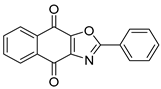
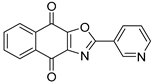
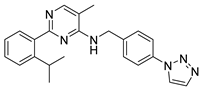
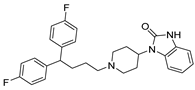
Abstract
Cancer management has traditionally depended on chemotherapy as the mainstay of treatment; however, recent advancements in targeted therapies and immunotherapies have offered new options. Ubiquitin-specific proteases (USPs) have emerged as promising therapeutic targets in cancer treatment due to their crucial roles in regulating protein homeostasis and various essential cellular processes. This review covers the following: (1) the structural and functional characteristics of USPs, highlighting their involvement in key cancer-related pathways, and (2) the discovery, chemical structures, mechanisms of action, and potential clinical implications of USP inhibitors in cancer therapy. Particular attention is given to the role of USP inhibitors in enhancing cancer immunotherapy, e.g., modulation of the tumor microenvironment, effect on regulatory T cell function, and influence on immune checkpoint pathways. Furthermore, this review summarizes the current progress and challenges of clinical trials involving USP inhibitors as cancer therapy. We also discuss the complexities of achieving target selectivity, the ongoing efforts to develop more specific and potent USP inhibitors, and the potential of USP inhibitors to overcome drug resistance and synergize with existing cancer treatments. We finally provide a perspective on future directions in targeting USPs, including the potential for personalized medicine based on specific gene mutations, underscoring their significant potential for enhancing cancer treatment. By elucidating their mechanisms of action, clinical progress, and potential future applications, we hope that this review could serve as a useful resource for both basic scientists and clinicians in the field of cancer therapeutics.
1. Introduction
1.1. Overview of the Ubiquitin–Proteasome Pathway
The ubiquitin–proteasome system (UPS) is a vital pathway regulating protein levels in eukaryotic cells, which is essential for maintaining cellular homeostasis. This pathway consists of several enzymatic steps that ensure the targeted degradation of substrate proteins. The UPS process begins with the activation of ubiquitin (Ub) by the E1 ubiquitin-activating enzyme in an ATP-dependent reaction, which is then transferred to an E2 ubiquitin-conjugating enzyme and finally transferred to a specific lysine residue on the target proteins by an E3 ubiquitin ligase (Figure 1). This process is repeated to form a polyubiquitin chain on the target proteins, which serves as a signal for recognition by the 26S proteasome [1]. The 26S proteasome, a multi-subunit protease complex consisting of a 20S core particle with proteolytic activity and two 19S regulatory particles that recognize ubiquitinated proteins, is then responsible for the degradation of the targeted substrate proteins into small peptides. Deubiquitinating enzymes (DUBs) play a significant role in the regulation of the UPS by removing ubiquitin molecules from ubiquitinated substrate proteins. The UPS is involved in multiple cellular processes, including DNA damage response (DDR), apoptosis, signal transduction, and drug resistance. Dysfunction of the UPS has been implicated in a range of disease types, including cancer [2]. Cancers with higher protein turnover are generally more sensitive to the inhibition of proteasome and deubiquitinases. The importance of the UPS in pathogenesis is underscored by the development of proteasome inhibitors as therapeutic agents. To date, the FDA has approved three 20S proteasome inhibitors, Borezomib, Carfilzomib, and Ixazomib, for the treatment of hematologic malignancies, namely multiple myeloma and mantle cell lymphoma. Bortezomib, which was discovered in 1995 and approved by the US FDA for the treatment of multiple myeloma in 2003 and later for the treatment of mantle cell lymphoma, validated the therapeutic potential of targeting the UPS, particularly in hematologic malignancies [3]. Carfilzomib was subsequently approved in 2012, followed by Ixazomib, both specifically for the treatment of multiple myeloma. These drugs have offered new therapeutic options for patients with these challenging blood cancers [4]. The UPS’s roles in a wide range of cellular processes make it an important target of ongoing research.
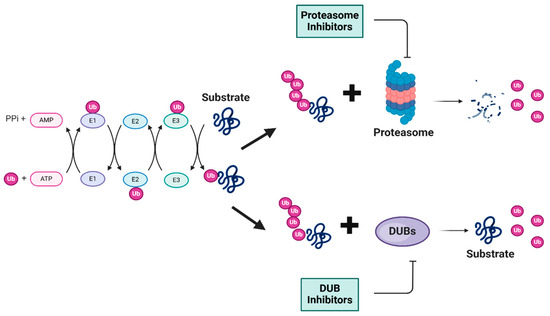
Figure 1.
Schematic representation of ubiquitin-tagged protein degradation. This schematic illustrates the UPS, a vital cellular protein degradation mechanism. The process begins with the ATP-dependent activation of ubiquitin, which leads to a cascade of enzymatic reactions that ultimately results in the degradation of the substrates into smaller peptides by the proteasome or stabilization of the substrates by the relevant DUB. In both cases, ubiquitin molecules are released for reuse. Ub: ubiquitin (Ub) molecules (purple); ATP/AMP: energy source for ubiquitin activation; PPi: inorganic pyrophosphate; proteasome: multi-subunit protein degradation complex (blue and pink); degradation products: released peptide fragments and free ubiquitin molecules; DUBs: deubiquitinating enzymes. (Created using BioRender. Bakkar, M. (2025)).
1.2. Role of Deubiquitinating Enzymes
Deubiquitinating enzymes (DUBs) are proteases that cleave ubiquitin from various substrate proteins, playing a crucial role in regulating the ubiquitin signaling pathway. The human genome encodes approximately 100 putative DUBs, which are classified into two main classes: metalloproteases and cysteine (Cys) proteases. These classes are further subclassed based on their domain structures. The first family belongs to the JAB1/MPN/Mov34 metalloenzyme (JAMM) domain zinc-dependent metalloprotease family, while the other five families—the ubiquitin C-terminal hydrolases (UCH), the ovarian tumor proteases (OTU), the Machado–Josephin domain proteases (MJDs), the ubiquitin-specific protease (USP/UBP), and the recently discovered motif interacting with ubiquitin (MIU)-containing novel DUB family (MINDY) and Zinc finger with UFM1-specific peptidase domain protein (ZUFSP)—are papain-like cysteine proteases.
1.2.1. Metalloprotease DUBs
Metalloproteases form one of the two main classes of DUBs, with the primary subclass of metalloproteases being JAMM (JAB1/MPN/MOV34 metalloenzyme) domain proteases. The catalytic site of this subclass of DUBs contains two histidine residues, an aspartate (Asp), and a catalytic serine (Ser). The main unique property of metalloproteases is that they use zinc in their catalytic mechanism, which distinguishes them from the cysteine protease DUBs. The zinc ion plays a role in the generation of a hydroxide ion from water, which acts as a nucleophile to hydrolyze the isopeptide bond between the protein substrate and ubiquitin. A key consequence of this mechanism is that this type of DUB does not form a covalent intermediate with the substrate during catalysis. In contrast to ubiquitin-specific proteases (USPs), which use their catalytic serine to form a covalent intermediate with the substrate during enzyme cleavage, metalloprotease DUBs utilize a nucleophilic hydroxide ion that directly attacks the peptide bond without forming a covalent bond with the enzyme. This characteristic makes them inherently resistant to DUB inhibitors, which often target the formation of such covalent intermediates [5].
1.2.2. Cysteine Protease DUBs
The six subfamilies of cysteine protease DUBs are organized based on their domain architecture. It is important to note that most DUBs in this class utilize a catalytic triad composed of a histidine (His), an active site cysteine (Cys), and in most cases, an asparagine (Asn) or aspartate (Asp).
Ubiquitin C-Terminal Hydrolases (UCHs)
This subfamily of cysteine proteases was one of the first types of DUBs identified with UCHL3 discovered in 1997 [6]. The UCH domain contains three conserved residues, namely His, Cys, and Asp, and there are four known members of this subfamily. These small enzymes preferentially remove small peptides from the C-terminus of ubiquitin. The loop structure covering the active site limits the size of the substrate with which they can interact, unlike USPs, which can handle larger substrates. UCHs have been implicated in playing a role in the oncogenesis of various cancers [7].
Ovarian Tumor Proteases (OTUs)
This subfamily displays specificity for the ubiquitin substrates with which it interacts by utilizing additional ubiquitin interaction sites that can bind to specific linkages on longer protein chains. Another important distinction is the lack of an asparagine or aspartate residue in some members of this subfamily [5]. OTUD5 is an important member of the OTU subfamily of cysteine proteases and has been implicated in both tumor progression and tumor suppression depending on the disease type. These DUBs play an important role in various cellular processes, including DNA repair and protein quality control [8].
Machado–Josephin Domain Proteases (MJDs)
These DUBs comprise four members in humans, and all contain a catalytic domain known as the Josephin domain. This important feature contains two conserved histidines, a catalytic cysteine, and two ubiquitin-binding sites. This subfamily is named after the Machado–Josephin disease, a neurological disorder caused by a CAG repeat expansion motif producing polyglutamine resulting in protein aggregation and misfolding [9]. Ataxin-3, a critical MJD in humans, has been shown to play a role in the proliferation of testicular cancer, gastric cancer, and lung cancer cells [10].
Motif Interacting with Ubiquitin (MIU)-Containing Novel DUB Family (MINDY) and Zinc Finger with UFM1-Specific Peptidase Domain Protein (ZUFSP)
These two subfamilies of cysteine proteases were the two most recently identified. The MINDY subfamily of DUBs has a unique catalytic triad consisting of cysteine, histidine, and threonine [11]. There are four known members of this subfamily, and these enzymes appear to preferentially cleave long ubiquitin chains starting from the direction of the distal end [12]. ZUFSP contains only one known member in humans, which utilizes multiple ubiquitin-binding domains and possesses highly specific cleavage of K63 ubiquitin linkage [13]. ZUFSP has a modular architecture, with its overall specificity and activity being influenced by its various modular ubiquitin-binding domains [14].
Ubiquitin-Specific Proteases (USPs)
USPs form the largest subclass of DUBs with 58 known members and are the focus of this review article. USPs are generally larger enzymes, ranging from 50 to 300 kDa and typically have N-terminal extensions involved in substrate recognition and protein–protein interactions. USPs can remove ubiquitin from protein conjugates, process ubiquitin precursors, and disassemble ubiquitin chains. Some types of USPs show specificity for certain substrates, while others have broader activity. They play crucial roles in various cellular processes including the cell cycle, protein degradation, signal transduction, and DNA repair. Their substrate specificities make them important potential targets for therapeutic development in the treatment of diseases such as cancer [5]. Given the crucial role USPs play in regulating protein homeostasis as well as their potential as therapeutic targets in cancer therapy, the remainder of this review will focus on the structural and functional characteristics of USPs, their involvement in key cancer-related pathways, and the development of USP inhibitors as promising anticancer agents.
2. USPs: Structure, Biological Function, and Role in Tumorigenesis and Cancer Progression
2.1. The Structure of USPs
USPs represent the most prominent family of DUBs. They represent a sophisticated class of enzymes whose function depends on the intricate interplay between three vital structural elements. These elements—the core catalytic domain, ubiquitin-like domains, and ancillary domains—work congruently to achieve precise protein regulation [15]. Their activities are fine-tuned through post-translational modifications (PTMs), creating a complex regulatory network [16]. Understanding this structural and regulatory framework is crucial for basic research and therapeutic development. As common with signaling proteins, most USP deubiquitinases have a modular architecture with a catalytic domain, putative site of interaction, and localization domains.
Most USP domains have at least two ubiquitin-binding sites, one for the proximal ubiquitin molecules and one for the distal ubiquitin molecules. These sites cleave the isopeptide bond linking two ubiquitin moieties or the proximal ubiquitin with a protein substrate [17]. The USP family is the largest, with more than 50 members. The USP catalytic domain may be flanked at its N- or C-terminus by several additional domains involved in differential substrate recognition, sub-cellular localization, and regulation of enzymatic activity [18]. Among the members of the USP domain family, the majority act upon larger protein substrates in contrast to other subclasses of DUBs [19].
2.1.1. Core Catalytic Domain
At the heart of USP function is the core catalytic domain, whose structure reveals the precise ingenuity of these enzymes. This domain features homolog sequences derived from papain-like cysteine proteases arranged in a distinctive “palm, thumb, and fingers” configuration (Figure 2) that orchestrates substrate binding and cleavage [20,21].
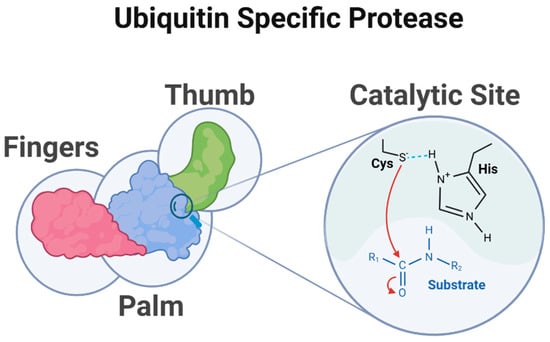
Figure 2.
Structure of ubiquitin-specific protease. This diagram depicts the structural organization and catalytic mechanism of a USP. The USP contains a catalytic core domain featuring the characteristic catalytic triad consisting of cysteine (Cys), histidine (His), and, in most cases, an asparagine (Asn) or aspartate (Asp), which interacts with histidine (the cysteine and histidine residues are illustrated in the figure) highlighted in the active site between the palm and thumb sub-domains. The isopeptide bond between ubiquitin and the substrate protein is cleaved resulting in the removal of ubiquitin modifications, which is crucial for protein regulation and cellular homeostasis. (Created using BioRender. Bakkar, M. (2025)).
The catalytic center of USPs is located at the interface between the palm and thumb regions of the USP domain. The USP catalytic core can be thought of as being divided into six conserved boxes (Figure 3).
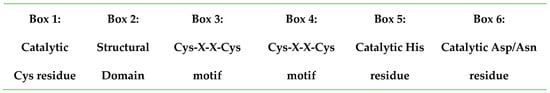
Figure 3.
Molecular basis of ubiquitin-specific protease catalytic core protein domains.
Box 1 includes a catalytic Cys residue, box 5 includes a catalytic His, and box 6 includes a catalytic Asp/Asn residue. Each of these boxes is characterized by several other conserved features and residues. Boxes 3 and 4 contain a Cys-X-X-Cys motif, which has been reported to form a functional zinc-binding motif in some USPs. It is speculated that zinc binding facilitates the folding of the USP core, thus allowing the interaction of sequence motifs that are several hundred residues apart. USP domains exhibit a common conserved fold. The catalytic triad resides between the thumb (Cys) and palm sub-domains (His/Asp) [17]. The principal site of the DUB has a strong interaction with the distal ubiquitin, mainly through the Ile44 patch, with different interacting surfaces within DUB subfamilies. The stretch of amino acids extending from the binding site of the C-terminus of the distal ubiquitin to the DUB catalytic center is responsible for distinguishing ubiquitin from other ubiquitin-like molecules (ULMs) [22,23]. The residues that are responsible for the difference in the C-terminal sequence of ubiquitin (Leu71, Arg72, Leu73, Arg74, Gly75, Gly76) when compared to that of ULMs are vital for identifying ubiquitin by DUBs [22,24]. The catalytic domain is responsible for the isopeptidase activity that cleaves the isopeptide bond between ubiquitin and the target substrate protein [18]. Most USPs contain a core catalytic domain with insertions and terminal extensions bearing additional protein interaction domains [20].
2.1.2. Ubiquitin-like Domains (UBL)
Building upon this catalytic foundation, UBL domains extend the functional repertoire of USPs. Strategically positioned near the distal ubiquitin-binding site, they play a crucial role in substrate recognition and regulation. Their integration within the catalytic core’s variable insertions facilitates essential intra-molecular interactions that maintain proper USP conformation. Many USPs contain UBL domains that can bind to ubiquitin or other UBLs, as well as compete with ubiquitin binding and regulate catalysis. These UBL domains may mediate substrate recognition and binding, as well as play a vital role in localization at the proteasome [18].
2.1.3. Ancillary Domains
Complementing these two core components mentioned above, ancillary domains add another layer of functional sophistication through specialized structural elements. These include zinc finger domains, ubiquitin-interacting motifs (UIMs), and SUMO-interacting motifs (SIMs) that regulate their activity, localization, and substrate specificity [18]. These domains have fine-tuned substrate specificity and mediate crucial protein–protein interactions, although the role of these ancillary domains in regulating USP activity is not always consistent [21]. Their structural adaptability, particularly in the substrate-induced ordering of the unstructured region, provides additional regulatory flexibility through PTM-mediated control [17].
2.2. The Roles of USPs in Tumorigenesis and Cancer Progression
USPs regulate several critical cancer-related pathways, including NF-κB, Wnt/β-catenin, JAK/STAT, p53 signaling, c-MYC, TGF-β, DNA repair, apoptosis, cell cycle regulation, MAPK, and hypoxia pathways (Table 1). These pathways are involved in various aspects of cancer biology, such as cellular proliferation, survival, metastasis, and therapy resistance. USPs modulate these pathways through deubiquitination of key substrate proteins, which affects their stability and, consequently, their functions. This regulation can either promote or inhibit cancer progression depending on the specific USP and pathway involved. The diverse roles of USPs in these pathways highlight their importance in cancer biology and their potential as therapeutic targets.

Table 1.
Cancer-related cellular pathways influenced by USPs.
2.3. Unique Structural and Functional Characteristics of Various USPs
USPs are a diverse family of DUBs that play crucial roles in protein homeostasis. A common theme among many USPs is their association with cancer pathogenesis, highlighting the importance of ubiquitin regulation in malignancies. USPs share some common structural features, but they also possess unique elements that contribute to their specific functions [53]. Table 2 provides a description of some of the similarities and differences of USPs.
The mechanism of action of the USP family of DUBs is generally conserved across members. The enzymatic activity relies on nucleophilic substitution of the catalytic cysteine on the isopeptide bond between ubiquitin and its protein substrate. The histidine residue acts as a general base and activates the catalytic cysteine for this reaction by lowering its pKa (shown in Figure 2). This mechanism is crucial for the deubiquitinating activity of USPs, allowing them to remove ubiquitin modifications from target proteins and thereby regulate their stability and functions [18].
Beyond the catalytic domain, USPs often have additional structural elements that regulate their activity and confer specificity. These include ubiquitin-binding domains, zinc finger domains, and other protein–protein interaction motifs that allow USPs to recognize specific substrates or participate in particular cellular pathways [18]. Despite their structural and functional similarities, the development of specific inhibitors for USPs has been uneven. Several USP inhibitors have been developed through various drug discovery approaches, including high-throughput screening and structure-based drug design. However, for many USPs, specific inhibitors have not yet been discovered [54]. This gap represents both a challenge and an opportunity in the field of cancer drug development and research, as targeting specific USPs could lead to novel cancer therapies that can potentially overcome drug resistance, improve efficacy, and reduce side effects.

Table 2.
Unique structural and functional characteristics of various USPs.
Table 2.
Unique structural and functional characteristics of various USPs.
| USP Name | Domains | Catalytic Site | Cancer Association | References |
|---|---|---|---|---|
| USP1 | Conserved USP catalytic domain (thumb, palm, and finger sub-domains). | Three catalytic residues: Cys90, His593, and Asp751. | Osteosarcoma, renal clear cell carcinoma, colorectal carcinoma, non-small cell lung cancer, and gastric cancers | [42] |
| USP3 | Zinc finger (ZnF) domain and catalytic domain. | Conserved catalytic domain containing key Cys, Hys, and Asp/Asn residues critical for USP activity. | Gastric cancer and breast cancer | [55,56] |
| USP5 | Catalytic domain flanked by two ubiquitin-associated (UBA) domains and two Zinc finger (ZnF) ubiquitin-binding protein (UBP) domains. | Papain-like proteases have a Cys and a His residue, which form an ion pair with the negatively charged cysteine thiolate functioning as a nucleophile. | Breast, lung, colorectal, hepatocellular, pancreatic, and non-small cell lung cancer | [57,58,59] |
| USP7 | N-terminal MATH domain, central catalytic domain, and five C-terminal tandem ubiquitin-like domains (UbL). | Cys223, His464, and Asp481 residues are critical for deubiquitination. | Colorectal cancer, osteosarcoma, acute myeloid leukemia, breast cancer, prostate cancer, multiple myeloma, ovarian cancer, bladder cancer, esophageal squamous cell carcinoma, chronic lymphocytic lymphoma, and medulloblastoma | [54,60,61] |
| USP10 | N-terminus region, USP catalytic structure domain, and a smaller C-terminus region. | Conserved catalytic domain containing key Cys, His, and Asp/Asn residues critical for USP activity. | Colorectal cancer, prostate cancer, hepatocellular carcinoma, glioblastoma multiforme, non-small-cell lung cancer, chronic myeloid leukemia, and acute myeloid leukemia | [62] |
| USP19 | Two CHORD-SGT1/P23 domains (CS1 and CS2 at the N-terminus), USP catalytic domain. | Contains Cys and His residues critical for enzymatic activity and auto-inhibition mechanism. | Breast cancer and osteosarcoma | [63,64] |
| USP20 | ZnF-UBP domain, catalytic USP domain, and two domains present in ubiquitin-specific protease (DUSP) domains. | Conserved Cys and His residues that catalyze proteolysis of the isopeptide bond between a target protein lysine residue and a glycine residue of ubiquitin. | Breast cancer, lung cancer, colon cancer, gastric cancer, and adult T-cell leukemia | [44,65] |
| USP32 | USP catalytic domain, Domain in USP (DUSP), two UbL domains, and calcium-binding EF-hand with a signal transduction mechanism. | Conserved catalytic domain containing key Cys and His residues critical for USP activity. | Small cell lung cancer, gastric cancer, breast cancer, epithelial ovarian cancer, glioblastoma, gastrointestinal stromal tumor, AML, and pancreatic adenocarcinoma | [66] |
| USP36 | Conserved USP catalytic domain (thumb, palm, and finger sub-domains). | Key residues: Cys223, His464, Asp481 in the catalytic domain. | Colon cancer and breast cancer | [36,67] |
2.4. History of the Development of USP Inhibitors
Over the past decade, the development of USP inhibitors has gained significant momentum as a promising area in cancer therapeutics. Discovery of USP inhibitors primarily relied on high-throughput screening prior to 2014. However, the field has since evolved with structure-guided drug design based on co-crystal structure complexes becoming a prominent approach [68,69]. The interest in USP inhibitors stems from the crucial role USPs play in various cellular processes and their involvement in multiple cancer-related pathways [54]. The shift in focus to targeting USPs was partly due to the limitations of existing UPS-targeted inhibitors such as Bortezomib, which showed efficacy primarily in a couple of hematologic malignancies but only marginal effects on solid tumors [68]. More than seventy USP inhibitors have been reported over the past 20 years with six reaching clinical trials, but despite these advancements, no USP inhibitor has yet been approved for clinical use [69]. The development of USP inhibitors faces the challenge of achieving target selectivity, but despite this, there is growing optimism that USPs represent a new reservoir of therapeutic targets [54].
2.5. Therapeutic Implications
The therapeutic significance of the USP structural organization becomes apparent in disease contexts, particularly in cancer biology. This integrated understanding of USP structure and regulation reveals a remarkable system where each domain contributes to a more extensive functional network. PTMs modulate the activity of signaling proteins further, forming an intricate regulatory network evident in several disease processes, particularly in cancer biology. Specifically, these modifications regulate essential cellular processes, including proliferation, immune responses, and apoptosis [70]. Drug discovery efforts over the last decade have capitalized on this knowledge for the development of adequate targeted therapeutic strategies.
3. Small Molecule Inhibitors Targeting USPs
3.1. Chemical Structure, Inhibition Potency, and Mechanism of Action of Small Molecule Inhibitors Targeting USPs
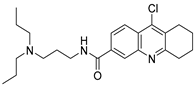
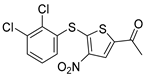
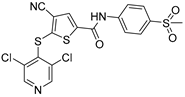
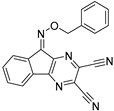
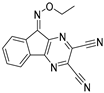
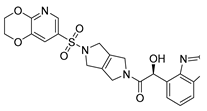
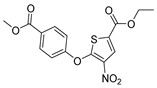
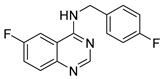
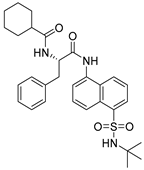
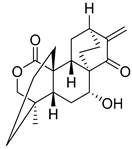
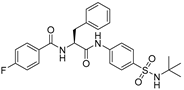
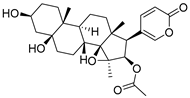
To date, more than seventy small-molecule USP inhibitors have been reported in the literature. The chemical structures, inhibition potency, and mechanisms of action of some selected USP inhibitors are summarized in Table 3. USP inhibitors that are currently being or were previously tested in cancer clinical trials will be discussed in Section 3.3. USP inhibitors can be broadly categorized based on their specificity and the USPs they target. Some USP inhibitors, such as SP-002 and Pimozide (PMZ), are highly selective for USP1, while others like PR619 are pan-inhibitors affecting multiple USPs. The mechanisms of action vary among these compounds, with some binding reversibly to their targets (e.g., IU1), while others form irreversible covalent bonds (e.g., Q29, XL177A). Many of these inhibitors demonstrate promising anticancer effects through various mechanisms. Common themes include inducing cell cycle arrest, enhancing DNA damage, promoting apoptosis, and modulating key signaling pathways, as described in Table 1. For instance, USP7 inhibitors HBX-19818 and P22077 work by stabilizing p53 and promoting the degradation of oncogenic proteins. USP14 inhibitors like IU1 and its derivatives have shown potential in treating neurodegenerative disorders by modulating the degradation of tau protein. Research on these USP inhibitors spans various stages, from in vitro studies to in vivo animal models, with some compounds showing efficacy in xenograft mouse models. Several inhibitors have been shown to exhibit synergistic effects when combined with other cancer therapies, such as PARP inhibitors or traditional chemotherapeutic agents. While these USP inhibitors show promise as a novel class of anticancer drugs in preclinical studies, the potential of these agents in clinical settings needs to be determined.

Table 3.
Small molecule inhibitors targeting USPs.
3.2. USP Inhibitors and Immunotherapy
As mentioned above, the development of USP inhibitors represents a significant advancement in cancer treatment, offering a multifaceted approach to improving patient outcomes. USP inhibitors work by various mechanisms to inhibit cancer cell growth and proliferation, such as destabilizing oncoproteins [181]. USP inhibitors thus offer a novel strategy to overcome drug resistance, a major challenge in cancer therapy, by targeting the mechanisms that cancer cells employ to evade chemotherapy and targeted treatments [182]. Some USP inhibitors also show promise in enhancing cancer immunotherapy by making cancer cells more vulnerable to immune attacks [183]. In this section, we focus on the potential of USP inhibitors in enhancing the immune system and immunotherapy against cancer.
3.2.1. Role of USP1 in Immune Response
USP1 plays a significant role in immunotherapy, particularly in the context of cancer treatment. USP1 has been identified as a critical regulator of T-cell differentiation, which is crucial for effective immunotherapy responses. By deubiquitinating and stabilizing the transcriptional co-activator with PDZ-binding motif (TAZ), USP1 leads to enhanced activity of RORγt, which is an important transcription factor for T helper type 17 (Th17) cell development. Deubiquitination of TAZ also causes decreased acetylation of Foxp3, which promotes its proteasomal degradation leading to reduced Treg cell differentiation [184]. USP1 has also been implicated in resistance to chemotherapy and Rituximab in DLBCL. USP1 deubiquitinates MAX, which is an important MYC-binding protein and promotes MYC gene transcription. High expression of USP1 in DLBCL has been found to be associated with a poorer prognosis, while inhibition with Pimozide or knockdown of USP1 has been shown to reduce cancer cell growth and induce cell cycle arrest. Inhibition of USP1 was also demonstrated to significantly reduce tumor burden in a therapy-resistant DLBCL-engrafted PDX mouse model. Importantly, Pimozide was shown to have evidence of synergy with Etoposide in therapy-resistant DLBCL [80]. The potential of USP1 inhibitors lies in their ability to modulate the tumor microenvironment and enhance the efficacy of immunotherapeutics. Combining USP1 inhibitors with existing immunotherapies may enhance treatment efficacy, particularly in cases of resistance to current therapies.
3.2.2. Role of USP7 in Immune Response
Recent research has substantially advanced our understanding of how USP7 modulates the immune response in cancer patients. Elevated levels of USP7 have been shown to facilitate tumor growth by enhancing the immunosuppressive functions of Foxp3+ Tregs [185,186]. Blocking USP7 hinders the activity of Tip60-dependent Foxp3+ Treg cells, decreasing their inhibitory function and enhancing the body’s ability to fight against tumors [186]. Histone acetyltransferase Tat-interactive protein (Tip60) is essential for the survival of Treg cells. Tip60 is a USP7 substrate, thus targeting USP7 could interfere with Tip60-mediated Foxp3 dimer formation, resulting in reduced activation of immunosuppressive molecules such as CTLA4 and IL-10 while promoting the expression of pro-inflammatory cytokines such as IL-2 and IFN-γ [186]. USP7 is important in regulating the equilibrium of M1 (which suppresses tumor growth and promotes an anti-tumor immune response) and M2 (which aids in tumor progression and inhibits the immune system) macrophages. Targeted inhibition of USP7 results in a change in the appearance and behavior of M2 macrophages, leading to a higher growth rate of differentiated CD8+ T cell groups in a laboratory setting [61]. In research on mice with Lewis lung carcinoma, inhibiting USP7 led to decreased tumor growth and increased levels of M1 macrophages and CD8+ T cells that expressed IFN-γ. USP7 also maintains the stability of PD-L1 by inhibiting its degradation [187].
3.2.3. Enhancing Immunotherapy by USP Inhibitors
USP inhibitors can make cancer cells more susceptible to immune attack by preventing them from evading the immune system and enhancing the effectiveness of immunotherapy [183]. Tumors have specific properties that allow them to evade the immune system, hindering an anti-tumor response and promoting their growth. Mechanisms of immune evasion include the production of immunosuppressive factors, down-regulation of major histocompatibility complex molecules, and recruitment of immunoregulatory cells. Additional studies have shown that USPs can influence the effectiveness of immunotherapy through the regulation of immune cell function and the immune response within the tumor microenvironment [157,188].
HBX-19818 and GNE-6776 are covalent USP7 inhibitors that have shown promise in stabilizing p53 and enhancing its activity, which can lead to improved anti-tumor responses [116]. USP22 inhibitors have also shown potential in improving responses to immunotherapy by increasing levels of CD8+ T cells and NK cells, transforming immune-desert tumors into immune-inflamed tumors [189]. In liver tumors, inhibiting USP22 has been shown to boost tumor immunogenicity, enhance T-cell infiltration, and increase responsiveness to anti-PD-L1 immunotherapy [183,190]. Recent research has identified potent macrocycle inhibitors of USP22, including compound S02, which binds tightly to the catalytic domain pocket of USP22. These USP22 inhibitors have demonstrated significant potential in reducing cancer cell growth, promoting apoptosis, and enhancing the efficacy of existing treatments [183]. USP8 inhibitors have shown potential in enhancing anti-tumor activity when combined with anti-PD-1/PD-L1 immunotherapy. USP13 inhibitors may enhance the antitumor effects of DNA damage response inhibitors as well as promote immune cell infiltration and innate immunity [191]. USP14 inhibitors have been shown to sensitize cancer cells to immune checkpoint inhibitors [192]. USP15 inhibitors can disrupt pathways controlling Toll-like receptor, RIG-I, and NF-kB signaling, enhancing the immune response against tumors [183]. USP9X inhibitors like G9 have shown promise in inactivating Notch signaling, reducing proinflammatory cytokines, and enhancing antitumor immune response [183,193].
In summary, USP inhibitors represent a promising avenue for enhancing cancer immunotherapy by modulating various aspects of the immune response as well as the tumor microenvironment. Their ability to target multiple pathways involved in immune evasion and tumor progression and to enhance the efficacy of existing treatments makes them a valuable addition to the arsenal of cancer therapeutics.
3.3. Development of USP Inhibitors for Cancer Therapy: Clinical Trials Status
Regarding the development of USP inhibitors for the treatment of cancer, most of these inhibitors are still in the preclinical stage. However, a few USP inhibitors are being explored in clinical trials for their potential therapeutic effects, given their mechanism of action and possible benefits to patients with resistant or refractory disease. Multiple USP1 inhibitors are currently being tested in phase 1 and phase 2 clinical trials, which aim to evaluate the safety, tolerability, and potential antitumor activity of these compounds. While there are no USP7 inhibitors currently in clinical trials, several compounds have shown promise in preclinical studies. Inhibition of USP7 has long been viewed as a promising anticancer target being the key DUB in regulating p53 levels. FT671 and FT827 are both small-molecule USP7 inhibitors that have shown high affinity and selectivity to USP7 in vitro. Both compounds appear to target the auto-inhibited apo form of USP7 near the catalytic center, which is distinct from other USPs [123]. Most recently, YCH2823 was discovered as a next-generation USP7 inhibitor with enhanced cellular activity compared to FT671, demonstrating about five times the potency. YCH2823 inhibited the growth of TP53-wild-type, TP53-mutant, and MYCN-amplified cell lines with exceptional efficacy by binding directly to the catalytic domain of USP7. A synergy between USP7 and mTOR inhibitors was also observed, demonstrating potential for novel therapeutic strategies [126]. Table 4 provides a comprehensive overview of the USP inhibitors that are currently or were previously investigated in clinical trials. The study design, preliminary/current results, and key findings from these clinical studies are also described where applicable in the table.
3.3.1. USP1 Inhibitors
USP1 inhibitors represent a promising class of drugs targeting DNA damage repair mechanisms, particularly in homologous recombination deficient (HRD) cancers. Several USP1 inhibitor compounds are currently being studied in various stages of clinical development. TNG348, developed by Tango Therapeutics, is an oral, potent, and highly selective allosteric USP1 inhibitor. It showed potential in preclinical models, demonstrating enhanced activity when combined with DNA repair pathway targeted therapies and efficacy in xenografts with both primary and acquired resistance to PARP inhibitors. Malignancies with HRD are generally more sensitive to therapies that target DNA repair. BRCA1-mutated tumor cells are known to be sensitive to PARP inhibitors due to defects in replication fork stability, and they have shown synergy with chemotherapy. HRD cancers represent up to 1 in 2 ovarian cancers, 1 in 4 breast cancers, 1 in 10 prostate cancers, and 1 in 20 pancreatic cancers. USP1 has been found to be upregulated in tumors with BRCA1 mutations, and its knockdown destabilizes replication forks resulting in cell death [194]. Inhibiting USP1 could be a potential therapeutic strategy for cancers with BRCA1-deficient cells, especially those resistant to PARP inhibitors [194]. However, the phase 1/2 trial (NCT06065059) was terminated due to significant liver toxicity, with grade 3/4 hepatotoxicity observed in patients who continued treatment for longer than eight weeks. These results demonstrate the challenges in developing novel therapies and highlight the importance of safety monitoring in early-phase clinical trials.
KSQ-4279, also known as RO7623066, was developed by Roche/KSQ Therapeutics, and it is a potent and highly selective oral USP1 inhibitor. It binds to a cryptic site in USP1, similar to ML323, but with subtle rearrangements in the site folding, accounting for differences in potency and selectivity [195]. Currently, in a phase 1 trial (NCT05240898) for advanced solid tumors, KSQ-4279 is being studied as a single agent and in combination with Carboplatin or Olaparib. The combination of a PARP inhibitor and KSQ-4279 was shown to induce regression of several PDX PARP-resistant tumors in the preclinical phase [196]. Preliminary results showed limited efficacy as monotherapy but suggested potential synergy with Olaparib, particularly in BRCA1-mutated tumors. Pharmacokinetic studies demonstrated that the tested drug becomes saturated at higher doses, suggesting the need for twice-daily dosing. Pharmacodynamic data also appeared to support the theorized mechanism of action of USP1 inhibition.
XL309, also known as ISM3091, was developed using AI technology by Insilico Medicine, Boston, Massachusetts, USA and licensed to Exelixis, Alameda, California, USA, and it is a highly selective and non-covalent oral inhibitor of USP1 [197]. Preclinical data demonstrated high efficacy in BRCA1 mutated triple-negative breast cancer cell lines and synergistic effects with Olaparib in vivo. Interestingly, it also showed efficacy against a lung adenocarcinoma cell line without HRD, suggesting potential applications beyond HRD cancers. This opens the door to consider potentially utilizing XL309 in HR-proficient cancers. A phase 1 trial (NCT05932862) is ongoing to investigate its safety and preliminary antitumor activity.
SIM0501, developed by Simcere Jiangsu Pharmaceutical Co., Nanjing, Jiangsu, China, is another highly selective and non-covalent oral USP1 inhibitor [195]. It has shown evidence of synergy with Olaparib both in vitro and in vivo across various HRD-positive cancers. In vivo efficacy studies have demonstrated a dose-dependent increase of Ub-PCNA, which may be a useful biomarker to track for response, as Ub-PCNA is a direct target of USP1 [196]. A phase 1 trial (NCT06331559) is evaluating its safety and preliminary efficacy in patients with advanced solid tumors.
HSK39775, developed by Tibet Haisco Pharmaceutical Co. Ltd., Zedang Town, Shannan, Tibet, is a highly selective and non-covalent oral USP1 inhibitor. It has been shown to inhibit the USP1/UAF1 complex and exhibit strong dose-dependent tumor response in xenograft-derived BRCA-mutated triple-negative breast cancer with synergy seen when given with a PARP inhibitor. HSK39775 monotherapy was also found to inhibit the growth of BRCA wild-type lung cancer [196]. A phase 1/2 trial (NCT06314373) is underway to evaluate its safety and efficacy in advanced solid tumors. The field of USP1 inhibitors is rapidly evolving, and these clinical trials will provide crucial data for this novel therapeutic approach in cancer management, particularly for patients with HRD-positive tumors and those resistant to PARP inhibitors.
3.3.2. USP14 Inhibitor
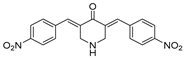
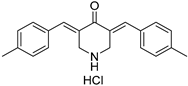
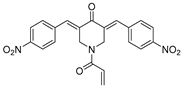
VLX1570, a small molecule DUB inhibitor derived from b-AP15 [(3E,5E)-3,5-bis[(4-nitrophenyl)methylidene]-1-(prop-2-enoyl)piperidin-4-one], selectively inhibits USP14 and UCHL5, which are associated with the 19S regulatory subunit of the proteasome. Both of these small molecule inhibitors are α,β-unsaturated carbonyl DUB inhibitors and have similar mechanisms of action. It was studied in a phase 1 clinical trial (NCT02372240) for relapsed or refractory multiple myeloma patients who had developed resistance to Bortezomib [198]. The trial, conducted by Eric K Rowinsky et al., aimed to determine the safety and tolerability of VLX1570. While the compound showed anti-myeloma effects at doses ≥ 0.6 mg/kg, the study was discontinued due to severe respiratory insufficiency in two patients that led to death [199]. Despite these setbacks, the unique mechanism of action of DUB inhibitors in Bortezomib-resistant multiple myeloma has prompted further research, including the development of a rat model to assess VLX1570-induced lung toxicity and the development of specific DUB inhibitors [199].

Table 4.
Clinical trials involving USP inhibitors.
Table 4.
Clinical trials involving USP inhibitors.
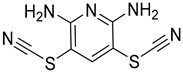
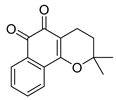
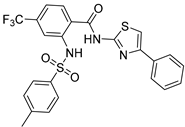
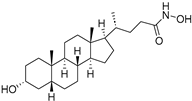
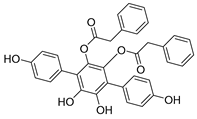
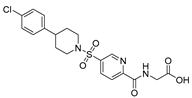
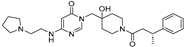
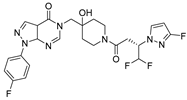
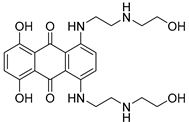
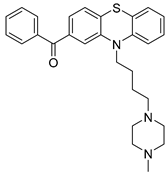
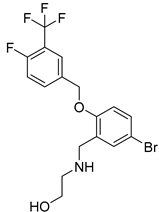
| Chemical Structure | Type of USP Inhibitor | Clinical Trials.gov ID | Trial Phase | Study Design | Results | References |
|---|---|---|---|---|---|---|
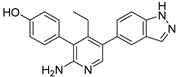
TNG348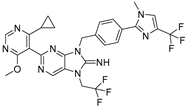 | USP1 IC50: 82 nM | NCT06065059 | Phase 1 dose escalation and Phase 2 dose expansion trial (terminated) |
|
| [194,200,201,202] |
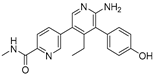
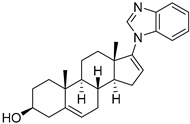
KSQ-4279, also known as RO7623066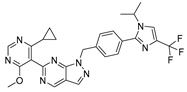 | USP1 IC50: 10 nM | NCT05240898 | Phase 1 (recruiting) |
|
| [71,196,203,,204] |
| XL309, also known as ISM3091 Chemical structure is not publicly disclosed | USP1 | NCT05932862 | Phase 1 (recruiting) |
|
| [197,205,206] |
| SIM0501 Chemical structure is not publicly disclosed. | USP1 | NCT06331559 | Phase 1 (recruiting) |
|
| [197,,207,208] |
| HSK39775 Chemical structure is not publicly disclosed. | USP1 | NCT06314373 | Phase 1 dose escalation and Phase 2 dose expansion trial (recruiting) |
|
| [197,209,210] |
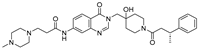
VLX1570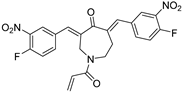 | USP14/UCHL5 IC50: ~10 µM | NCT02372240 | Phase 1 dose escalation and Phase 2 dose expansion trial (terminated) |
|
| [198,199,211,212] |
4. Summary and Future Directions
USP inhibitors have shown significant potential in combating drug resistance and enhancing the effectiveness of existing cancer therapies. The UPS is involved in a vast array of cellular functions; thus, developing more potent and selective USP inhibitors remains a primary focus. Current inhibitors often face challenges related to specificity and potency, given the possible off-target effects these USP inhibitors may have. Researchers are currently exploring novel allosteric binding sites within USPs to design inhibitors with improved selectivity and efficacy. Combination therapies involving USP inhibitors are being studied to overcome drug resistance in cancer treatment. Integrating USP inhibitors with existing anticancer therapies, such as chemotherapy and targeted therapy, shows potential for enhancing treatment outcomes [182]. USP inhibitors also have the key benefit of enhancing the effectiveness of immunotherapy by modulating the tumor microenvironment. Tailored approaches to treatment may lead to more effective and personalized therapy. As research progresses, expanding our understanding of the complex roles of USPs in various signaling pathways and cellular processes will be crucial [54]. Ongoing clinical trials suggest that USP inhibitors, particularly those targeting USP1, may have the potential to become valuable components of cancer therapy. While no USP inhibitors have yet been approved for clinical use, they have shown significant promise in preclinical studies. Advancing USP inhibitors from preclinical studies to clinical trials remains a significant goal, with the aim of bringing these promising therapeutic agents closer to clinical application. As these trials continue to expand and progress, more information about their safety and efficacy in patients will emerge, potentially leading to novel treatment strategies for patients with cancer.
5. Conclusions
In conclusion, this review provides an in-depth analysis of USPs’ structural and functional characteristics, their roles in various cancer-related pathways, and the mechanisms of action of over seventy different small molecule USP inhibitors. We also discuss how these inhibitors can enhance cancer immunotherapy, overcome drug resistance, and synergize with existing cancer treatments. Additionally, we explore the current progress and challenges in clinical trials involving USP inhibitors, highlighting their growing potential.
Research on USP inhibitors has shown their potential in targeting oncogenic pathways, modulating immune responses, and mitigating neurodegenerative protein aggregation. Targeting USPs by small-molecule inhibitors has demonstrated significant potential in tackling drug resistance and enhancing the effectiveness of existing cancer therapies. Several USP inhibitors, such as VLX1570 and b-AP15, have demonstrated efficacy in preclinical and early clinical trials, particularly in multiple myeloma and other malignancies.
However, challenges remain in developing selective and potent USP inhibitors due to the highly conserved catalytic domains among DUBs and potential off-target effects. The USPs are involved in multiple pathways, and additional studies are required to understand their complex mechanisms. Further advancements in structure-based drug design, high-throughput screening, and combination therapies may enhance their clinical applicability.
Author Contributions
M.B.: conceptualization; writing—original draft; writing—review and editing of the manuscript. S.K.: writing—original draft of USP inhibitors and immunotherapy section; writing—review and editing of the manuscript. K.B.: writing—original draft of USP inhibitor mechanisms of action section included in Table 3; writing—review and editing of the manuscript. N.D.K. and A.S.: writing—original draft of Table 3 and Table 4 structures and IC50 values; writing—review and editing of the manuscript. S.D.: writing—original draft of USP structure, biological function, and role in tumorigenesis and progression section; writing—review and editing of the manuscript. H.E.: writing—review and editing of the manuscript. Q.P.D.: writing—review and editing of the manuscript. Y.G.: conceptualization; supervision; writing—original draft; writing—review and editing of the manuscript. N.S.G.: conceptualization; supervision; writing—original draft; writing—review and editing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The research work in the Gavande laboratory is supported by the National Institutes of Health (R01CA247370 and R01AI161570), the Department of Defense (W81XWH-22-1-0369), the VA, KCI’s Michigan SPORE, Richard Barber Interdisciplinary Research Program, DMC Foundation, WSU Applebaum Faculty Research Award (FRAP), and Wayne State University. Sadaf Dorandish is supported by the NIH-funded Wayne State University Chemistry Biology Interface (CBI) Program T32GM142519-01.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Y.G. and N.S.G. highly appreciate the Karmanos Cancer Institute’s Molecular Therapeutics Program’s support for our initial studies on developing USP inhibitors for cancer therapy. The research work in the Ge laboratory is supported by the U Can-Cer Vive Foundation, Kids without Cancer, Children’s Foundation, and the Molecular Therapeutics Program of the Karmanos Cancer Institute.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bedford, L.; Lowe, J.; Dick, L.R.; Mayer, R.J.; Brownell, J.E. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat. Rev. Drug Discov. 2011, 10, 29–46. [Google Scholar] [CrossRef]
- Du, W.; Mei, Q.B. Ubiquitin-proteasome system, a new anti-tumor target. Acta Pharmacol. Sin. 2013, 34, 187–188. [Google Scholar] [CrossRef] [PubMed]
- Arkwright, R.; Pham, T.M.; Zonder, J.A.; Dou, Q.P. The preclinical discovery and development of bortezomib for the treatment of mantle cell lymphoma. Expert. Opin. Drug Discov. 2017, 12, 225–235. [Google Scholar] [CrossRef]
- Teicher, B.A.; Tomaszewski, J.E. Proteasome inhibitors. Biochem. Pharmacol. 2015, 96, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Snyder, N.A.; Silva, G.M. Deubiquitinating enzymes (DUBs): Regulation, homeostasis, and oxidative stress response. J. Biol. Chem. 2021, 297, 101077. [Google Scholar] [CrossRef]
- Johnston, S.C.; Larsen, C.N.; Cook, W.J.; Wilkinson, K.D.; Hill, C.P. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution. EMBO J. 1997, 16, 3787–3796. [Google Scholar] [CrossRef]
- Fang, Y.; Fu, D.; Shen, X.Z. The potential role of ubiquitin c-terminal hydrolases in oncogenesis. Biochim. Biophys. Acta 2010, 1806, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Lu, K.; Jiao, Q.; Chen, X.; Jia, F. The Regulation and Double-Edged Roles of the Deubiquitinase OTUD5. Cells 2023, 12, 1161. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, Y.; Okamoto, T.; Taniwaki, M.; Aizawa, M.; Inoue, M.; Katayama, S.; Kawakami, H.; Nakamura, S.; Nishimura, M.; Akiguchi, I.; et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat. Genet. 1994, 8, 221–228. [Google Scholar] [CrossRef]
- Shi, Z.; Chen, J.; Zhang, X.; Chu, J.; Han, Z.; Xu, D.; Gan, S.; Pan, X.; Ye, J.; Cui, X. Ataxin-3 promotes testicular cancer cell proliferation by inhibiting anti-oncogene PTEN. Biochem. Biophys. Res. Commun. 2018, 503, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Abdul Rehman, S.A.; Armstrong, L.A.; Lange, S.M.; Kristariyanto, Y.A.; Grawert, T.W.; Knebel, A.; Svergun, D.I.; Kulathu, Y. Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2. Mol. Cell 2021, 81, 4176–4190.e6. [Google Scholar] [CrossRef]
- Abdul Rehman, S.A.; Kristariyanto, Y.A.; Choi, S.Y.; Nkosi, P.J.; Weidlich, S.; Labib, K.; Hofmann, K.; Kulathu, Y. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol. Cell 2016, 63, 146–155. [Google Scholar] [CrossRef]
- Trulsson, F.; Akimov, V.; Robu, M.; van Overbeek, N.; Berrocal, D.A.P.; Shah, R.G.; Cox, J.; Shah, G.M.; Blagoev, B.; Vertegaal, A.C.O. Deubiquitinating enzymes and the proteasome regulate preferential sets of ubiquitin substrates. Nat. Commun. 2022, 13, 2736. [Google Scholar] [CrossRef] [PubMed]
- Hermanns, T.; Pichlo, C.; Woiwode, I.; Klopffleisch, K.; Witting, K.F.; Ovaa, H.; Baumann, U.; Hofmann, K. A family of unconventional deubiquitinases with modular chain specificity determinants. Nat. Commun. 2018, 9, 799. [Google Scholar] [CrossRef]
- Harper, S.; Gratton, H.E.; Cornaciu, I.; Oberer, M.; Scott, D.J.; Emsley, J.; Dreveny, I. Structure and catalytic regulatory function of ubiquitin specific protease 11 N-terminal and ubiquitin-like domains. Biochemistry 2014, 53, 2966–2978. [Google Scholar] [CrossRef] [PubMed]
- Cruz, L.; Soares, P.; Correia, M. Ubiquitin-Specific Proteases: Players in Cancer Cellular Processes. Pharmaceuticals 2021, 14, 848. [Google Scholar] [CrossRef]
- Ye, Y.; Scheel, H.; Hofmann, K.; Komander, D. Dissection of USP catalytic domains reveals five common insertion points. Mol. Biosyst. 2009, 5, 1797–1808. [Google Scholar] [CrossRef]
- Harrigan, J.A.; Jacq, X.; Martin, N.M.; Jackson, S.P. Deubiquitylating enzymes and drug discovery: Emerging opportunities. Nat. Rev. Drug Discov. 2018, 17, 57–78. [Google Scholar] [CrossRef]
- Reyes-Turcu, F.E.; Wilkinson, K.D. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem. Rev. 2009, 109, 1495–1508. [Google Scholar] [CrossRef]
- Reyes-Turcu, F.E.; Ventii, K.H.; Wilkinson, K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009, 78, 363–397. [Google Scholar] [CrossRef]
- Komander, D. Mechanism, specificity and structure of the deubiquitinases. Subcell. Biochem. 2010, 54, 69–87. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Zhou, Z.; Shah, A.A.; Zou, H.; Tao, J.; Chen, Q.; Wan, Y. The emerging role of deubiquitinating enzymes in genomic integrity, diseases, and therapeutics. Cell Biosci. 2016, 6, 62. [Google Scholar] [CrossRef] [PubMed]
- Dikic, I.; Wakatsuki, S.; Walters, K.J. Ubiquitin-binding domains—From structures to functions. Nat. Rev. Mol. Cell Biol. 2009, 10, 659–671. [Google Scholar] [CrossRef]
- Drag, M.; Mikolajczyk, J.; Bekes, M.; Reyes-Turcu, F.E.; Ellman, J.A.; Wilkinson, K.D.; Salvesen, G.S. Positional-scanning fluorigenic substrate libraries reveal unexpected specificity determinants of DUBs (deubiquitinating enzymes). Biochem. J. 2008, 415, 367–375. [Google Scholar] [CrossRef]
- Fan, Y.H.; Yu, Y.; Mao, R.F.; Tan, X.J.; Xu, G.F.; Zhang, H.; Lu, X.B.; Fu, S.B.; Yang, J. USP4 targets TAK1 to downregulate TNFalpha-induced NF-kappaB activation. Cell Death Differ. 2011, 18, 1547–1560. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Tan, X.; Wang, H.; Sun, W.; Shi, Y.; Burlingame, S.; Gu, X.; Cao, G.; Zhang, T.; Qin, J.; et al. Ubiquitin-specific peptidase 21 inhibits tumor necrosis factor alpha-induced nuclear factor kappaB activation via binding to and deubiquitinating receptor-interacting protein 1. J. Biol. Chem. 2010, 285, 969–978. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Gaynor, R.B. Therapeutic potential of inhibition of the NF-kappaB pathway in the treatment of inflammation and cancer. J. Clin. Investig. 2001, 107, 135–142. [Google Scholar] [CrossRef]
- Yun, S.I.; Kim, H.H.; Yoon, J.H.; Park, W.S.; Hahn, M.J.; Kim, H.C.; Chung, C.H.; Kim, K.K. Ubiquitin specific protease 4 positively regulates the WNT/beta-catenin signaling in colorectal cancer. Mol. Oncol. 2015, 9, 1834–1851. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, N.; Kurzrock, R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat. Rev. 2018, 62, 50–60. [Google Scholar] [CrossRef]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.; Liu, C.; Guan, X.; Wang, B.; Yao, Y.; Su, D.; Ma, Y.; Fang, L.; Zhang, Y. Ubiquitin specific peptidase 5 enhances STAT3 signaling and promotes migration and invasion in Pancreatic Cancer. J. Cancer 2020, 11, 6802–6811. [Google Scholar] [CrossRef] [PubMed]
- Qureshy, Z.; Johnson, D.E.; Grandis, J.R. Targeting the JAK/STAT pathway in solid tumors. J. Cancer Metastasis Treat 2020, 6, 21. [Google Scholar] [CrossRef]
- Lee, J.T.; Gu, W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010, 17, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, T.; Su, W.; Dou, Z.; Zhao, D.; Jin, X.; Lei, H.; Wang, J.; Xie, X.; Cheng, B.; et al. Mutant p53 in cancer: From molecular mechanism to therapeutic modulation. Cell Death Dis. 2022, 13, 974. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Yu, W.; Li, J.; Ma, H.; Wang, P.; Zhou, Y.; Wang, Y.; Zhang, J.; Shi, G. USP16 regulates castration-resistant prostate cancer cell proliferation by deubiquitinating and stablizing c-Myc. J. Exp. Clin. Cancer Res. 2021, 40, 59. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Luo, M. Cinobufotalin regulates the USP36/c-Myc axis to suppress malignant phenotypes of colon cancer cells in vitro and in vivo. Aging 2024, 16, 5526–5544. [Google Scholar] [CrossRef]
- Ruiz, E.J.; Pinto-Fernandez, A.; Turnbull, A.P.; Lan, L.; Charlton, T.M.; Scott, H.C.; Damianou, A.; Vere, G.; Riising, E.M.; Da Costa, C.; et al. USP28 deletion and small-molecule inhibition destabilizes c-MYC and elicits regression of squamous cell lung carcinoma. Elife 2021, 10, e71596. [Google Scholar] [CrossRef]
- Guo, J.; Zhao, J.; Sun, L.; Yang, C. Role of ubiquitin specific proteases in the immune microenvironment of prostate cancer: A new direction. Front. Oncol. 2022, 12, 955718. [Google Scholar] [CrossRef]
- Ji, M.; Shi, H.; Xie, Y.; Zhao, Z.; Li, S.; Chang, C.; Cheng, X.; Li, Y. Ubiquitin specific protease 22 promotes cell proliferation and tumor growth of epithelial ovarian cancer through synergy with transforming growth factor beta1. Oncol. Rep. 2015, 33, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xiang, H.; Lu, Y.; Wu, T. Role and clinical significance of TGF-beta1 and TGF-betaR1 in malignant tumors (Review). Int. J. Mol. Med. 2021, 47, 55. [Google Scholar] [CrossRef]
- Liu, J.; Kruswick, A.; Dang, H.; Tran, A.D.; Kwon, S.M.; Wang, X.W.; Oberdoerffer, P. Ubiquitin-specific protease 21 stabilizes BRCA2 to control DNA repair and tumor growth. Nat. Commun. 2017, 8, 137. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.M.; Kim, S.; Seo, S.U.; Kim, S.; Park, J.W.; Kim, G.; Choi, Y.R.; Hur, K.; Kwon, T.K. Inhibition of USP1 enhances anticancer drugs-induced cancer cell death through downregulation of survivin and miR-216a-5p-mediated upregulation of DR5. Cell Death Dis. 2022, 13, 821. [Google Scholar] [CrossRef]
- Foster, B.M.; Wang, Z.; Schmidt, C.K. DoUBLing up: Ubiquitin and ubiquitin-like proteases in genome stability. Biochem. J. 2024, 481, 515–545. [Google Scholar] [CrossRef]
- Li, Q.; Ye, C.; Tian, T.; Jiang, Q.; Zhao, P.; Wang, X.; Liu, F.; Shan, J.; Ruan, J. The emerging role of ubiquitin-specific protease 20 in tumorigenesis and cancer therapeutics. Cell Death Dis. 2022, 13, 434. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, S.; Narita, M.; Tsujimoto, Y. Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature 1999, 399, 483–487. [Google Scholar] [CrossRef]
- Trivigno, D.; Essmann, F.; Huber, S.M.; Rudner, J. Deubiquitinase USP9x confers radioresistance through stabilization of Mcl-1. Neoplasia 2012, 14, 893–904. [Google Scholar] [CrossRef]
- Ma, C.; Wang, D.; Tian, Z.; Gao, W.; Zang, Y.; Qian, L.; Xu, X.; Jia, J.; Liu, Z. USP13 deubiquitinates and stabilizes cyclin D1 to promote gastric cancer cell cycle progression and cell proliferation. Oncogene 2023, 42, 2249–2262. [Google Scholar] [CrossRef]
- Li, G.; Yang, T.; Chen, Y.; Bao, J.; Wu, D.; Hu, X.; Feng, C.; Xu, L.; Li, M.; Li, G.; et al. USP5 Sustains the Proliferation of Glioblastoma Through Stabilization of CyclinD1. Front. Pharmacol. 2021, 12, 720307. [Google Scholar] [CrossRef]
- Zhang, Z.; Cui, Z.; Xie, Z.; Li, C.; Xu, C.; Guo, X.; Yu, J.; Chen, T.; Facchinetti, F.; Bohnenberger, H.; et al. Deubiquitinase USP5 promotes non-small cell lung cancer cell proliferation by stabilizing cyclin D1. Transl. Lung Cancer Res. 2021, 10, 3995–4011. [Google Scholar] [CrossRef]
- Sun, H.; Ou, B.; Zhao, S.; Liu, X.; Song, L.; Liu, X.; Wang, R.; Peng, Z. USP11 promotes growth and metastasis of colorectal cancer via PPP1CA-mediated activation of ERK/MAPK signaling pathway. EBioMedicine 2019, 48, 236–247. [Google Scholar] [CrossRef]
- Brahimi-Horn, C.; Pouyssegur, J. When hypoxia signalling meets the ubiquitin-proteasomal pathway, new targets for cancer therapy. Crit. Rev. Oncol. Hematol. 2005, 53, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.C.; Zhao, C.J.; Jin, Z.F.; Zheng, J.; Ma, L.T. Targeted therapy based on ubiquitin-specific proteases, signalling pathways and E3 ligases in non-small-cell lung cancer. Front. Oncol. 2023, 13, 1120828. [Google Scholar] [CrossRef]
- Keijzer, N.; Priyanka, A.; Stijf-Bultsma, Y.; Fish, A.; Gersch, M.; Sixma, T.K. Variety in the USP deubiquitinase catalytic mechanism. Life Sci. Alliance 2024, 7, e202302533. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, Y.; Zhou, H. Advances in the Development Ubiquitin-Specific Peptidase (USP) Inhibitors. Int. J. Mol. Sci. 2021, 22, 4546. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, Y.; Niu, K.; Yang, R.; Lv, Q.; Zhang, W.; Feng, K.; Zhang, Y. Ubiquitin specific peptidase 3: An emerging deubiquitinase that regulates physiology and diseases. Cell Death Discov. 2024, 10, 243. [Google Scholar] [CrossRef]
- Wu, X.; Wang, H.; Zhu, D.; Chai, Y.; Wang, J.; Dai, W.; Xiao, Y.; Tang, W.; Li, J.; Hong, L.; et al. USP3 promotes gastric cancer progression and metastasis by deubiquitination-dependent COL9A3/COL6A5 stabilisation. Cell Death Dis. 2021, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.; Layfield, R.; Oldham, N.J. Structural insights into interactions between ubiquitin specific protease 5 and its polyubiquitin substrates by mass spectrometry and ion mobility spectrometry. Protein Sci. 2015, 24, 1257–1263. [Google Scholar] [CrossRef] [PubMed]
- Novinec, M.; Lenarcic, B. Papain-like peptidases: Structure, function, and evolution. Biomol. Concepts 2013, 4, 287–308. [Google Scholar] [CrossRef]
- Yan, B.; Guo, J.; Deng, S.; Chen, D.; Huang, M. A pan-cancer analysis of the role of USP5 in human cancers. Sci. Rep. 2023, 13, 8972. [Google Scholar] [CrossRef]
- Nie, L.; Wang, C.; Liu, X.; Teng, H.; Li, S.; Huang, M.; Feng, X.; Pei, G.; Hang, Q.; Zhao, Z.; et al. USP7 substrates identified by proteomics analysis reveal the specificity of USP7. Genes. Dev. 2022, 36, 1016–1030. [Google Scholar] [CrossRef] [PubMed]
- Korenev, G.; Yakukhnov, S.; Druk, A.; Golovina, A.; Chasov, V.; Mirgayazova, R.; Ivanov, R.; Bulatov, E. USP7 Inhibitors in Cancer Immunotherapy: Current Status and Perspective. Cancers 2022, 14, 5539. [Google Scholar] [CrossRef]
- Tao, L.; Liu, X.; Jiang, X.; Zhang, K.; Wang, Y.; Li, X.; Jiang, S.; Han, T. USP10 as a Potential Therapeutic Target in Human Cancers. Genes. 2022, 13, 831. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.A.; Rossi, M. Emerging Role of Ubiquitin-Specific Protease 19 in Oncogenesis and Cancer Development. Front. Cell Dev. Biol. 2022, 10, 889166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; van Dinther, M.; Thorikay, M.; Gourabi, B.M.; Kruithof, B.P.T.; Ten Dijke, P. Opposing USP19 splice variants in TGF-beta signaling and TGF-beta-induced epithelial-mesenchymal transition of breast cancer cells. Cell Mol. Life Sci. 2023, 80, 43. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Clague, M.J.; Urbe, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Song, Y.; Wang, K.; Liu, G.; Dong, X.; Yang, F.; Chen, G.; Cao, C.; Zhang, H.; Wang, M.; et al. USP32 deubiquitinase: Cellular functions, regulatory mechanisms, and potential as a cancer therapy target. Cell Death Discov. 2023, 9, 338. [Google Scholar] [CrossRef]
- O’Dea, R.; Kazi, N.; Hoffmann-Benito, A.; Zhao, Z.; Recknagel, S.; Wendrich, K.; Janning, P.; Gersch, M. Molecular basis for ubiquitin/Fubi cross-reactivity in USP16 and USP36. Nat. Chem. Biol. 2023, 19, 1394–1405. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Young, M.A.; Donato, N.J. Emerging potential of therapeutic targeting of ubiquitin-specific proteases in the treatment of cancer. Cancer Res. 2014, 74, 4955–4966. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.L.; Shen, G.T.; Li, N.L. Emerging potential of ubiquitin-specific proteases and ubiquitin-specific proteases inhibitors in breast cancer treatment. World J. Clin. Cases 2022, 10, 11690–11701. [Google Scholar] [CrossRef]
- Clague, M.J.; Urbe, S.; Komander, D. Breaking the chains: Deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 2019, 20, 338–352. [Google Scholar] [CrossRef]
- Zhou, F.; Li, Z.; Zhou, L.; Zhu, W.; Zhang, J.; Yang, W.; Xue, L.; Qin, X.; Chen, P.; Tang, R. Abstract 6201: Identification of SP-002, a highly selective USP1 inhibitor effectively inhibits HRD tumor growth and displays low hematotoxicity risk. Cancer Res. 2023, 83, 6201. [Google Scholar] [CrossRef]
- Gore, S.D.; Carducci, M.A. Modifying histones to tame cancer: Clinical development of sodium phenylbutyrate and other histone deacetylase inhibitors. Expert. Opin. Investig. Drugs 2000, 9, 2923–2934. [Google Scholar] [CrossRef] [PubMed]
- Mistry, H.; Hsieh, G.; Buhrlage, S.J.; Huang, M.; Park, E.; Cuny, G.D.; Galinsky, I.; Stone, R.M.; Gray, N.S.; D’Andrea, A.D.; et al. Small-molecule inhibitors of USP1 target ID1 degradation in leukemic cells. Mol. Cancer Ther. 2013, 12, 2651–2662. [Google Scholar] [CrossRef] [PubMed]
- Shang, K.; Zhang, L.; Yu, Y.; Xiao, H.; Gao, Y.; Yang, L.; Huang, J.; Song, H.; Han, H. Disulfide-containing polymer delivery of C527 and a Platinum(IV) prodrug selectively inhibited protein ubiquitination and tumor growth on cisplatin resistant and patient-derived liver cancer models. Mater. Today Bio 2023, 18, 100548. [Google Scholar] [CrossRef]
- Das, D.S.; Das, A.; Ray, A.; Song, Y.; Samur, M.K.; Munshi, N.C.; Chauhan, D.; Anderson, K.C. Blockade of Deubiquitylating Enzyme USP1 Inhibits DNA Repair and Triggers Apoptosis in Multiple Myeloma Cells. Clin. Cancer Res. 2017, 23, 4280–4289. [Google Scholar] [CrossRef]
- Dexheimer, T.S.; Rosenthal, A.S.; Luci, D.K.; Liang, Q.; Villamil, M.A.; Chen, J.; Sun, H.; Kerns, E.H.; Simeonov, A.; Jadhav, A.; et al. Synthesis and structure-activity relationship studies of N-benzyl-2-phenylpyrimidin-4-amine derivatives as potent USP1/UAF1 deubiquitinase inhibitors with anticancer activity against nonsmall cell lung cancer. J. Med. Chem. 2014, 57, 8099–8110. [Google Scholar] [CrossRef] [PubMed]
- Rennie, M.L.; Arkinson, C.; Chaugule, V.K.; Walden, H. Cryo-EM reveals a mechanism of USP1 inhibition through a cryptic binding site. Sci. Adv. 2022, 8, eabq6353. [Google Scholar] [CrossRef] [PubMed]
- Dexheimer, T.S.; Rosenthal, A.S.; Liang, Q.; Chen, J.; Villamil, M.A.; Kerns, E.H.; Simeonov, A.; Jadhav, A.; Zhuang, Z.; Maloney, D.J. Discovery of ML323 as a Novel Inhibitor of the USP1/UAF1 Deubiquitinase Complex. In Probe Reports from the NIH Molecular Libraries Program; National Center for Biotechnology Information: Bethesda, MD, USA, 2010. [Google Scholar]
- Chen, J.; Dexheimer, T.S.; Ai, Y.; Liang, Q.; Villamil, M.A.; Inglese, J.; Maloney, D.J.; Jadhav, A.; Simeonov, A.; Zhuang, Z. Selective and cell-active inhibitors of the USP1/ UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chem. Biol. 2011, 18, 1390–1400. [Google Scholar] [CrossRef]
- Li, X.Y.; Wu, J.C.; Liu, P.; Li, Z.J.; Wang, Y.; Chen, B.Y.; Hu, C.L.; Fei, M.Y.; Yu, P.C.; Jiang, Y.L.; et al. Inhibition of USP1 reverses the chemotherapy resistance through destabilization of MAX in the relapsed/refractory B-cell lymphoma. Leukemia 2023, 37, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Dakir, E.H.; Pickard, A.; Srivastava, K.; McCrudden, C.M.; Gross, S.R.; Lloyd, S.; Zhang, S.D.; Margariti, A.; Morgan, R.; Rudland, P.S.; et al. The anti-psychotic drug pimozide is a novel chemotherapeutic for breast cancer. Oncotarget 2018, 9, 34889–34910. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Xia, Y.; Gao, T.; Xu, F.; Lei, Q.; Peng, C.; Yang, Y.; Xue, Q.; Hu, X.; Wang, Q.; et al. The antipsychotic agent trifluoperazine hydrochloride suppresses triple-negative breast cancer tumor growth and brain metastasis by inducing G0/G1 arrest and apoptosis. Cell Death Dis. 2018, 9, 1006. [Google Scholar] [CrossRef] [PubMed]
- Valacchi, G.; Pecorelli, A.; Sticozzi, C.; Torricelli, C.; Muscettola, M.; Aldinucci, C.; Maioli, E. Rottlerin exhibits antiangiogenic effects in vitro. Chem. Biol. Drug Des. 2011, 77, 460–470. [Google Scholar] [CrossRef]
- Kumar, D.; Shankar, S.; Srivastava, R.K. Rottlerin-induced autophagy leads to the apoptosis in breast cancer stem cells: Molecular mechanisms. Mol. Cancer 2013, 12, 171. [Google Scholar] [CrossRef] [PubMed]
- McClurg, U.L.; Summerscales, E.E.; Harle, V.J.; Gaughan, L.; Robson, C.N. Deubiquitinating enzyme Usp12 regulates the interaction between the androgen receptor and the Akt pathway. Oncotarget 2014, 5, 7081–7092. [Google Scholar] [CrossRef] [PubMed]
- Altun, M.; Kramer, H.B.; Willems, L.I.; McDermott, J.L.; Leach, C.A.; Goldenberg, S.J.; Kumar, K.G.; Konietzny, R.; Fischer, R.; Kogan, E.; et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 2011, 18, 1401–1412. [Google Scholar] [CrossRef]
- Lin, W.C.; Chiu, Y.L.; Kuo, K.L.; Chow, P.M.; Hsu, C.H.; Liao, S.M.; Dong, J.R.; Chang, S.C.; Liu, S.H.; Liu, T.J.; et al. Anti-tumor effects of deubiquitinating enzyme inhibitor PR-619 in human chondrosarcoma through reduced cell proliferation and endoplasmic reticulum stress-related apoptosis. Am. J. Cancer Res. 2023, 13, 3055–3066. [Google Scholar] [PubMed]
- Ohayon, S.; Refua, M.; Hendler, A.; Aharoni, A.; Brik, A. Harnessing the oxidation susceptibility of deubiquitinases for inhibition with small molecules. Angew. Chem. Int. Ed. Engl. 2015, 54, 599–603. [Google Scholar] [CrossRef]
- Zhang, S.; Guo, Y.; Zhang, S.; Wang, Z.; Zhang, Y.; Zuo, S. Targeting the deubiquitinase USP2 for malignant tumor therapy (Review). Oncol. Rep. 2023, 50, 176. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.I.; Pragani, R.; Fox, J.T.; Shen, M.; Parmar, K.; Gaudiano, E.F.; Liu, L.; Tanega, C.; McGee, L.; Hall, M.D.; et al. Small Molecule Inhibition of the Ubiquitin-specific Protease USP2 Accelerates cyclin D1 Degradation and Leads to Cell Cycle Arrest in Colorectal Cancer and Mantle Cell Lymphoma Models. J. Biol. Chem. 2016, 291, 24628–24640. [Google Scholar] [CrossRef] [PubMed]
- Magiera, K.; Tomala, M.; Kubica, K.; De Cesare, V.; Trost, M.; Zieba, B.J.; Kachamakova-Trojanowska, N.; Les, M.; Dubin, G.; Holak, T.A.; et al. Lithocholic Acid Hydroxyamide Destabilizes Cyclin D1 and Induces G(0)/G(1) Arrest by Inhibiting Deubiquitinase USP2a. Cell Chem. Biol. 2017, 24, 458–470.e18. [Google Scholar] [CrossRef]
- Tomala, M.D.; Magiera-Mularz, K.; Kubica, K.; Krzanik, S.; Zieba, B.; Musielak, B.; Pustula, M.; Popowicz, G.M.; Sattler, M.; Dubin, G.; et al. Identification of small-molecule inhibitors of USP2a. Eur. J. Med. Chem. 2018, 150, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Chuang, S.J.; Cheng, S.C.; Tang, H.C.; Sun, C.Y.; Chou, C.Y. 6-Thioguanine is a noncompetitive and slow binding inhibitor of human deubiquitinating protease USP2. Sci. Rep. 2018, 8, 3102. [Google Scholar] [CrossRef]
- Lin, H.C.; Kuan, Y.; Chu, H.F.; Cheng, S.C.; Pan, H.C.; Chen, W.Y.; Sun, C.Y.; Lin, T.H. Disulfiram and 6-Thioguanine synergistically inhibit the enzymatic activities of USP2 and USP21. Int. J. Biol. Macromol. 2021, 176, 490–497. [Google Scholar] [CrossRef]
- Issaenko, O.A.; Amerik, A.Y. Chalcone-based small-molecule inhibitors attenuate malignant phenotype via targeting deubiquitinating enzymes. Cell Cycle 2012, 11, 1804–1817. [Google Scholar] [CrossRef]
- Coughlin, K.; Anchoori, R.; Iizuka, Y.; Meints, J.; MacNeill, L.; Vogel, R.I.; Orlowski, R.Z.; Lee, M.K.; Roden, R.B.; Bazzaro, M. Small-molecule RA-9 inhibits proteasome-associated DUBs and ovarian cancer in vitro and in vivo via exacerbating unfolded protein responses. Clin. Cancer Res. 2014, 20, 3174–3186. [Google Scholar] [CrossRef] [PubMed]
- Mirzapoiazova, T.; Pozhitkov, A.; Nam, A.; Mambetsariev, I.; Nelson, M.S.; Tan, Y.C.; Zhang, K.; Raz, D.; Singhal, S.; Nasser, M.W.; et al. Effects of selected deubiquitinating enzyme inhibitors on the proliferation and motility of lung cancer and mesothelioma cell lines. Int. J. Oncol. 2020, 57, 80–86. [Google Scholar] [CrossRef]
- Nicholson, B.; Leach, C.A.; Goldenberg, S.J.; Francis, D.M.; Kodrasov, M.P.; Tian, X.; Shanks, J.; Sterner, D.E.; Bernal, A.; Mattern, M.R.; et al. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci. 2008, 17, 1035–1043. [Google Scholar] [CrossRef]
- Vamisetti, G.B.; Meledin, R.; Gopinath, P.; Brik, A. Halogen Substituents in the Isoquinoline Scaffold Switches the Selectivity of Inhibition between USP2 and USP7. Chembiochem 2019, 20, 282–286. [Google Scholar] [CrossRef]
- Okada, K.; Ye, Y.Q.; Taniguchi, K.; Yoshida, A.; Akiyama, T.; Yoshioka, Y.; Onose, J.; Koshino, H.; Takahashi, S.; Yajima, A.; et al. Vialinin A is a ubiquitin-specific peptidase inhibitor. Bioorg. Med. Chem. Lett. 2013, 23, 4328–4331. [Google Scholar] [CrossRef]
- Xu, J.; Chen, D.; Jin, L.; Chen, Z.; Tu, Y.; Huang, X.; Xue, F.; Xu, J.; Chen, M.; Wang, X.; et al. Ubiquitously specific protease 4 inhibitor-Vialinin A attenuates inflammation and fibrosis in S100-induced hepatitis mice through Rheb/mTOR signalling. J. Cell Mol. Med. 2021, 25, 1140–1150. [Google Scholar] [CrossRef]
- Bailly, C.; Vergoten, G. Binding of Vialinin A and p-Terphenyl Derivatives to Ubiquitin-Specific Protease 4 (USP4): A Molecular Docking Study. Molecules 2022, 27, 5909. [Google Scholar] [CrossRef]
- Mann, M.K.; Zepeda-Velázquez, C.A.; González-Álvarez, H.; Dong, A.; Kiyota, T.; Aman, A.M.; Loppnau, P.; Li, Y.; Wilson, B.; Arrowsmith, C.H.; et al. Structure-Activity Relationship of USP5 Inhibitors. J. Med. Chem. 2021, 64, 15017–15036. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.N.; Zhou, Y.B.; Gao, A.H.; Cao, J.Y.; Gao, L.X.; Sheng, L.; Xu, L.; Su, M.B.; Cao, X.C.; Han, M.M.; et al. Curcusone D, a novel ubiquitin-proteasome pathway inhibitor via ROS-induced DUB inhibition, is synergistic with bortezomib against multiple myeloma cell growth. Biochim. Biophys. Acta 2014, 1840, 2004–2013. [Google Scholar] [CrossRef]
- Kapuria, V.; Peterson, L.F.; Fang, D.; Bornmann, W.G.; Talpaz, M.; Donato, N.J. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010, 70, 9265–9276. [Google Scholar] [CrossRef]
- Kim, S.; Woo, S.M.; Min, K.J.; Seo, S.U.; Lee, T.J.; Kubatka, P.; Kim, D.E.; Kwon, T.K. WP1130 Enhances TRAIL-Induced Apoptosis through USP9X-Dependent miR-708-Mediated Downregulation of c-FLIP. Cancers 2019, 11, 344. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liu, H.M. Recent advances in the development of ubiquitin-specific-processing protease 7 (USP7) inhibitors. Eur. J. Med. Chem. 2020, 191, 112107. [Google Scholar] [CrossRef]
- D’Arcy, P.; Wang, X.; Linder, S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol. Ther. 2015, 147, 32–54. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.F.; Sun, H.; Liu, Y.; Potu, H.; Kandarpa, M.; Ermann, M.; Courtney, S.M.; Young, M.; Showalter, H.D.; Sun, D.; et al. Targeting deubiquitinase activity with a novel small-molecule inhibitor as therapy for B-cell malignancies. Blood 2015, 125, 3588–3597. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, L.; Niu, X.; Guo, Y.; Zhao, J.; Li, L.; Zhao, J. EOAI, a ubiquitin-specific peptidase 5 inhibitor, prevents non-small cell lung cancer progression by inducing DNA damage. BMC Cancer 2023, 23, 28. [Google Scholar] [CrossRef] [PubMed]
- Colland, F.; Formstecher, E.; Jacq, X.; Reverdy, C.; Planquette, C.; Conrath, S.; Trouplin, V.; Bianchi, J.; Aushev, V.N.; Camonis, J.; et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol. Cancer Ther. 2009, 8, 2286–2295. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Donato, N.J. Ubiquitin-specific proteases as therapeutic targets for the treatment of breast cancer. Breast Cancer Res. 2014, 16, 461. [Google Scholar] [CrossRef] [PubMed]
- Reverdy, C.; Conrath, S.; Lopez, R.; Planquette, C.; Atmanene, C.; Collura, V.; Harpon, J.; Battaglia, V.; Vivat, V.; Sippl, W.; et al. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem. Biol. 2012, 19, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.M.; Cheng, G.; Cheng, X.D.; Xu, Z.; Xu, B.; Zhang, W.D.; Qin, J.J. Targeting USP7-Mediated Deubiquitination of MDM2/MDMX-p53 Pathway for Cancer Therapy: Are We There Yet? Front. Cell Dev. Biol. 2020, 8, 233. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Miyazaki, M.; Kodrasov, M.P.; Rotinsulu, H.; Losung, F.; Mangindaan, R.E.; de Voogd, N.J.; Yokosawa, H.; Nicholson, B.; Tsukamoto, S. Spongiacidin C, a pyrrole alkaloid from the marine sponge Stylissa massa, functions as a USP7 inhibitor. Bioorg Med. Chem. Lett. 2013, 23, 3884–3886. [Google Scholar] [CrossRef] [PubMed]
- Kategaya, L.; Di Lello, P.; Rouge, L.; Pastor, R.; Clark, K.R.; Drummond, J.; Kleinheinz, T.; Lin, E.; Upton, J.P.; Prakash, S.; et al. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature 2017, 550, 534–538. [Google Scholar] [CrossRef]
- Kategaya, L.; Di Lello, P.; Rougé, L.; Pastor , R.; Clark, K.R.; Drummond, J. Identified Selective USP7 Inhibitors Compete with Ubiquitin Binding. Cancer Discov. 2017, 7, 1365. [Google Scholar] [CrossRef]
- Wang, S.A.; Young, M.J.; Wang, Y.C.; Chen, S.H.; Liu, C.Y.; Lo, Y.A.; Jen, H.H.; Hsu, K.C.; Hung, J.J. USP24 promotes drug resistance during cancer therapy. Cell Death Differ. 2021, 28, 2690–2707. [Google Scholar] [CrossRef]
- Lamberto, I.; Liu, X.; Seo, H.S.; Schauer, N.J.; Iacob, R.E.; Hu, W.; Das, D.; Mikhailova, T.; Weisberg, E.L.; Engen, J.R.; et al. Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7. Cell Chem. Biol. 2017, 24, 1490–1500 e1411. [Google Scholar] [CrossRef] [PubMed]
- Schauer, N.J.; Liu, X.; Magin, R.S.; Doherty, L.M.; Chan, W.C.; Ficarro, S.B.; Hu, W.; Roberts, R.M.; Iacob, R.E.; Stolte, B.; et al. Selective USP7 inhibition elicits cancer cell killing through a p53-dependent mechanism. Sci. Rep. 2020, 10, 5324. [Google Scholar] [CrossRef] [PubMed]
- Gavory, G.; O’Dowd, C.R.; Helm, M.D.; Flasz, J.; Arkoudis, E.; Dossang, A.; Hughes, C.; Cassidy, E.; McClelland, K.; Odrzywol, E.; et al. Discovery and characterization of highly potent and selective allosteric USP7 inhibitors. Nat. Chem. Biol. 2018, 14, 118–125. [Google Scholar] [CrossRef]
- O’Dowd, C.R.; Helm, M.D.; Rountree, J.S.S.; Flasz, J.T.; Arkoudis, E.; Miel, H.; Hewitt, P.R.; Jordan, L.; Barker, O.; Hughes, C.; et al. Identification and Structure-Guided Development of Pyrimidinone Based USP7 Inhibitors. ACS Med. Chem. Lett. 2018, 9, 238–243. [Google Scholar] [CrossRef]
- Turnbull, A.P.; Ioannidis, S.; Krajewski, W.W.; Pinto-Fernandez, A.; Heride, C.; Martin, A.C.L.; Tonkin, L.M.; Townsend, E.C.; Buker, S.M.; Lancia, D.R.; et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature 2017, 550, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Benavente, C. Abstract PR04: USP7 inhibitors prevent triple negative breast cancer metastasis by inducing UHRF1 protein degradation. Cancer Res. 2024, 84, PR04. [Google Scholar] [CrossRef]
- Li, X.; Kong, L.; Yang, Q.; Duan, A.; Ju, X.; Cai, B.; Chen, L.; An, T.; Li, Y. Parthenolide inhibits ubiquitin-specific peptidase 7 (USP7), Wnt signaling, and colorectal cancer cell growth. J. Biol. Chem. 2020, 295, 3576–3589. [Google Scholar] [CrossRef]
- Cheng, Y.J.; Zhuang, Z.; Miao, Y.L.; Song, S.S.; Bao, X.B.; Yang, C.H.; He, J.X. Identification of YCH2823 as a novel USP7 inhibitor for cancer therapy. Biochem. Pharmacol. 2024, 222, 116071. [Google Scholar] [CrossRef] [PubMed]
- Weisberg, E.L.; Schauer, N.J.; Yang, J.; Lamberto, I.; Doherty, L.; Bhatt, S.; Nonami, A.; Meng, C.; Letai, A.; Wright, R.; et al. Inhibition of USP10 induces degradation of oncogenic FLT3. Nat. Chem. Biol. 2017, 13, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Javaid, S.; Zadi, S.; Awais, M.; Wahab, A.T.; Zafar, H.; Maslennikov, I.; Choudhary, M.I. Identification of new leads against ubiquitin specific protease-7 (USP7): A step towards the potential treatment of cancers. RSC Adv. 2024, 14, 33080–33093. [Google Scholar] [CrossRef]
- Lee, G.; Oh, T.I.; Um, K.B.; Yoon, H.; Son, J.; Kim, B.M.; Kim, H.I.; Kim, H.; Kim, Y.J.; Lee, C.S.; et al. Small-molecule inhibitors of USP7 induce apoptosis through oxidative and endoplasmic reticulum stress in cancer cells. Biochem. Biophys. Res. Commun. 2016, 470, 181–186. [Google Scholar] [CrossRef]
- Fan, Y.H.; Cheng, J.; Vasudevan, S.A.; Dou, J.; Zhang, H.; Patel, R.H.; Ma, I.T.; Rojas, Y.; Zhao, Y.; Yu, Y.; et al. USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis. 2013, 4, e867. [Google Scholar] [CrossRef]
- Cartel, M.; Mouchel, P.L.; Gotanegre, M.; David, L.; Bertoli, S.; Mansat-De Mas, V.; Besson, A.; Sarry, J.E.; Manenti, S.; Didier, C. Inhibition of ubiquitin-specific protease 7 sensitizes acute myeloid leukemia to chemotherapy. Leukemia 2021, 35, 417–432. [Google Scholar] [CrossRef] [PubMed]
- Pozhidaeva, A.; Valles, G.; Wang, F.; Wu, J.; Sterner, D.E.; Nguyen, P.; Weinstock, J.; Kumar, K.G.S.; Kanyo, J.; Wright, D.; et al. USP7-Specific Inhibitors Target and Modify the Enzyme’s Active Site via Distinct Chemical Mechanisms. Cell Chem. Biol. 2017, 24, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, D.; Tian, Z.; Nicholson, B.; Kumar, K.G.; Zhou, B.; Carrasco, R.; McDermott, J.L.; Leach, C.A.; Fulcinniti, M.; Kodrasov, M.P.; et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell 2012, 22, 345–358. [Google Scholar] [CrossRef]
- An, T.; Gong, Y.; Li, X.; Kong, L.; Ma, P.; Gong, L.; Zhu, H.; Yu, C.; Liu, J.; Zhou, H.; et al. USP7 inhibitor P5091 inhibits Wnt signaling and colorectal tumor growth. Biochem. Pharmacol. 2017, 131, 29–39. [Google Scholar] [CrossRef]
- Granieri, L.; Marocchi, F.; Melixetian, M.; Mohammadi, N.; Nicoli, P.; Cuomo, A.; Bonaldi, T.; Confalonieri, S.; Pisati, F.; Giardina, G.; et al. Targeting the USP7/RRM2 axis drives senescence and sensitizes melanoma cells to HDAC/LSD1 inhibitors. Cell Rep. 2022, 40, 111396. [Google Scholar] [CrossRef]
- Weinstock, J.; Wu, J.; Cao, P.; Kingsbury, W.D.; McDermott, J.L.; Kodrasov, M.P.; McKelvey, D.M.; Suresh Kumar, K.G.; Goldenberg, S.J.; Mattern, M.R.; et al. Selective Dual Inhibitors of the Cancer-Related Deubiquitylating Proteases USP7 and USP47. ACS Med. Chem. Lett. 2012, 3, 789–792. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Vallese, S.; Peretto, I.; Jacq, X.; Rain, J.C.; Colland, F.; Guedat, P. Synthesis and biological evaluation of 9-oxo-9H-indeno[1,2-b]pyrazine-2,3-dicarbonitrile analogues as potential inhibitors of deubiquitinating enzymes. ChemMedChem 2010, 5, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Clancy, A.; Heride, C.; Pinto-Fernandez, A.; Elcocks, H.; Kallinos, A.; Kayser-Bricker, K.J.; Wang, W.; Smith, V.; Davis, S.; Fessler, S.; et al. The deubiquitylase USP9X controls ribosomal stalling. J. Cell Biol. 2021, 220, e202004211. [Google Scholar] [CrossRef]
- Yu, M.; Fang, Z.X.; Wang, W.W.; Zhang, Y.; Bu, Z.L.; Liu, M.; Xiao, X.H.; Zhang, Z.L.; Zhang, X.M.; Cao, Y.; et al. Wu-5, a novel USP10 inhibitor, enhances crenolanib-induced FLT3-ITD-positive AML cell death via inhibiting FLT3 and AMPK pathways. Acta Pharmacol. Sin. 2021, 42, 604–612. [Google Scholar] [CrossRef]
- Yang, J.; Meng, C.; Weisberg, E.; Case, A.; Lamberto, I.; Magin, R.S.; Adamia, S.; Wang, J.; Gray, N.; Liu, S.; et al. Inhibition of the deubiquitinase USP10 induces degradation of SYK. Br. J. Cancer 2020, 122, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xia, H.; Kim, M.; Xu, L.; Li, Y.; Zhang, L.; Cai, Y.; Norberg, H.V.; Zhang, T.; Furuya, T.; et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 2011, 147, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Ndubaku, C.; Tsui, V. Inhibiting the deubiquitinating enzymes (DUBs). J. Med. Chem. 2015, 58, 1581–1595. [Google Scholar] [CrossRef] [PubMed]
- Kunimasa, K.; Ikeda-Ishikawa, C.; Tani, Y.; Tsukahara, S.; Sakurai, J.; Okamoto, Y.; Koido, M.; Dan, S.; Tomida, A. Spautin-1 inhibits mitochondrial complex I and leads to suppression of the unfolded protein response and cell survival during glucose starvation. Sci. Rep. 2022, 12, 11533. [Google Scholar] [CrossRef]
- Burkhart, R.A.; Peng, Y.; Norris, Z.A.; Tholey, R.M.; Talbott, V.A.; Liang, Q.; Ai, Y.; Miller, K.; Lal, S.; Cozzitorto, J.A.; et al. Mitoxantrone targets human ubiquitin-specific peptidase 11 (USP11) and is a potent inhibitor of pancreatic cancer cell survival. Mol. Cancer Res. 2013, 11, 901–911. [Google Scholar] [CrossRef]
- McClurg, U.L.; Azizyan, M.; Dransfield, D.T.; Namdev, N.; Chit, N.; Nakjang, S.; Robson, C.N. The novel anti-androgen candidate galeterone targets deubiquitinating enzymes, USP12 and USP46, to control prostate cancer growth and survival. Oncotarget 2018, 9, 24992–25007. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Lee, M.J.; Park, S.; Oh, D.C.; Elsasser, S.; Chen, P.C.; Gartner, C.; Dimova, N.; Hanna, J.; Gygi, S.P.; et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 2010, 467, 179–184. [Google Scholar] [CrossRef]
- Xu, L.; Wang, J.; Yuan, X.; Yang, S.; Xu, X.; Li, K.; He, Y.; Wei, L.; Zhang, J.; Tian, Y. IU1 suppresses proliferation of cervical cancer cells through MDM2 degradation. Int. J. Biol. Sci. 2020, 16, 2951–2963. [Google Scholar] [CrossRef] [PubMed]
- Boselli, M.; Lee, B.H.; Robert, J.; Prado, M.A.; Min, S.W.; Cheng, C.; Silva, M.C.; Seong, C.; Elsasser, S.; Hatle, K.M.; et al. An inhibitor of the proteasomal deubiquitinating enzyme USP14 induces tau elimination in cultured neurons. J. Biol. Chem. 2017, 292, 19209–19225. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, Y.; Ding, S.; Li, J.; Song, N.; Ren, Y.; Hong, D.; Wu, C.; Li, B.; Wang, F.; et al. Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade. Cell Res. 2018, 28, 1186–1194. [Google Scholar] [CrossRef]
- Li, P.; Zhu, X.; Qu, H.; Han, Z.; Yao, X.; Wei, Y.; Li, B.; Chen, H. Synergistic Effect of Ubiquitin-Specific Protease 14 and Poly(ADP-Ribose) Glycohydrolase Co-Inhibition in BRCA1-Mutant, Poly(ADP-Ribose) Polymerase Inhibitor-Resistant Triple-Negative Breast Cancer Cells. Onco Targets Ther. 2024, 17, 741–753. [Google Scholar] [CrossRef]
- Chen, X.; Yang, Q.; Xiao, L.; Tang, D.; Dou, Q.P.; Liu, J. Metal-based proteasomal deubiquitinase inhibitors as potential anticancer agents. Cancer Metastasis Rev. 2017, 36, 655–668. [Google Scholar] [CrossRef]
- Liu, N.; Li, X.; Huang, H.; Zhao, C.; Liao, S.; Yang, C.; Liu, S.; Song, W.; Lu, X.; Lan, X.; et al. Clinically used antirheumatic agent auranofin is a proteasomal deubiquitinase inhibitor and inhibits tumor growth. Oncotarget 2014, 5, 5453–5471. [Google Scholar] [CrossRef]
- Liu, N.; Liu, C.; Li, X.; Liao, S.; Song, W.; Yang, C.; Zhao, C.; Huang, H.; Guan, L.; Zhang, P.; et al. A novel proteasome inhibitor suppresses tumor growth via targeting both 19S proteasome deubiquitinases and 20S proteolytic peptidases. Sci. Rep. 2014, 4, 5240. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Chen, X.; Yang, C.; Zang, D.; Lan, X.; Liao, S.; Zhang, P.; Wu, J.; Li, X.; Liu, N.; et al. Repurposing an antidandruff agent to treating cancer: Zinc pyrithione inhibits tumor growth via targeting proteasome-associated deubiquitinases. Oncotarget 2017, 8, 13942–13956. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Chen, X.; Zang, D.; Lan, X.; Liao, S.; Yang, C.; Zhang, P.; Wu, J.; Li, X.; Liu, N.; et al. Platinum-containing compound platinum pyrithione is stronger and safer than cisplatin in cancer therapy. Biochem. Pharmacol. 2016, 116, 22–38. [Google Scholar] [CrossRef]
- Zhao, C.; Chen, X.; Zang, D.; Lan, X.; Liao, S.; Yang, C.; Zhang, P.; Wu, J.; Li, X.; Liu, N.; et al. A novel nickel complex works as a proteasomal deubiquitinase inhibitor for cancer therapy. Oncogene 2016, 35, 5916–5927. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Ning, S.; Yu, B.; Wang, Y. USP14: Structure, Function, and Target Inhibition. Front. Pharmacol. 2021, 12, 801328. [Google Scholar] [CrossRef]
- Wang, X.; Stafford, W.; Mazurkiewicz, M.; Fryknas, M.; Brjnic, S.; Zhang, X.; Gullbo, J.; Larsson, R.; Arner, E.S.; D’Arcy, P.; et al. The 19S Deubiquitinase inhibitor b-AP15 is enriched in cells and elicits rapid commitment to cell death. Mol. Pharmacol. 2014, 85, 932–945. [Google Scholar] [CrossRef]
- Ward, J.A.; Pinto-Fernandez, A.; Cornelissen, L.; Bonham, S.; Diaz-Saez, L.; Riant, O.; Huber, K.V.M.; Kessler, B.M.; Feron, O.; Tate, E.W. Re-Evaluating the Mechanism of Action of alpha, beta-Unsaturated Carbonyl DUB Inhibitors b-AP15 and VLX1570: A Paradigmatic Example of Unspecific Protein Cross-linking with Michael Acceptor Motif-Containing Drugs. J. Med. Chem. 2020, 63, 3756–3762. [Google Scholar] [CrossRef]
- Cai, J.; Xia, X.; Liao, Y.; Liu, N.; Guo, Z.; Chen, J.; Yang, L.; Long, H.; Yang, Q.; Zhang, X.; et al. A novel deubiquitinase inhibitor b-AP15 triggers apoptosis in both androgen receptor-dependent and -independent prostate cancers. Oncotarget 2017, 8, 63232–63246. [Google Scholar] [CrossRef]
- Gubat, J.; Selvaraju, K.; Sjostrand, L.; Kumar Singh, D.; Turkina, M.V.; Schmierer, B.; Sabatier, P.; Zubarev, R.A.; Linder, S.; D’Arcy, P. Comprehensive Target Screening and Cellular Profiling of the Cancer-Active Compound b-AP15 Indicate Abrogation of Protein Homeostasis and Organelle Dysfunction as the Primary Mechanism of Action. Front. Oncol. 2022, 12, 852980. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Gao, Q.; Jia, Y.; Wei, J.; Chaudhuri, S.M.; Wang, S.; Tang, A.; Mani, N.L.; Iyer, R.; Cheng, Y.; et al. Ubiquitin-specific peptidase 22 controls integrin-dependent cancer cell stemness and metastasis. iScience 2024, 27, 110592. [Google Scholar] [CrossRef]
- Prokakis, E.; Dyas, A.; Grun, R.; Fritzsche, S.; Bedi, U.; Kazerouni, Z.B.; Kosinsky, R.L.; Johnsen, S.A.; Wegwitz, F. USP22 promotes HER2-driven mammary carcinoma aggressiveness by suppressing the unfolded protein response. Oncogene 2021, 40, 4004–4018. [Google Scholar] [CrossRef] [PubMed]
- Montauti, E.; Weinberg, S.E.; Chu, P.; Chaudhuri, S.; Mani, N.L.; Iyer, R.; Zhou, Y.; Zhang, Y.; Liu, C.; Xin, C.; et al. A deubiquitination module essential for T(reg) fitness in the tumor microenvironment. Sci. Adv. 2022, 8, eabo4116. [Google Scholar] [CrossRef] [PubMed]
- Prokakis, E.; Bamahmoud, H.; Jansari, S.; Fritsche, L.; Dietz, A.; Boshnakovska, A.; Rehling, P.; Johnsen, S.A.; Gallwas, J.; Wegwitz, F. USP22 supports the aggressive behavior of basal-like breast cancer by stimulating cellular respiration. Cell Commun. Signal 2024, 22, 120. [Google Scholar] [CrossRef]
- Wang, S.A.; Wu, Y.C.; Yang, F.M.; Hsu, F.L.; Zhang, K.; Hung, J.J. NCI677397 targeting USP24-mediated induction of lipid peroxidation induces ferroptosis in drug-resistant cancer cells. Mol. Oncol. 2024, 18, 2255–2276. [Google Scholar] [CrossRef] [PubMed]
- Young, M.J.; Wang, S.A.; Chen, Y.C.; Liu, C.Y.; Hsu, K.C.; Tang, S.W.; Tseng, Y.L.; Wang, Y.C.; Lin, S.M.; Hung, J.J. USP24-i-101 targeting of USP24 activates autophagy to inhibit drug resistance acquired during cancer therapy. Cell Death Differ. 2024, 31, 574–591. [Google Scholar] [CrossRef] [PubMed]
- Wrigley, J.D.; Gavory, G.; Simpson, I.; Preston, M.; Plant, H.; Bradley, J.; Goeppert, A.U.; Rozycka, E.; Davies, G.; Walsh, J.; et al. Identification and Characterization of Dual Inhibitors of the USP25/28 Deubiquitinating Enzyme Subfamily. ACS Chem. Biol. 2017, 12, 3113–3125. [Google Scholar] [CrossRef] [PubMed]
- Patzke, J.V.; Sauer, F.; Nair, R.K.; Endres, E.; Proschak, E.; Hernandez-Olmos, V.; Sotriffer, C.; Kisker, C. Structural basis for the bi-specificity of USP25 and USP28 inhibitors. EMBO Rep. 2024, 25, 2950–2973. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Xu, Z.; Huang, Y.; Wang, H.; Zhu, X.; Zhang, W.; Song, W.; Gao, T.; Liu, T.; Wang, M.; et al. Structure-based discovery of potent USP28 inhibitors derived from Vismodegib. Eur. J. Med. Chem. 2023, 254, 115369. [Google Scholar] [CrossRef]
- Aditya, S.; Rattan, A. Vismodegib: A smoothened inhibitor for the treatment of advanced basal cell carcinoma. Indian. Dermatol. Online J. 2013, 4, 365–368. [Google Scholar] [CrossRef]
- Wang, H.; Meng, Q.; Ding, Y.; Xiong, M.; Zhu, M.; Yang, Y.; Su, H.; Gu, L.; Xu, Y.; Shi, L.; et al. USP28 and USP25 are downregulated by Vismodegib in vitro and in colorectal cancer cell lines. FEBS J. 2021, 288, 1325–1342. [Google Scholar] [CrossRef]
- Kluge, A.F.; Lagu, B.R.; Maiti, P.; Jaleel, M.; Webb, M.; Malhotra, J.; Mallat, A.; Srinivas, P.A.; Thompson, J.E. Novel highly selective inhibitors of ubiquitin specific protease 30 (USP30) accelerate mitophagy. Bioorg. Med. Chem. Lett. 2018, 28, 2655–2659. [Google Scholar] [CrossRef]
- Zhang, X.; Han, Y.; Liu, S.; Guo, B.; Xu, S.; He, Y.; Liu, L. MF-094 nanodelivery inhibits oral squamous cell carcinoma by targeting USP30. Cell Mol. Biol. Lett. 2022, 27, 107. [Google Scholar] [CrossRef]
- Li, X.; Wang, T.; Tao, Y.; Wang, X.; Li, L.; Liu, J. MF-094, a potent and selective USP30 inhibitor, accelerates diabetic wound healing by inhibiting the NLRP3 inflammasome. Exp. Cell Res. 2022, 410, 112967. [Google Scholar] [CrossRef]
- Wang, W.; Liu, H.; Wang, S.; Hao, X.; Li, L. A diterpenoid derivative 15-oxospiramilactone inhibits Wnt/beta-catenin signaling and colon cancer cell tumorigenesis. Cell Res. 2011, 21, 730–740. [Google Scholar] [CrossRef] [PubMed]
- Yue, W.; Chen, Z.; Liu, H.; Yan, C.; Chen, M.; Feng, D.; Yan, C.; Wu, H.; Du, L.; Wang, Y.; et al. A small natural molecule promotes mitochondrial fusion through inhibition of the deubiquitinase USP30. Cell Res. 2014, 24, 482–496. [Google Scholar] [CrossRef]
- Waters, C.S.; Angenent, S.B.; Altschuler, S.J.; Wu, L.F. A PINK1 input threshold arises from positive feedback in the PINK1/Parkin mitophagy decision circuit. Cell Rep. 2023, 42, 113260. [Google Scholar] [CrossRef]
- Rusilowicz-Jones, E.V.; Barone, F.G.; Lopes, F.M.; Stephen, E.; Mortiboys, H.; Urbe, S.; Clague, M.J. Benchmarking a highly selective USP30 inhibitor for enhancement of mitophagy and pexophagy. Life Sci. Alliance 2022, 5, e202101287. [Google Scholar] [CrossRef]
- Li, M.; Wang, X.; Qiu, Y.; Zhang, Y.; Pan, X.; Tang, N.; Chen, T.; Ruan, B.; Shao, S.; He, L.; et al. Safety reassessment of cinobufotalin injection: New findings into cardiotoxicity. Toxicol. Res. 2020, 9, 390–398. [Google Scholar] [CrossRef]
- Kim, Y.J.; Lee, Y.; Shin, H.; Hwang, S.; Park, J.; Song, E.J. Ubiquitin-proteasome system as a target for anticancer treatment-an update. Arch. Pharm. Res. 2023, 46, 573–597. [Google Scholar] [CrossRef]
- Gao, H.; Xi, Z.; Dai, J.; Xue, J.; Guan, X.; Zhao, L.; Chen, Z.; Xing, F. Drug resistance mechanisms and treatment strategies mediated by Ubiquitin-Specific Proteases (USPs) in cancers: New directions and therapeutic options. Mol. Cancer 2024, 23, 88. [Google Scholar] [CrossRef]
- Gao, H.; Yin, J.; Ji, C.; Yu, X.; Xue, J.; Guan, X.; Zhang, S.; Liu, X.; Xing, F. Targeting ubiquitin specific proteases (USPs) in cancer immunotherapy: From basic research to preclinical application. J. Exp. Clin. Cancer Res. 2023, 42, 225. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, P.; Zhan, X.; Zhang, Y.; Sheng, J.; He, S.; Chen, Y.; Nie, D.; You, X.; Mai, H.; et al. USP1-regulated reciprocal differentiation of Th17 cells and Treg cells by deubiquitinating and stabilizing TAZ. Cell Mol. Immunol. 2023, 20, 252–263. [Google Scholar] [CrossRef] [PubMed]
- van Loosdregt, J.; Fleskens, V.; Fu, J.; Brenkman, A.B.; Bekker, C.P.; Pals, C.E.; Meerding, J.; Berkers, C.R.; Barbi, J.; Grone, A.; et al. Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg-cell-suppressive capacity. Immunity 2013, 39, 259–271. [Google Scholar] [CrossRef]
- Wang, L.; Kumar, S.; Dahiya, S.; Wang, F.; Wu, J.; Newick, K.; Han, R.; Samanta, A.; Beier, U.H.; Akimova, T.; et al. Ubiquitin-specific Protease-7 Inhibition Impairs Tip60-dependent Foxp3+ T-regulatory Cell Function and Promotes Antitumor Immunity. EBioMedicine 2016, 13, 99–112. [Google Scholar] [CrossRef]
- Wang, Z.; Kang, W.; Li, O.; Qi, F.; Wang, J.; You, Y.; He, P.; Suo, Z.; Zheng, Y.; Liu, H.M. Abrogation of USP7 is an alternative strategy to downregulate PD-L1 and sensitize gastric cancer cells to T cells killing. Acta Pharm. Sin. B 2021, 11, 694–707. [Google Scholar] [CrossRef]
- Selvaraju, K.; Mazurkiewicz, M.; Wang, X.; Gullbo, J.; Linder, S.; D’Arcy, P. Inhibition of proteasome deubiquitinase activity: A strategy to overcome resistance to conventional proteasome inhibitors? Drug Resist. Updat. 2015, 21–22, 20–29. [Google Scholar] [CrossRef]
- Li, J.; Yuan, S.; Norgard, R.J.; Yan, F.; Yamazoe, T.; Blanco, A.; Stanger, B.Z. Tumor Cell-Intrinsic USP22 Suppresses Antitumor Immunity in Pancreatic Cancer. Cancer Immunol. Res. 2020, 8, 282–291. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, Q.; Lou, Y.; Wang, J.; Zhao, X.; Wang, L.; Zhang, X.; Li, S.; Zhao, Y.; Chen, Q.; et al. USP22 Deubiquitinates CD274 to Suppress Anticancer Immunity. Cancer Immunol. Res. 2019, 7, 1580–1590. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Yu, H.; Yao, J.; Li, J.; Li, Z.; Jiang, Z. ncRNA-mediated overexpression of ubiquitin-specific proteinase 13 contributes to the progression of prostate cancer via modulating AR signaling, DNA damage repair and immune infiltration. BMC Cancer 2022, 22, 1350. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Wu, X.; Jian, Y.; Wang, J.; Huang, C.; Mo, S.; Li, Y.; Li, F.; Zhang, C.; Zhang, D.; et al. USP14 promotes tryptophan metabolism and immune suppression by stabilizing IDO1 in colorectal cancer. Nat. Commun. 2022, 13, 5644. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, A.; Murakami, K.; Elia, A.; Shibahara, Y.; Done, S.J.; Wood, S.A.; Donato, N.J.; Ohashi, P.S.; Reedijk, M. Therapeutic inhibition of USP9x-mediated Notch signaling in triple-negative breast cancer. Proc. Natl. Acad. Sci. USA 2021, 118, e2101592118. [Google Scholar] [CrossRef]
- Lim, K.S.; Li, H.; Roberts, E.A.; Gaudiano, E.F.; Clairmont, C.; Sambel, L.A.; Ponnienselvan, K.; Liu, J.C.; Yang, C.; Kozono, D.; et al. USP1 Is Required for Replication Fork Protection in BRCA1-Deficient Tumors. Mol. Cell 2018, 72, 925–941 e924. [Google Scholar] [CrossRef] [PubMed]
- Rennie, M.L.; Gundogdu, M.; Arkinson, C.; Liness, S.; Frame, S.; Walden, H. Structural and Biochemical Insights into the Mechanism of Action of the Clinical USP1 Inhibitor, KSQ-4279. J. Med. Chem. 2024, 67, 15557–15568. [Google Scholar] [CrossRef] [PubMed]
- Cadzow, L.; Brenneman, J.; Tobin, E.; Sullivan, P.; Nayak, S.; Ali, J.A.; Shenker, S.; Griffith, J.; McGuire, M.; Grasberger, P.; et al. The USP1 Inhibitor KSQ-4279 Overcomes PARP Inhibitor Resistance in Homologous Recombination-Deficient Tumors. Cancer Res. 2024, 84, 3419–3434. [Google Scholar] [CrossRef]
- Li, H.; Liu, B.J.; Xu, J.; Song, S.S.; Ba, R.; Zhang, J.; Huan, X.J.; Wang, D.; Miao, Z.H.; Liu, T.; et al. Design, synthesis, and biological evaluation of pyrido[2,3-d]pyrimidin-7(8H)-one derivatives as potent USP1 inhibitors. Eur. J. Med. Chem. 2024, 275, 116568. [Google Scholar] [CrossRef]
- Wang, X.; Mazurkiewicz, M.; Hillert, E.K.; Olofsson, M.H.; Pierrou, S.; Hillertz, P.; Gullbo, J.; Selvaraju, K.; Paulus, A.; Akhtar, S.; et al. The proteasome deubiquitinase inhibitor VLX1570 shows selectivity for ubiquitin-specific protease-14 and induces apoptosis of multiple myeloma cells. Sci. Rep. 2016, 6, 26979. [Google Scholar] [CrossRef]
- Rowinsky, E.K.; Paner, A.; Berdeja, J.G.; Paba-Prada, C.; Venugopal, P.; Porkka, K.; Gullbo, J.; Linder, S.; Loskog, A.; Richardson, P.G.; et al. Phase 1 study of the protein deubiquitinase inhibitor VLX1570 in patients with relapsed and/or refractory multiple myeloma. Investig. New Drugs 2020, 38, 1448–1453. [Google Scholar] [CrossRef]
- Simoneau, A.; Pratt, C.; Comer, G.; Wu, H.-J.; Choi, A.; Khendu, T.; Meier, S.; Yu, Y.; Liu, S.; Zhang, W.; et al. Abstract B054: TNG348, a selective USP1 inhibitor, shows strong preclinical combination activity with PARP inhibitors and other agents targeting DNA repair. Mol. Cancer Ther. 2023, 22, B054. [Google Scholar] [CrossRef]
- Simoneau, A.; Wu, H.-J.; Bandi, M.; Lazarides, K.; Sun, S.; Liu, S.; Meier, S.; Choi, A.; Zhang, H.; Shen, B.; et al. Abstract 4968: Characterization of the clinical development candidate TNG348 as a potent and selective inhibitor of USP1 for the treatment of BRCA1/2mut cancers. Cancer Res. 2023, 83, 4968. [Google Scholar] [CrossRef]
- Phase 1/2, Multi-Center, Open-Label Study to Evaluate the Safety, Tolerability, and Preliminary Antitumor Activity of TNG348 Single Agent and in Combination With a PARP Inhibitor in Patients With BRCA 1/2 Mutant or Other HRD+ Solid Tumors. 2024. Available online: https://www.dana-farber.org/clinical-trials/23-643 (accessed on 13 August 2024).
- A Phase 1 Study of RO7623066 Alone and in Combination in Patients With Advanced Solid Tumors. 2022. Available online: https://www.cancer.gov/research/participate/clinical-trials-search/v?id=NCI-2022-01620 (accessed on 4 February 2025).
- Yap, T.A.; Lakhani, N.J.; Patnaik, A.; Lee, E.K.; Gutierrez, M.; Moore, K.N.; Carneiro, B.A.; Hays, J.L.; Huang, M.; LoRusso, P.; et al. First-in-human phase I trial of the oral first-in-class ubiquitin specific peptidase 1 (USP1) inhibitor KSQ-4279 (KSQi), given as single agent (SA) and in combination with olaparib (OLA) or carboplatin (CARBO) in patients (pts) with advanced solid tumors, enriched for deleterious homologous recombination repair (HRR) mutations. J. Clin. Oncol. 2024, 42, 3005. [Google Scholar] [CrossRef]
- Li, Y.; Wu, J.; Liu, J.; Qin, L.; Cai, X.; Qiao, J.; Wang, L.; Rao, S.; Ren, F.; Zhavoronkov, A. Abstract 502: ISM3091, a novel selective USP1 inhibitor as a targeted anticancer therapy. Cancer Res. 2023, 83, 502. [Google Scholar] [CrossRef]
- An Open-Label, Multicenter Study to Investigate the Safety, Tolerability, Pharmacokinetics, and Preliminary Efficacy of XL309 (ISM3091) as Single-Agent and Combination Therapy in Patients With Advanced Solid Tumors. 2025. Available online: https://clinicaltrials.gov/study/NCT05932862 (accessed on 24 January 2025).
- A Phase I First-in-human, Open-label Trial to Investigate the Safety, Pharmacokinetics and Antitumor Activity of SIM0501 as Monotherapy and in Combination in Participants With Advanced Solid Tumors. 2024. Available online: https://www.medifind.com/articles/clinical-trial/507724029 (accessed on 4 February 2025).
- Yang, S.; Li, Z.; Zhou, L.; Zhou, F.; Qin, X.; Xue, L.; Xu, P.; Tang, J.; Wang, W.; Tang, R. Abstract LB271: Synergistic anti-tumor efficacy in olaparib-sensitive and -resistant models via simultaneously inhibition of USP1 and PARP. Cancer Res. 2024, 84, LB271. [Google Scholar] [CrossRef]
- Xu, J.; Li, Y.; Wang, J.; Tang, P.; Wang, J.; Zhang, C.; Yan, P. Abstract 7146: HSK39775: A USP1 inhibitor for the treatment of cancers with homologous recombination deficiencies. Cancer Res. 2024, 84, 7146. [Google Scholar] [CrossRef]
- A Phase I/II Study to Assess the Safety, Tolerability, PK/PD, and Preliminary Efficacy of HSK39775 Tablet Monotherapy in Participants With Advanced Solid Malignancies. 2024. Available online: https://adisinsight.springer.com/trials/700372061 (accessed on 22 March 2024).
- VLX1570 and Low-Dose Dexamethasone in Relapsed or Relapsed and Refractory Multiple Myeloma: A Clinical and Correlative Phase 1/2 Study. 2018. Available online: https://clinicaltrials.gov/study/NCT02372240 (accessed on 11 May 2018).
- Kharel, P.; Uprety, D.; Chandra, A.B.; Hu, Y.; Belur, A.A.; Dhakal, A. Bortezomib-Induced Pulmonary Toxicity: A Case Report and Review of Literature. Case Rep. Med. 2018, 2018, 2913124. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).