R.; Vergano, G.; Me, G.; Vallania, R. Floral biology and embryo development in chestnut (Castanea sativa Mill.). HortScience 1995, 30, 1283–1286. [Google Scholar] [CrossRef] [Green Version]
- Feijó, J.A.; Certal, A.C.; Boavida, L.; Van Nerum, I.; Valdiviesso, T.; Oliveira, M.M.; Broothaerts, W. Advances on the Study of Sexual Reproduction in the Cork-Tree (Quercus suber L.), Chestnut (Castanea sativa Mill.) and in Rosaceae (Apple and Almond). Fertil. High. Plants 1999, 377–396. [Google Scholar] [CrossRef]
- Soylu, A.; Ayfer, M. Floral biology and fruit set of some chestnut cultivars (C. sativa Mill.). Proc. Int. Congr. Chestnut Spoleto Italy 1993, 28, 125–130. [Google Scholar]
- Dinis, L.T.; Ramos, S.; Gomes-Laranjo, J.; Peixoto, F.; Costa, R.; Vallania, R.; Botta, R. Phenology and reproductive biology in cultivar “Judia” (Castanea sativa Mill.). Acta Hortic. 2010, 866, 169–174. [Google Scholar] [CrossRef]
- Fang, G.C.; Blackmon, B.P.; Staton, M.E.; Nelson, C.D.; Kubisiak, T.L.; Olukolu, B.A.; Henry, D.; Zhebentyayeva, T.; Saski, C.A.; Cheng, C.H.; et al. A physical map of the Chinese chestnut (Castanea mollissima) genome and its integration with the genetic map. Tree Genet. Genomes 2013, 9, 525–537. [Google Scholar] [CrossRef] [Green Version]
- Xing, Y.; Liu, Y.; Zhang, Q.; Nie, X.; Sun, Y.; Zhang, Z.; Li, H.; Fang, K.; Wang, G.; Huang, H.; et al. Hybrid de novo genome assembly of Chinese chestnut (Castanea mollissima). Gigascience 2019, 8, 1–7. [Google Scholar] [CrossRef]
- Ainsworth, C.; Rahman, A.; Parker, J.; Edwards, G. Intersex inflorescences of Rumex acetosa demonstrate that sex determination is unique to each flower. New Phytol. 2005, 165, 711–720. [Google Scholar] [CrossRef]
- Di Stilio, V.S.; Kramer, E.M.; Baum, D.A. Floral MADS box genes and homeotic gender dimorphism in Thalictrum dioicum (Ranunculaceae)—A new model for the study of dioecy. Plant J. 2005, 41, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Sather, D.N.; Jovanovic, M.; Golenberg, E.M. Functional analysis of B and C class floral organ genes in spinach demonstrates their role in sexual dimorphism. BMC Plant Biol. 2010, 10, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coutinho, X.P. Esboço de uma flora lenhosa Portuguesa. Publ. Serv. Florest. Port. 1936, 3, 60–62. [Google Scholar]
- Guerreiro, M.G. Alguns estudos no género Castanea. Publ. Serv. Florest. Port. 1948, 15, 10–15. [Google Scholar]
- Kientzler, L. Cas d’hermaphroditisme chez le châtaignier. Bull. Soc. Bot. France 1959, 106, 211–212. [Google Scholar] [CrossRef]
- Solignat, G. Recherches sur le châtaignier à la St. de Brive. INRA 1952, 3, 15–35. [Google Scholar]
- Solignat, G. Un renouveau de la châtaigneraie fruitière. INRA Cent. Rech. Agron. Bordeaux. Extr. Du Boll. Tech. D’Inf. 1973, 280, 280–281. [Google Scholar]
- Shi, Z.; Stösser, R. Reproductive biology of Chinese chestnut (Castanea mollissima Blume). Eur. J. Hortic. Sci. 2005, 70, 96–103. [Google Scholar]
- Guo, S.J.; Zou, F. Observation on the pistillate differentiation of Chestnut (Castanea) cultivar “Yanshanzaofeng”. J. Chem. Pharm. Res. 2014, 6, 686–690. [Google Scholar]
- Fan, X.; Yuan, D.; Tian, X.; Zhu, Z.; Liu, M.; Cao, H. Comprehensive Transcriptome Analysis of Phytohormone Biosynthesis and Signaling Genes in the Flowers of Chinese Chinquapin (Castanea henryi). J. Agric. Food Chem. 2017, 65, 10332–10349. [Google Scholar] [CrossRef] [PubMed]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 Gene of Arabidopsis thaliana Functions in Floral Organ and Meristem Identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunner, A.M.; Rottmann, W.H.; Sheppard, L.A.; Krutovskii, K.; DiFazio, S.P.; Leonardi, S.; Strauss, S.H. Structure and expression of duplicate AGAMOUS orthologues in poplar. Plant Mol. Biol. 2000, 44, 619–634. [Google Scholar] [CrossRef]
- Lemmetyinen, J.; Hassinen, M.; Elo, A.; Porali, I.; Keinonen, K.; Mäkelä, H.; Sopanen, T. Functional characterization of SEPALLATA3 and AGAMOUS orthologues in silver birch. Physiol. Plant. 2004, 121, 149–162. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, Z.; Yong, X.; Ahmad, S.; Yang, W.; Cheng, T.; Wang, J.; Zhang, Q. SEP-class genes in Prunus mume and their likely role in floral organ development. BMC Plant Biol. 2017, 17, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Favaro, R.; Pinyopich, A.; Battaglia, R.; Kooiker, M.; Borghi, L.; Ditta, G.; Yanofsky, M.F.; Kater, M.M.; Colombo, L. MADS-Box Protein Complexes Control Carpel and Ovule Development in Arabidopsis. Plant Cell 2003, 15, 2603–2611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Z.; Zhuo, S.; Liu, X.; Che, G.; Wang, Z.; Gu, R.; Shen, J.; Song, W.; Zhou, Z.; Han, D.; et al. The MADS-Box Gene CsSHP Participates in Fruit Maturation and Floral Organ Development in Cucumber. Front. Plant Sci. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fourquin, C.; Ferrándiz, C. Functional analyses of AGAMOUS family members in Nicotiana benthamiana clarify the evolution of early and late roles of C-function genes in eudicots. Plant J. 2012, 71, 990–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, Y.; Liu, W.; Chen, X.; Xu, Y.; Lu, W.; Hou, J.; Ni, J.; Wang, Y.; Wu, L. Flower development and sex determination between male and female flowers in Vernicia fordii. Front. Plant Sci. 2017, 8, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Li, W.; Zhao, G.; Fan, X.; Long, H.; Fan, Y.; Shi, M.; Tan, X.; Zhang, L. New Insights of Salicylic Acid into Stamen Abortion of Female Flowers in Tung Tree (Vernicia fordii). Front. Genet. 2019, 10, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Wang, Y.; Chen, Y.; Yin, H.; Wu, L.; Zhao, Y.; Wang, M.; Gao, M. A model of hormonal regulation of stamen abortion during pre-meiosis of Litsea cubeba. Genes 2020, 11, 48. [Google Scholar] [CrossRef] [Green Version]
- Ooi, S.E.; Sarpan, N.; Aziz, N.A.; Nuraziyan, A.; Nor-Azwani, A.B.; Ong-Abdullah, M. Transcriptomics of microdissected staminodes and early developing carpels from female inflorescences of Elaeis guineensis. J. Oil Palm Res. 2020, 32, 559–568. [Google Scholar] [CrossRef]
- Ferreira-cardoso, J.; Portela, E.; Abreu, C.G. Castanheiros; Universidade de Trás-os-Montes e Alto Douro: Vila Real, Portugal, 2007; ISBN 9789726698449. [Google Scholar]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Le Provost, G.; Herrera, R.; Paiva, J.A.; Chaumeil, P.; Salin, F.; Plomion, C. A micromethod for high throughput RNA extraction in forest trees. Biol. Res. 2007, 40, 291–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serrazina, S.; Santos, C.; Machado, H.; Pesquita, C.; Vicentini, R.; Pais, M.S.; Sebastiana, M.; Costa, R. Castanea root transcriptome in response to Phytophthora cinnamomi challenge. Tree Genet. Genomes 2015, 11, 6. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinform.babraham.ac.uk/proj./Fastqc/2010 (accessed on 26 July 2021).
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner. In Proceedings of the 9th Annual Genomics of Energy & Environment Meeting, Walnut Creek, CA, USA, 17–20 March 2014. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Smith-Unna, R.; Boursnell, C.; Patro, R.; Hibberd, J.M.; Kelly, S. TransRate: Reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 2016, 26, 1134–1144. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Simao, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO online supplementary information: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [Green Version]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Marum, L.; Miguel, A.; Ricardo, C.P.; Miguel, C. Reference gene selection for quantitative real-time PCR normalization in Quercus suber. PLoS ONE 2012, 7, e35113. [Google Scholar] [CrossRef]
- Coen, E.S.; Romero, J.M.; Doyle, S.; Elliott, R.; Murphy, G.; Carpenter, R. floricaula: A homeotic gene required for flower development in antirrhinum majus. Cell 1990, 63, 1311–1322. [Google Scholar] [CrossRef]
- Viejo, M.; Rodríguez, R.; Valledor, L.; Pérez, M.; Cañal, M.J.; Hasbún, R. DNA methylation during sexual embryogenesis and implications on the induction of somatic embryogenesis in Castanea sativa Miller. Sex Plant Reprod. 2010, 23, 315–323. [Google Scholar] [CrossRef]
- Causier, B.; Davies, B. Analysing protein-protein interactions with the yeast two-hybrid system. Plant Mol. Biol. 2002, 50, 855–870. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).