The New Nematicide Cyclobutrifluram Targets the Mitochondrial Succinate Dehydrogenase Complex in Caenorhabditis elegans
Abstract
:1. Introduction
2. Materials and Methods
2.1. C. elegans Strains and Culture
2.2. Survival Assay
2.3. Brood Size and Embryonic Viability
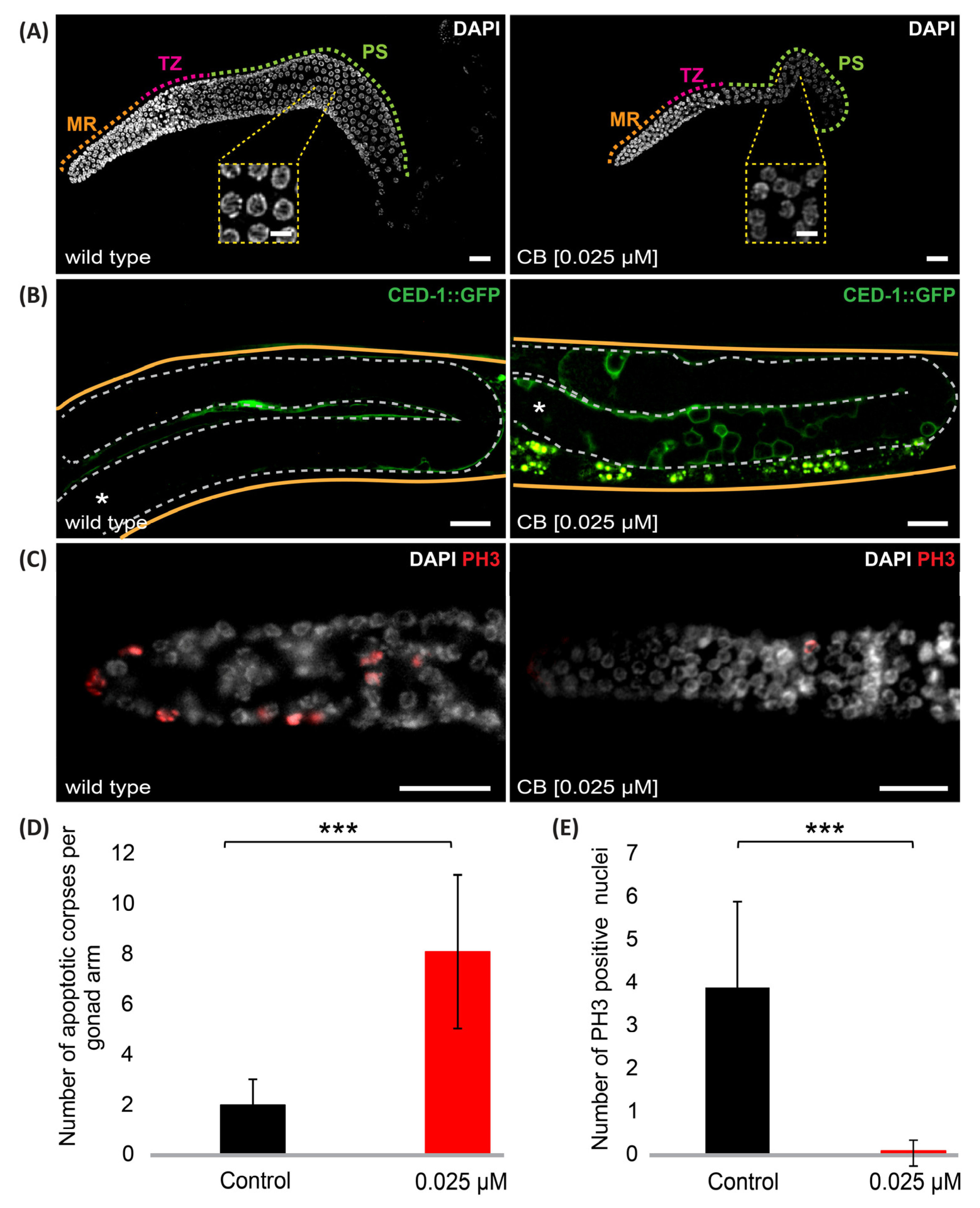
2.4. Germline Apoptosis
2.5. Immunostaining and DAPI Staining
2.6. Microscopy and Image Processing
2.7. RNA Extraction, cDNA Library Preparation and Sequencing
2.8. RNA-seq Data Analysis
3. Results
3.1. Decreased Survival Rate of Worms Exposed to Cyclobutrifluram
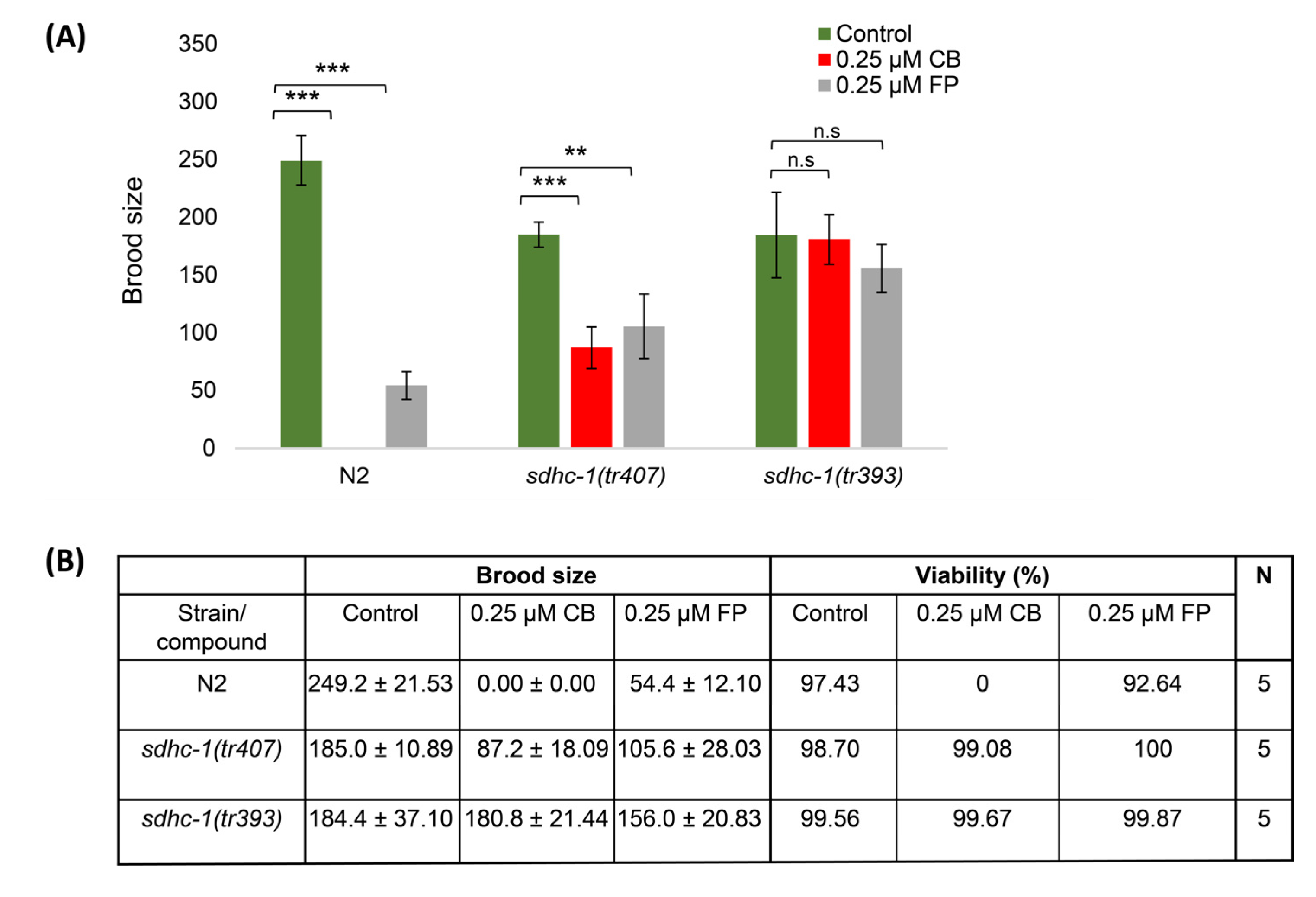
3.2. Impact of Cyclobutrifluram on Brood Size and Embryo Viability
3.3. Cyclobutrifluram Treatment Leads to a Decreased Number of Germ Cells
3.4. Cyclobutrifluram Inhibits C. elegans Mitochondrial Complex II
3.5. Transcriptional Response of C. elegans Treated with Cyclobutrifluram
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Den Hoogen, J.; Geisen, S.; Routh, D.; Ferris, H.; Traunspurger, W.; Wardle, D.A.; De Goede, R.G.; Adams, B.J.; Ahmad, W.; Andriuzzi, W.S. Soil nematode abundance and functional group composition at a global scale. Nature 2019, 572, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Gamalero, E.; Glick, B.R. The use of plant growth-promoting bacteria to prevent nematode damage to plants. Biology 2020, 9, 381. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.T.; Haegeman, A.; Danchin, E.G.; Gaur, H.S.; Helder, J.; Jones, M.G.; Kikuchi, T.; Manzanilla-López, R.; Palomares-Rius, J.E.; Wesemael, W.M. Top 10 plant—Parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Desaeger, J.; Wram, C.; Zasada, I. New reduced-risk agricultural nematicides-rationale and review. J. Nematol. 2020, 52, e2020-91. [Google Scholar] [CrossRef] [PubMed]
- Zasada, I.A.; Halbrendt, J.M.; Kokalis-Burelle, N.; LaMondia, J.; McKenry, M.V.; Noling, J.W. Managing nematodes without methyl bromide. Annu. Rev. Phytopathol. 2010, 48, 311–328. [Google Scholar] [CrossRef]
- Costa, L.G.; Giordano, G.; Guizzetti, M.; Vitalone, A. Neurotoxicity of pesticides: A brief review. Front. Biosci.-Landmark 2008, 13, 1240–1249. [Google Scholar] [CrossRef]
- Opperman, C.; Chang, S. Plant-parasitic nematode acetylcholinesterase inhibition by carbamate and organophosphate nematicides. J. Nematol. 1990, 22, 481. [Google Scholar]
- Umetsu, N.; Shirai, Y. Development of novel pesticides in the 21st century. J. Pestic. Sci. 2020, 45, 54–74. [Google Scholar] [CrossRef]
- Burns, A.R.; Luciani, G.M.; Musso, G.; Bagg, R.; Yeo, M.; Zhang, Y.; Rajendran, L.; Glavin, J.; Hunter, R.; Redman, E. Caenorhabditis elegans is a useful model for anthelmintic discovery. Nat. Commun. 2015, 6, 7485. [Google Scholar] [CrossRef]
- Schleker, A.S.S.; Rist, M.; Matera, C.; Damijonaitis, A.; Collienne, U.; Matsuoka, K.; Habash, S.S.; Twelker, K.; Gutbrod, O.; Saalwächter, C. Mode of action of fluopyram in plant-parasitic nematodes. Sci. Rep. 2022, 12, 11954. [Google Scholar] [CrossRef]
- Oka, Y.; Shuker, S.; Tkachi, N. Nematicidal efficacy of MCW-2, a new nematicide of the fluoroalkenyl group, against the root-knot nematode Meloidogyne javanica. Pest Manag. Sci. Former. Pestic. Sci. 2009, 65, 1082–1089. [Google Scholar] [CrossRef] [PubMed]
- Oka, Y.; Shuker, S.; Tkachi, N. Systemic nematicidal activity of fluensulfone against the root-knot nematode Meloidogyne incognita on pepper. Pest Manag. Sci. 2012, 68, 268–275. [Google Scholar] [CrossRef]
- Thoden, T.C.; Alkader, M.A.; Markakis, E.A.; Yum, M.Y.; Wiles, J.A. Biological attributes of Salibro™, a novel sulfonamide nematicide. Part 3: Biocompatibility with beneficial soil fungi. Nematology 2022, 24, 915–924. [Google Scholar] [CrossRef]
- Hunt, P. The C. elegans model in toxicity testing. J. Appl. Toxicol. 2017, 37, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Meneely, P.M.; Dahlberg, C.L.; Rose, J.K. Working with worms: Caenorhabditis elegans as a model organism. Curr. Protoc. Essent. Lab. Tech. 2019, 19, e35. [Google Scholar] [CrossRef]
- Holden-Dye, L.; Walker, R. Anthelmintic drugs and nematicides: Studies in Caenorhabditis elegans. In WormBook: The Online Review of C. elegans Biology; WormBook: Pasadena, CA, USA, 2014; pp. 1–29. [Google Scholar] [CrossRef]
- Kaminsky, R.; Ducray, P.; Jung, M.; Clover, R.; Rufener, L.; Bouvier, J.; Weber, S.S.; Wenger, A.; Wieland-Berghausen, S.; Goebel, T. A new class of anthelmintics effective against drug-resistant nematodes. Nature 2008, 452, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, W.; Wang, Y.; Liu, H.; Zhang, S.; Ji, X.; Qiao, K. Oxidative stress, intestinal damage, and cell apoptosis: Toxicity induced by fluopyram in Caenorhabditis elegans. Chemosphere 2022, 286, 131830. [Google Scholar] [CrossRef] [PubMed]
- Kearn, J.; Ludlow, E.; Dillon, J.; O’Connor, V.; Holden-Dye, L. Fluensulfone is a nematicide with a mode of action distinct from anticholinesterases and macrocyclic lactones. Pestic. Biochem. Physiol. 2014, 109, 44–57. [Google Scholar] [CrossRef]
- Donkin, S.G.; Dusenbery, D.B. A soil toxicity test using the nematode Caenorhabditis elegans and an effective method of recovery. Arch. Environ. Contam. Toxicol. 1993, 25, 145–151. [Google Scholar] [CrossRef]
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef]
- Park, H.-E.H.; Jung, Y.; Lee, S.-J.V. Survival assays using Caenorhabditis elegans. Mol. Cells 2017, 40, 90. [Google Scholar] [CrossRef]
- Lant, B.; Derry, W.B. Analysis of apoptosis in Caenorhabditis elegans. Cold Spring Harb. Protoc. 2014, pdb.top070458. [Google Scholar] [CrossRef] [PubMed]
- Shaham, S. Methods in Cell Biology. In WormBook: The Online Review of C. elegans Biology; The C. elegans Research Community, Ed.; WormBook: Pasadena, CA, USA, 2006. [Google Scholar]
- Phillips, C.M.; McDonald, K.L.; Dernburg, A.F. Cytological analysis of meiosis in Caenorhabditis elegans. In Meiosis: Volume 2, Cytological Methods; Humana Press: Totowa, NJ, USA, 2009; pp. 171–195. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Van den Beek, M.; Blankenberg, D.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Eberhard, C. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016, 44, W3–W10. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
- Angeles-Albores, D.; Lee, R.Y.N.; Chan, J.; Sternberg, P.W. Tissue enrichment analysis for C. elegans genomics. BMC Bioinform. 2016, 17, 366. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Larigot, L.; Mansuy, D.; Borowski, I.; Coumoul, X.; Dairou, J. Cytochromes P450 of Caenorhabditis elegans: Implication in biological functions and metabolism of xenobiotics. Biomolecules 2022, 12, 342. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Flemming, A. TYMIRIUM® Technology: The Mechanism of Action of Cyclobutrifluram in Nematodes. In Proceedings of the 15th IUPAC International Congress of Crop Protection Chemistry, New Delhi, India, 14–17 March 2023. [Google Scholar]
- Gaberthueel, M.; Morsello, S.; Hamill, J.; Ireland, D.; Simmons, J.; Bachiega, A. Biological activity of Tymirium™ molecule as a soil- and seed-applied nematicide. In Proceedings of the 7th International Nematode Conference, Antibes Juan-Les-Pins, France, 1–6 May 2022. [Google Scholar]
- Slaats, B.; Gaberthueel, M.; Loiseleur, O.; Flemming, A.; Luksch, T. Tymirium™: The story of a powerful new molecule. In Proceedings of the 7th International Nematode Conference, Antibes Juan-Les-Pins, France, 1–6 May 2022. [Google Scholar]
- Wram, C.L.; Hesse, C.N.; Zasada, I.A. Transcriptional response of Meloidogyne incognita to non-fumigant nematicides. Sci. Rep. 2022, 12, 9814. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heydari, F.; Rodriguez-Crespo, D.; Wicky, C. The New Nematicide Cyclobutrifluram Targets the Mitochondrial Succinate Dehydrogenase Complex in Caenorhabditis elegans. J. Dev. Biol. 2023, 11, 39. https://doi.org/10.3390/jdb11040039
Heydari F, Rodriguez-Crespo D, Wicky C. The New Nematicide Cyclobutrifluram Targets the Mitochondrial Succinate Dehydrogenase Complex in Caenorhabditis elegans. Journal of Developmental Biology. 2023; 11(4):39. https://doi.org/10.3390/jdb11040039
Chicago/Turabian StyleHeydari, Fariba, David Rodriguez-Crespo, and Chantal Wicky. 2023. "The New Nematicide Cyclobutrifluram Targets the Mitochondrial Succinate Dehydrogenase Complex in Caenorhabditis elegans" Journal of Developmental Biology 11, no. 4: 39. https://doi.org/10.3390/jdb11040039
APA StyleHeydari, F., Rodriguez-Crespo, D., & Wicky, C. (2023). The New Nematicide Cyclobutrifluram Targets the Mitochondrial Succinate Dehydrogenase Complex in Caenorhabditis elegans. Journal of Developmental Biology, 11(4), 39. https://doi.org/10.3390/jdb11040039






