Characterization of Skeletal Muscle Regeneration Revealed a Novel Growth Network Induced by Molecular Acupuncture-like Transfection
Abstract
1. Introduction
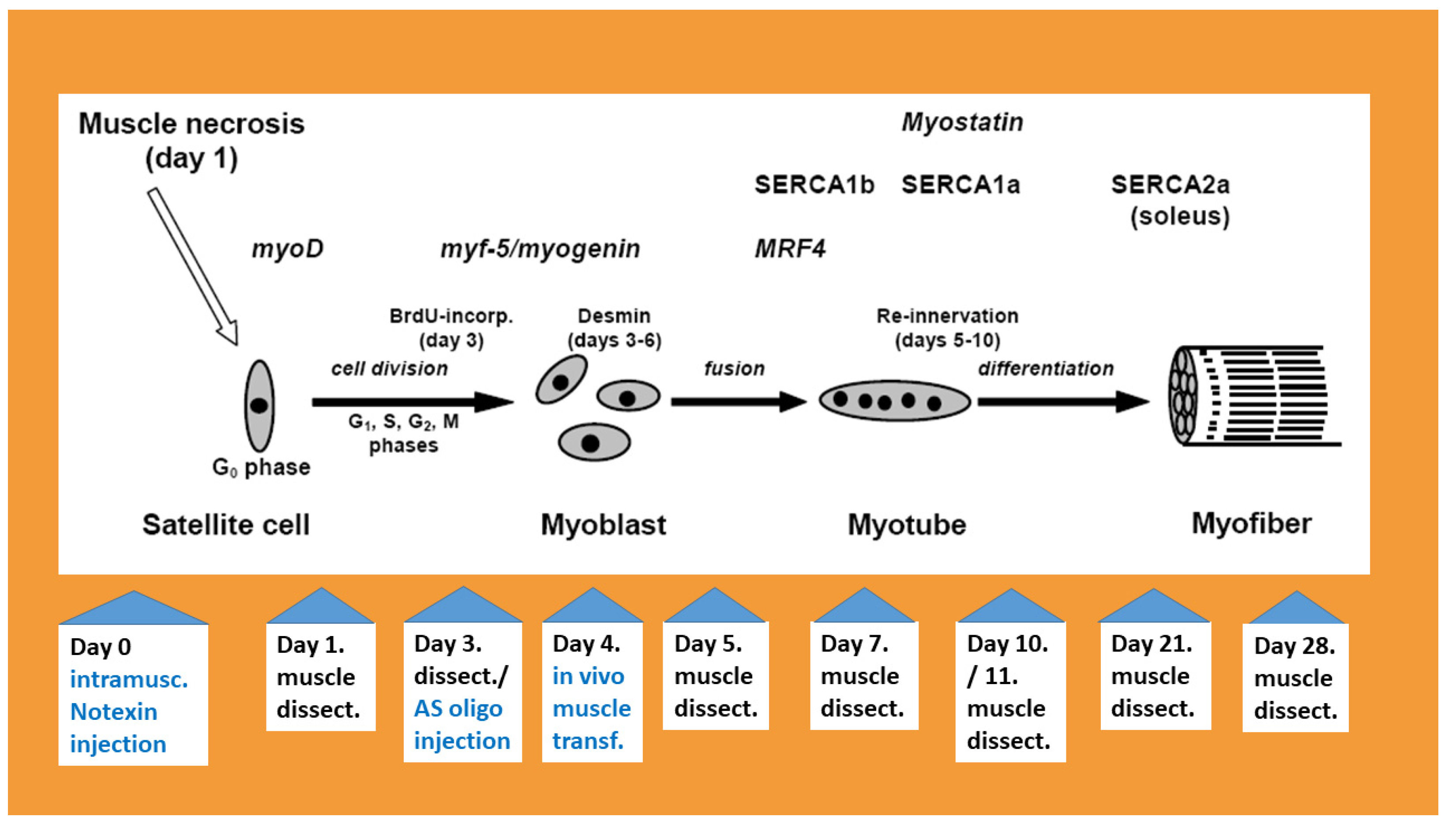
2. Muscle Degradation, Inflammation and Increase in TNF-α Levels
3. Myogenic Regulatory Factors
4. The Expression of Sarco/Endoplasmic Reticulum Ca2+ ATPases (SERCAs) during Regeneration
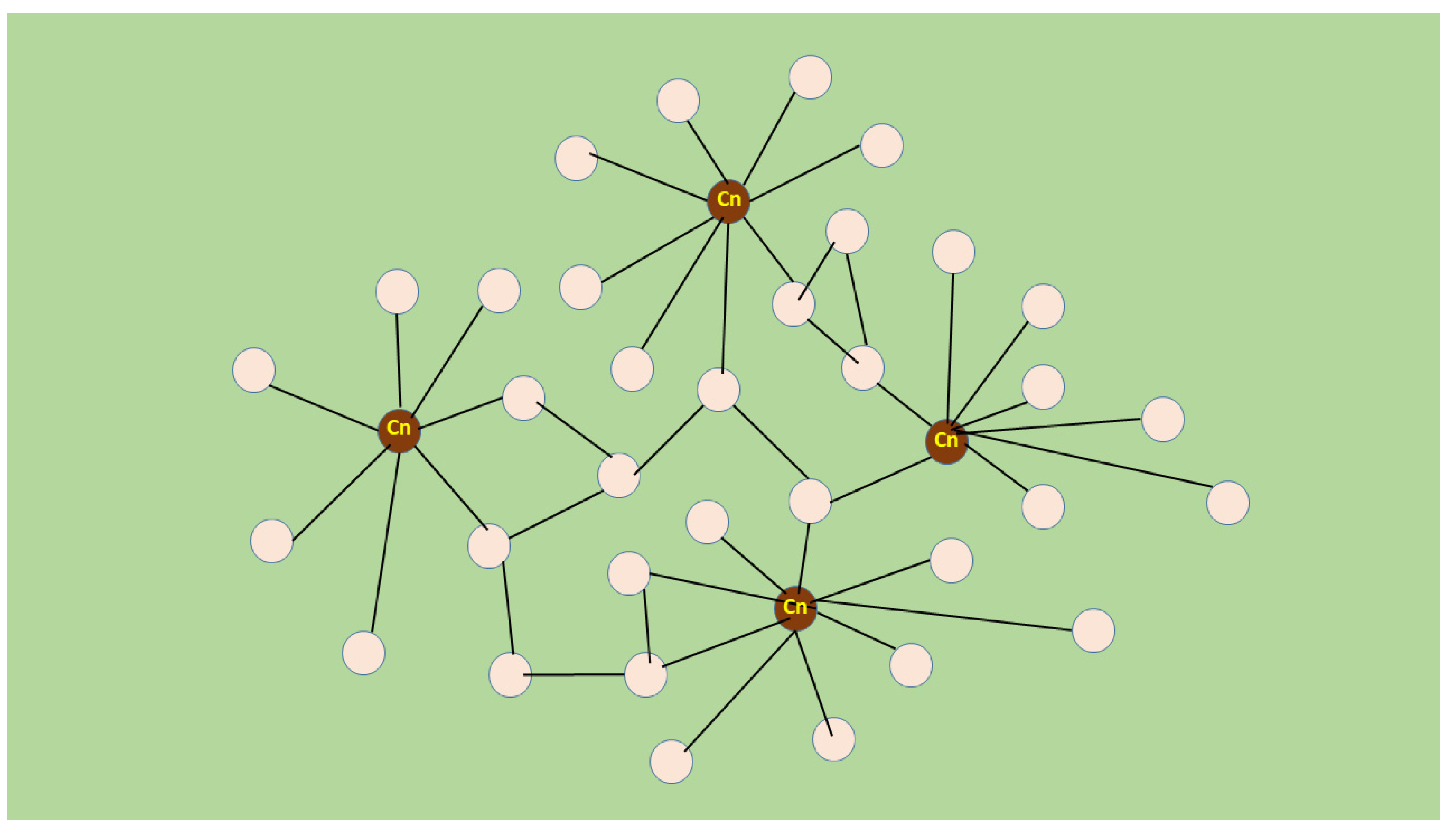
5. Novel Muscle Growth Regulation Revealed Using Molecular Acupuncture during Regeneration Takes after a Scale-Free Network
6. Conclusions
Funding
Conflicts of Interest
References
- Kim, K.M.; Jang, H.C.; Lim, S. Differences among skeletal muscle mass indices derived from height-, weight-, and body mass index-adjusted models in assessing sarcopenia. Korean J. Intern. Med. 2016, 31, 643–650. [Google Scholar] [CrossRef]
- Henrot, P.; Blervaque, L.; Dupin, I.; Zysman, M.; Esteves, P.; Gouzi, F.; Hayot, M.; Pomiès, P.; Berger, P. Cellular interplay in skeletal muscle regeneration and wasting: Insights from animal models. J. Cachexia Sarcopenia Muscle 2023, 14, 745–757. [Google Scholar] [CrossRef]
- Lieber, R.L.; Roberts, T.J.; Blemker, S.S.; Lee, S.S.M.; Herzog, W. Skeletal muscle mechanics, energetics and plasticity. J. Neuroeng. Rehabil. 2017, 14, 108. [Google Scholar] [CrossRef]
- Baldwin, K.M.; Haddad, F. The Evolution of Skeletal Muscle Plasticity in Response to Physical Activity and Inactivity. In Muscle and Exercise Physiology; Zoladz, J.A., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 347–377. [Google Scholar]
- Laumonier, T.; Menetrey, J. Muscle injuries and strategies for improving their repair. J. Exp. Orthop. 2016, 3, 15. [Google Scholar] [CrossRef]
- Mauro, A. Satellite cells of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 1961, 9, 493–495. [Google Scholar] [CrossRef]
- Seale, P.; Rudnicki, M.A. A new look at the origin, function, and “stem-cell” status of muscle satellite cells. Dev. Biol. 2000, 218, 115–124. [Google Scholar] [CrossRef]
- Morrissey, J.B.; Cheng, R.Y.; Davoudi, S.; Gilbert, P.M. Biomechanical Origins of Muscle Stem Cell Signal Transduction. J. Mol. Biol. 2016, 428, 1441–1454. [Google Scholar] [CrossRef]
- Fukada, S.I.; Akimoto, T.; Sotiropoulos, A. Role of damage and management in muscle hypertrophy: Different behaviors of muscle stem cells in regeneration and hypertrophy. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118742. [Google Scholar] [CrossRef]
- Ferrari, G.; Cusella-De Angelis, G.; Coletta, M.; Paolucci, E.; Stornaiuolo, A.; Cossu, G.; Mavilio, F. Muscle regeneration by bone marrow-derived myogenic progenitors. Science 1998, 279, 1528–1530. [Google Scholar] [CrossRef]
- Schulze, M.; Belema-Bedada, F.; Technau, A.; Braun, T. Mesenchymal stem cells are recruited to striated muscle by NFAT/IL-4-mediated cell fusion. Genes. Dev. 2005, 19, 1787–1798. [Google Scholar] [CrossRef][Green Version]
- Andrade, B.M.; Baldanza, M.R.; Ribeiro, K.C.; Porto, A.; Peçanha, R.; Fortes, F.S.; Zapata-Sudo, G.; Campos-de-Carvalho, A.C.; Goldenberg, R.C.; Werneck-de-Castro, J.P. Bone marrow mesenchymal cells improve muscle function in a skeletal muscle re-injury model. PLoS ONE 2015, 10, e0127561. [Google Scholar] [CrossRef]
- Gibson, A.J.; Karasinski, J.; Relvas, J.; Moss, J.; Sherratt, T.G.; Strong, P.N.; Watt, D.J. Dermal fibroblasts convert to a myogenic lineage in mdx mouse muscle. J. Cell Sci. 1995, 108, 207–214. [Google Scholar] [CrossRef]
- Chapman, M.A.; Meza, R.; Lieber, R.L. Skeletal muscle fibroblasts in health and disease. Differentiation 2016, 92, 108–115. [Google Scholar] [CrossRef]
- Giuliani, G.; Rosina, M.; Reggio, A. Signaling pathways regulating the fate of fibro/adipogenic progenitors (FAPs) in skeletal muscle regeneration and disease. FEBS J. 2022, 289, 6484–6517. [Google Scholar] [CrossRef]
- Molina, T.; Fabre, P.; Dumont, N.A. Fibro-adipogenic progenitors in skeletal muscle homeostasis, regeneration and diseases. Open Biol. 2021, 11, 210110. [Google Scholar] [CrossRef]
- Carlson, B.M. Muscle regeneration in animal models. In Skeletal Muscle Repair and Regeneration; Schiaffino, S., Partridge, T., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 163–180. [Google Scholar]
- Mendler, L.; Zádor, E.; Dux, L.; Wuytack, F. mRNA levels of myogenic regulatory factors in rat slow and fast muscles regenerating from notexin-induced necrosis. Neuromuscul. Disord. 1998, 8, 533–541. [Google Scholar] [CrossRef]
- Bernard, C.; Jomard, C.; Chazaud, B.; Gondin, J. Kinetics of skeletal muscle regeneration after mild and severe muscle damage induced by electrically-evoked lengthening contractions. FASEB J. 2023, 37, e23107. [Google Scholar] [CrossRef]
- Tidball, J.G.; Dorshkind, K.; Wehling-Henricks, M. Shared signaling systems in myeloid cell-mediated muscle regeneration. Development 2014, 141, 1184–1196. [Google Scholar] [CrossRef]
- Khuu, S.; Fernandez, J.W.; Handsfield, G.G. Delayed skeletal muscle repair following inflammatory damage in simulated agent-based models of muscle regeneration. PLoS Comput. Biol. 2023, 19, e1011042. [Google Scholar] [CrossRef]
- Bordon, K.C.F.; Cologna, C.T.; Fornari-Baldo, E.C.; Pinheiro-Júnior, E.L.; Cerni, F.A.; Amorim, F.G.; Anjolette, F.A.P.; Cordeiro, F.A.; Wiezel, G.A.; Cardoso, I.A.; et al. From Animal Poisons and Venoms to Medicines: Achievements, Challenges and Perspectives in Drug Discovery. Front. Pharmacol. 2020, 11, 1132. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Lomonte, B. Phospholipases A2: Unveiling the secrets of a functionally versatile group of snake venom toxins. Toxicon 2013, 62, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Benoit, P.W.; Belt, W.D. Destruction and regeneration of skeletal muscle after treatment with a local anaesthetic, bupivacaine (Marcaine). J. Anat. 1970, 107, 547–556. [Google Scholar] [PubMed]
- Chen, Y.; Li, X.; Huo, Z.; Chen, H.; Zhang, L. An overview of bupivacaine-induced morphological changes: A novel animal model of skeletal muscle injury. Int. J. Clin. Exp. Med. 2020, 13, 7–15. [Google Scholar]
- Harris, J.B.; Johnson, M.A.; Karlsson, E. Pathological responses of rat skeletal muscle to a single subcutaneous injection of a toxin isolated from the venom of the Australian tiger snake, Notechis scutatus scutatus. Clin. Exp. Pharmacol. Physiol. 1975, 2, 383–404. [Google Scholar] [CrossRef]
- Plant, D.R.; Colarossi, F.E.; Lynch, G.S. Notexin causes greater myotoxic damage and slower functional repair in mouse skeletal muscles than bupivacaine. Muscle Nerve 2006, 34, 577–585. [Google Scholar] [CrossRef]
- Zádor, E.; Mendler, L.; Ver Heyen, M.; Dux, L.; Wuytack, F. Changes in mRNA levels of the sarcoplasmic/endoplasmic-reticulum Ca2+-ATPase isoforms in the rat soleus muscle regenerating from notexin-induced necrosis. Biochem. J. 1996, 320, 107–113. [Google Scholar] [CrossRef]
- Murgia, M.; Serrano, A.L.; Calabria, E.; Pallafacchina, G.; Lomo, T.; Schiaffino, S. Ras is involved in nerve-activity-dependent regulation of muscle genes. Nat. Cell Biol. 2000, 2, 142–147. [Google Scholar] [CrossRef]
- Zhou, K.; Luo, W.; Liu, T.; Ni, Y.; Qin, Z. Neurotoxins Acting at Synaptic Sites: A Brief Review on Mechanisms and Clinical Applications. Toxins 2022, 15, 18. [Google Scholar] [CrossRef]
- Montecucco, C.; Gutiérrez, J.M.; Lomonte, B. Cellular pathology induced by snake venom phospholipase A2 myotoxins and neurotoxins: Common aspects of their mechanisms of action. Cell Mol. Life Sci. 2008, 65, 2897–2912. [Google Scholar] [CrossRef]
- Cintra-Francischinelli, M.; Pizzo, P.; Rodrigues-Simioni, L.; Ponce-Soto, L.A.; Rossetto, O.; Lomonte, B.; Gutiérrez, J.M.; Pozzan, T.; Montecucco, C. Calcium imaging of muscle cells treated with snake myotoxins reveals toxin synergism and presence of acceptors. Cell Mol. Life Sci. 2009, 66, 1718–1728. [Google Scholar] [CrossRef]
- Whalen, R.G.; Harris, J.B.; Butler-Browne, G.S.; Sesodia, S. Expression of myosin isoforms during notexin-induced regeneration of rat soleus muscles. Dev. Biol. 1990, 141, 24–40. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Rucavado, A.; Escalante, T.; Herrera, C.; Fernández, J.; Lomonte, B.; Fox, J.W. Unresolved issues in the understanding of the pathogenesis of local tissue damage induced by snake venoms. Toxicon 2018, 148, 123–131. [Google Scholar] [CrossRef]
- Harris, J.B.; Scott-Davey, T. Secreted phospholipases A2 of snake venoms: Effects on the peripheral neuromuscular system with comments on the role of phospholipases A2 in disorders of the CNS and their uses in industry. Toxins 2013, 5, 2533–2571. [Google Scholar] [CrossRef]
- Harris, J.B. Myotoxic phospholipases A2 and the regeneration of skeletal muscles. Toxicon 2003, 42, 933–945. [Google Scholar] [CrossRef]
- Gasanov, S.E.; Dagda, R.K.; Rael, E.D. Snake Venom Cytotoxins, Phospholipase A2s, and Zn2+-dependent Metalloproteinases: Mechanisms of Action and Pharmacological Relevance. J. Clin. Toxicol. 2014, 4, 1000181. [Google Scholar] [CrossRef]
- Bickler, P.E. Amplification of Snake Venom Toxicity by Endogenous Signaling Pathways. Toxins 2020, 12, 68. [Google Scholar] [CrossRef]
- Delp, M.D.; Duan, C. Composition and size of type I, IIA, IID/X, and IIB fibers and citrate synthase activity of rat muscle. J. Appl. Physiol. 1996, 80, 261–270. [Google Scholar] [CrossRef]
- Zádor, E.; Wuytack, F. Expression of SERCA2a is independent of innervation in regenerating soleus muscle. Am. J. Physiol.-Cell Physiol. 2003, 285, C853–C861. [Google Scholar] [CrossRef]
- Zádor, E.; Szakonyi, G.; Rácz, G.; Mendler, L.; Ver Heyen, M.; Lebacq, J.; Dux, L.; Wuytack, F. Expression of the sarco/endoplasmic reticulum Ca2+-transport ATPase protein isoforms during regeneration from notexin induced necrosis of rat muscle. Acta Histochem. 1998, 100, 355–369. [Google Scholar] [CrossRef]
- Zádor, E.; Mendler, L.; Takács, V.; De Bleecker, J.; Wuytack, F. Regenerating soleus and EDL muscles of the rat show elevated levels of TNF-α and its receptors, TNFR-60 and TNFR-80. Muscle Nerve 2001, 24, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Tidball, J.G.; Villalta, S.A. Regulatory interactions between muscle and the immune system during muscle regeneration. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 298, R1173–R1187. [Google Scholar] [CrossRef] [PubMed]
- Mourkioti, F.; Rosenthal, N. NF-kappaB signaling in skeletal muscle: Prospects for intervention in muscle diseases. J. Mol. Med. 2008, 86, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Webster, J.D.; Vucic, D. The Balance of TNF Mediated Pathways Regulates Inflammatory Cell Death Signaling in Healthy and Diseased Tissues. Front. Cell Dev. Biol. 2020, 8, 365. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, S.; Panguluri, S.K.; Gupta, S.K.; Dahiya, S.; Lundy, R.F.; Kumar, A. Tumor necrosis factor-α regulates distinct molecular pathways and gene networks in cultured skeletal muscle cells. PLoS ONE 2010, 5, e13262. [Google Scholar] [CrossRef] [PubMed]
- Langen, R.C.; Van der Velden, J.L.; Schols, A.M.; Kelder, M.C.; Wouters, E.F.; Janssen-Heininger, Y.M. Tumor necrosis factor-alpha inhibits myogenic differentiation through MyoD protein destabilization. FASEB J. 2004, 18, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.P. TNF-α is mitogen in skeletal muscle. Am. J. Physiol. Cell Physiol. 2003, 285, C370–C376. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, M.E.; Londino, J.; McGinnis, M.; Weathington, N.; Adair, J.; Suber, T.; Kagan, V.; Chen, K.; Zou, C.; Chen, B.; et al. Tumor Necrosis Factor Alpha Regulates Skeletal Myogenesis by Inhibiting SP1 Interaction with cis-Acting Regulatory Elements within the Fbxl2 Gene Promoter. Mol. Cell Biol. 2020, 40, e00040-20. [Google Scholar] [CrossRef]
- Chen, S.E.; Jin, B.; Li, Y.P. TNF-alpha regulates myogenesis and muscle regeneration by activating p38 MAPK. Am. J. Physiol. Cell Physiol. 2007, 292, C1660–C1671. [Google Scholar] [CrossRef]
- Careccia, G.; Mangiavini, L.; Cirillo, F. Regulation of Satellite Cells Functions during Skeletal Muscle Regeneration: A Critical Step in Physiological and Pathological Conditions. Int. J. Mol. Sci. 2023, 25, 512. [Google Scholar] [CrossRef]
- Ceafalan, L.C.; Popescu, B.O.; Hinescu, M.E. Cellular players in skeletal muscle regeneration. Biomed. Res. Int. 2014, 2014, 957014. [Google Scholar] [CrossRef]
- Tu, H.; Li, Y.L. Inflammation balance in skeletal muscle damage and repair. Front. Immunol. 2023, 14, 1133355. [Google Scholar] [CrossRef] [PubMed]
- Kovács, E.; Zádor, E. The effect of a TNF-α inhibiting drug on skeletal muscle regeneration. XXXIV. In Proceedings of the European Muscle Conference, Hungary, Hortobágy, 17–21 September 2005. Abstract. J. Muscle Res. Cell Motil. 2005, 26, 57–89. [Google Scholar]
- Ermolova, N.V.; Martinez, L.; Vetrone, S.A.; Jordan, M.C.; Roos, K.P.; Sweeney, H.L.; Spencer, M.J. Long-term administration of the TNF blocking drug Remicade (cV1q) to mdx mice reduces skeletal and cardiac muscle fibrosis, but negatively impacts cardiac function. Neuromuscul. Disord. 2014, 24, 583–595. [Google Scholar] [CrossRef]
- McCroskery, S.; Thomas, M.; Maxwell, L.; Sharma, M.; Kambadur, R. Myostatin negatively regulates satellite cell activation and self-renewal. J. Cell Biol. 2003, 162, 1135–1147. [Google Scholar] [CrossRef]
- Mendler, L.; Zador, E.; Ver Heyen, M.; Dux, L.; Wuytack, F. Myostatin in regenerating rat muscles and in myogenic cell cultures. J. Muscle Res. Cell Mot. 2000, 21, 551–563. [Google Scholar] [CrossRef]
- Rodriguez-Outeiriño, L.; Hernandez-Torres, F.; Ramírez-de Acuña, F.; Matías-Valiente, L.; Sanchez-Fernandez, C.; Franco, D.; Aranega, A.E. Muscle Satellite Cell Heterogeneity: Does Embryonic Origin Matter? Front. Cell Dev. Biol. 2021, 9, 750534. [Google Scholar] [CrossRef]
- Vaidya, T.B.; Rhodes, S.J.; Moore, J.L.; Sharman, D.A.; Konieczny, S.F.; Taparowsky, E.J. Isolation and structural analysis of the rat myoD gene. Gene 1992, 116, 223–230. [Google Scholar] [CrossRef]
- Miller, J.B.; Everitt, E.A.; Smith, T.H.; Block, N.E.; Dominov, J.A. Cellular and Molecular Diversity in Skeletal Muscle Development: News from in vitro and in vivo. BioEssays 1993, 15, 191–195. [Google Scholar] [CrossRef]
- Olson, E.N.; Klein, W.H. bHLH factors in muscle development: Dead lines and commitments, what to leave in and what to leave out. Genes. Dev. 1994, 8, 1–8. [Google Scholar] [CrossRef]
- Motohashi, N.; Asakura, A. Muscle satellite cell heterogeneity and self-renewal. Front. Cell Dev. Bio. 2014, 2, 1. [Google Scholar] [CrossRef] [PubMed]
- Ludolph, D.C.; Konieczny, S.F. Transcription factor families: Muscling in on the myogenic program. FASEB J. 1995, 9, 1595–1604. [Google Scholar] [CrossRef] [PubMed]
- Maione, R.; Amati, P. Interdependence between muscle differentiation and cell cycle control. Biochim. Biophys. Acta 1997, 1322, M19–M30. [Google Scholar] [CrossRef] [PubMed]
- Tapscott, S.J. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 2005, 132, 2685–2695. [Google Scholar] [CrossRef] [PubMed]
- Lowe, D.A.; Lund, T.; Alway, S.E. Hypertrophy-stimulated myogenic regulatory factor mRNA increases are attenuated in fast muscle of aged quails. Am. J. Physiol. Cell Physiol. 1998, 275, C155–C166. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Dilworth, F.J. Differential modulation of cell cycle progression distinguishes members of the myogenic regulatory factor family of transcription factors. FEBS J. 2013, 280, 3991–4003. [Google Scholar] [CrossRef] [PubMed]
- Zammit, P.S. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin. Cell Dev. Biol. 2017, 72, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Rudnicki, M.A.; Schnegelsberg, P.N.J.; Stead, R.H.; Braun, T.; Arnold, H.-H.; Jaenisch, R. MyoD and myf-5 is required for the formation of skeletal muscle. Cell 1993, 75, 1351–1359. [Google Scholar] [CrossRef]
- Gerber, A.N.; Klesert, T.R.; Bergstrom, D.A.; Tapscott, S.J. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: A mechanism for lineage determination in myogenesis. Genes. Dev. 1997, 11, 436–450. [Google Scholar] [CrossRef]
- Londhe, P.; Davie, J.K. Sequential association of myogenic regulatory factors and E proteins at muscle-specific genes. Skelet. Muscle 2011, 1, 14. [Google Scholar] [CrossRef]
- de Martin, X.; Sodaei, R.; Santpere, G. Mechanisms of Binding Specificity among bHLH Transcription Factors. Int. J. Mol. Sci. 2021, 22, 9150. [Google Scholar] [CrossRef]
- Sabourin, L.A.; Rudnicki, M.A. The molecular regulation of myogenesis. Clin. Genet. 2000, 57, 16–25. [Google Scholar] [CrossRef]
- Zammit, P.S. All muscle satellite cells are equal, but are some more equal than others? J. Cell Sci. 2008, 121, 2975–2982. [Google Scholar] [CrossRef]
- Günther, S.; Kim, J.; Kostin, S.; Lepper, C.; Fan, C.M.; Braun, T. Myf5-positive satellite cells contribute to Pax7-dependent long-term maintenance of adult muscle stem cells. Cell Stem Cell 2013, 13, 590–601, Erratum in: Cell Stem Cell 2013, 13, 769. [Google Scholar] [CrossRef]
- Zádor, E.; Bottka, S.; Wuytack, F. Antisense inhibition of myoD expression in regenerating rat soleus muscle is followed by an increase in the mRNA levels of myoD, myf-5 and myogenin and by a retarded regeneration. Biochim. Biophys. Acta Mol. Cell Res. 2002, 1590, 52–63. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Grubb, B.; Harris, J.; Schofield, I. Neuromuscular transmission of newly formed neuromuscular junctions in the regenerating soleus muscle. J. Physiol. 1991, 441, 405–421. [Google Scholar] [CrossRef] [PubMed]
- Abu Hatoum, O.; Gross-Mesilaty, S.; Breitschop, K.; Hoffman, A.; Gonen, H.; Ciechanover, A.; Bengal, E. Degradation of myogenic transcription factor myoD by the ubiquitin pathway in vivo and in vitro: Regulation by specific DNA binding. Mol. Cell Biol. 1998, 18, 5670–5677. [Google Scholar] [CrossRef][Green Version]
- Bisbal, C.; Silhol, M.; Laubenthal, H.; Kaluza, T.; Carnac, G.; Milligan, L.; Le Roy, F.; Salehzada, T. The 2V–5Voligoadenylate/RNase L/RNase L inhibitor pathway regulates both myoD mRNA stability and muscle cell differentiation. Mol. Cell Biol. 2000, 20, 4959–4969. [Google Scholar] [CrossRef]
- Phillis, M.I.; Gyurko, R. Antisense oligonucleotides: New tools for physiology. News Physiol. Sci. 1997, 12, 99–105. [Google Scholar]
- Tu, G.-C.; Cao, Q.-N.; Zhou, F.; Yedy, I. Tetranucleotide GGGA motif in primary RNA transcripts. Novel target site for antisense design. J. Biol. Chem. 1998, 273, 25125–25131. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Dennis, R.G.; Baar, K. Cultured slow vs. fast skeletal muscle cells differ in physiology and responsiveness to stimulation. Am. J. Physiol. Cell Physiol. 2006, 291, C11–C17. [Google Scholar] [CrossRef]
- Gorbe, A.; Becker, D.L.; Dux, L.; Stelkovics, E.; Krenacs, L.; Bagdi, E.; Krenacs, T. Transient upregulation of connexin43 gap junctions and synchronized cell cycle control precede myoblast fusion in regenerating skeletal muscle in vivo. Histochem. Cell Biol. 2005, 123, 573–583. [Google Scholar] [CrossRef]
- Gorbe, A.; Becker, D.L.; Dux, L.; Krenacs, L.; Krenacs, T. In differentiating prefusion myoblasts connexin43 gap junction coupling is upregulated before myoblast alignment then reduced in post-mitotic cells. Histochem. Cell Biol. 2006, 125, 705–716. [Google Scholar] [CrossRef]
- Gorbe, A.; Krenacs, T.; Cook, J.E.; Becker, D.L. Myoblast proliferation and syncytial fusion both depend on connexin43 function in transfected skeletal muscle primary cultures. Exp. Cell Res. 2007, 313, 1135–1148. [Google Scholar] [CrossRef]
- Ishido, M.; Kasuga, N. Characteristics of the Localization of Connexin 43 in Satellite Cells during Skeletal Muscle Regeneration In Vivo. Acta Histochem. Cytochem. 2015, 48, 53–60. [Google Scholar] [CrossRef]
- Gash, M.C.; Kandle, P.F.; Murray, I.V.; Varacallo, M. Physiology, Muscle Contraction. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Xu, H.; Van Remmen, H. The SarcoEndoplasmic Reticulum Calcium ATPase (SERCA) pump: A potential target for intervention in aging and skeletal muscle pathologies. Skelet. Muscle 2021, 11, 25. [Google Scholar] [CrossRef]
- Brandl, C.J.; Green, N.M.; Korczak, B.; MacLennan, D.H. Two Ca2+ ATPase genes: Homologies and mechanistic implications of deduced amino acid sequences. Cell 1986, 44, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Brandl, C.J.; DeLeon, S.; Martin, D.R.; MacLennan, D.H. Adult forms of the Ca2+ ATPase of sarcoplasmic reticulum. Expression in developing skeletal muscle. J. Biol. Chem. 1987, 262, 3768–3774. [Google Scholar] [CrossRef] [PubMed]
- Korczak, B.; Zarain-Herzberg, A.; Brandl, C.J.; Ingles, C.J.; Green, M.N.; MacLennan, D.H. Structure of the rabbit fast-twitch skeletal muscle Ca2+ ATPase gene. J. Biol. Chem. 1988, 263, 4813–4819. [Google Scholar] [CrossRef]
- Zádor, E.; Vangheluwe, P.; Wuytack, F. The expression of the neonatal sarcoplasmic reticulum Ca2+ pump (SERCA1b) hints to a role in muscle growth and development. Cell Calcium 2007, 41, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Zador, E.; Dux, L.; Wuytack, F. Prolonged passive stretch of rat soleus muscle provokes an increase in the mRNA levels of the muscle regulatory factors distributed along the entire length of the fibers. J. Muscle Res. Cell Mot. 1999, 20, 395–402. [Google Scholar] [CrossRef]
- Szabó, A.; Wuytack, F.; Zádor, E. The effect of passive movement on denervated soleus highlights a differential nerve control on SERCA and MyHC isoforms. J. Histochem. Cytochem. 2008, 56, 1013–1022. [Google Scholar] [CrossRef]
- Zádor, E.; Kósa, M. The neonatal sarcoplasmic/endoplasmic reticulum calcium ATPase (SERCA1b): A neglected pump in scope. Pflugers Arch. 2015, 467, 1395–1401. [Google Scholar] [CrossRef]
- Kósa, M.; Brinyiczki, K.; van Damme, P.; Goemans, N.; Hancsák, K.; Mendler, L.; Zádor, E. The neonatal sarcoplasmic reticulum Ca2+-ATPase gives a clue to development and pathology in human muscles. J. Muscle Res. Cell Motil. 2015, 36, 195–203. [Google Scholar] [CrossRef]
- Fodor, J.; Gomba-Tóth, A.; Oláh, T.; Zádor, E.; Tóth, Z.C.; Ioannis, I.; Molnár, B.; Kovács, I.; Csernoch, L. Alteration of sarcoplasmic reticulum Ca2+ ATPase expression in lower limb ischemia caused by atherosclerosis obliterans. Physiol. Int. 2017, 104, 183–192. [Google Scholar] [CrossRef]
- Zhao, Y.; Ogawa, H.; Yonekura, S.; Mitsuhashi, H.; Mitsuhashi, S.; Nishino, I.; Toyoshima, C.; Ishiura, S. Functional analysis of SERCA1b, a highly expressed SERCA1 variant in myotonic dystrophy type 1 muscle. Biochim. Biophys. Acta 2015, 1852, 2042–2047. [Google Scholar] [CrossRef]
- Guglielmi, V.; Oosterhof, A.; Voermans, N.C.; Cardani, R.; Molenaar, J.P.; van Kuppevelt, T.H.; Meola, G.; van Engelen, B.G.; Tomelleri, G.; Vattemi, G. Characterization of sarcoplasmic reticulum Ca2+ ATPase pumps in muscle of patients with myotonic dystrophy and with hypothyroid myopathy. Neuromuscul. Disord. 2016, 26, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Mendler, L.; Szakonyi, G.; Zádor, E.; Görbe, A.; Dux, L.; Wuytack, F. Expression of sarcoplasmic/endoplasmic reticulum Ca2+ ATPases in the rat extensor digitorum longus (EDL) muscle regenerating from notexin-induced necrosis. J. Muscle Res. Cell Mot. 1998, 19, 777–785. [Google Scholar] [CrossRef]
- Kiss, G.; Zádor, E.; Szalay, J.; Somogyi, F.; Vér, A. Molecular forms of acetylcholinesterase in rat extensor digitorum longus and soleus muscles regenerating from notexin induced necrosis. J. Muscle Res. Cell Motil. 2004, 25, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Fenyvesi, R.; Rácz, G.; Wuytack, F.; Zádor, E. The calcineurin activity and MCIP1.4 mRNA levels are increased by innervation in regenerating soleus muscle. Biochem. Biophys. Res. Commun. 2004, 320, 599–605. [Google Scholar] [CrossRef]
- Zádor, E.; Fenyvesi, R.; Wuytack, F. Expression of SERCA2a is not regulated by calcineurin or upon mechanical unloading in skeletal muscle regeneration. FEBS Lett. 2005, 579, 749–752. [Google Scholar] [CrossRef] [PubMed]
- Serrano, A.L.; Murgia, M.; Pallafacchina, G.; Calabria, E.; Coniglio, P.; Lømo, T.; Schiaffino, S. Calcineurin controls nerve activity-dependent specification of slow skeletal muscle fibers but not muscle growth. Proc. Natl. Acad. Sci. USA 2001, 98, 13108–13113. [Google Scholar] [CrossRef] [PubMed]
- McCullagh, K.J.; Calabria, E.; Pallafacchina, G.; Ciciliot, S.; Serrano, A.L.; Argentini, C.; Kalhovde, J.M.; Lømo, T.; Schiaffino, S. NFAT is a nerve activity sensor in skeletal muscle and controls activity-dependent myosin switching. Proc. Natl. Acad. Sci. USA 2004, 101, 10590–10595. [Google Scholar] [CrossRef] [PubMed]
- Launay, T.; Noirez, P.; Butler-Browne, G.; Agbulut, O. Expression of slow myosin heavy chain during muscle regeneration is not always dependent on muscle innervation and calcineurin phosphatase activity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 290, R1508–R1514. [Google Scholar] [CrossRef] [PubMed]
- Misquitta, C.M.; Chen, T.; Grover, A.K. Control of protein expression through mRNA stability in calcium signalling. Cell Calcium 2006, 40, 329–346. [Google Scholar] [CrossRef] [PubMed]
- Zádor, E. dnRas stimulates autocrine-paracrine growth of regenerating muscle via calcineurin-NFAT-IL-4 pathway. Biochem. Biophys. Res. Commun. 2008, 375, 265–270. [Google Scholar] [CrossRef]
- Kósa, M.; Zádor, E. Transfection efficiency along the regenerating soleus muscle of the rat. Mol. Biotechnol. 2013, 54, 220–227. [Google Scholar] [CrossRef]
- Horsley, V.; Jansen, K.M.; Mills, S.T.; Pavlath, G.K. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell 2003, 113, 483–494. [Google Scholar] [CrossRef]
- Shaikh, S.; Lee, E.; Ahmad, K.; Ahmad, S.S.; Chun, H.; Lim, J.; Lee, Y.; Choi, I. Cell Types Used for Cultured Meat Production and the Importance of Myokines. Foods 2021, 10, 2318. [Google Scholar] [CrossRef]
- Horsley, V.; Friday, B.B.; Matteson, S.; Kegley, K.M.; Gephart, J.; Pavlath, G.K. Regulation of the growth of multinucleated muscle cells by an NFATC2-dependent pathway. J. Cell Biol. 2001, 153, 329–338. [Google Scholar] [CrossRef]
- Ichida, M.; Finkel, T. Ras regulates NFAT3 activity in cardiac myocytes. J. Biol. Chem. 2001, 276, 3524–3530. [Google Scholar] [CrossRef] [PubMed]
- Sanna, B.; Bueno, O.F.; Dai, Y.S.; Wilkins, B.J.; Molkentin, J.D. Direct and indirect interactions between calcineurin-NFAT and MEK1-extracellular signal-regulated kinase 1/2 signaling pathways regulate cardiac gene expression and cellular growth. Mol. Cell Biol. 2005, 25, 865–878. [Google Scholar] [CrossRef]
- Zádor, E.; Owsianik, G.; Wuytack, F. Silencing SERCA1b in a few fibers stimulates growth in the entire regenerating soleus muscle. Histochem. Cell Biol. 2011, 135, 11–20. [Google Scholar] [CrossRef]
- Heredia, J.E.; Mukundan, L.; Chen, F.M.; Mueller, A.A.; Deo, R.C.; Locksley, R.M.; Rando, T.A.; Chawla, A. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell 2013, 153, 376–388. [Google Scholar] [CrossRef]
- Pan, Y.; Zvaritch, E.; Tupling, A.R.; Rice, W.J.; de Leon, S.; Rudnicki, M.; McKerlie, C.; Banwell, B.L.; MacLennan, D.H. Targeted disruption of the ATP2A1 gene encoding the sarco(endo)plasmic reticulum Ca2+ ATPase isoform 1 (SERCA1) impairs diaphragm function and is lethal in neonatal mice. J. Biol. Chem. 2003, 278, 13367–13375. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Anderson, K.M.; Chang, C.L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Hansson, K.A.; Eftestøl, E.; Bruusgaard, J.C.; Juvkam, I.; Cramer, A.W.; Malthe-Sørenssen, A.; Millay, D.P.; Gundersen, K. Myonuclear content regulates cell size with similar scaling properties in mice and humans. Nat. Commun. 2020, 11, 6288. [Google Scholar] [CrossRef]
- Bagley, J.R.; Denes, L.T.; McCarthy, J.J.; Wang, E.T.; Murach, K.A. The myonuclear domain in adult skeletal muscle fibres: Past, present and future. J. Physiol. 2023, 601, 723–741. [Google Scholar] [CrossRef]
- Lin, J.G.; Kotha, P.; Chen, Y.H. Understandings of acupuncture application and mechanisms. Am. J. Transl. Res. 2022, 14, 1469–1481. [Google Scholar]
- Rajula, H.S.R.; Mauri, M.; Fanos, V. Scale-free networks in metabolomics. Bioinformation 2018, 14, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Barabasi, A.L.; Albert, R. Emergence of scaling in random networks. Science 1999, 286, 509–512. [Google Scholar] [CrossRef]
- Newlands, S.; Levitt, L.K.; Robinson, C.S.; Karpf, A.B.; Hodgson, V.R.; Wade, R.P.; Hardeman, E.C. Transcription occurs in pulses in muscle fibers. Genes Dev. 1998, 12, 2748–2758. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zádor, E. Characterization of Skeletal Muscle Regeneration Revealed a Novel Growth Network Induced by Molecular Acupuncture-like Transfection. Biomolecules 2024, 14, 363. https://doi.org/10.3390/biom14030363
Zádor E. Characterization of Skeletal Muscle Regeneration Revealed a Novel Growth Network Induced by Molecular Acupuncture-like Transfection. Biomolecules. 2024; 14(3):363. https://doi.org/10.3390/biom14030363
Chicago/Turabian StyleZádor, Ernő. 2024. "Characterization of Skeletal Muscle Regeneration Revealed a Novel Growth Network Induced by Molecular Acupuncture-like Transfection" Biomolecules 14, no. 3: 363. https://doi.org/10.3390/biom14030363
APA StyleZádor, E. (2024). Characterization of Skeletal Muscle Regeneration Revealed a Novel Growth Network Induced by Molecular Acupuncture-like Transfection. Biomolecules, 14(3), 363. https://doi.org/10.3390/biom14030363





