Abstract
Estrogens, which bind to estrogen receptor alpha (ERα), are important for proper bone mineral density. When women go through menopause, estrogen levels decrease, and there is a decrease in bone quality, along with an increased risk for fractures. We previously identified an enhancer near FOXC1 as the most significantly enriched binding site for estrogen receptor alpha (ERα) in osteoblasts. FOXC1 is a transcription factor belonging to a large group of proteins known as forkhead box genes and is an important regulator of bone formation. Here, we demonstrate that 17β-estradiol (E2) increases the mRNA and protein levels of FOXC1 in primary mouse and human osteoblasts. GATA4 is a pioneer factor for ERα and it is also recruited to enhancers near Foxc1. Knockdown of Gata4 in mouse osteoblasts in vitro decreases Foxc1 expression as does knockout of Gata4 in vivo. Functionally, GATA4 and FOXC1 interact and regulate osteoblast proteins such as RUNX2, as demonstrated by ChIP-reChIP and luciferase assays. The most enriched motif in GATA4 binding sites from ChIP-seq is for FOXC1, supporting the notion that GATA4 and FOXC1 cooperate in regulating osteoblast differentiation. Together, these data demonstrate the interactions of the transcription factors ERα, GATA4, and FOXC1 to regulate each other’s expression and other osteoblast differentiation genes.
1. Introduction
Transcription factors bind to enhancers and promoters to regulate gene expression. Tissue-specific gene regulation is controlled in part by the spatiotemporal and combinatorial binding of transcription factors. There are many known transcription factors in bone, including RUNX2, estrogen receptor alpha (ERα), GATA4, and FOXC1. Herein, we describe the transcriptional regulation of Foxc1 and Runx2 in bone that is mediated by ERα, GATA4, and FOXC1.
Estrogens are important for maintaining bone mineral density in both mice and humans via estrogen receptor alpha (ERα) []. Estrogen signaling regulates osteoblasts, osteoclasts, and the immune system to maintain bone mass. Specifically in osteoblasts, ERα inhibits apoptosis and induces the expression of differentiation genes such as BMP2, osteoprotegerin (OPG), and alkaline phosphatase [,].
GATA transcription factors, so named because they bind to the consensus DNA sequence (A/T)GATA(A/G), have been identified as important regulators of tissue-specific gene expression during development. GATA4 is necessary for heart, liver, and osteoblast differentiation, among other tissues [,]. Bone-specific deletion of GATA4 leads to embryonic lethality []. GATA4 recruits ERα to estrogen-regulated genes including alkaline phosphatase []. In addition, there are estrogen-independent functions of GATA4. Knockout of Gata4 in osteoblasts using the Prx1, Runx2, or OCN (Bglap) promoters driving Cre-recombinase leads to trabecular bone loss [,,]. GATA4 regulates many osteoblast differentiation genes, including Runx2, bone sialoprotein, and osteocalcin [].
A pioneer factor is a protein that can bind to the genome in sites of condensed chromatin, leading to nucleosome remodeling and the opening of chromatin, the recruitment of additional transcription factors, and the activation of transcription []. GATA4 has been shown to be a pioneer factor in the liver, heart, and osteoblasts [,,]. In the liver, this activity depends on FOXA proteins for stable nucleosome binding [], as FOXA proteins can bind to closed chromatin and displace histones, leading to open and active enhancers. The binding of FOXA proteins to the DNA allows for GATA4 recruitment in liver cells []. However, FOXA1 is not expressed in osteoblasts [], suggesting a role for a different forkhead protein to recruit GATA4 to chromatin in osteoblasts.
There are 50 human and 44 mouse forkhead proteins, categorized from FoxA to FoxS []. FOXC1 has been shown to be important for skeletal development. Mutations in FOXC1 in humans cause Axenfeld–Rieger Syndrome, characterized by ocular defects but also skeletal abnormalities []. A spontaneous loss-of-FOXC1-function mutant mouse (named Foxc1ch/ch because it has congenital hydrocephalus) dies at birth and has skull and axial and appendicular bone defects [,], demonstrating a role for FOXC1 in intramembranous and endochondral bone formation. Chondrocyte-specific deletion of Foxc1 and Foxc2 leads to disrupted endochondral ossification and skeletal dysplasia []. Mechanistically, FOXC1 has been shown to regulate osteoblast differentiation by regulating Runx2, the master transcription factor for osteoblastogenesis [,].
For the first time, we identified key regulators of Foxc1 regulation and functions for FOXC1 in osteoblasts. Importantly, GATA4 and FOXC1 interact in osteoblasts to directly regulate Runx2.
2. Material and Methods
2.1. Mice
Animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Tennessee Health Science Center. Animals were maintained in a specific pathogen-free environment at 20–26 °C with a relative humidity of 30–70% and a 12 h light/dark cycle. Commercial rodent chow (LM-485, Teklad, Madison, WI, USA) and drinking water were available ad libitum.
GATA4-Flag-biotin mice (Flag-bio, Gt(ROSA)26Sortm1(birA)Mejr Gata4tm3.1 Wtp/J) [] were obtained from Jackson Labs (stock #018121). GATA4 Prx-cKO mice were previously described [].
Six-week-old female BALB/c mice were sham-operated or ovariectomized and then treated with 50 mg/kg body weight of E2 or sesame oil (as a control) by intraperitoneal injection for 24 h.
ERα, ERβ, and ERαβ heterozygous mice were kindly provided by Dr. Pierre Chambon []. ERα+/−ERβ+/− mice were bred to produce WT, ERαKO, and ERαβKO littermates.
Mouse calvarial osteoblasts were isolated from 2-day-old CD1 or Flag-bio mice by sequential collagenase digestion []. The cells were incubated for 40 min in α-MEM with 1.0 mg/mL collagenase P and 1.25% trypsin at 37 °C. The cells were then washed in α-MEM and then incubated in α-MEM-1.0 mg/mL collagenase P-1.25% trypsin for 1 h at 37 °C. Collagenase digestion was stopped by the addition of complete α-MEM media containing 10% FBS. The cells from the second digest were obtained and allowed to proliferate in α-MEM media containing 10% FBS. Differentiation was induced in α-MEM media containing 5 mM β-glycerophosphate and 100 mg/mL ascorbic acid (mineralization medium) for 14 days.
2.2. Human Cells
The human study was approved by the Institutional Review Board at the University of Tennessee Health Science Center, and all individuals provided informed written consent before participation. Primary human osteoblasts were isolated from the trabecular bone in the femoral or humeral heads of individuals who underwent total joint replacement surgery, as described previously [].
2.3. Cell Lines
Human osteosarcoma U2OS-ERα cells were kindly provided by Dr. Thomas Spelsberg and were maintained as described []. ERα expression was induced by treatment with 100 ng/mL doxycycline (DOX; Sigma-Aldrich Co., St. Louis, MO, USA) for 24 h after culture in phenol-red-free media containing 5% CDT-FBS for three days. Cells were treated with 10 nM E2 or vehicle control (ethanol, EtOH). The cell line was verified each year by STR profiling and tested for mycoplasma.
2.4. shGATA4
Lentivirus shC (a short hairpin that does not recognize any mammalian DNA) and shGATA4 were purchased from Sigma Aldrich. Two different shRNAs from the RNAi Consortium (TRC) in the pLKO vector were used to knockdown mouse Gata4 (TRCN0000095215:
CCGGCCCAATCTCGATATGTTTGATCTCGAGATCAAACATATCGAGATTGGGTTTTTG and TRCN0000095217: CCGGCATCTCCTGTCACTCAGACATCTCGAGATGTCTGAGTGACAGGAGATGTTTTTG). The knockdown of Gata4 was confirmed by qPCR.
2.5. RNA and qPCR
RNA and qPCR were performed as previously described []. The specific primers used for SYBR Green assays are listed in Supplemental Table S1. For analysis of the cDNA data, the values were normalized to β-actin (Actb) values.
2.6. Immunoprecipitation and Immunoblotting
Immunoprecipitation (IP) reactions were conducted on whole-cell lysates prepared in standard RIPA buffer extracted from calvarial osteoblasts. IPs were performed with antibodies to GATA4 (Clone G-4, Santa Cruz Biotechnology, Dallas, TX, USA) or normal mouse IgG, along with Protein G magnetic beads (Invitrogen, ThermoFisher, Waltham, MA, USA). Immunoblotting was performed with antibodies to FOXC1 (Cell Signaling Technology, clone D8A6, Danvers, MA, USA) and β-actin (Cell Signaling Technology, clone 8H10D10). Whole blots are shown in Supplemental Figure S1.
2.7. Chromatin Immunoprecipitation (ChIP)
ChIP was performed as previously described for U2OS cells [] and Flag-biotin-tagged GATA4 []. ChIP-sequencing was previously described []. Motif analysis was performed using Meme Suite []. FOXC1 was immunoprecipitated with a goat polyclonal antibody (Abcam, ab5079). The specific primers for SYBR Green assays used for qPCR are listed in Supplemental Table S1. All data were compared to the percent input and a negative control region [].
2.8. ChIP-reChIP
ChIP was performed as described above, except that after the first overnight incubation with the first antibody, the magnetic beads were washed 3 times with PBS/BSA and then resuspended in TE with 10 mM DTT for 30 min. The eluate was resuspended in ChIP dilution buffer, and the second antibody was added overnight.
2.9. Immunohistochemistry (IHC)
Formalin-fixed, paraffin-embedded samples were processed through standard deparaffinization protocols. Antigen retrieval was performed by placing the slides in 90 °C citrate buffer and leaving them at room temperature until the temperature reached 55 °C. The tissue was then incubated in blocking buffer (5% normal goat serum and 2.5% bovine serum albumin [BSA] in PBS at pH 7.5) for 30 min. Anti-FOXC1 (LSBio catalog #LS-B1800, Shirley, MA, USA) antibodies were incubated overnight at 4 °C in a humidified chamber, followed by the DAKO Envision Visualization system (Agilent, Santa Clara, CA, USA) and counterstaining with hematoxylin. Immunohistochemistry samples were scored without bias by three independent people using a bone-specific immune reactive scoring index of 0 (negative) -12 (strongly positive), multiplying the percent of positive cells by the intensity of staining [].
2.10. Luciferase Assay
U2OS osteosarcoma cells were plated in 24-well tissue culture dishes and transfected with the rat 0.6 kb Runx2-promoter-luciferase (a kind gift from Dr. Gary S. Stein) [], pcDNA3-Gata4 (a kind gift from Dr. Michael Parmacek []), and pRL-SV40 (Promega, Madison, WI, USA) as a transfection control. pcDNA3.1-DYK-Foxc1 was purchased from GenScript.
2.11. Cistrome Data Browser
The Cistrome Data Browser [,] was used to search for factors 100 kb from the transcriptional start site of Foxc1. The datasets that were used are listed below in the Data Availability section.
2.12. FOXC1 Expression in Osteoporotic Patients
The expression of FOXC1 in Human Mesenchymal Stem Cells from old, age-matched osteoporotic and non-osteoporotic individuals was obtained in the GEO dataset GSE35959 [].
2.13. Statistical Analysis
All experiments represent both biological and experimental triplicates. Unless otherwise stated, error bars represent the mean ± 1 standard deviation. Statistical analyses, including Student’s t-test, were performed using GraphPad Prism® (version 9, Boston, MA, USA) software.
3. Results
3.1. The Transcriptional Regulation of Foxc1
To search for estrogen-regulated genes in osteoblasts, we analyzed our ERα binding sites in U2OS-ERα cells after 45 min. of 17β-estradiol (E2) treatment []. U2OS-ERα cells are an osteosarcoma cell line that has an inducible expression of ERα. Overexpression of ERα in U2OS cells leads to an osteoblast phenotype that serves as a model for normal osteoblasts [,,,]. The ERα binding site with the highest enrichment was near FOXC1 and FOXC1 has been shown to be important for osteoblast biology [,,]. Therefore, we first searched the literature for transcription factors that could regulate FOXC1 in bone cells. All of the papers investigated murine Foxc1 regulation. SOX9 was shown to bind 43 kb upstream of Foxc1 (site C, Figure 1A,B) by ChIP-sequencing in chondrocytes []. In addition, phosphorylated SMAD1,5,9 was shown to bind 839 to 384 bp upstream of the Foxc1 transcriptional start site [] (site D, Figure 1A,B). (ChIP-sequencing was not performed to look for additional sites for pSMAD1,5,9 recruitment).
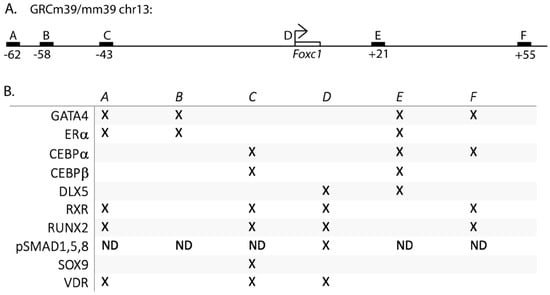
Figure 1.
Transcription factors regulating Foxc1. (A) Schematic of enhancers near Foxc1. (B) “X” indicates the presence of the transcription factor at the indicated enhancer or promoter by ChIP-sequencing and/or ChIP-qPCR. ND = not determined.
Next, we retrospectively searched our ERα (translated to the murine genome) and GATA4 ChIP-sequencing data in combination with publicly available ChIP-seq data obtained from the Cistrome Data Browser for additional transcription factors that bind near Foxc1 in MSCs, osteoblasts, and chondrocytes. We identified additional enhancers near Foxc1 bound by CEBPα, CEBPβ, DLX5, RUNX2, RXR, and VDR (Figure 1A,B). Each of these proteins has been shown to have important roles in bone development. Interestingly, the location and number of binding sites are different for each transcription factor, demonstrating complex regulation involving over 100 kb.
3.2. Estrogen Regulates Foxc1 in Osteoblasts
To verify the binding sites for ERα near Foxc1, primary mouse calvarial osteoblasts were treated with 10 nM E2 for 45 min., and ChIP was performed. qPCR primers were designed for these predicted enhancers and the promoter of Foxc1. Indeed, after E2 treatment, ERα was enriched at enhancers A, B, and E (Figure 2A), compared to untreated cells or a negative control (NC) genomic region.
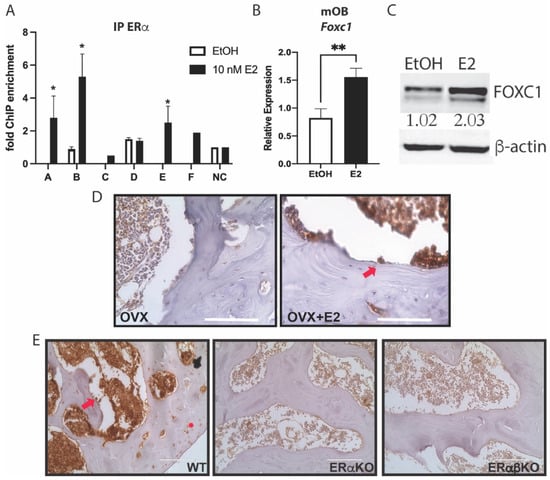
Figure 2.
ERα regulates Foxc1. (A) Primary calvarial osteoblasts were treated with ethanol (EtOH) as a vehicle control or 10 nM E2 for 45 min before being fixed with 4% PFA. ChIP was performed with an antibody to ERα. qPCR was performed at the indicated regions near Foxc1 or a negative control (NC) region. (B) Primary calvarial osteoblasts were treated with EtOH or 10 nM E2 for 24 h. RNA was obtained, and qPCR was performed with primers for Foxc1 and normalized to Actb mRNA. (C) Primary calvarial osteoblasts were treated with 10 nM E2 for 24 h. Protein was obtained, and immunoblotting was performed with antibodies to FOXC1 and β-actin. The bands were quantified and normalized to β-actin and are displayed under the FOXC1 band. (D) Immunohistochemistry was performed with an antibody to FOXC1 (brown) on femurs from ovariectomized mice treated with or without 50 mg/kg body weight of E2 for 24 h. The slides were co-stained with hematoxylin. The arrow indicates an example of a FOXC1+ osteoblast. Bar = 100 μm. (E) Immunohistochemistry was performed with an antibody to FOXC1 (brown) on femurs from wild-type, ERαKO, and ERαβKO mice and co-stained with hematoxylin. The arrow indicates an example of a FOXC1+ osteoblast. * indicates an example of a FOXC1+ chondrocyte. Bar = 100 μm. * = p value < 0.05, ** = p value < 0.01.
To determine if E2 induced expression of Foxc1 after ERα binding, calvarial osteoblasts were treated with 10 nM E2 for 24 h. RNA and protein were obtained and analyzed for the expression of Foxc1. qPCR (Figure 2B) and immunoblotting (Figure 2C) both demonstrate that E2 induced a 2-fold increase in Foxc1 expression.
Next, we analyzed the effect of E2 on FOXC1 in bone in vivo. Mice were ovariectomized (OVX) or sham-operated and then treated with vehicle or E2 for 24 h. Immunohistochemistry was performed with an antibody to FOXC1. FOXC1 is increased in the E2-treated osteoblasts found along the trabecular bone (Figure 2D). To demonstrate that ERα is necessary for the regulation of Foxc1, immunohistochemistry for FOXC1 was also performed on the femurs from WT, ERα knockout (ERαKO), and ERα/ERβ double knockout (ERαβKO) mice. FOXC1 staining was quantified by the number of positive cells and the intensity of staining. FOXC1 was markedly reduced in the ERαKO and ERαβKO mice (Figure 2E and Supplemental Figure S2).
Together, these experiments demonstrate that E2 and ERα directly up-regulate Foxc1 expression by binding to Foxc1 enhancers.
3.3. GATA4 Regulates Foxc1 Expression
We have previously demonstrated that GATA4 is a pioneer for ERα in osteoblasts []. To determine if GATA4 also regulates Foxc1, ChIP was performed to detect GATA4 at the enhancers near Foxc1. Because of inferior antibodies to GATA4, a mouse with GATA4 tagged with the Flag epitope and a biotinylation site at the endogenous locus was created (GATA4 Flag-bio mice) and has been successful for GATA4 ChIP [,,]. Calvariae from these mice were obtained, and ChIP was performed with streptavidin beads. GATA4 was enriched at enhancers A, B, E, and F in the GATA4 Flag-bio mice (Figure 3A). GATA4 has ERα-independent functions [], as evidenced by enhancer F, which is bound by GATA4 but not ERα.
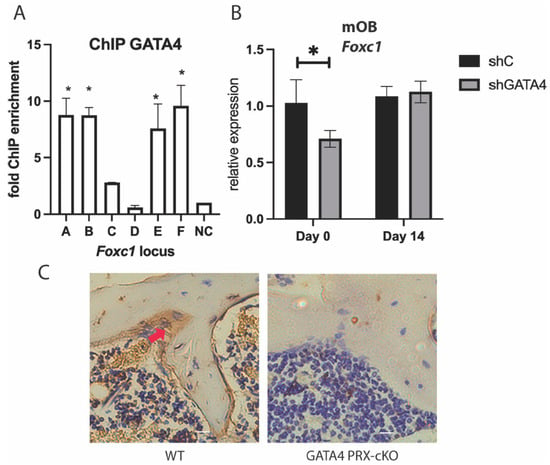
Figure 3.
GATA4 regulates Foxc1. (A) Primary calvarial osteoblasts from GATA4 Flag-bio mice or WT CD1 mice were fixed, and ChIP was performed with streptavidin beads. qPCR was performed at a region upstream and downstream of the Foxc1 gene. (B) Primary calvarial osteoblasts were exposed to lentivirus encoding shRNA for Gata4 or a negative control. RNA was obtained, and qPCR was performed with primers for Foxc1 and normalized to Actb mRNA. (C) Immunohistochemistry on femurs from wild-type or GATA4 PRX1-cKO was performed with an antibody to FOXC1 (brown) and co-stained with hematoxylin. The arrow indicates an example of a FOXC1+ osteoblast. * = p value < 0.05.
To further demonstrate that GATA4 regulates Foxc1, wild-type calvarial cells were cultured and knockdown of Gata4 was performed with lentiviral shRNA to Gata4 []. Knockdown of Gata4 correlated with a decrease in Foxc1 mRNA at day 0 of differentiation, but not at day 14, (Figure 3B), which is consistent with a role for GATA4 early in differentiation []. Conditional knockout (cKO) of Gata4 in osteoblast progenitors (using PRX1-CRE) causes osteopenia []. Immunohistochemistry with an antibody to FOXC1 revealed a significant decrease in FOXC1 protein in the cKO femurs (Figure 3C and Supplemental Figure S2). WT mice express FOXC1 along the trabecular bone and in the bone marrow, whereas the Gata4 cKO mice have lost nearly all expression of FOXC1.
Together, these experiments demonstrate that GATA4 in vivo and in vitro directly up-regulates Foxc1 expression by binding to Foxc1 enhancers.
3.4. GATA4 and FOXC1 Interact in Osteoblasts to Regulate Osteoblast Differentiation
Analysis of our GATA4 ChIP-seq data [] using Analysis of Enriched Motifs (AME) [] software revealed that the most highly enriched motif (p value < 3.95 × 10−100) in the 5983 identified GATA4 binding sites is a canonical forkhead motif. Simple Enrichment Analysis (SEA) [] also identified a forkhead motif (MA0481.1, p value < 3.15 × 10−15, Figure 4A). 79.8% of the GATA4 binding sites contained the motif MA0481.1 (Figure 4B), suggesting that FOXC1 binds with or near GATA4 to regulate osteoblast-specific genes. Because the GATA4 ChIP-seq data predicts a forkhead protein/FOXC1 at the same binding sites, we sought to determine if GATA4 and FOXC1 physically interact through direct or indirect protein–protein interactions. Co-immunoprecipitations were performed with an antibody to GATA4, FOXC1, or normal IgG and demonstrated that GATA4 and FOXC1 interact in calvarial osteoblasts (Figure 4C).
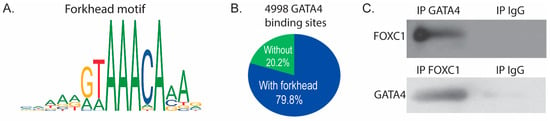
Figure 4.
FOXC1 and GATA4 interact to regulate bone genes. (A) ChIP sequencing for GATA4 was performed on calvarial osteoblasts obtained from GATA4 Flag-bio mice. GATA4 binding sites were analyzed for enriched motifs, which included the depicted motif for FOXC1. (B) 79.8% of the GATA4 binding sites contained the motif MA0481.1 (the motif depicted in part (A)). (C) Equal amounts of wild-type calvarial osteoblast protein were immunoprecipitated with FOXC1, GATA4, or normal IgG. Then immunoblots were performed to detect FOXC1 or GATA4.
We have previously shown that GATA4 binds to both promoters of Runx2 and to a downstream enhancer (Figure 5A) to regulate its expression and osteoblast differentiation []. In primary osteoblast cells, FOXC1 and GATA4 are recruited to these genomic regions, as detected by ChIP on the Runx2 enhancer 1 and on both promoters 1 and 2 (Figure 5B,C). ChIP-reChIP showed that FOXC1 and GATA4 are at the Runx2 promoters and an enhancer at the same time (Figure 5D). In addition, GATA4 enhances the transcription of a Runx2-luciferase plasmid containing the osteoblast-specific P1 promoter [], and the addition of FOXC1 enhances this transcriptional regulation (p < 0.01) (Figure 5E). These results support the notion that FOXC1 is directly enhancing GATA4 activation of Runx2 expression.
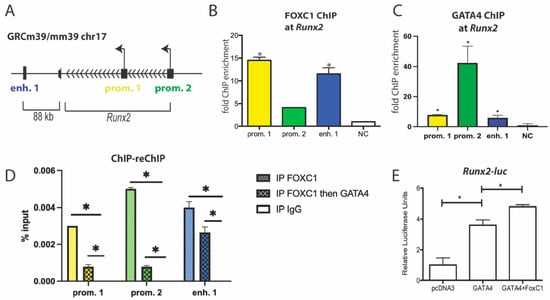
Figure 5.
GATA4 and FOXC1 regulate Runx2. (A) Schematic diagram of the mouse Runx2 genomic locus. The arrowheads indicate the direction of gene transcription. The transcriptional start sites are shown by the arrow. GATA4 binding sites are denoted at the promoters and enhancer 1. (B) ChIP was performed in primary calvarial osteoblasts with an antibody to FOXC1. qPCR was performed with primers for the indicated regions near Runx2 or a negative control genomic region. (C) ChIP was performed using calvaria from GATA4 Flag-bio mice or wild-type (WT) mice and streptavidin beads to detect GATA4. qPCR was performed with primers for the indicated regions near Runx2 or a negative control genomic region. (D) ChIP was performed on whole-cell lysate from U2OS cells with an antibody to FOXC1. ChIP-reChIP was performed first with an antibody to FOXC1 and then GATA4. IgG was used as a negative control. (E) The Runx2-promoter luciferase construct was transfected into U2OS cells along with pcDNA3-GATA4 or FOXC1, where indicated. Luciferase values were normalized to Renilla luciferase. n = 3. * = p value < 0.05.
3.5. Clinical Relevance of FOXC1 and Osteoporosis
FOXC1 has been mostly studied in murine models of bone, and little work has been conducted in human models to determine whether it is clinically important. However, we show that E2 induces FOXC1 expression not only in murine calvarial osteoblasts (Figure 2) but also in primary human osteoblasts (Figure 6A) and the U2OS-ERα model system (Supplemental Figure S3).
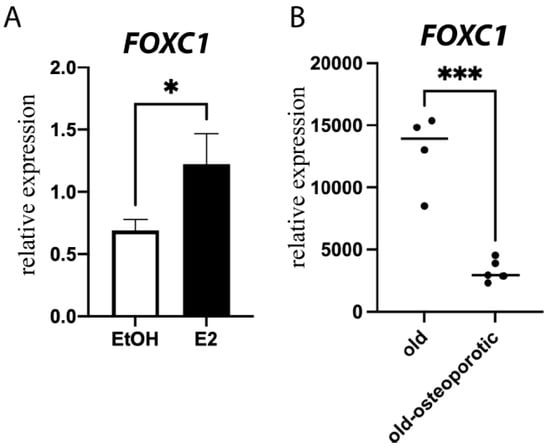
Figure 6.
FOXC1 in human cells. (A) Primary human osteoblasts were treated with vehicle control (Ethanol, EtOH) or 10 nM E2 for 24 h. RNA was obtained, and qPCR was performed for FOXC1 and normalized to ACTB. (B) GEO dataset GSE35959 was analyzed for the expression of FOXC1 in age-matched elderly individuals with or without osteoporosis. * = p value < 0.05; *** = p value < 0.001.
Because we saw a decrease in FOXC1 in ovariectomized mice, we sought to determine if there is a decrease in FOXC1 in osteoporosis. Publicly available expression arrays of mesenchymal stem cells from osteoporotic individuals demonstrate significantly lower expression of FOXC1 in comparison with age-matched elderly (79–94 years old) individuals (Figure 6B). These data suggest that FOXC1 expression is clinically relevant for osteoporosis.
4. Discussion
ERα, GATA4, and FOXC1 are important transcription factors in osteoblast differentiation. Here we show that the transcription factors ERα and GATA4, along with SOX9, pSMAD1,5,9, CEBPα, CEBPβ, DLX5, RUNX2, RXR, and VDR, regulate Foxc1 expression. Furthermore, we demonstrated that GATA4 and FOXC1 physically interact to regulate the master osteoblast transcription factor Runx2.
FOXC1 is critical to bone development as it controls endochondral and intramembranous ossification [,,,]. Loss-of-function of FOXC1 in mice and humans leads to developmental abnormalities in the appendicular and axial skeletons. Mechanistically, FOXC1 has been shown to regulate Runx2 [], β-catenin [,], and Msx2 [], all important regulators of skeletogenesis. In osteoblasts, FOXC1 binds to the Runx2 promoter and activates the expression of Runx2 in response to intermittent parathyroid hormone in MC3T3-E1 cells []. FOX proteins can function as either classical transcription factors, pioneer factors, or cofactors []. We show that FOXC1 interacts with GATA4 and upregulates Runx2 by binding to an enhancer and its promoters in mouse osteoblasts. FOXC1 also binds to the DKK1 promoter to suppress expression [], while GATA4 is also recruited to the DKK1 promoter and enhancer to activate WNT signaling in osteoblasts []. Moreover, we find that 79.8% of the GATA4 binding sites contained the motif MA0481.1, a forkhead recognition site. This suggests a coregulation pattern between FOXC1 and GATA4 in osteoblasts. Other osteogenic markers such as Osx, Alp, and Bglap (osteocalcin) are upregulated by FOXC1 [,]. Thus, ChIP-sequencing of FOXC1 would provide many additional gene targets in osteoblast differentiation. An analysis of histone modifications and the timing of FOXC1 and GATA4 binding would help determine if they are pioneer factors.
GATA4 and ERα show synchronized actions on osteoblast differentiation by activating the expression of RUNX2, ALPL, and BGLAP [,]. GATA4 and ERα interact and bind at either estrogen response elements or GATA binding sites for the cis-regulation of many genes []. We elucidate that GATA4 and ERα control the expression of Foxc1 by binding to enhancers in mouse osteoblasts. Mechanistically, activation of ERα increases its binding and the expression of FOXC1, while GATA4 knockdown abolishes the binding of ERα at the FOXC1 enhancer in U2OS-ERα cells []. We show multiple transcription factors, in addition to ERα and GATA4 bind upstream and downstream of Foxc1. Binding alone is not sufficient for transcriptional regulation [], so functional experiments could be done to show up- or down-regulation of Foxc1 mRNA. Furthermore, chromatin conformation assays would reveal additional complexity of the Foxc1 locus.
Osteosarcoma is a bone cancer that can be caused by osteoblast-committed cells with differentiation defects []. FOXC1 shows high expression in osteosarcoma and promotes cancer progression and metastasis [,]. RUNX2 also has high expression in osteosarcoma and correlates with metastasis and poor survival [,]. This suggests a correlation between FOXC1 and RUNX2 in osteosarcoma. Thus, understanding the function of FOXC1 in normal osteoblast differentiation may help find the role of FOXC1 in osteosarcoma and osteoporosis.
In conclusion, we show that GATA4 and FOXC1 directly promote Runx2 expression by interacting with and binding to the Runx2 enhancer and its two promoters. Furthermore, ERα and GATA4 control the expression of Foxc1, in addition to other transcription factors and cofactors.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jdb11030038/s1, Figure S1: Whole immunoblots from Figure 2 and Figure 4; Figure S2: Immunoreactivity scoring; Figure S3: Foxc1 regulation by E2 in human cells; Table S1: Primers.
Author Contributions
Conceptualization, S.A.K.; Methodology, S.S., J.L., R.S.P., G.A.M.-C. and S.A.K.; Writing—Original Draft Preparation, S.S. and S.A.K.; Writing—Review and Editing, S.S., G.A.M.-C. and S.A.K.; Funding Acquisition, S.A.K. All authors have read and agreed to the published version of the manuscript.
Funding
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number AR-064354 to SAK. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Institutional Review Board Statement
Animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Tennessee Health Science Center. The human study was approved by the Institutional Review Board at The University of Tennessee Health Science Center.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The following data was obtained from NCBI GEO: The expression of FOXC1 in Human Mesenchymal Stem Cells from old age-matched osteoporotic and not-osteoporotic individuals was obtained in the GEO dataset GSE35959.
| ChIP-seq | |
| ERα | GSE28918 |
| CEBPα | GSM2104127 |
| CEBPβ | GSM2104228 |
| DLX5 | GSM1976254 |
| RXR | GSM2104118 |
| RUNX2 | GSM2104159 |
| SOX9 | GSM826703 |
| VDR | GSM2104116 |
Conflicts of Interest
The authors declare no conflict of interest.
References
- Khalid, A.B.; Krum, S.A. Estrogen receptors alpha and beta in bone. Bone 2016, 87, 130–135. [Google Scholar] [CrossRef]
- Krum, S.A. Direct transcriptional targets of sex steroid hormones in bone. J. Cell. Biochem. 2011, 112, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Guemes, M.; Garcia, A.J.; Rigueur, D.; Runke, S.; Wang, W.; Zhao, G.; Mayorga, V.H.; Atti, E.; Tetradis, S.; Peault, B.; et al. GATA4 is essential for bone mineralization via ERalpha and TGFbeta/BMP pathways. J. Bone Miner. Res. 2014, 29, 2676–2687. [Google Scholar] [CrossRef]
- Patient, R.K.; McGhee, J.D. The GATA family (vertebrates and invertebrates). Curr. Opin. Genet. Dev. 2002, 12, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Khalid, A.B.; Slayden, A.V.; Kumpati, J.; Perry, C.D.; Lillo, M.A.; Arroyo, S.R.; Miranda-Carboni, G.; Krum, S.A. GATA4 directly regulates Runx2 expression and osteoblast differentiation. JBMR Plus 2018, 2, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Carboni, G.A.; Guemes, M.; Bailey, S.; Anaya, E.; Corselli, M.; Peault, B.; Krum, S.A. GATA4 regulates estrogen receptor-alpha-mediated osteoblast transcription. Mol. Endocrinol. 2011, 25, 1126–1136. [Google Scholar] [CrossRef]
- Khalid, A.B.; Pence, J.; Suthon, S.; Lin, J.; Miranda-Carboni, G.A.; Krum, S.A. GATA4 regulates mesenchymal stem cells via direct transcriptional regulation of the WNT signalosome. Bone 2021, 144, 115819. [Google Scholar] [CrossRef]
- Khalid, A.B.; Slayden, A.V.; Kumpati, J.; Perry, C.D.; Berryhill, S.B.; Crawford, J.A.; Fatima, I.; Morselli, M.; Pellegrini, M.; Miranda-Carboni, G.A.; et al. GATA4 represses RANKL in osteoblasts via multiple long-range enhancers to regulate osteoclast differentiation. Bone 2018, 116, 78–86. [Google Scholar] [CrossRef]
- Zaret, K.S.; Carroll, J.S. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 2011, 25, 2227–2241. [Google Scholar] [CrossRef]
- Zhou, P.; Zhang, Y.; Sethi, I.; Ye, L.; Trembley, M.A.; Cao, Y.; Akerberg, B.N.; Xiao, F.; Zhang, X.; Li, K.; et al. GATA4 Regulates Developing Endocardium Through Interaction With ETS1. Circ. Res. 2022, 131, e152–e168. [Google Scholar] [CrossRef]
- Cirillo, L.A.; Zaret, K.S. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell 1999, 4, 961–969. [Google Scholar] [CrossRef]
- Krum, S.A.; Miranda-Carboni, G.A.; Lupien, M.; Eeckhoute, J.; Carroll, J.S.; Brown, M. Unique ERα cistromes control cell type-specific gene regulation. Mol. Endocrinol. 2008, 22, 2393–2406. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.C.; Carpenter, C.; Nebert, D.W.; Vasiliou, V. Update of human and mouse forkhead box (FOX) gene families. Hum. Genom. 2010, 4, 345–352. [Google Scholar] [CrossRef]
- Gokce, G.; Oren, N.C.; Ozgonul, C. Axenfeld-Rieger syndrome associated with severe maxillofacial and skeletal anomalies. J. Oral Maxillofac. Pathol. JOMFP 2015, 19, 109. [Google Scholar] [CrossRef]
- Machida, A.; Okuhara, S.; Harada, K.; Iseki, S. Difference in apical and basal growth of the frontal bone primordium in Foxc1ch/ch mice. Congenit. Anom. 2014, 54, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.K.; Lass, J.H.; Chakravarti, A. Pleiotropic skeletal and ocular phenotypes of the mouse mutation congenital hydrocephalus (ch/Mf1) arise from a winged helix/forkhead transcriptionfactor gene. Hum. Mol. Genet. 1999, 8, 625–637. [Google Scholar] [CrossRef]
- Almubarak, A.; Lavy, R.; Srnic, N.; Hu, Y.; Maripuri, D.P.; Kume, T.; Berry, F.B. Loss of Foxc1 and Foxc2 function in chondroprogenitor cells disrupts endochondral ossification. J. Biol. Chem. 2021, 297, 101020. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, N.; Li, H.; Wang, M.; Shen, H.; Si, J.; Shen, G. The Transcription Factor Foxc1 Promotes Osteogenesis by Directly Regulating Runx2 in Response of Intermittent Parathyroid Hormone (1-34) Treatment. Front. Pharmacol. 2020, 11, 592. [Google Scholar] [CrossRef]
- Mirzayans, F.; Lavy, R.; Penner-Chea, J.; Berry, F.B. Initiation of early osteoblast differentiation events through the direct transcriptional regulation of Msx2 by FOXC1. PLoS ONE 2012, 7, e49095. [Google Scholar] [CrossRef]
- He, A.; Shen, X.; Ma, Q.; Cao, J.; von Gise, A.; Zhou, P.; Wang, G.; Marquez, V.E.; Orkin, S.H.; Pu, W.T. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012, 26, 37–42. [Google Scholar] [CrossRef]
- Dupont, S.; Krust, A.; Gansmuller, A.; Dierich, A.; Chambon, P.; Mark, M. Effect of single and compound knockouts of estrogen receptors alpha (ERalpha) and beta (ERbeta) on mouse reproductive phenotypes. Development 2000, 127, 4277–4291. [Google Scholar] [CrossRef]
- Krum, S.A.; Miranda-Carboni, G.A.; Hauschka, P.V.; Carroll, J.S.; Lane, T.F.; Freedman, L.P.; Brown, M. Estrogen protects bone by inducing Fas ligand in osteoblasts to regulate osteoclast survival. EMBO J. 2008, 27, 535–545. [Google Scholar] [CrossRef]
- Suthon, S.; Lin, J.; Perkins, R.S.; Crockarell, J.R., Jr.; Miranda-Carboni, G.A.; Krum, S.A. Estrogen receptor alpha and NFATc1 bind to a bone mineral density-associated SNP to repress WNT5B in osteoblasts. Am. J. Hum. Genet. 2022, 109, 97–115. [Google Scholar] [CrossRef]
- Monroe, D.G.; Getz, B.J.; Johnsen, S.A.; Riggs, B.L.; Khosla, S.; Spelsberg, T.C. Estrogen receptor isoform-specific regulation of endogenous gene expression in human osteoblastic cell lines expressing either ERalpha or ERbeta. J. Cell. Biochem. 2003, 90, 315–326. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Fedchenko, N.; Reifenrath, J. Different approaches for interpretation and reporting of immunohistochemistry analysis results in the bone tissue—A review. Diagn. Pathol. 2014, 9, 221. [Google Scholar] [CrossRef]
- Zhang, Y.; Hassan, M.Q.; Xie, R.L.; Hawse, J.R.; Spelsberg, T.C.; Montecino, M.; Stein, J.L.; Lian, J.B.; van Wijnen, A.J.; Stein, G.S. Co-stimulation of the bone-related Runx2 P1 promoter in mesenchymal cells by SP1 and ETS transcription factors at polymorphic purine-rich DNA sequences (Y-repeats). J. Biol. Chem. 2009, 284, 3125–3135. [Google Scholar] [CrossRef] [PubMed]
- Morrisey, E.E.; Ip, H.S.; Lu, M.M.; Parmacek, M.S. GATA-6: A zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev. Biol. 1996, 177, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Mei, S.; Qin, Q.; Wu, Q.; Sun, H.; Zheng, R.; Zang, C.; Zhu, M.; Wu, J.; Shi, X.; Taing, L.; et al. Cistrome Data Browser: A data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 2017, 45, D658–D662. [Google Scholar] [CrossRef]
- Zheng, R.; Wan, C.; Mei, S.; Qin, Q.; Wu, Q.; Sun, H.; Chen, C.H.; Brown, M.; Zhang, X.; Meyer, C.A.; et al. Cistrome Data Browser: Expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019, 47, D729–D735. [Google Scholar] [CrossRef]
- Benisch, P.; Schilling, T.; Klein-Hitpass, L.; Frey, S.P.; Seefried, L.; Raaijmakers, N.; Krug, M.; Regensburger, M.; Zeck, S.; Schinke, T.; et al. The transcriptional profile of mesenchymal stem cell populations in primary osteoporosis is distinct and shows overexpression of osteogenic inhibitors. PLoS ONE 2012, 7, e45142. [Google Scholar] [CrossRef]
- Suthon, S.; Perkins, R.S.; Lin, J.; Crockarell, J.R., Jr.; Miranda-Carboni, G.A.; Krum, S.A. GATA4 and estrogen receptor alpha bind at SNPs rs9921222 and rs10794639 to regulate AXIN1 expression in osteoblasts. Hum. Genet. 2022, 141, 1849–1861. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.; Mirzayans, F.; Berry, F. Foxc1 Expression in Early Osteogenic Differentiation Is Regulated by BMP4-SMAD Activity. J. Cell. Biochem. 2016, 117, 1707–1717. [Google Scholar] [CrossRef] [PubMed]
- McLeay, R.C.; Bailey, T.L. Motif Enrichment Analysis: A unified framework and an evaluation on ChIP data. BMC Bioinform. 2010, 11, 165. [Google Scholar] [CrossRef] [PubMed]
- Barutcu, A.R.; Tai, P.W.; Wu, H.; Gordon, J.A.; Whitfield, T.W.; Dobson, J.R.; Imbalzano, A.N.; Lian, J.B.; van Wijnen, A.J.; Stein, J.L.; et al. The bone-specific Runx2-P1 promoter displays conserved three-dimensional chromatin structure with the syntenic Supt3h promoter. Nucleic Acids Res. 2014, 42, 10360–10372. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Li, J.; Yao, W.; Wang, W.; Shi, B.; Yuan, F.; Dong, J.; Zhang, H. FOXC1 Negatively Regulates DKK1 Expression to Promote Gastric Cancer Cell Proliferation Through Activation of Wnt Signaling Pathway. Front. Cell Dev. Biol. 2021, 9, 662624. [Google Scholar] [CrossRef]
- Liu, K.; Ni, J.D.; Li, W.Z.; Pan, B.Q.; Yang, Y.T.; Xia, Q.; Huang, J. The Sp1/FOXC1/HOTTIP/LATS2/YAP/beta-catenin cascade promotes malignant and metastatic progression of osteosarcoma. Mol. Oncol. 2020, 14, 2678–2695. [Google Scholar] [CrossRef]
- Lam, E.W.; Brosens, J.J.; Gomes, A.R.; Koo, C.Y. Forkhead box proteins: Tuning forks for transcriptional harmony. Nat. Rev. Cancer 2013, 13, 482–495. [Google Scholar] [CrossRef]
- Lin, Y.H.; Jewell, B.E.; Gingold, J.; Lu, L.; Zhao, R.; Wang, L.L.; Lee, D.F. Osteosarcoma: Molecular Pathogenesis and iPSC Modeling. Trends Mol. Med. 2017, 23, 737–755. [Google Scholar] [CrossRef]
- Qiu, Z.; Dan, Z.; Che, X. FoxC1 promotes osteosarcoma cell proliferation and metastasis through the activation of EZH2. Int. J. Clin. Exp. Med. 2017, 10, 376–384. [Google Scholar]
- Won, K.Y.; Park, H.R.; Park, Y.K. Prognostic implication of immunohistochemical Runx2 expression in osteosarcoma. Tumori 2009, 95, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Sadikovic, B.; Thorner, P.; Chilton-Macneill, S.; Martin, J.W.; Cervigne, N.K.; Squire, J.; Zielenska, M. Expression analysis of genes associated with human osteosarcoma tumors shows correlation of RUNX2 overexpression with poor response to chemotherapy. BMC Cancer 2010, 10, 202. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).