Abstract
The OVATE gene family is a class of conserved transcription factors that play significant roles in plant growth, development, and abiotic stress, and also affect fruit shape in vegetable crops. Bottle gourd (Lagenaria siceraria), commonly known as calabash or gourd, is an annual climber belonging to the Cucurbitaceae family. Studies on bottle gourd OVATE genes are limited. In this study, we performed genome-wide identification of the OVATE gene family in bottle gourd, and identified a total of 20 OVATE family genes. The identified genes were unevenly distributed across 11 bottle gourd chromosomes. We also analyzed the gene homology, amino acid sequence conservation, and three-dimensional protein structure (via prediction) of the 20 OVATE family genes. We used RNA-seq data to perform expression analysis, which found 20 OVATE family genes to be differentially expressed based on spatial and temporal characteristics, suggesting that they have varying functions in the growth and development of bottle gourd. In situ hybridization and subcellular localization analysis showed that the expression characteristics of the LsOVATE1 gene, located on chromosome 7 homologous to OVATE, is a candidate gene for affecting the fruit shape of bottle gourd. In addition, RT-qPCR data from bottle gourd roots, stems, leaves, and flowers showed different spatial expression of the LsOVATE1 gene. The ectopic expression of LsOVATE1 in tomato generated a phenotype with a distinct fruit shape and development. Transgenic-positive plants that overexpressed LsOVATE1 had cone-shaped fruit, calyx hypertrophy, petal degeneration, and petal retention after flowering. Our results indicate that LsOVATE1 could serve important roles in bottle gourd development and fruit shape determination, and provide a basis for future research into the function of LsOVATE1.
1. Introduction
Bottle gourd [Lagenaria siceraria (Mol.) Standl.] (2n = 2x = 22), a member of the Cucurbitaceae family, also known as calabash or long melon, is a cultivated vegetable, medicinal plant, and grafting rootstock [1]. It originated in Africa and was independently domesticated in Asia [2]. With a cultivation history of 7000 years in China, it is one of the characteristic summer vegetables in southern China. Bottle gourd exhibits high genetic variability, especially in fruit shape, which can be round, oblate, pyriform, Hulu (double gourd), dipper, slender straight, tubby, etc. [3,4]. The diverse morphology of the bottle gourd fruit makes it a good candidate for studying variations in fruit shape. Fruit shape is a major horticultural trait for many fruit and vegetable crops that can influence their yield and quality from a crop-breeding perspective [5,6]. It is usually evaluated in terms of fruit diameter (FD), fruit length (FL), and fruit shape index (FSI, the ratio of FL to FD) [7,8]. Changes in fruit shape are not only the result of natural selection but also of artificial domestication to adapt to diverse environments or consumer preferences [9].
Fruit shape has historically been a research hotspot [10,11,12], and many fruit shape-determining genes have been identified or cloned in various crops [13,14,15,16,17,18,19,20]. In tomato, OVATE is expressed during flower and fruit development. A single mutation in OVATE leads to a premature stop codon, causing tomato fruits to transition from round to pear-shaped [14]. SUN, one of the major genes causing elongated fruit shapes in tomato, was found to encode a member of the IQ67 domain-containing family [15]. Research shows that extreme fruit sizes are the result of a regulatory change of a transcription factor (FASCIATED) that controls carpel number during flower or fruit development [16]. In addition to FASCIATED, a single nucleotide mutation of LC will also cause changes in the number of locules [17]. Regarding cucurbit crops, studies have generally focused on cucumber. SF1 induces larger but fewer fruit cells, which leads to a short fruit phenotype. In addition, transcriptomic analysis revealed that SF1 might control fruit length via fine-tuning of cytokinin and auxin signaling, gibberellin biosynthesis, and signal transduction in cucumber fruits [18]. CsCLV3 is a homolog of the Arabidopsis CLAVATA3 gene. CsCLV3 induces carpel number variations in cucumber [19]. Further research also showed that CsCLV3 and CsWUS function as a negative and positive regulator for carpel number variation, respectively. Biochemical analyses indicated that CsWUS directly binds to the promoter of CsCLV3 to activate its expression [20].
Among the known fruit shape-related tomato genes, OVATE is a representative gene that is also known to affect fruit shape in other crops. Eighteen OVATE family transcription factors were identified in Arabidopsis thaliana. AtOFP1 overexpression leads to silique shortening. Furthermore, this gene inhibits cell elongation by inhibiting AtGA20ox1 expression, which encodes gibberellin synthase [21,22]. Rice also contains multiple OVATE family transcription factors. The overexpression of OsOFP1 and OsOFP2 results in dwarf plants with thicker leaves, anther abortions, and low seed-setting rates [23,24]. In addition, the expression of OVATE homologs has been confirmed to be closely related to fruit shape in pepper, peach, and melon [6,25,26,27]. The mechanism by which OVATE regulates fruit shape is related to its interaction with TONNEAU1 Recruiting Motif (TRM) family proteins, which are involved in regulating microtubule arrangement in order to control cell division patterns and growth [26,28,29]. In Arabidopsis thaliana, TRM proteins (LONGIFOLIA1 (LNG1) and LONGIFOLIA2 (LNG2)) regulate the longitudinal elongation of cells. Overexpression of these two proteins causes the aerial portions of plants, such as leaves, floral organs, and siliques, to become longer, while the cotyledons and pods become shorter if loss-of-function mutations of these proteins occur [30,31]. In tomato, SlTRM5 knockout in OVATE/SOV1 double mutants induced fruit shapes to change from long pear shapes to round [32].
In previous studies, we performed RAD-Seq genotyping on a natural population of bottle gourd comprising 80 accessions, based on which two sub-gene pools were suggested to be associated with fruit shape. Outlier testing suggested a locus on LG7, which harbors an ortholog of the tomato fruit shape gene OVATE, to be a candidate site for selection [4]. In this study, bottle gourd OVATE gene family members were first identified at the whole-genome level. A phylogenetic tree was constructed to assess the evolutionary relationships of 20 OVATE family genes between bottle gourd and other plants. Then, we analyzed the conserved amino acid sequences and three-dimensional protein structures of 20 OVATE family genes. In addition, the expression patterns of 20 OVATE family genes in the ovaries were also analyzed across various developmental stages. We cloned the OVATE ortholog named LsOVATE1. In situ hybridization, subcellular localization, and RT-qPCR were used to analyze the temporal and spatial expression characteristics of LsOVATE1. RNA in situ hybridization analysis revealed that LsOVATE1 transcripts were detectable in young ovaries, which were largely restricted to the placental area. Finally, ectopic expression of LsOVATE1 in tomato-induced phenotypes which included cone-shaped fruit, calyx hypertrophy, petal degeneration, and petal retention after flowering. Function analyses suggested a potential regulatory mechanism of LsOVATE1 during bottle gourd development.
2. Materials and Methods
2.1. Plant Materials
In this study, plant materials included the ‘Hangzhou gourd’ (hereafter ‘HZ’) and YD-4 (Figure S1). HZ is a local cultivar with slender straight fruit, which originated from Southeast China. YD-4 is a landrace with near-round fruit. For OVATE gene family member expression profiling, roots, stems, leaves, male flowers, female flowers, immature fruits, and ovaries (8 days before anthesis [DBA], 6 DBA, 4 DBA, and 0 DBA) from HZ and YD-4 were used. Ten individuals of each accession were grown in 30 m rows, spaced 0.5 m apart, at the Haining experimental station (30° N, 120° E).
2.2. Identification and Phylogenetic Analysis of the OVATE Gene Family
For identification of the OVATE gene family, the sequences of tomato OVATE proteins were obtained from the Sol Genomics Network (https://www.sgn.cornell.edu/, accessed on 20 March 2020). These sequences were used as queries to search against bottle gourd proteins using the BLASTP program with a threshold e-value of 1 × e−50. Data on OVATE family genes in bottle gourd were downloaded from the Cucurbit Genomics Database (http://cucurbitgenomics.org/, accessed on 20 March 2020) and Gourdbase (http://www.gourdbase.cn/, accessed on 20 March 2020). The genome data of Arabidopsis thaliana were downloaded from TAIR (https://www.arabidopsis.org/, accessed on 20 March 2020). Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 7, based on OVATE protein alignment [33,34]. The phylogenetic tree was constructed in Toolbox for Biologists software (TBtools, Ver. 1.098693) using the maximum likelihood (ML) method with 10,000 bootstrap number.
2.3. Sequence Alignment, Chromosomal Location, and Three-Dimensional Protein Structure Prediction
OVATE gene family members were mapped on bottle gourd chromosomes according to positional information from Gourdbase. BioEdit software [35] was used to compare the homologous proteins between bottle gourd and tomatoes (using amino acid sequence information), and data of the conserved domain were analyzed. The online SWISS-MODEL software (https://swissmodel.expasy.org/, accessed on 25 March 2020), which can predict the three-dimensional structure of proteins upon input of the amino acid sequence of related proteins, was used to predict the three-dimensional structures of homologs in bottle gourd. These tools were used to compare the differences in genes in the OVATE family.
2.4. RNA Extraction and RT-qPCR Analysis
Total RNA was extracted using TRIzol Reagent (Plant RNA Purification Reagent for plant tissue) according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). Reverse transcription was performed using SynScipt™ III cDNA Synthesis Mix (Tsingke, Beijing, China) with 1 µg total RNA. The cDNA samples were stored at −20 °C before being used. Gene expression was examined via RT-qPCR using the SYBR Green method on a StepOneTM real-time PCR System. PCR amplifications were performed on a StepOneTM real-time thermal cycler in a final volume of 20 µL containing 1.0 µL of cDNA, 0.8 µL of each primer (10 µM), 7.0 µL of sterile water, and 10 µL (2×) of TSINGKE Master qPCR Mix (SYBR Green I) (Tsingke). The amplification conditions were as follows: 1 min of denaturation at 95 °C, followed by 40 cycles at 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 30 s, after which a melt curve was produced at 60 °C. Relative gene expressions were estimated according to the 2−∆∆Ct method [36,37]. The bottle gourd TuB-α (tubulin alpha chain-like) gene (HG_GLEAN_10019204) was used as a reference gene. Primer sequences of the reference and target genes are listed in Table S1. Three replicates were performed for each reaction. The relationship between gene expression and phenotype was further obtained by analyzing gene expression using RT-qPCR.
2.5. Transcriptional Profiling
To intuitively compare the expression differences of 20 OVATE family genes in different fruit types and different periods, an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Rosa, CA, USA) was used to assess the quality of the total RNAs and RNA samples with RNA integrity numbers > 7 were selected for library preparation. Multiplexed libraries for next-generation sequencing were prepared using the TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Paired-end sequencing was performed using an Illumina HiSeq 4000 platform. The raw paired-end reads were trimmed and quality controlled using SeqPrep (https://github.com/jstjohn/SeqPrep, accessed on 1 July 2020) and Sickle (https://github.com/najoshi/sickle, accessed on 1 July 2020) with the default parameters. TopHat (v2.1.0) was used to align the high-quality reads to the reference genome. The count of the mapped reads from each sample was derived and normalized to reads per kilobase of exon per million mapped reads (RPKM) for each predicted transcript using Cufflinks (v2.2.1). [38,39].
2.6. In Situ Hybridization
Young ovaries at 8, 4, and 0 DBA from HZ and YD-4 were fixed in 3.7% formol-acetic-alcohol and stored at 4 °C until use. In situ hybridization was performed as previously described [40]. The LsOVATE1 probe was designed according to the specific gene fragments. Sense and antisense probes were generated via PCR amplification with specific primers using SP6 and T7 polymerase, respectively. The primers for probe synthesis are listed in Table S1.
2.7. Subcellular Localization
The LsOVATE1 probe was inserted into the N-terminus of green fluorescent protein (GFP) in a pCAMBIA1305-GFP vector, generating the fusion construct p35S:LsOVATE1-GFP. The fusion expression and empty vectors were transformed into DH5α (Tsingke) competent cells, and after PCR amplification and verification by sequencing, these plasmids were transformed into competent Agrobacterium tumefaciens GV3101 (Tsingke) cells using the freezing method. The A. tumefaciens cells containing the recombinant plasmid and blank control were injected into the abaxial side of tobacco leaf explants using the vacuum infiltration method. The fusion expression vector was transiently expressed in N. benthamiana leaves. Fluorescent signals were detected using a laser confocal scanning microscope (ZEISS Microsystems LSM 700) [41].
2.8. Construction of Overexpression Plasmid and Phenotypic Evaluation of Transgenic Plants
After the study of gene expression characteristics, the LsOVATE1 function was further verified through transgenic analysis. The full-length CDS of LsOVATE1 was PCR-amplified using a cDNA sample prepared from an ‘Hangzhou gourd’ ovary with gene-specific primers (Table S1). The PCR reaction was carried out using kodakarensis (KOD) and DNA polymerase (ToYoBo, Shanghai, China) in a total volume of 50 µL. The procedure was as follows: an initial denaturing step at 95 °C for 5 min, 34 cycles of 95 °C for 30 s, 58 °C for 30 s, and 68 °C for 90 s, and a final extension step at 68 °C for 7 min. The LsOVATE1 CDS was inserted into the BamHI and SpeI sites of the pCAMBIA1305-GFP vector. The primer sequences used to obtain the target gene and overexpression vector are listed in Table S1. The constructs were then transformed into tomato using A. tumefaciens GV3101. Tomato transformation was performed as previously described [42]. Transgenic tomatoes were verified with PCR using the vector- and gene-specific primers listed in Table S1. Wild tomatoes were used as control plants.
3. Results
3.1. Identification of OVATE Family Genes in Bottle Gourd
To identify OVATE family genes in the bottle gourd genome, BLAST searches were performed using tomato OVATE proteins as query sequences. The sequences of candidate proteins identified from the BLAST searches were compared. Sequences encoding very short polypeptides or those that did not contain the conserved motifs (characteristic of OVATE family proteins) were excluded after phylogenetic and conserved domain analysis. After multiple screening and validation steps of the conserved domains, we finally identified 20 putative OVATE genes (Table 1). To determine the chromosomal distribution of the identified bottle gourd genes, their physical locations on the bottle gourd chromosomes were investigated. As result, the 20 genes were mapped on 11 chromosomes. Chromosomes 1, 2, 6, 7, 9, 10, and 11 contained one gene each; chromosomes 8 contained two genes each; and chromosome 4 and 5 contained three genes. Five OVATE homologs were located on chromosome 3, which is the chromosome with the maximum number of homologous genes.

Table 1.
Twenty OVATE family genes in bottle gourd.
3.2. Phylogenetic Relationship of the OVATE Family Genes in Bottle Gourd
To evaluate the evolutionary relationships among OVATE proteins, a phylogenetic tree was constructed using the protein sequences from the 20 putative bottle gourd OVATE gene families, 19 melon OVATE gene families, 20 Arabidopsis OVATE gene families, and 27 tomato OVATE gene families, including OVATE (Figure 1). Specific information on the other OVATE gene families is provided in Table S2. The evolutionary tree classified the OVATE genes into four clades with well-supported bootstrap values. The results show that bottle gourd OVATE genes were unevenly distributed in three clades, with the gene related to bottle gourd shape (chromosome 7) having high homology with the tomato OVATE gene. The corresponding GO analysis is shown in Table S3.
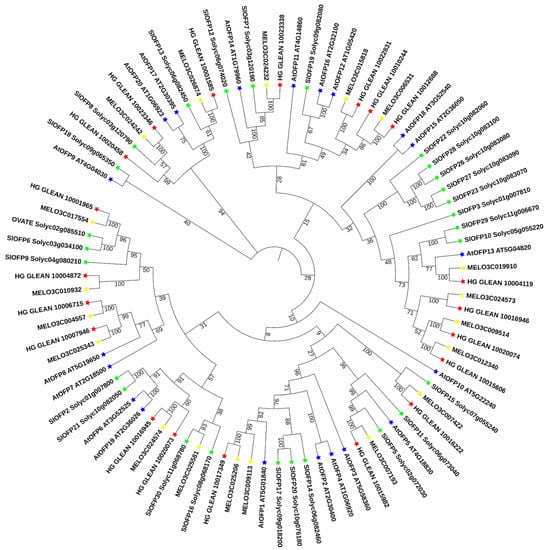
Figure 1.
Phylogenetic relationships of the OVATE family in plants. The phylogenetic tree was constructed using full-length amino acid sequences of the 86 OVATE gene families and MEGA 7.0. Green, yellow, red, and blue stars represent tomato, melon, bottle gourd and Arabidopsis genes, respectively.
3.3. Structural Characterization of Bottle Gourd OVATE Family Genes
The structures of the proteins encoded by the 20 OVATE family genes in bottle gourd were predicted, and the amino acid sequences of the OVATE proteins were aligned with the tomato OVATE protein (Figure 2). An analysis of the conserved domains of the bottle gourd OVATE family genes revealed that corresponding identical amino acids were found at multiple sites. This suggests that the 20 identified genes have similar structures and functions, and are also similar to the tomato OVATE gene. This result indicates that the OVATE gene family is highly conserved among these species. The three-dimensional protein structure predictions showed that bottle gourd OVATE family proteins were mainly composed of α-helices and random coils (Figure 3). In terms of spatial configuration, HG_GLEAN_10017349, HG_GLEAN_10016946, HG_GLEAN_10015982, HG_GLEAN_10004872, HG_GLEAN_10007946, and HG_GLEAN_10001965 showed high similarity. Among the 20 genes, only HG_GLEAN_10023346 contains introns, likely because the three-dimensional structure of its encoded protein has the highest complexity.
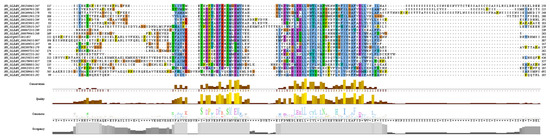
Figure 2.
Sequence alignment of OVATE homologous proteins between bottle gourd and tomato.
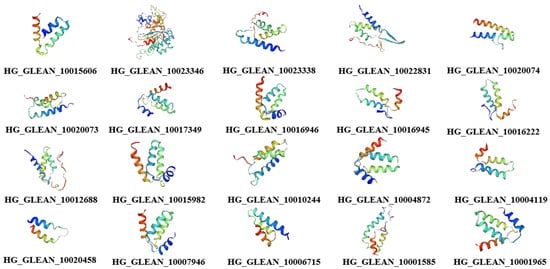
Figure 3.
Predicted three-dimensional structures of bottle gourd OVATE family proteins.
3.4. Expression Profiles of the OVATE Gene Family in the Bottle Gourd
RNA-seq data were used to observe the expression patterns at five stages of ovarian development in both HZ and YD-4 bottle gourd (8, 6, 4, 2, and 0 DBA), in order to assess the possible roles of the 20 OVATE family genes in the growth and development of bottle gourd (Table S4). A heat map was constructed according to the Log10 values (tpm + 0.0001) of the genes (Figure 4). The levels of expression varied widely between different bottle gourd OVATE family genes, and between different stages in individual bottle gourd. Some OVATE family genes in bottle gourd demonstrated opposing expression patterns; for example, HG_GLEAN_10020458 is highly expressed in YD-4 at 0 DBA, but HG_GLEAN_10022831 and HG_GLEAN_10010244 are the opposite. HG_GLEAN_10016946 is highly expressed in YD-4 at 2 DBA, but HG_GLEAN_10001965 shows the opposite expression trend. These varying expression levels suggest that these genes may play different significant roles during different developmental stages.
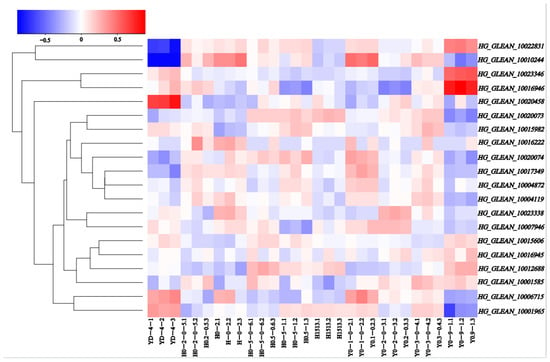
Figure 4.
OVATE gene family members expression profiles in the ovaries of bottle gourd. Blue or red indicates lower or higher levels of expression, respectively, of each transcript in each sample.
3.5. Expression Characteristics of LsOVATE1 Gene
To provide new insights into the expression characteristics of the LsOVATE1 gene, we first cloned LsOVATE1 (720 bp) (Figure S2). Next, we performed RT-qPCR and in situ hybridization to analyze LsOVATE1 expression. Specific RNA probes were used to perform in situ hybridization on HZ and YD-4 samples at 8, 4, and 0 DBA (Figure 5). The results showed that LsOVATE1 is expressed in tissues around the embryonic seat of young ovaries at the 8, 4, and 0 DBA growth stages. LsOVATE1 expression at different stages of growth was consistent between HZ and YD-4 specimens. When hybridization was performed with the sense probe (Figure 5C), no signal was detected. In addition, RT-qPCR analysis showed that LsOVATE1 was most highly expressed in leaves, followed by roots and young fruits, while male flowers had the lowest expression levels (Figure 6). The gene was expressed differently at different growth stages, but no significant expression trend was evident. To analyze the subcellular localization of LsOVATE1, p35S:CmOFP13-GFP constructs were transfected into N. benthamiana epidermal cells for transient expression. We found that the LsOVATE1-GFP fusion protein was localized both in the cytoplasm and nucleus (Figure 7).
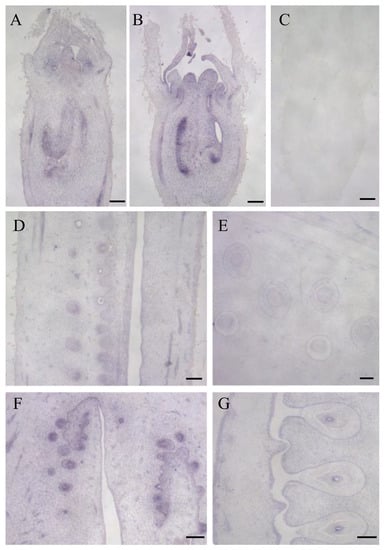
Figure 5.
In situ hybridization of LsOVATE1. (A,B) show longitudinal sections of young ovaries at 8 days before anthesis in HZ and YD-4, respectively; (D,E) show longitudinal sections of young ovaries at 4 days before anthesis in HZ and YD-4, respectively. (F,G) show longitudinal sections of ovaries on the day of anthesis in HZ and YD-4, respectively. (C) indicates that negative controls successfully hybridized with the sense probe. Scale bar = 200 µM.

Figure 6.
LsOVATE1 expression levels in different organs and growth stages, including expression of LsOVATE1 in roots, stems, leaves, male flowers, female flowers, young fruits, and ovaries at 8 days before anthesis (8 DBA) and 0 days before anthesis (0 DBA) from HZ plants. Values are means ± SD of three replicates.

Figure 7.
Subcellular localization of LsOVATE1 in N. benthamiana epidermal cells. Empty GFP vectors were used as controls, and showed green fluorescence. The tobacco nuclei showed red fluorescence. C and F are digitally merged images of both bright-field and fluorescent signals. (A–C) show the localization of empty GFP control vector; (D–F) show the localization of LsOVATE1. Scale bar = 50 µM.
3.6. Functional Validation of LsOVATE1
To further validate the function of LsOVATE1, we used the CaMV35S promoter to overexpress the coding sequence. Since transformation technology is commonly difficult to use in bottle gourd, we could not confirm the function of LsOVATE1 by generating transgenic bottle gourd lines. Instead, we transformed the LsOVATE1 overexpression vector (p35S:LsOVATE1) into wild-type tomato. The transgenic tomato plants were successfully detected using PCR, and the transcript levels of representative transgenic lines were confirmed with RT-qPCR. Compared with wild-type tomato, the fruit of the transgenic plants showed significant changes and a cone-shaped morphology. In addition to the fruit phenotype, phenotypic changes including calyx hypertrophy, petal degeneration, and petal retention after anthesis were observed (Figure 8). Our results indicate that LsOVATE1 not only affected the fruit shape, but also played a role in petal development. The relative expression levels of LsOVATE1 in the four transgenic plants with respect to the wild-type control were determined. It was found that LsOVATE1 expression in transgenic plants correlated positively with the phenotype (Figure 9).

Figure 8.
Phenotypes of transgenic plants overexpressing the LsOVATE1 gene compared to wild-type (WT) tomato. (A) shows the flowers of the representative transgenic line (right) and that of the WT (left); (B) shows the young fruits of the representative transgenic line (right) and that of the WT (left).

Figure 9.
LsOVATE1 expression levels in overexpressing and wild-type plants. Relative expression of LsOVATE1 in wild-type (WT) tomato and four overexpressing tomatoes (OE➀, OE➁, OE➂, OE➃). Values are means ± SD of three replicates.
4. Discussion
The OVATE protein family is unique to plants and includes specific transcription factors that regulate plant growth and development [43]. OVATE was first found to be a major QTL in controlling the development of pear-shaped tomato fruit [44]. OVATE was first cloned in tomato, and the OVATE domain was found to be conserved across tomato, Arabidopsis, and rice [14,22]. It was further confirmed that OVATE overexpression can induce the transition from pear-to round-shaped tomato [45].
To date, many OVATE family genes, in addition to OVATE, have been described. Their functions are related to the regulation of fruit shape; however, different phenotypes arise based on which genes are active. When studying the expression characteristics of tomato OVATE, subcellular localization revealed that the gene was localized in the nucleus [46]. In addition, SlOFP20 was shown to be localized in the nucleus and cytoplasm. Phenotypic observation of SlOFP20-overexpressing transgenic tomato plants showed that the tomato fruit shape reverted from pear- to round-shaped [26]. Many OVATE family genes have also been found in the model crop Arabidopsis. Nine AtOFPs have been found to regulate the development of Arabidopsis meristems and leaves. AtOFP1 transgenic Arabidopsis plants were found to develop slowly, with shorter leaves and anthers, thicker filaments, and fewer seeds. Subcellular localization analysis showed that AtOFP1 was localized in the nucleus [21]. A total of 31 transcription factors of the OVATE family have been detected in rice [24]. The internodes of rice stems are shortened and the leaves become shorter and wider when OsOFP22 is overexpressed. OsOFP1 and OsOFP2 overexpressed transgenic lines were dwarf in size, with thicker leaves and a lower seed setting rate [23]. In melon crops, CmFSI8/CmOFP13 (a fruit shape-related gene), has been characterized as a homolog to SlOFP20 and AtOFP1 [6]. The overexpression of this gene in Arabidopsis led to significantly smaller and curly kidney-shaped leaves, as well as significantly shorter plants. Similar to SlOFP20, CmFSI8/CmOFP13 is localized both in the cytoplasm and nucleus, but its expression is relatively weaker. The protein encoded by OVATE in grapes is also localized in the nucleus. The differential expression of VvOVATE in different varieties may be an important factor in fruit shape [47]. In round fruit and long fruit pepper, CaOVATE was found to negatively regulate the expression of CaGA20ox1, resulting in alterations in the gibberellin synthesis pathway, which led to changes in fruit shape from round to oval [25].
In this study, 20 OVATE homologs were successfully identified in bottle gourd. These OVATE homologs were highly conserved with tomato OVATE, which is consistent with previous findings regarding OVATE family genes in tomato, rice, and other crops. LsOVATE1, a homolog with the highest homology to tomato OVATE, was successfully cloned. Subcellular localization and in situ hybridization studies indicated that LsOVATE1 might be involved in the regulation of nuclear gene transcription. In addition, subcellular localization results were also consistent with those of OVATE family genes in other crops [48], and the expressed LsOVATE1 was localized in the cytoplasm and nucleus. The function of LsOVATE1 was verified using a transgenic approach, which also proved that LsOVATE1 could affect fruit shape. LsOVATE1-overexpressing transgenic seedlings showed obvious changes in fruit shape and flowering period. The transgenic plants exhibited phenotypic characteristics such as calyx hypertrophy, petal degeneration, and petal retainment after anthesis. During further ripening of young fruits, the transgenic fruit dehisced in the pericarp and flowered further at dehiscence. No mature seeds were found in ripe fruits upon opening. This transgenic phenotype is similar to that of Arabidopsis thaliana overexpressing AtOFP1 [43], which affects petals and pollen, as well as the seed-setting rate of mature seeds. Therefore, it is speculated that this gene affects the pollen activity and reproductive mode of plants. Recently, VvMADS39 was discovered in grapes [49], and is homologous to the SEP2 gene of Arabidopsis. The heterologous overexpression of VvMADS39 reduced the fruit and seed size, as well as the seed number in tomato. Research on VvMADS39 also found that MADS39 is required for the normal development of the inner three whorls of floral organs as well as the maintenance of floral meristem identity. Further research found that SlMADS39, which is homologous to VvMADS39, is expressed in tomatoes, and used CRISPER technology to knock out SlMADS39. It was observed that the SlMADS39-knockout phenotype was similar to that of LsOVATE1 overexpression, which was characterized in this study. Whether there is an interaction between the LsOVATE1 protein and MAD proteins needs further verification.
Homologs of the tomato OVATE, SUN, FASCIATED, and LC family genes have been suggested as candidate genes regulating fruit shape [9,50,51]. Apart from the above-mentioned “classical” genes, recently an ethylene biosynthesis gene (ACS2) [52] and FRUITFULL-like MADS-box gene (FUL1) [53] in cucumber, an AP2/ERF transcription factor gene (AP2a) gene and TONNEAU1 Recruiting Motif protein (TRM5) in tomato [26,54], as well as an LRR-RLK family gene (CAD1) in peach [55], have also been associated with fruit shape. The CsFUL1 gene directly depresses the expression of the auxin transporters CsPIN1 and CsPIN7, resulting in decreases in auxin accumulation in fruits [53]. In the bottle gourd, despite the number of fsQTLs mapped [56], no fruit shape gene has been identified thus far. Our results also suggest certain OVATE family members as candidate fruit shape genes in the bottle gourd.
5. Conclusions
We identified a total of 20 OVATE family genes in bottle gourd and divided them into four clades to help understand the evolutionary relationships between these genes, which all had at least one conserved domain. Our chromosomal distribution, sequence alignment, three-dimensional protein structure prediction, and exon/intron analyses of the OVATE gene family provide a useful basis for understanding the function of the OVATE gene family. The expression pattern analysis demonstrated that OVATE family genes in bottle gourd were differentially expressed, indicating that they played different roles in the growth and development of bottle gourd. We studied the candidate gene LsOVATE1, and found that it is homologous to OVATE and affects fruit shape. We also analyzed the expression characteristics of LsOVATE1 based on in situ hybridization, expression level, and subcellular localization analyses. In situ hybridization further revealed that LsOVATE1 transcripts were detectable in young ovaries, which were largely restricted to the placental area. Additionally, LsOVATE1 overexpression in tomatoes caused fruit shape changes, as well as secondary flower and non-seed phenotypes, highlighting LsOVATE1’s role in the regulation of plant development (Figure S3). Further studies on fruit shape-regulating genes should provide new gene information that would allow the generation of new bottle gourd varieties with desirable phenotypes. By comparing with other genes that have similar effects on fruit shape, it is speculated that there may be some regulation mechanism, which ultimately led to the effect of LsOVATE1 on the fruit shape of bottle gourd. This will guide further study on the regulation mechanism of fruit shape in bottle gourd.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biom13010085/s1, Figure S1: Fruit shape pictures of bottle gourd HZ and YD-4; Figure S2: Cloning of LsOVATE1 and the pCAMBIA1305.1-GFP vector; Figure S3: Framework figure; Table S1: Primer list summary; Table S2: Composition and chromosomal location of the plants; Table S3: GO analysis of OVATE proteins; Table S4: List of RNA-Seq data.
Author Contributions
Data curation, Formal analysis, Writing—original draft, Writing—review & editing, Z.F.; Project supervision, Conceptualization, X.W. (Xiaohua Wu); Investigation, Resources, J.W. and X.W. (Xiaohua Wu); Investigation, Methodology, X.W. (Xinyi Wu) and B.W.; Supervision, Project administration, Z.L.; Project administration, Supervision, Funding acquisition, Z.Y. and G.L.; Project supervision, Conceptualization, Funding acquisition, Writing—original draft, Writing—review & editing, Y.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31801865), the Key Research and Development Project of Zhejiang Province (2021C02052), and the Key Science Project of Vegetable Breeding in Zhejiang (2021C02065).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Beevy, S.S.; Kuriachan, P. Chromosome numbers of south Indian Cucurbitaceae and a note on the cytological evolution in the family. J. Cytol. Genet. 1996, 31, 65–71. [Google Scholar]
- Erickson, D.L.; Smith, B.D.; Clarke, A.C.; Sandweiss, D.H.; Tuross, N. An Asian origin for a 10,000-year-old domesticated plant in the Americas. Proc. Natl. Acad. Sci. USA 2005, 102, 18315–18320. [Google Scholar] [CrossRef] [PubMed]
- Heiser, C.B. The Gourd Book: A Thorough and Fascinating Account of Gourds from throughout the World; University of Oklahoma Press: Norman, OK, USA, 1979. [Google Scholar]
- Xu, P.; Xu, S.; Wu, X.; Tao, Y.; Wang, B.; Wang, S.; Qin, D.; Lu, Z.; Li, G. Population genomic analyses from low-coverage RAD-Seq data: A case study on the non-model cucurbit bottle gourd. Plant J. 2014, 77, 430–442. [Google Scholar] [CrossRef]
- Chen, S.; Wang, X.; Tan, G.; Zhou, W.; Wang, G. Gibberellin and the plant growth retardant paclobutrazol altered fruit shape and ripening in tomato. Protoplasma 2020, 257, 853–861. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, C.; Zong, M.; Qiu, Y.; Liu, Y.; Huang, Y.; Xie, Y.; Zhang, H.; Wang, J. CmFSI8/CmOFP13 gene encoding an OFP family protein controls fruit shape in melon (Cucumis melo L.). J. Exp. Bot. 2021, 5, 1370–1384. [Google Scholar] [CrossRef]
- Pan, Y.; Liang, X.; Gao, M.; Liu, H.; Meng, H.; Weng, Y.; Cheng, Z. Round fruit shape in WI7239 cucumber is controlled by two interacting quantitative trait loci with one putatively encoding a tomato SUN homolog. Theor. Appl. Genet. 2017, 130, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.; Ruggieri, V.; Pérez, S.; Alexiou, K.G.; Fernández, M.; Jahrmann, T.; Pujol, M.; Garcia-Mas, J. QTL mapping of melon fruit quality traits using a high-density GBS-based genetic map. BMC Plant Biol. 2018, 18, 324. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Wang, Y.; McGregor, C.; Liu, S.; Luan, F.; Gao, M.; Weng, Y. Genetic architecture of fruit size and shape variation in cucurbits: A comparative perspective. Theor. Appl. Genet. 2020, 133, 1–21. [Google Scholar] [CrossRef]
- Ercisli, S.; Esitken, A.; Turkkal, C.; Orhan, E. The allelopathic effects of juglone and walnut leaf extracts on yield, growth, chemical and PNE composition of strawberry cv. Fern. Plant Soil Environ. 2005, 51, 283–387. [Google Scholar] [CrossRef]
- Bolaric, S.; Müller, I.D.; Vokurka, A.; Cepo, D.V.; Ruscic, M.; Srecec, S.; Kremer, D. Morphological and molecular characterization of Croatian carob tree (Ceratonia siliqua L.) germplasm. Turk. J. Agric. For. 2021, 45, 807–818. [Google Scholar] [CrossRef]
- Milosevic, T.; Milosevic, N.; Glisic, I. Early tree performances, precocity and fruit quality attributes of newly introduced apricot cultivars grown under western Serbian conditions. Turk. J. Agric. For. 2021, 45, 819–833. [Google Scholar] [CrossRef]
- Han, J.L. Functional Identification of Arabidopsis Transcription Factor AtOFP19. Master’s Thesis, Northeast Agricultural University, Harbin, China, 2021. [Google Scholar] [CrossRef]
- Liu, J.; Van Eck, J.; Cong, B.; Tanksley, S.D. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. USA 2002, 99, 13302. [Google Scholar] [CrossRef]
- Xiao, H.; Jiang, N.; Schaffner, E.; Stockinger, E.J.; Knaap, E.V.D. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 2018, 319, 1527–1530. [Google Scholar] [CrossRef] [PubMed]
- Cong, B.; Barrero, L.S.; Tanksley, S.D. Regulatory change in yabby-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 2008, 40, 800–804. [Google Scholar] [CrossRef] [PubMed]
- Munos, S.; Ranc, N.; Botton, E.; Berard, A.; Rolland, S.; Duffe, P.; Carretero, Y.; Le Paslier, M.; Delalande, C.; Bouzayen, M.; et al. Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near wuschel. Plant Physiol. 2011, 156, 2244–2254. [Google Scholar] [CrossRef]
- Wang, L.; Cao, C.; Zheng, S.; Zhang, H.; Liu, P.; Ge, Q.; Li, J.; Ren, Z. Transcriptomic analysis of short-fruit 1 (SF1) reveals new insights into the variation of fruit-related traits in cucumis sativus. Sci. Rep. 2017, 7, 2950. [Google Scholar] [CrossRef]
- Li, S.; Pan, Y.; Wen, C.; Li, Y.; Liu, X.; Zhang, X.; Behera, T.; Xing, G.; Weng, Y. Integrated analysis in bi-parental and natural populations reveals Csclavata3 (CsCLV3) underlying carpel number variations in cucumber. Theor. Appl. Genet. 2016, 129, 1007–1022. [Google Scholar] [CrossRef] [PubMed]
- Che, G.; Gu, R.; Zhao, J.; Liu, X.; Song, X.; Zi, H.; Cheng, Z.; Shen, J.; Wang, Z.; Liu, R.; et al. Gene regulatory network of carpel number variation in cucumber. Development 2020, 147, dev.184788. [Google Scholar] [CrossRef] [PubMed]
- Hackbusch, J.; Richter, K.; Müller, J.; Salamini, F.; Uhrig, J.F. A central role of Arabidopsis thaliana OVATE family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 4908–4912. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chang, Y.; Guo, J.; Chen, J. Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 2007, 50, 858–872. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, A.J.; Walia, H.; Begcy, K.; Sarath, G. Rice OVATE family protein 2 (OFP2) alters hormonal homeostasis and vasculature development. Plant Sci. 2015, 241, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Hui, Y.; Jiang, W.; Liu, Q.; Zhang, H.; Mingxin, P.; Chen, Z.; Bian, M. Expression pattern and subcellular localization of the OVATE protein family in rice. PLoS ONE 2017, 10, e0118966. [Google Scholar] [CrossRef]
- Tsaballa, A.; Pasentsis, K.; Darzentas, N.; Tsaftaris, A.S. Multiple evidence for the role of an OVATE-like gene in determining fruit shape in pepper. BMC Plant Biol. 2011, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, B.; Keyhaninejad, N.; Rodríguez, G.; Kim, H.J.; Chakrabarti, M.; Illa-Berenguer, E.; Taitano, N.K.; Gonzalo, M.J.; Díaz, A.; et al. A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nat. Commun. 2018, 9, 4734. [Google Scholar] [CrossRef]
- Guan, J.; Xu, Y.; Yu, Y.; Fu, J.; Ren, F.; Guo, J.; Zhao, J.; Jiang, Q.; Wei, J.; Xie, H. Genome structure variation analyses of peach reveal population dynamics and a 1.67 Mb causal inversion for fruit shape. Genome Biology. 2021, 22, 13. [Google Scholar] [CrossRef]
- Spinner, L.; Gadeyne, A.; Belcram, K.; Goussot, M.; Moison, M.; Duroc, Y.; Eeckhout, D.; De Winne, N.; Schaefer, E.; Van De Slijke, E.; et al. A protein phosphatase 2A complex spatially controls plant cell division. Nat. Commun. 2013, 4, 1863. [Google Scholar] [CrossRef]
- Schaefer, E.; Belcram, K.; Uyttewaal, M.; Duroc, Y.; Goussot, M.; Legland, D.; Laruelle, E.; de Tauzia-Moreau, M.-L.; Pastuglia, M.; Bouchez, D. The preprophase band of microtubules controls the robustness of division orientation in plants. Science 2017, 356, 186–189. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, G.; Kim, I.J.; Park, J.; Kwak, S.; Choi, G.; Chung, W. LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development 2006, 133, 4305–4314. [Google Scholar] [CrossRef]
- Drevensek, S.; Goussot, M.; Duroc, Y.; Christodoulidou, A.; Steyaert, S.; Schaefer, E.; Duvernois, E.; Grandjean, O.; Vantard, M.; Bouchez, D.; et al. The Arabidopsis TRM1-TON1 interaction reveals a recruitment network common to plant cortical microtubule arrays and eukaryotic centrosomes. Plant Cell 2012, 24, 178–191. [Google Scholar] [CrossRef]
- Lazzaro, M.D.; Wu, S.; Snouffer, A.; Wang, Y.; van der Knaap, E. Plant organ shapes are regulated by protein interactions and associations with microtubules. Front. Plant Sci. 2018, 9, 1766. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Ahmed, M.A. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Wang, G.; Lovato, A.; Polverari, A.; Wang, M.; Liang, Y.H.; Ma, Y.C.; Cheng, Z.M. Genome-wide identification and analysis of mitogen activated protein kinase kinase kinase gene family in grapevine (Vitis vinifera). BMC Plant Biol. 2014, 14, 219. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−∆∆Ct) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- De Hoon, M.J.; Imoto, S.; Nolan, J.; Miyano, S. Open source clustering software. Bioinformatics 2004, 20, 1453–1454. [Google Scholar] [CrossRef]
- Saldanha, A.J. Java Treeview–extensible visualization of microarray data. Bioinformatics 2004, 20, 3246–3248. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Ding, L.; Zhang, X. RNA in situ hybridization technology for vegetable crops. J. Shanxi Agric. Univ. (Nat. Sci. Ed.) 2013, 33, 42–45. [Google Scholar] [CrossRef]
- Haseloff, J.; Siemering, K.R.; Prasher, D.C.; Hodge, S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 1997, 94, 2122–2127. [Google Scholar] [CrossRef]
- Sun, S.; Kang, X.; Xing, X.; Xu, X.; Cheng, J.; Zheng, S.; Xing, G. Agrobacterium-mediated transformation of tomato (Lycopersicon esculentum L. cv. Hezuo 908) with improved efficiency. Biotechnol. Biotechnol. Equip. 2015, 29, 861–868. [Google Scholar] [CrossRef]
- Wang, S.; Chang, Y.; Guo, J.; Zeng, Q.; Ellis, B.E.; Chen, J. Arabidopsis OVATE Family Proteins, a Novel Transcriptional Repressor Family, Control Multiple Aspects of Plant Growth and Development. PLoS ONE 2011, 6, e23896. [Google Scholar] [CrossRef] [PubMed]
- Copp, A.J.; Fisher, E.M.C. Abstracts of papers presented at the sixteenth Genetics Society’s Mammalian Genetics and Development Workshop held at the Institute of Child Health, University College London on 21 and 22 November 2005. Genet. Res. 2006, 88, 67–76. [Google Scholar] [CrossRef][Green Version]
- Guo, Y.; Li, N.; Li, S.; Han, Y.; Duan, M.; Ma, F. Identification and Expression Analysis of OFP Gene Family in Setavia italica. J. Shan Xi Agric. Sci. 2020, 48, 467–476. [Google Scholar] [CrossRef]
- Rodríguez, G.R.; Muños, S.; Anderson, C.; Sim, S.-C.; Michel, A.; Causse, M.; Gardener, B.B.M.; Francis, D.; van der Knaap, E. Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 2011, 156, 275–285. [Google Scholar] [CrossRef]
- Song, X. Cloning and functional analysis of fruit shape-related gene in grape. Nanjing Agric. Univ. 2014, 7, 82. [Google Scholar]
- Han, J.; Wang, H.; Zhang, D.; Chang, Y. Preliminary study on the function of Arabidopsis transcription factor AtOFP19. North China J. Agric. 2021, 36, 56–63. [Google Scholar] [CrossRef]
- Zhang, S.; Yao, J.; Wang, L.; Wu, N.; van Nocker, S.; Li, Z.; Gao, M.; Wang, X. Role of grapevine SEPALLATA-related MADS-box gene VvMADS39 in flower and ovule development. Plant J. Cell Mol. Biol. 2022, 111, 1565–1579. [Google Scholar] [CrossRef]
- Kim, K.; Hwang, J.; Han, D.; Park, M.; Kim, S.; Choi, D.; Kim, Y.; Lee, G.; Kim, S.; Park, Y. Major Quantitative Trait Loci and Putative Candidate Genes for Powdery Mildew Resistance and Fruit-Related Traits Revealed by an Intraspecific Genetic Map for Watermelon (Citrullus lanatus var. lanatus). PLoS ONE 2015, 10, e0145665. [Google Scholar] [CrossRef]
- Wei, Q.; Fu, W.X.; Wang, Y.Z.; Qin, X.D.; Wang, J.; Li, J.; Lou, Q.F.; Chen, J.F. Rapid identification of fruit length loci in cucumber (Cucumis sativus L.) using next-generation sequencing (NGS)-based QTL analysis. Sci. Rep. 2016, 6, 27496. [Google Scholar] [CrossRef]
- Tan, J.Y.; Tao, Q.Y.; Niu, H.H.; Zhang, Z.; Li, D.D.; Gong, Z.H.; Weng, Y.Q.; Li, Z. A novel allele of monoecious (m) locus is responsible for elongated fruit shape and perfect flowers in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2015, 128, 2483–2493. [Google Scholar] [CrossRef]
- Zhao, J.; Jiang, L.; Che, G.; Pan, Y.; Li, Y.; Hou, Y.; Zhao, W.; Zhong, Y.; Ding, L.; Yan, S.; et al. A Functional Allele of CsFUL1 Regulates Fruit Length through Repressing CsSUP and Inhibiting Auxin Transport in Cucumber. Plant Cell 2019, 31, 1289–1307. [Google Scholar] [CrossRef] [PubMed]
- Karlova, R.; Rosin, F.M.; Busscher-Lange, J.; Parapunova, V.; Do, P.T.; Fernie, A.R.; Fraser, P.D.; Baxter, C.; Angenent, G.C.; de Maagd, R.A. Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 2011, 23, 923–941. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Zhou, Z.; Wang, Q.; Guo, J.; Zhao, P.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wang, X.; et al. Genome-wide association study of 12 agronomic traits in peach. Nat. Commun. 2016, 7, 13246. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Wang, Y.; Sun, F.; Wu, R.; Du, H.; Wang, Y.; Jiang, L.; Wu, X.; Wu, X.; Yang, L.; et al. Long-read genome assembly and genetic architecture of fruit shape in the bottle gourd. Plant J. Cell Mol. Biol. 2021, 107, 956–968. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).