Comprehensive Analysis of the Putative Substratome of FAM20C, the Master Serine Kinase of the Secretory Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Analysis of Conserved Residues in Peptide Sequences
2.3. Heatmap Generation
2.4. GO Annotation
2.5. Sequence Retrieval, Alignment, and Filtering of Orthologues
2.6. Phylogenetic Inference and Network Construction
2.7. Identification and In Silico Analysis of Pathogenic Variants in FAM20C Substrate Phosphorylation Motifs
3. Results
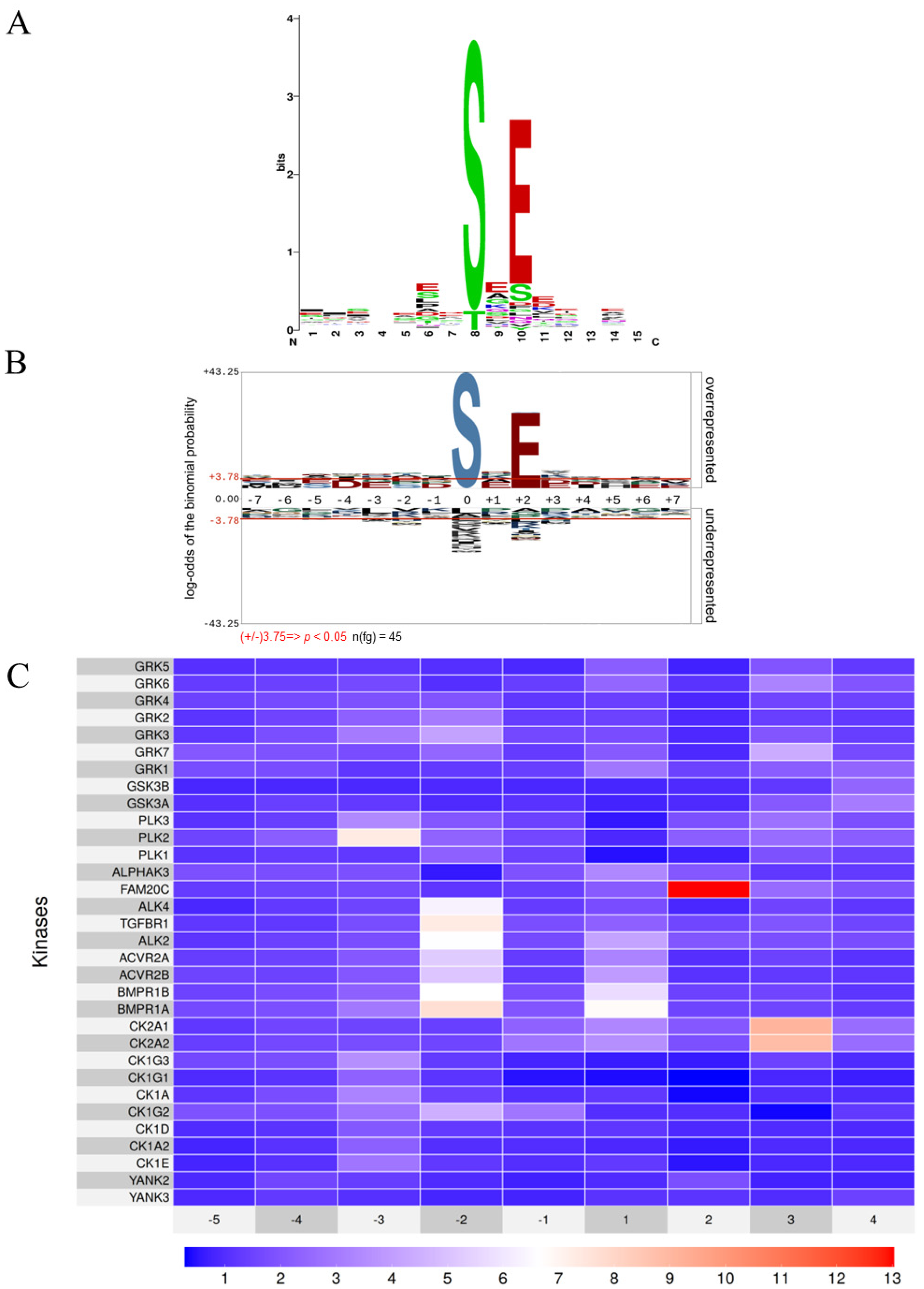
3.1. Is the FAM20C Kinase Consensus Sequence Sufficiently Distinctive to Serve as a Characteristic Signature of the Kinase?
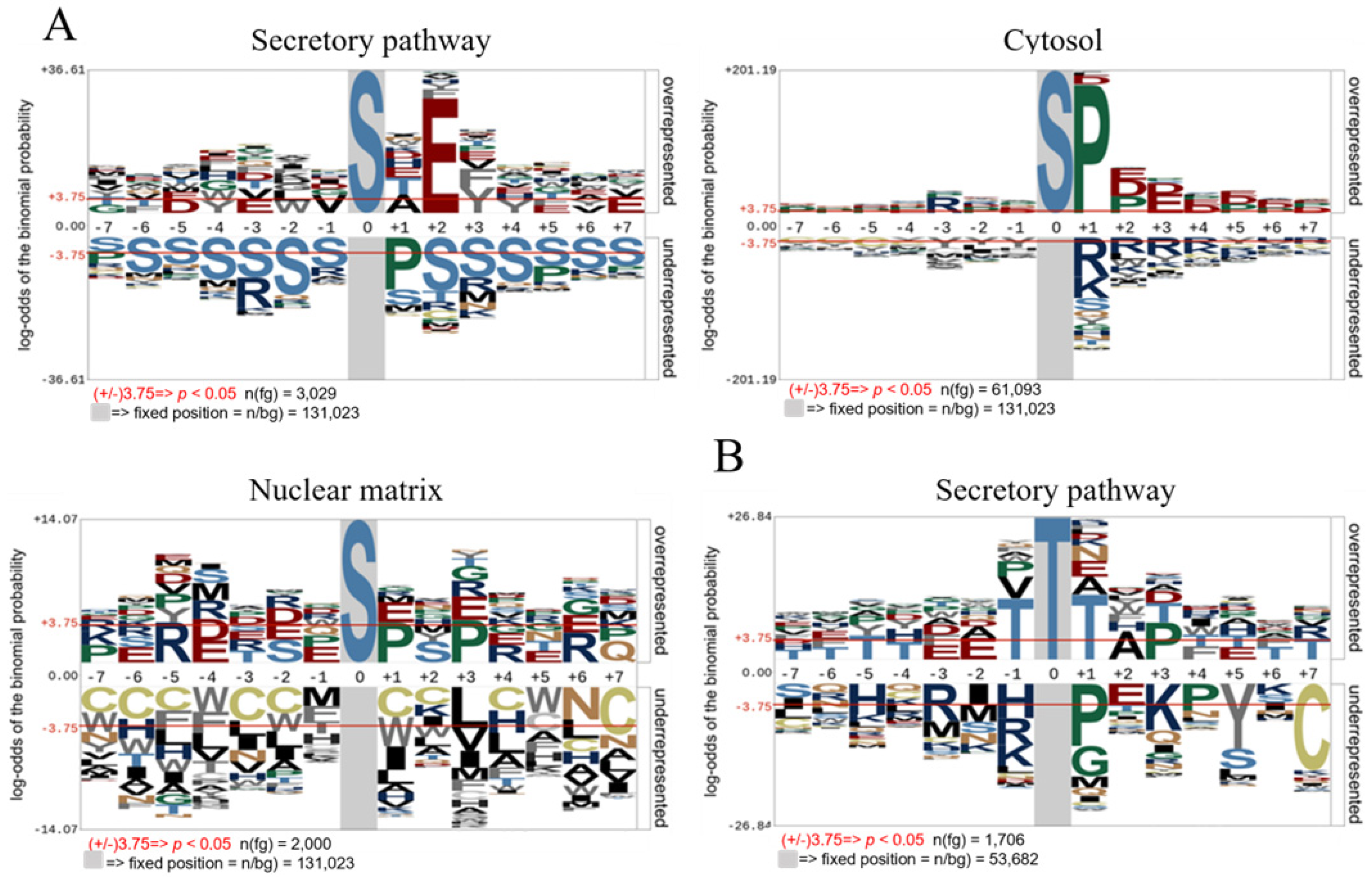
3.2. FAM20C Motif in the Secretory Pathway
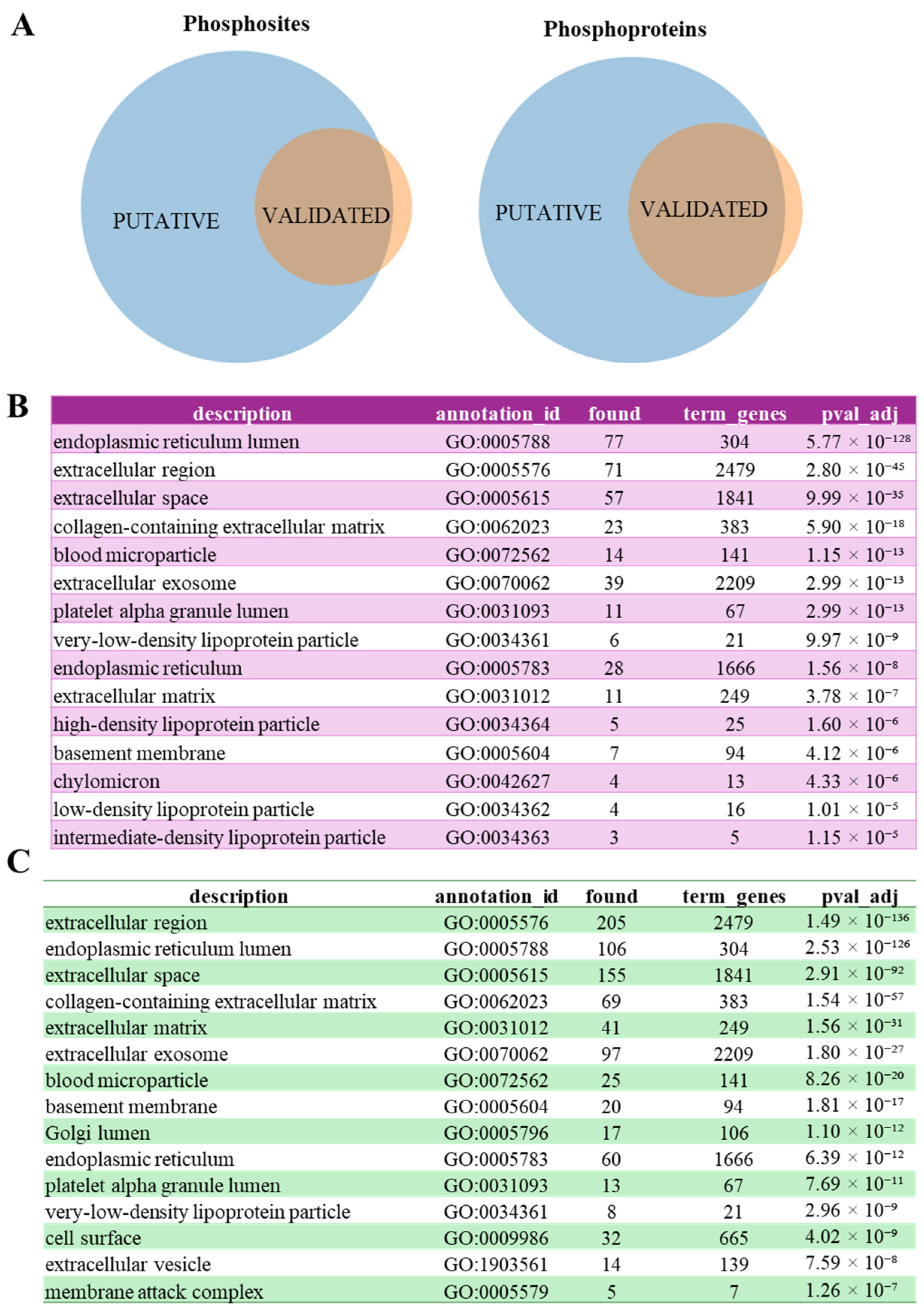
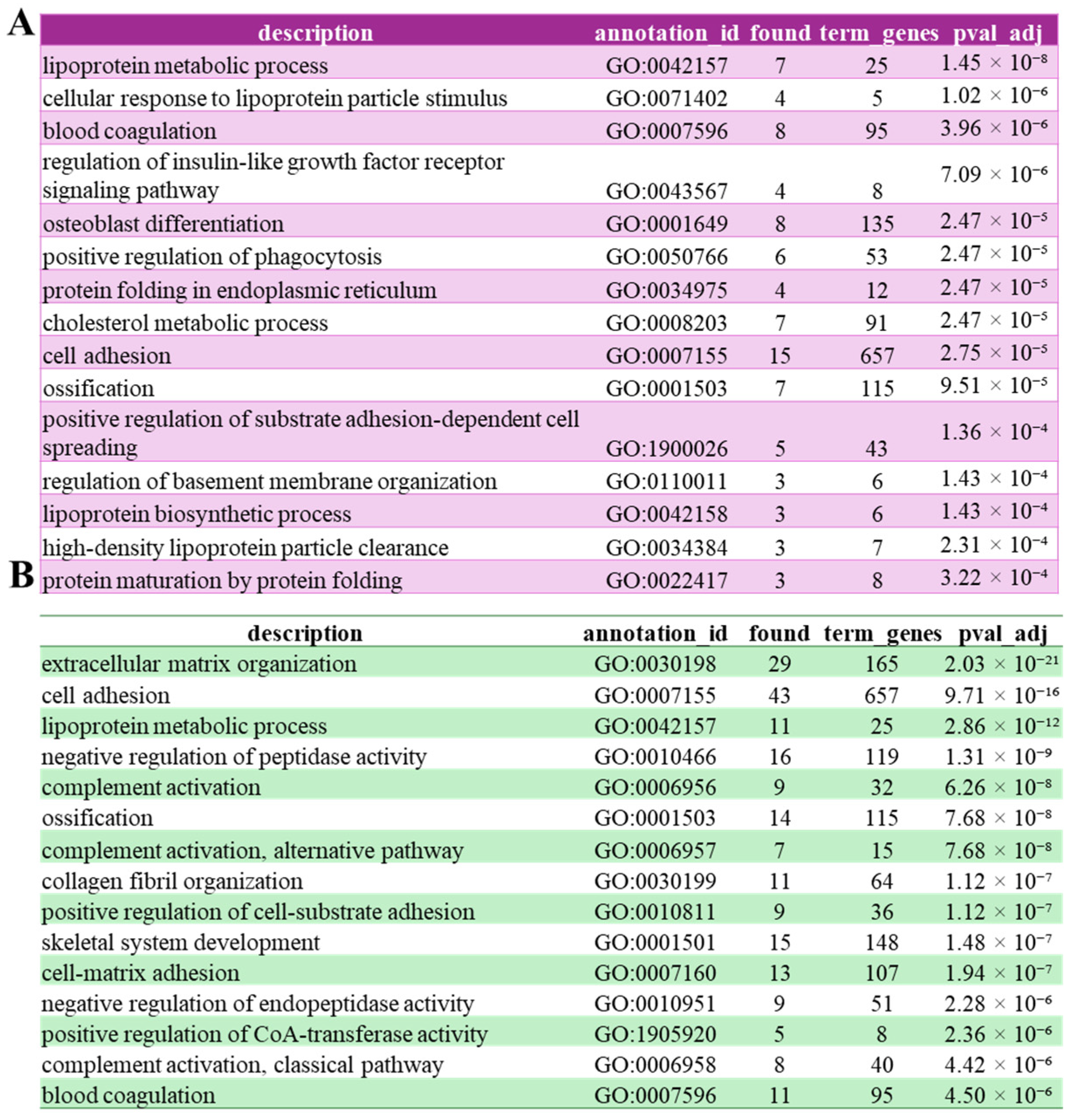
3.3. FAM20C Putative Substratome
3.4. Phylogenetic Structure and Network Signals of SxE-Containing Proteins in the Complement and Coagulation Cascades
3.5. Pathogenic Variants Affecting FAM20C Substrates Phosphorylation Motifs in Human Diseases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hammarsten, O. Zur Frage, Ob Das Caseïn Ein Einheitlicher Stoff Sei. Biol. Chem. 1883, 7, 227–273. [Google Scholar] [CrossRef]
- Burnett, G.; Kennedy, E.P. The Enzymatic Phosphorylation of Proteins. J. Biol. Chem. 1954, 211, 969–980. [Google Scholar] [CrossRef]
- Venerando, A.; Ruzzene, M.; Pinna, L.A. Casein Kinase: The Triple Meaning of a Misnomer. Biochem. J. 2014, 460, 141–156. [Google Scholar] [CrossRef]
- Ew, B.; Hm, F.; Jj, B. Phosphorylation of Casein. Role of the Golgi Apparatus. J. Biol. Chem. 1972, 247, 8193–8194. [Google Scholar]
- Lasa-Benito, M.; Marin, O.; Meggio, F.; Pinna, L.A. Golgi Apparatus Mammary Gland Casein Kinase: Monitoring by a Specific Peptide Substrate and Definition of Specificity Determinants. FEBS Lett. 1996, 382, 149–152. [Google Scholar] [CrossRef]
- Lasa, M.; Marin, O.; Pinna, L.A. Rat Liver Golgi Apparatus Contains a Protein Kinase Similar to the Casein Kinase of Lactating Mammary Gland. Eur. J. Biochem. 1997, 243, 719–725. [Google Scholar] [CrossRef]
- Salvi, M.; Cesaro, L.; Tibaldi, E.; Pinna, L.A. Motif Analysis of Phosphosites Discloses a Potential Prominent Role of the Golgi Casein Kinase (GCK) in the Generation of Human Plasma Phospho-Proteome. J. Proteome Res. 2010, 9, 3335–3338. [Google Scholar] [CrossRef]
- Ishikawa, H.O.; Xu, A.; Ogura, E.; Manning, G.; Irvine, K.D. The Raine Syndrome Protein FAM20C Is a Golgi Kinase That Phosphorylates Bio-Mineralization Proteins. PLoS ONE 2012, 7, e42988. [Google Scholar] [CrossRef]
- Tagliabracci, V.S.; Engel, J.L.; Wen, J.; Wiley, S.E.; Worby, C.A.; Kinch, L.N.; Xiao, J.; Grishin, N.V.; Dixon, J.E. Secreted Kinase Phosphorylates Extracellular Proteins That Regulate Biomineralization. Science 2012, 336, 1150–1153. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The Protein Kinase Complement of the Human Genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Tagliabracci, V.S.; Wiley, S.E.; Guo, X.; Kinch, L.N.; Durrant, E.; Wen, J.; Xiao, J.; Cui, J.; Nguyen, K.B.; Engel, J.L.; et al. A Single Kinase Generates the Majority of the Secreted Phosphoproteome. Cell 2015, 161, 1619–1632. [Google Scholar] [CrossRef]
- Johnson, J.L.; Yaron, T.M.; Huntsman, E.M.; Kerelsky, A.; Song, J.; Regev, A.; Lin, T.-Y.; Liberatore, K.; Cizin, D.M.; Cohen, B.M.; et al. An Atlas of Substrate Specificities for the Human Serine/Threonine Kinome. Nature 2023, 613, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Tan, H.; Zhang, J.; Yuan, Z.; Xie, Q.; Zhang, L. Fam20C in Human Diseases: Emerging Biological Functions and Therapeutic Implications. Front. Mol. Biosci. 2021, 8, 790172. [Google Scholar] [CrossRef]
- Simpson, M.A.; Hsu, R.; Keir, L.S.; Hao, J.; Sivapalan, G.; Ernst, L.M.; Zackai, E.H.; Al-Gazali, L.I.; Hulskamp, G.; Kingston, H.M.; et al. Mutations in FAM20C Are Associated with Lethal Osteosclerotic Bone Dysplasia (Raine Syndrome), Highlighting a Crucial Molecule in Bone Development. Am. J. Hum. Genet. 2007, 81, 906–912. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, J.; Liu, P.; Wei, Y.; Wang, X.; Xiao, J.; Wang, C.-C.; Wang, L. Proteolytic Processing of Secretory Pathway Kinase Fam20C by Site-1 Protease Promotes Biomineralization. Proc. Natl. Acad. Sci. USA 2021, 118, e2100133118. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, Q.; Wang, X.; Yu, J.; Chen, X.; Wang, J.; Wang, X.; Xiao, J.; Wang, C.-C.; Wang, L. Secretory Kinase Fam20C Tunes Endoplasmic Reticulum Redox State via Phosphorylation of Ero1α. EMBO J. 2018, 37, e98699. [Google Scholar] [CrossRef]
- Pollak, A.J.; Haghighi, K.; Kunduri, S.; Arvanitis, D.A.; Bidwell, P.A.; Liu, G.-S.; Singh, V.P.; Gonzalez, D.J.; Sanoudou, D.; Wiley, S.E.; et al. Phosphorylation of Serine96 of Histidine-Rich Calcium-Binding Protein by the Fam20C Kinase Functions to Prevent Cardiac Arrhythmia. Proc. Natl. Acad. Sci. USA 2017, 114, 9098–9103. [Google Scholar] [CrossRef]
- Pollak, A.J.; Liu, C.; Gudlur, A.; Mayfield, J.E.; Dalton, N.D.; Gu, Y.; Chen, J.; Heller Brown, J.; Hogan, P.G.; Wiley, S.E.; et al. A Secretory Pathway Kinase Regulates Sarcoplasmic Reticulum Ca2+ Homeostasis and Protects against Heart Failure. eLife 2018, 7, e41378. [Google Scholar] [CrossRef]
- Yu, J.; Li, T.; Liu, Y.; Wang, X.; Zhang, J.; Wang, X.; Shi, G.; Lou, J.; Wang, L.; Wang, C.-C.; et al. Phosphorylation Switches Protein Disulfide Isomerase Activity to Maintain Proteostasis and Attenuate ER Stress. EMBO J. 2020, 39, e103841. [Google Scholar] [CrossRef]
- Mayfield, J.E.; Dixon, J.E. Emerging Mechanisms of Regulation for Endoplasmic/Sarcoplasmic Reticulum Ca2+ Stores by Secretory Pathway Kinase FAM20C. Curr. Opin. Chem. Biol. 2023, 74, 102279. [Google Scholar] [CrossRef]
- Noventa, F.; Salvi, M. Protein Kinase FAM20C-When Subcellular Localization Matters. FEBS Lett. 2025; early view. [Google Scholar] [CrossRef]
- Tagliabracci, V.S.; Pinna, L.A.; Dixon, J.E. Secreted Protein Kinases. Trends Biochem. Sci. 2013, 38, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Xiao, J.; Tagliabracci, V.S.; Wen, J.; Rahdar, M.; Dixon, J.E. A Secretory Kinase Complex Regulates Extracellular Protein Phosphorylation. eLife 2015, 4, e06120. [Google Scholar] [CrossRef]
- Cui, J.; Zhu, Q.; Zhang, H.; Cianfrocco, M.A.; Leschziner, A.E.; Dixon, J.E.; Xiao, J. Structure of Fam20A Reveals a Pseudokinase Featuring a Unique Disulfide Pattern and Inverted ATP-Binding. eLife 2017, 6, e23990. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, J.; Bitu, C.C.; Daly, S.B.; Urquhart, J.E.; Barron, M.J.; Bhaskar, S.S.; Martelli-Júnior, H.; dos Santos Neto, P.E.; Mansilla, M.A.; Murray, J.C.; et al. Whole-Exome Sequencing Identifies FAM20A Mutations as a Cause of Amelogenesis Imperfecta and Gingival Hyperplasia Syndrome. Am. J. Hum. Genet. 2011, 88, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and Recalibrations. Nucleic Acids Res 2015, 43, D512–D520. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. Advantages of Combined Transmembrane Topology and Signal Peptide Prediction--the Phobius Web Server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef]
- O’Shea, J.P.; Chou, M.F.; Quader, S.A.; Ryan, J.K.; Church, G.M.; Schwartz, D. pLogo: A Probabilistic Approach to Visualizing Sequence Motifs. Nat. Methods 2013, 10, 1211–1212. [Google Scholar] [CrossRef]
- Garcia-Moreno, A.; López-Domínguez, R.; Villatoro-García, J.A.; Ramirez-Mena, A.; Aparicio-Puerta, E.; Hackenberg, M.; Pascual-Montano, A.; Carmona-Saez, P. Functional Enrichment Analysis of Regulatory Elements. Biomedicines 2022, 10, 590. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res 2025, 53, D609–D617. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Ostell, J.; Pruitt, K.D.; Sayers, E.W. GenBank. Nucleic Acids Res. 2018, 46, D41–D47. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. The SplitsTree App: Interactive Analysis and Visualization Using Phylogenetic Trees and Networks. Nat. Methods 2024, 21, 1773–1774. [Google Scholar] [CrossRef]
- Huson, D.H. SplitsTree: Analyzing and Visualizing Evolutionary Data. Bioinformatics 1998, 14, 68–73. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. aLeaves Facilitates On-Demand Exploration of Metazoan Gene Family Trees on MAFFT Sequence Alignment Server with Enhanced Interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar: Public Archive of Relationships among Sequence Variation and Human Phenotype. Nucleic Acids Res. 2014, 42, D980–D985. [Google Scholar] [CrossRef]
- Rehm, H.L.; Berg, J.S.; Brooks, L.D.; Bustamante, C.D.; Evans, J.P.; Landrum, M.J.; Ledbetter, D.H.; Maglott, D.R.; Martin, C.L.; Nussbaum, R.L.; et al. ClinGen—The Clinical Genome Resource. N. Engl. J. Med. 2015, 372, 2235–2242. [Google Scholar] [CrossRef]
- Cozza, G.; Salvi, M. The Acidophilic Kinases PLK2 and PLK3: Structure, Substrate Targeting and Inhibition. Curr. Protein Pept. Sci. 2018, 19, 728–745. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Cesaro, L.; Zuliani, A.M.; Bosello Travain, V.; Salvi, M. Exploring Protein Kinase CK2 Substrate Recognition and the Dynamic Response of Substrate Phosphorylation to Kinase Modulation. Kinases Phosphatases 2023, 1, 251–264. [Google Scholar] [CrossRef]
- Salvi, M.; Cesaro, L.; Pinna, L.A. Variable Contribution of Protein Kinases to the Generation of the Human Phosphoproteome: A Global Weblogo Analysis. Biomol. Concepts 2010, 1, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Cozza, G.; Moro, E.; Black, M.; Marin, O.; Salvi, M.; Venerando, A.; Tagliabracci, V.S.; Pinna, L.A. The Golgi “casein Kinase” Fam20C Is a Genuine “Phosvitin Kinase” and Phosphorylates Polyserine Stretches Devoid of the Canonical Consensus. FEBS J. 2018, 285, 4674–4683. [Google Scholar] [CrossRef]
- Brunati, A.M.; Marin, O.; Bisinella, A.; Salviati, A.; Pinna, L.A. Novel Consensus Sequence for the Golgi Apparatus Casein Kinase, Revealed Using Proline-Rich Protein-1 (PRP1)-Derived Peptide Substrates. Biochem. J. 2000, 351 Pt 3, 765–768. [Google Scholar] [CrossRef]
- Liu, P.; Ma, S.; Zhang, H.; Liu, C.; Lu, Y.; Chen, L.; Qin, C. Specific Ablation of Mouse Fam20C in Cells Expressing Type I Collagen Leads to Skeletal Defects and Hypophosphatemia. Sci. Rep. 2017, 7, 3590. [Google Scholar] [CrossRef]
- Da, Q.; Han, H.; Valladolid, C.; Fernández, M.; Khatlani, T.; Pradhan, S.; Nolasco, J.; Matsunami, R.K.; Engler, D.A.; Cruz, M.A.; et al. In Vitro Phosphorylation of von Willebrand Factor by FAM20c Enhances Its Ability to Support Platelet Adhesion. J. Thromb. Haemost. 2019, 17, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Mameli, C.; Zichichi, G.; Mahmood, N.; Elalaoui, S.C.; Mirza, A.; Dharmaraj, P.; Burrone, M.; Cattaneo, E.; Sheth, J.; Gandhi, A.; et al. Natural History of Non-Lethal Raine Syndrome during Childhood. Orphanet J. Rare Dis. 2020, 15, 93. [Google Scholar] [CrossRef]
- Simpson, M.; Scheuerle, A.; Hurst, J.; Patton, M.; Stewart, H.; Crosby, A. Mutations in FAM20C Also Identified in Non-Lethal Osteosclerotic Bone Dysplasia. Clin. Genet. 2009, 75, 271–276. [Google Scholar] [CrossRef]
- Fradin, M.; Stoetzel, C.; Muller, J.; Koob, M.; Christmann, D.; Debry, C.; Kohler, M.; Isnard, M.; Astruc, D.; Desprez, P.; et al. Osteosclerotic Bone Dysplasia in Siblings with a Fam20C Mutation. Clin. Genet. 2011, 80, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Ren, Y.; Ju, Y.; Zhang, Y.; Zhang, Y.; Wang, Y. FAM20C: A Key Protein Kinase in Multiple Diseases. Genes. Dis. 2025, 12, 101179. [Google Scholar] [CrossRef] [PubMed]
- Ben Djoudi Ouadda, A.; Gauthier, M.-S.; Susan-Resiga, D.; Girard, E.; Essalmani, R.; Black, M.; Marcinkiewicz, J.; Forget, D.; Hamelin, J.; Evagelidis, A.; et al. Ser-Phosphorylation of PCSK9 (Proprotein Convertase Subtilisin-Kexin 9) by Fam20C (Family With Sequence Similarity 20, Member C) Kinase Enhances Its Ability to Degrade the LDLR (Low-Density Lipoprotein Receptor). Arterioscler. Thromb. Vasc. Biol. 2019, 39, 1996–2013. [Google Scholar] [CrossRef] [PubMed]
- Rallapalli, P.M.; Orengo, C.A.; Studer, R.A.; Perkins, S.J. Positive Selection during the Evolution of the Blood Coagulation Factors in the Context of Their Disease-Causing Mutations. Mol. Biol. Evol. 2014, 31, 3040–3056. [Google Scholar] [CrossRef]
- Zakas, P.M.; Coyle, C.W.; Brehm, A.; Bayer, M.; Solecka-Witulska, B.; Radford, C.E.; Brown, C.; Nesbitt, K.; Dwyer, C.; Kannicht, C.; et al. Molecular Coevolution of Coagulation Factor VIII and von Willebrand Factor. Blood Adv. 2021, 5, 812–822. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cesaro, L.; Noventa, F.; De Los Angeles Cordero, T.; Molon, B.; Bosello Travain, V.; Aspromonte, M.C.; Salvi, M. Comprehensive Analysis of the Putative Substratome of FAM20C, the Master Serine Kinase of the Secretory Pathway. Biomolecules 2025, 15, 1582. https://doi.org/10.3390/biom15111582
Cesaro L, Noventa F, De Los Angeles Cordero T, Molon B, Bosello Travain V, Aspromonte MC, Salvi M. Comprehensive Analysis of the Putative Substratome of FAM20C, the Master Serine Kinase of the Secretory Pathway. Biomolecules. 2025; 15(11):1582. https://doi.org/10.3390/biom15111582
Chicago/Turabian StyleCesaro, Luca, Francesca Noventa, Trinidad De Los Angeles Cordero, Barbara Molon, Valentina Bosello Travain, Maria Cristina Aspromonte, and Mauro Salvi. 2025. "Comprehensive Analysis of the Putative Substratome of FAM20C, the Master Serine Kinase of the Secretory Pathway" Biomolecules 15, no. 11: 1582. https://doi.org/10.3390/biom15111582
APA StyleCesaro, L., Noventa, F., De Los Angeles Cordero, T., Molon, B., Bosello Travain, V., Aspromonte, M. C., & Salvi, M. (2025). Comprehensive Analysis of the Putative Substratome of FAM20C, the Master Serine Kinase of the Secretory Pathway. Biomolecules, 15(11), 1582. https://doi.org/10.3390/biom15111582








