Methyltransferases of Riboviria
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. The Search for Methyltransferases Encoded by Riboviria, Their Multiple Alignment and Identification of Conserved Sequence Motifs
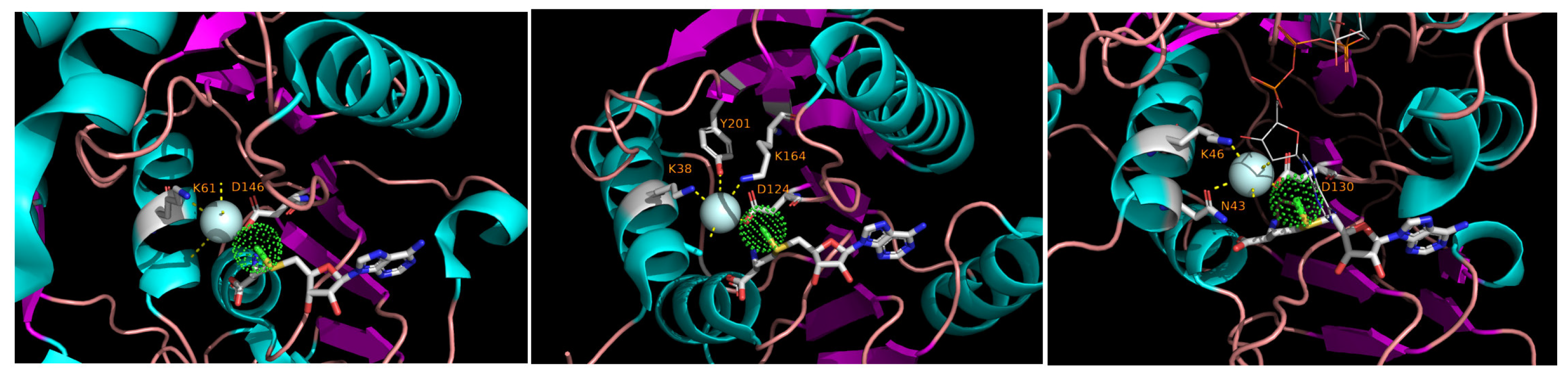
3.2. Functional Roles of the Conserved Motifs and the Mechanism of Methyltransferase Reaction
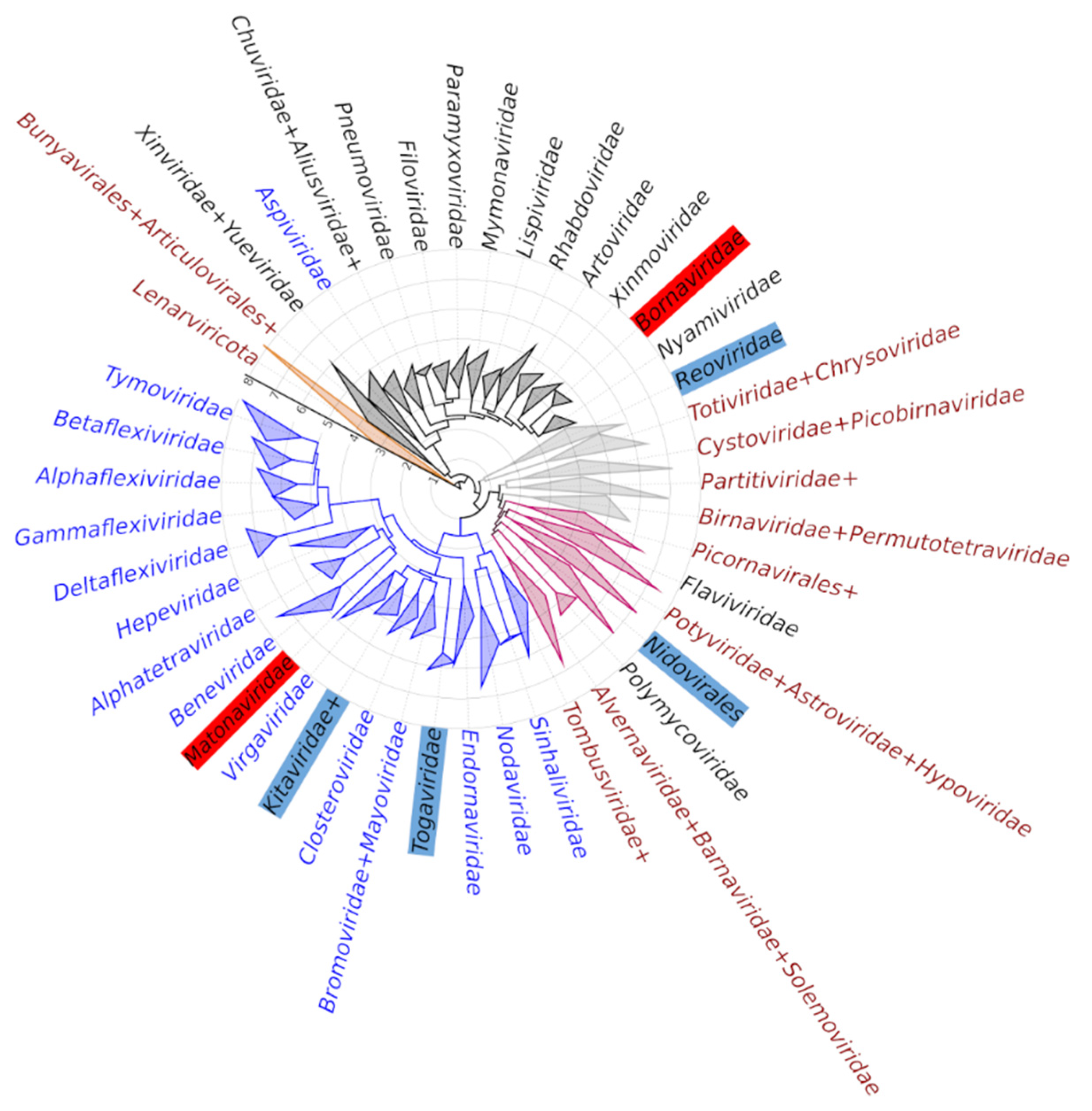
3.3. Gain and Loss of Methyltransferases in the Evolution of Riboviria
4. Discussion
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iwanowski, D. Über Die Mosaikkrankheit Der Tabakspflanze; Bulletin Scientifique Publié Par l’Académie Impériale des Sciences de Saint-Pétersbourg/Nouvelle Serie III; Glagoslav Publications: Oosterhout, The Netherlands, 1892; pp. 65–70. [Google Scholar]
- Keith, J.; Fraenkel-Conrat, H. Tobacco mosaic virus RNA carries 5′-terminal triphosphorylated guanosine blocked by 5′-linked 7-methylguanosine. FEBS Lett. 1975, 57, 31–33. [Google Scholar] [CrossRef]
- Zimmern, D. The 5′ end group of tobacco mosaic virus RNA is m7G5′ppp5′Gp. Nucleic Acids Res. 1975, 2, 1189–1202. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Byszewska, M.; Śmietański, M.; Purta, E.; Bujnicki, J.M. RNA methyltransferases involved in 5′ cap biosynthesis. RNA Biol. 2014, 11, 1597–1607. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.Z.; Dai, R.M.; Shen, X.R.; Sun, Y.K. The Location and Function of the 5′-Cap Structure of the RNA of Tobacco Mosaic Virus in the Virion. In Proceedings of the Proceedings of the Sixth International Congress of Virology, Sendai, Japan, 1–7 September 1984; p. 231. [Google Scholar]
- Wilson, T.M.A. Nucleocapsid Disassembly and Early Gene Expression by Positive-strand RNA Viruses. J. Gen. Virol. 1985, 66, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Plaskitt, K.; Wilson, T. Evidence that tobacco mosaic virus particles disassemble contranslationally in vivo. Virology 1986, 148, 326–336. [Google Scholar] [CrossRef]
- Wilson, T.M.A. Structural Interactions between Plant RNA Viruses and Cells. Oxf. Surv. Plant Mol. Cell Biol. 1988, 5, 89–144. [Google Scholar]
- Rozanov, M.N.; Koonin, E.V.; Gorbalenya, A. Conservation of the putative methyltransferase domain: A hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J. Gen. Virol. 1992, 73, 2129–2134. [Google Scholar] [CrossRef]
- Merits, A.; Kettunen, R.; Mäkinen, K.; Lampio, A.; Auvinen, P.; Kääriäinen, L.; Ahola, T. Virus-specific capping of tobacco mosaic virus RNA: Methylation of GTP prior to formation of covalent complex p126-m7GMP. FEBS Lett. 1999, 455, 45–48. [Google Scholar] [CrossRef]
- Martin, S.; Moss, B. Modification of RNA by mRNA guanylyltransferase and mRNA (guanine-7-)methyltransferase from vaccinia virions. J. Biol. Chem. 1975, 250, 9330–9335. [Google Scholar] [CrossRef]
- Wei, C.-M.; Gershowitz, A.; Moss, B. N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 1975, 257, 251–253. [Google Scholar] [CrossRef]
- Wei, C.M.; Moss, B. Methylated nucleotides block 5’-terminus of vaccinia virus messenger RNA. Proc. Natl. Acad. Sci. USA 1975, 72, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Krug, R.M.; Morgan, M.A.; Shatkin, A.J. Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. J. Virol. 1976, 20, 45–53. [Google Scholar] [CrossRef]
- Banerjee, A.K. 5’-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol. Rev. 1980, 44, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Fraser, R. TMV-RNA is not methylated and does not contain a polyadenylic acid sequence. Virology 1973, 56, 379–382. [Google Scholar] [CrossRef]
- Dubin, D.T.; Stollar, V. Methylation of sindbis virus “26S” messenger RNA. Biochem. Biophys. Res. Commun. 1975, 66, 1373–1379. [Google Scholar] [CrossRef]
- Hefti, E.; Bishop, D.H.L.; Dubin, D.T.; Stollar, V. 5′ Nucleotide Sequence of Sindbis Viral RNA. J. Virol. 1976, 17, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Durbin, R.; Huang, H.V.; Rice, C.M.; Stollar, V. Association of the sindbis virus RNA methyltransferase activity with the nonstructural protein nsP1. Virology 1989, 170, 385–391. [Google Scholar] [CrossRef]
- Wang, H.-L.; O’Rear, J.; Stollar, V. Mutagenesis of the Sindbis Virus nsP1 Protein: Effects on Methyltransferase Activity and Viral Infectivity. Virology 1996, 217, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Haseloff, J.; Goelet, P.; Zimmern, D.; Ahlquist, P.; Dasgupta, R.; Kaesberg, P. Striking similarities in amino acid sequence among nonstructural proteins encoded by RNA viruses that have dissimilar genomic organization. Proc. Natl. Acad. Sci. USA 1984, 81, 4358–4362. [Google Scholar] [CrossRef]
- Koonin, E.V. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and 2 protein of reovirus. J. Gen. Virol. 1993, 74, 733–740. [Google Scholar] [CrossRef]
- Bujnicki, J.M.; Rychlewski, L. Reassignment of specificities of two cap methyltransferase domains in the reovirus lambda2 protein. Genome Biol. 2001, 2, research0038.1. [Google Scholar] [CrossRef]
- Anantharaman, V.; Koonin, E.V.; Aravind, L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002, 30, 1427–1464. [Google Scholar] [CrossRef] [PubMed]
- Bujnicki, J.M.; Rychlewski, L. In silico identification, structure prediction and phylogenetic analysis of the 2′-O-ribose (cap 1) methyltransferase domain in the large structural protein of ssRNA negative-strand viruses. Protein Eng. Des. Sel. 2002, 15, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Egloff, M.; Benarroch, D.; Selisko, B.; Romette, J.; Canard, B. An RNA cap (nucleoside-2’-O-)-methyltransferase in the flavivirus RNA polymerase NS5: Crystal structure and functional characterization. EMBO J. 2002, 21, 2757–2768. [Google Scholar] [CrossRef] [PubMed]
- Hager, J.; Staker, B.L.; Bügl, H.; Jakob, U. Active Site in RrmJ, a Heat Shock-induced Methyltransferase. J. Biol. Chem. 2002, 277, 41978–41986. [Google Scholar] [CrossRef] [PubMed]
- Caldas, T.; Binet, E.; Bouloc, P.; Costa, A.; Desgres, J.; Richarme, G. The FtsJ/RrmJ Heat Shock Protein of Escherichia coli Is a 23 S Ribosomal RNA Methyltransferase. J. Biol. Chem. 2000, 275, 16414–16419. [Google Scholar] [CrossRef]
- Ma, Y.; Wei, Y.; Zhang, X.; Zhang, Y.; Cai, H.; Zhu, Y.; Shilo, K.; Oglesbee, M.; Krakowka, S.; Whelan, S.P.J.; et al. mRNA Cap Methylation Influences Pathogenesis of Vesicular Stomatitis Virus In Vivo. J. Virol. 2014, 88, 2913–2926. [Google Scholar] [CrossRef]
- Lin, S.; Chen, H.; Chen, Z.; Yang, F.; Ye, F.; Zheng, Y.; Yang, J.; Lin, X.; Sun, H.; Wang, L.; et al. Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity. Nucleic Acids Res. 2021, 49, 5382–5392. [Google Scholar] [CrossRef]
- Russo, A.T.; White, M.A.; Watowich, S.J. The Crystal Structure of the Venezuelan Equine Encephalitis Alphavirus nsP2 Protease. Structure 2006, 14, 1449–1458. [Google Scholar] [CrossRef]
- Decroly, E.; Ferron, F.; Lescar, J.; Canard, B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2012, 10, 51–65. [Google Scholar] [CrossRef]
- Malone, T.; Blumenthal, R.; Cheng, X. Structure-guided Analysis Reveals Nine Sequence Motifs Conserved among DNA Amino-methyl-transferases, and Suggests a Catalytic Mechanism for these Enzymes. J. Mol. Biol. 1995, 253, 618–632. [Google Scholar] [CrossRef] [PubMed]
- Kozbial, P.Z.; Mushegian, A.R. Natural history of S-adenosylmethionine-binding proteins. BMC Struct. Biol. 2005, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Ferron, F.; Subissi, L.; De Morais, A.T.S.; Le, N.T.T.; Sevajol, M.; Gluais, L.; Decroly, E.; Vonrhein, C.; Bricogne, G.; Canard, B.; et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. USA 2018, 115, E162–E171. [Google Scholar] [CrossRef]
- Ferron, F.; Debat, H.J.; Shannon, A.; Decroly, E.; Canard, B. A N7-guanine RNA cap methyltransferase signature-sequence as a genetic marker of large genome, non-mammalian Tobaniviridae. NAR Genom. Bioinform. 2019, 2, lqz022. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, R.D.; Liao, Y.; Cheng, H.; Grishin, N.V. ECOD: New developments in the evolutionary classification of domains. Nucleic Acids Res. 2016, 45, D296–D302. [Google Scholar] [CrossRef]
- Medvedev, K.E.; Kinch, L.N.; Grishin, N.V. Functional and evolutionary analysis of viral proteins containing a Rossmann-like fold. Protein Sci. 2018, 27, 1450–1463. [Google Scholar] [CrossRef]
- Feder, M.; Pas, J.; Wyrwicz, L.; Bujnicki, J.M. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene 2003, 302, 129–138. [Google Scholar] [CrossRef]
- Pei, J.; Kim, B.-H.; Grishin, N.V. PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008, 36, 2295–2300. [Google Scholar] [CrossRef]
- Schafer, A.A.; Aravind, L.; Madden, T.L.; Shavirin, S.; Spouge, J.L.; Wolf, Y.I.; Koonin, E.V.; Altschul, S.F. Improving the Accuracy of PSI-BLAST Protein Database Searches with Composition-Based Statistics and Other Refinements. Nucleic Acids Res. 2001, 29, 2994–3005. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Holm, L. Dali server: Structural unification of protein families. Nucleic Acids Res. 2022, 50, W210–W215. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, F.; Entfellner, J.-B.D.; Wilkinson, E.; Correia, D.; Felipe, M.D.; De Oliveira, T.; Gascuel, O. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature 2018, 556, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- PyMOL. The PyMOL Molecular Graphics System; Schroedinger LLC: New York, NY, USA, 2006. [Google Scholar]
- Cheng, H.; Liao, Y.; Schaeffer, R.D.; Grishin, N.V. Manual classification strategies in the ECOD database. Proteins: Struct. Funct. Bioinform. 2015, 83, 1238–1251. [Google Scholar] [CrossRef]
- Schaeffer, R.D.; Liao, Y.; Grishin, N.V. Searching ECOD for Homologous Domains by Sequence and Structure. Curr. Protoc. Bioinform. 2018, 61, e45. [Google Scholar] [CrossRef] [PubMed]
- Tkaczuk, K.L.; Dunin-Horkawicz, S.; Purta, E.; Bujnicki, J.M. Structural and evolutionary bioinformatics of the SPOUT superfamily of methyltransferases. BMC Bioinform. 2007, 8, 73. [Google Scholar] [CrossRef]
- Aravind, L.; Iyer, L.M. Provenance of SET-Domain Histone Methyltransferases Through Duplication of a Simple Structural Unit. Cell Cycle 2003, 2, 366–373. [Google Scholar] [CrossRef][Green Version]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Murphy, A.M.; Moerdyk-Schauwecker, M.; Mushegian, A.; Grdzelishvili, V.Z. Sequence–function analysis of the Sendai virus L protein domain VI. Virology 2010, 405, 370–382. [Google Scholar] [CrossRef][Green Version]
- Li, C.; Xia, Y.; Gao, X.; Gershon, P.D. Mechanism of RNA 2‘-O-Methylation: Evidence that the Catalytic Lysine Acts To Steer Rather than Deprotonate the Target Nucleophile. Biochemistry 2004, 43, 5680–5687. [Google Scholar] [CrossRef]
- Yang, W.; Lee, J.Y.; Nowotny, M. Making and Breaking Nucleic Acids: Two-Mg2+-Ion Catalysis and Substrate Specificity. Mol. Cell 2006, 22, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Bruice, T.C. Reaction mechanism of guanidinoacetate methyltransferase, concerted or step-wise. Proc. Natl. Acad. Sci. USA 2006, 103, 16141–16146. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.-M.; Perona, J.J. Stereochemical mechanisms of tRNA methyltransferases. FEBS Lett. 2009, 584, 278–286. [Google Scholar] [CrossRef]
- Liu, L.; Dong, H.; Chen, H.; Zhang, J.; Ling, H.; Li, Z.; Shi, P.-Y.; Li, H. Flavivirus RNA cap methyltransferase: Structure, function, and inhibition. Front. Biol. 2010, 5, 286–303. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Fink, K.; Züst, R.; Lim, S.P.; Qin, C.-F.; Shi, P.-Y. Flavivirus RNA methylation. J. Gen. Virol. 2014, 95, 763–778. [Google Scholar] [CrossRef]
- Sevajol, M.; Subissi, L.; Decroly, E.; Canard, B.; Imbert, I. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014, 194, 90–99. [Google Scholar] [CrossRef]
- Paesen, G.C.; Collet, A.; Sallamand, C.; Debart, F.; Vasseur, J.-J.; Canard, B.; Decroly, E.; Grimes, J.M. X-ray structure and activities of an essential Mononegavirales L-protein domain. Nat. Commun. 2015, 6, 8749. [Google Scholar] [CrossRef]
- Viswanathan, T.; Arya, S.; Chan, S.-H.; Qi, S.; Dai, N.; Misra, A.; Park, J.-G.; Oladunni, F.; Kovalskyy, D.; Hromas, R.A.; et al. Structural basis of RNA cap modification by SARS-CoV-2. Nat. Commun. 2020, 11, 3718. [Google Scholar] [CrossRef]
- Wilamowski, M.; Sherrell, D.A.; Minasov, G.; Kim, Y.; Shuvalova, L.; Lavens, A.; Chard, R.; Maltseva, N.; Jedrzejczak, R.; Rosas-Lemus, M.; et al. 2′-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography. Proc. Natl. Acad. Sci. USA 2021, 118, e2100170118. [Google Scholar] [CrossRef]
- Nencka, R.; Silhan, J.; Klima, M.; Otava, T.; Kocek, H.; Krafcikova, P.; Boura, E. Coronaviral RNA-methyltransferases: Function, structure and inhibition. Nucleic Acids Res. 2022, 50, 635–650. [Google Scholar] [CrossRef]
- Ramdhan, P.; Li, C. Targeting Viral Methyltransferases: An Approach to Antiviral Treatment for ssRNA Viruses. Viruses 2022, 14, 379. [Google Scholar] [CrossRef]
- Pogolotti, A.L.; Ono, A.; Subramaniam, R.; Santi, D.V. On the mechanism of DNA-adenine methylase. J. Biol. Chem. 1988, 263, 7461–7464. [Google Scholar] [CrossRef]
- Kealey, J.T.; Lee, S.; Floss, H.G.; Santi, D.V. Stereochemistry of methly transfer catalyzed by tRNA (m5U54)-methyltransferase—evidence for a single displacement mechanism. Nucleic Acids Res. 1991, 19, 6465–6468. [Google Scholar] [CrossRef]
- Viswanathan, T.; Misra, A.; Chan, S.-H.; Qi, S.; Dai, N.; Arya, S.; Martinez-Sobrido, L.; Gupta, Y.K. A metal ion orients SARS-CoV-2 mRNA to ensure accurate 2′-O methylation of its first nucleotide. Nat. Commun. 2021, 12, 3287. [Google Scholar] [CrossRef]
- Moura, M.; Conde, C. Phosphatases in Mitosis: Roles and Regulation. Biomolecules 2019, 9, 55. [Google Scholar] [CrossRef]
- Hubrich, F.; Müller, M.; Andexer, J.N. Chorismate- and isochorismate converting enzymes: Versatile catalysts acting on an important metabolic node. Chem. Commun. 2021, 57, 2441–2463. [Google Scholar] [CrossRef]
- O’Hagan, D.; Schmidberger, J.W. Enzymes that catalyse SN2 reaction mechanisms. Nat. Prod. Rep. 2010, 27, 900–918. [Google Scholar] [CrossRef]
- Neri, U.; Wolf, Y.I.; Roux, S.; Camargo, A.P.; Lee, B.; Kazlauskas, D.; Chen, I.M.; Ivanova, N.; Zeigler Allen, L.; Paez-Espino, D.; et al. A five-fold expansion of the global RNA virome reveals multiple new clades of RNA bacteriophages. bioRxiv 2022. [Google Scholar] [CrossRef]
- Wolf, Y.I.; Kazlauskas, D.; Iranzo, J.; Lucía-Sanz, A.; Kuhn, J.H.; Krupovic, M.; Dolja, V.V.; Koonin, E.V. Origins and Evolution of the Global RNA Virome. mBio 2018, 9, e02329-18. [Google Scholar] [CrossRef]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M.; Varsani, A.; Wolf, Y.I.; Yutin, N.; Zerbini, F.M.; Kuhn, J.H. Global Organization and Proposed Megataxonomy of the Virus World. Microbiol. Mol. Biol. Rev. 2020, 84, e00061-19. [Google Scholar] [CrossRef]
- Cross, S.T.; Michalski, D.; Miller, M.R.; Wilusz, J. RNA regulatory processes in RNA virus biology. Wiley Interdiscip. Rev. RNA 2019, 10, e1536. [Google Scholar] [CrossRef]
- Courtney, D. Post-Transcriptional Regulation of Viral RNA through Epitranscriptional Modification. Cells 2021, 10, 1129. [Google Scholar] [CrossRef]
- Tong, J.; Zhang, W.; Chen, Y.; Yuan, Q.; Qin, N.-N.; Qu, G. The Emerging Role of RNA Modifications in the Regulation of Antiviral Innate Immunity. Front. Microbiol. 2022, 13, 845625. [Google Scholar] [CrossRef]
- Born, E.V.D.; Omelchenko, M.V.; Bekkelund, A.; Leihne, V.; Koonin, E.V.; Dolja, V.V.; Falnes, P. Viral AlkB proteins repair RNA damage by oxidative demethylation. Nucleic Acids Res. 2008, 36, 5451–5461. [Google Scholar] [CrossRef]
- Martínez-Pérez, M.; Aparicio, F.; López-Gresa, M.P.; Bellés, J.M.; Sanchez-Navarro, J.A.; Pallás, V. Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. USA 2017, 114, 10755–10760. [Google Scholar] [CrossRef]
- Yue, J.; Wei, Y.; Sun, Z.; Chen, Y.; Wei, X.; Wang, H.; Pasin, F.; Zhao, M. AlkB RNA demethylase homologues and N 6 -methyladenosine are involved in Potyvirus infection. Mol. Plant Pathol. 2022. [Google Scholar] [CrossRef]
- Kao, Y.-T.; Lai, M.M.C.; Yu, C.-Y. How Dengue Virus Circumvents Innate Immunity. Front. Immunol. 2018, 9, 2860. [Google Scholar] [CrossRef]
- Martin, B.; Coutard, B.; Guez, T.; Paesen, G.C.; Canard, B.; Debart, F.; Vasseur, J.-J.; Grimes, J.M.; Decroly, E. The methyltransferase domain of the Sudan ebolavirus L protein specifically targets internal adenosines of RNA substrates, in addition to the cap structure. Nucleic Acids Res. 2018, 46, 7902–7912. [Google Scholar] [CrossRef]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef]
- Daffis, S.; Szretter, K.J.; Schriewer, J.; Li, J.; Youn, S.; Errett, J.; Lin, T.-Y.; Schneller, S.; Zust, R.; Dong, H.; et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 2010, 468, 452–456. [Google Scholar] [CrossRef]
- Hyde, J.L.; Diamond, M.S. Innate immune restriction and antagonism of viral RNA lacking 2׳-O methylation. Virology 2015, 479–480, 66–74. [Google Scholar] [CrossRef]
- Göertz, G.P.; McNally, K.L.; Robertson, S.J.; Best, S.M.; Pijlman, G.P.; Fros, J.J. The Methyltransferase-Like Domain of Chikungunya Virus nsP2 Inhibits the Interferon Response by Promoting the Nuclear Export of STAT1. J. Virol. 2018, 92, e01008-18. [Google Scholar] [CrossRef]
- Mandadi, K.K.; Scholthof, K.-B.G. Plant Immune Responses Against Viruses: How Does a Virus Cause Disease? Plant Cell 2013, 25, 1489–1505. [Google Scholar] [CrossRef]
- Meier, N.; Hatch, C.; Nagalakshmi, U.; Dinesh-Kumar, S.P. Perspectives on intracellular perception of plant viruses. Mol. Plant Pathol. 2019, 20, 1185–1190. [Google Scholar] [CrossRef]
- Wang, L.-Y.; Lin, S.-S.; Hung, T.-H.; Li, T.-K.; Lin, N.-C.; Shen, T.-L. Multiple Domains of the Tobacco mosaic virus p126 Protein Can Independently Suppress Local and Systemic RNA Silencing. Mol. Plant-Microbe Interact.® 2012, 25, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: Common ancestry vs convergence. Biol. Direct 2017, 12, 5. [Google Scholar] [CrossRef]
- Mathonnet, G.; Fabian, M.R.; Svitkin, Y.V.; Parsyan, A.; Huck, L.; Murata, T.; Biffo, S.; Merrick, W.C.; Darzynkiewicz, E.; Pillai, R.S.; et al. MicroRNA Inhibition of Translation Initiation in Vitro by Targeting the Cap-Binding Complex eIF4F. Science 2007, 317, 1764–1767. [Google Scholar] [CrossRef]
- Gregory, B.D.; O’Malley, R.C.; Lister, R.; Urich, M.A.; Tonti-Filippini, J.; Chen, H.; Millar, A.H.; Ecker, J.R. A Link between RNA Metabolism and Silencing Affecting Arabidopsis Development. Dev. Cell 2008, 14, 854–866. [Google Scholar] [CrossRef]
- Gruber, J.J.; Zatechka, D.S.; Sabin, L.R.; Yong, J.; Lum, J.J.; Kong, M.; Zong, W.-X.; Zhang, Z.; Lau, C.-K.; Rawlings, J.; et al. Ars2 Links the Nuclear Cap-Binding Complex to RNA Interference and Cell Proliferation. Cell 2009, 138, 328–339. [Google Scholar] [CrossRef]
- Ryu, I.; Park, J.H.; An, S.; Kwon, O.S.; Jang, S.K. eIF4GI Facilitates the MicroRNA-Mediated Gene Silencing. PLoS ONE 2013, 8, e55725. [Google Scholar] [CrossRef]
- Zhang, X.; Chapat, C.; Wang, P.; Choi, J.-H.; Li, Q.; Luo, J.; Wiebe, S.; Kim, S.-H.; Robichaud, N.; Karam, I.F.; et al. microRNA-induced translational control of antiviral immunity by the cap-binding protein 4EHP. Mol. Cell 2021, 81, 1187–1199.e5. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mushegian, A. Methyltransferases of Riboviria. Biomolecules 2022, 12, 1247. https://doi.org/10.3390/biom12091247
Mushegian A. Methyltransferases of Riboviria. Biomolecules. 2022; 12(9):1247. https://doi.org/10.3390/biom12091247
Chicago/Turabian StyleMushegian, Arcady. 2022. "Methyltransferases of Riboviria" Biomolecules 12, no. 9: 1247. https://doi.org/10.3390/biom12091247
APA StyleMushegian, A. (2022). Methyltransferases of Riboviria. Biomolecules, 12(9), 1247. https://doi.org/10.3390/biom12091247





