Abstract
Irritable bowel syndrome is a prototypical disorder of the brain–gut–microbiome axis, although the underlying pathogenesis and mechanisms remain incompletely understood. With the recent advances in ‘omics’ technologies, studies have attempted to uncover IBS-specific variations in the host–microbiome profile and function. However, no biomarker has been identified to date. Given the high inter-individual and day-to-day variability of the gut microbiota, and a lack of agreement across the large number of microbiome studies, this review focused on omics studies that had sampling at more than one time point. A systematic literature search was performed using various combinations of the search terms “Irritable Bowel Syndrome” and “Omics” in the Medline, EMBASE, and Cochrane Library up to 1 December 2022. A total of 16 original studies were reviewed. These multi-omics studies have implicated Bacteroides, Faecalibacterium prausnitzii, Ruminococcus spp., and Bifidobacteria in IBS and treatment response, found altered metabolite profiles in serum, faecal, or urinary samples taken from IBS patients compared to the healthy controls, and revealed enrichment in the immune and inflammation-related pathways. They also demonstrated the possible therapeutic mechanisms of diet interventions, for example, synbiotics and low fermentable oligosaccharides, disaccharides, monosaccharides, and polyol (FODMAP) diets on microbial metabolites. However, there was significant heterogeneity among the studies and no uniform characteristics of IBS-related gut microbiota. There is a need to further study these putative mechanisms and also ensure that they can be translated to therapeutic benefits for patients with IBS.
1. Introduction
Irritable bowel syndrome (IBS) is the most commonly diagnosed gastrointestinal disorder [1], characterized by recurrent abdominal pain or discomfort and a change in the frequency or form of one’s stools [2]. It is thought to affect around 12% of the global population [1] and is associated with a significant burden of illness as it impacts an individual’s health-related quality of life and work productivity [3].
Despite the prevalence of IBS and the enormous economic disease burden (totalling more than USD 20 billion in the United States alone [4]), the current state of IBS research into the etiopathogenesis and clinical phenotypes of IBS suggest that the condition is heterogenous, multifactorial, and remain incompletely understood [5,6]. There is also no cure or targeted therapy for IBS, and treatments are primarily aimed at providing symptom relief. Burgeoning research and experimental evidence have suggested IBS to be a disorder of the brain–gut–microbiome axis, driven by an altered intestinal and colonic microbiota, abnormal gut immune activation, and increased gut permeability [7]. However, traditional laboratory research is hampered by the lack of a reliable animal model for IBS with poor clinical translation [8], while clinical studies have yielded inconsistent results and may suffer from suboptimal design and power [9].
To overcome these limitations, in recent years, scientific advances in ‘omics’ technologies have enabled precise and accurate molecular measurements (proteins, genes, and metabolite etc.) within a tissue or cell, emerging as a powerful tool to unravel new mechanistic insights beyond the expressed phenotype [10,11]. Omics technologies such as genomics, transcriptomics, proteomics, and metabolomics are increasingly being used to study IBS and improve our understanding of its underlying molecular mechanisms: genomics studies have identified genetic variations associated with IBS and its subtypes. Transcriptomics can provide information on the gene expression changes that occur in response to IBS-related stimuli, while proteomics can identify changes in protein levels and post-translational modifications. Finally, metabolomics is being used to identify changes in the gut microbiota and metabolic pathways that may contribute to IBS symptoms. Alterations in metabolites and metabolite signatures have been associated with central sensitivity pain syndromes including IBS [12]. By integrating the results from different omics platforms, researchers can theoretically gain a more comprehensive view of the complex and multifactorial nature of IBS to inform the development of novel therapeutic strategies for the management of this disorder.
To the best of our knowledge, there has not been a review focusing on the contributions of longitudinal omics studies to our modern understanding of IBS. This review therefore aimed to systematically synthesise the current body of evidence from omics studies in humans as well as outline possible directions for future omics research and clinical applications. Given the known high inter-individual and day-to-day variability of the gut microbiota due to genetic, diet, environmental, and other factors [13], and a lack of agreement across the large number of faecal microbiome studies [14], this review focused only on studies that had sampling at more than one time point.
2. Methods
A systematic literature search was performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [15]. The review protocol was registered under the International Prospective Register of Systematic Reviews (PROSPERO), registration number CRD42022360859.
Various combinations of the key search terms “Irritable Bowel Syndrome” and “Omics” were used in the search strategy for Medline, EMBASE, and Cochrane Library, and searched from database inception through to 1 December 2022. No restrictions on date, language, or subject were implemented on the database search. The detailed search strategy can be found in Table S1. Abstracts were imported into Microsoft Excel and screened by five independent researchers (C.E.Y., C.Y.L.Y, R.I.H.C., N.Z.-Y.C., and S.E.T.). Full texts were obtained for all abstracts of relevance and their respective reference lists were hand-searched to identify additional relevant articles. Forward searching of prospective citations of the relevant full texts was also performed and authors of the respective articles were contacted if necessary to provide additional data.
Each article was reviewed by at least two researchers blinded to each other’s decision. Disputes were resolved through consensus from the senior author (Q.X.N. or Y.L.L.). The criterion for inclusion were: (1) human studies; (2) utilising omics technology (e.g., proteomics, transcriptomics, genomics, or metabolomics); (3) original published articles; (4) longitudinal study design (with sampling at more than one time point); (5) written or translated into the English language. Animal studies were excluded from this review. A cross-sectional study of the gut microbiome falls short in adequately capturing and reflecting the highly diverse gut ecosystem and dynamic microbiota–gut–brain axis interactions, moreover, the gut microbiome profile tends to vary significantly between individuals from different geographical regions, populations, and even development stages [13]. Studies have also shown that the commonly employed ‘omics’ methods lack accuracy when measuring a single time point [16] and it is more reliable to investigate microbiota changes over time or repeat metabolite analyses; hence, cross-sectional studies were excluded from the present review.
Data from the relevant studies were extracted using a standardized data form in Microsoft Excel including information on the study population, country of origin, type of sample(s) collected and analysed, study methods, time points for taking measurements, and the key study findings. These information were extracted by five independent researchers (C.E.Y., C.Y.L.Y, R.I.H.C., N.Z.-Y.C. and S.E.T.) and cross-checked by a senior author (Q.X.N. or Y.L.L.) for accuracy.
3. Results
A total of 16 studies were eligible for inclusion after a systematic literature search (Figure 1).
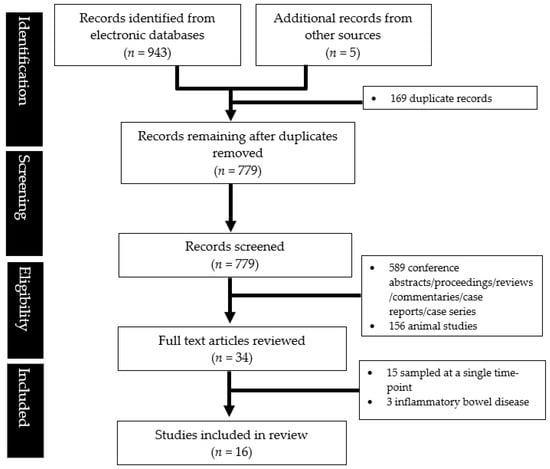
Figure 1.
PRISMA flowchart showing the study abstraction process.
Table 1 outlines the key characteristics and findings of the studies reviewed. A total of 16 studies were included in this review [16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31]. The studies mostly comprised intervention studies that took measurements at the baseline and after intervention (typically four to six weeks) [18,19,21,22,24,25,26,27,28,29,30]. The majority of studies had small sample sizes, with only two having more than 100 patients [20,30]. The studies tended to focus on patients with IBS-D and made use of healthy controls for comparison [16,17,18,20,23,26,27,31], and were generally aimed to identify the microbiota changes and cellular mediators underlying IBS via 16S rRNA gene sequencing, metabolomics, or transcriptomics analyses.

Table 1.
The characteristics and key findings of the studies reviewed (arranged alphabetically by the first author’s last name).
In terms of the sample type, six studies used stool samples [16,18,19,23,24,25], five used gastrointestinal mucosal samples [16,17,20,25,26], four analysed blood samples [22,27,29,30], and four analysed urine samples [21,27,28,31].
Findings of studies that utilised metagenomics were further elaborated in Table 2. Metagenomics and RNA sequencing are more sensitive, have greater resolution, and provide a more comprehensive picture regarding the structure and function of host microbial communities compared to traditional 16S rRNA sequencing. However, differences in taxa abundances between individuals with IBS and the healthy controls at the baseline and post-intervention appeared rather variable within and inconsistent across the studies (Table 2).

Table 2.
Comparisons in terms of taxa for metagenomics studies.
In terms of the metabolomics changes, reduced levels of short-chain fatty acids (SCFAs) have been associated with an altered gut microbiome in IBS [25], while elevated levels of branched-chain amino acids and certain gut peptides have also been observed in IBS patients [23].
4. Discussion
Traditional clinical trials involving IBS patients have been confounded by a heterogeneous patient population, highly variable symptoms, and a large placebo effect [9]. IBS consists of a constellation of gut symptoms, and burgeoning research into the gut microbiome has attempted to uncover uniform mechanisms underlying the microbiota–gut–brain axis interactions, especially at the level of metabolite changes and differential gene expressions. The intestinal microbiota comprises billions of diverse bacteria, viruses, fungi, and archaea, and their metabolites and by-products are probably a part of the complex bi-directional microbiota–gut–brain axis [7]. Alterations in the gut microbiome may contribute to the development of IBS symptoms.
As enabled by multi-omics studies, we have some hypotheses on the abnormal alterations in the gut microbiota and microbial metabolites underlying patients with IBS and their symptom flares. Several studies have implicated Bacteroides, Faecalibacterium prausnitzii, Ruminococcus spp., and Bifidobacteria in IBS and treatment response [19,24,26]. Similar to the findings of a 2019 review that examined case-control studies detecting gut microbiota in IBS patients [14], increased Firmicutes and decreased relative abundance of Bacteriodetes were common among faecal microbiota studies, but results for mucosal microbiota were more variable. Metabolomics studies have revealed alterations in the levels of specific metabolites such as SCFAs, bile acids, and amino acids [23,25,27,29], which are the end products of cellular metabolism and can reflect changes in the gut microbiome and other aspects of the gut environment. These changes may contribute to the development of gut symptoms; SCFAs are produced by the gut microbiome and are the primary energy source for intestinal epithelial cells [32]. Reduced levels of SCFAs, particularly butyrate, have been observed in IBS [33] and are thought to reflect alterations in the gut microbiome.
Several studies in this review also found altered metabolite profiles in serum, faecal, or urinary samples taken from IBS patients compared to healthy controls [27,28,29,31], and revealed an enrichment in the immune and inflammation-related pathways [16,22], although the results have been inconsistent. The intestinal mucosa is part of an intricate enteric immune system and comprises a large variety of immune cells [5]; low-grade inflammation and the effects of proinflammatory cytokines and tumour necrosis factor alpha (TNF-alpha) in the colonic mucosa may at least in part explain IBS symptoms and flares [34].
These multi-omics studies have also demonstrated the possible therapeutic mechanisms of diet interventions (e.g., synbiotics and low FODMAP diets) on microbial metabolites. It is known that the human gut microbiome can rapidly respond to dietary interventions and an altered diet [35]. Based on the findings of the studies reviewed, oral synbiotic yogurt normalized metabolites are involved in the one-carbon metabolism pathway [27]. Probiotic supplementation increased the counts of presumptive lactic acid bacteria (Lactobacillus and Bifidobacteria) [19]. Low FODMAP diets resulted in increased 2-hydroxybutyrate in serum and decreased pantothenate in urine [28], while starch- and sucrose-restriction led to increased alpha-linoleic acid and linoleic acid levels in the blood plasma [29], which were likely the direct results of an increased intake of specific foods rich in these essential metabolites. In a double-blind, crossover trial involving paediatric IBS patients compared to non-responders to a low FODMAP diet, responders were characterised to be enriched at the baseline in gut microbes with greater saccharolytic metabolic capacity (family Bacteroidaceae, e.g., Bacteroides, order Clostridiales, e.g., Ruminococcaceae, Dorea, and Faecalibacterium prausnitzii, and family Erysipilotrichaceae, e.g., cc_115), while non-responders were enriched at the baseline in Turicibacter (from the family Turicibacteraceae). [36]. Similarly, in adult IBS patients, non-responders had at the baseline gut dysbiosis, with an overrepresentation of Streptococcus and Dorea species [37]. This implies that gut microbiota may predict the treatment response. However, the differences in taxa abundances observed at the individual time points can be highly variable and inconsistent when comparing the different time points, and it may not overlap with changes observed in the averaged data [16]. The gut microbiome likely plays a role in the development of IBS symptoms, and patients with certain alterations in the gut microbiome diversity or composition may be more or less likely to respond to particular treatments, however, there are no firm conclusions yet. Metabolomics capture valuable information on metabolites that are either produced endogenously or from the digestion and metabolism of foods. The effects of dietary interventions may be transient and the correlation between the impact on the metabolite signature and long-term symptom control remains unclear. In particular, the effects of probiotic supplementation are likely to be dependent on the baseline host microbiome features and are not sustained [38,39].
To elucidate the effect of microbial metabolism on host function, one study also compared the transcriptional and epigenetic changes [16], hinting at alterations in purine nucleoside phosphorylase and increased purine degradation by gut microbiota in the IBS-D and IBS-C patients. There are complex metabolic changes that occur in IBS and this may have implications for new diagnostic and therapeutic approaches.
The present methods are not without limitations. First, in the majority of studies, 16S rRNA gene sequencing technology was used to detect the faecal microbiota of IBS patients and healthy controls, however, 16S rRNA sequencing provides taxa resolution up to the genus level and is unable to yield information on the functional characteristics compared to newer techniques such as shotgun-metagenome sequencing, which was used in a few studies [16,18,24]. Metagenomics and RNA sequencing are more sensitive, have greater resolution, and provide a more comprehensive picture regarding the structure and function of host microbial communities [40]. Second, at present, the gut microbiome is also primarily studied by the use of stool bacterial communities as a proxy. Stool samples are broadly representative of colonic luminal bacteria; however, some communities of bacteria may be overlooked including those found in the small intestine or embedded within intestinal mucosa [41], which may also explain the difference seen between studies that utilised mucosal as opposed to faecal samples. A study into human gastrointestinal and faecal microbiome found the two to be only partially correlated, and faecal microbiome was a limited indicator of gut mucosa-associated microbiome composition and metagenomic function [38]. As different parts of the intestinal tract contain different luminal and mucosal commensal microbiota, we should collect gut microbiota at different sites. Last but not least, although this review focused on the time-series feature of available studies, most studies only took measurements at two time points, and it is necessary to have more studies with greater longitudinal sampling to reliably investigate microbiota and metabolite changes over time. Longitudinal sampling would be particularly useful to compare different disease states such as IBS flare versus remission or the effects of interventions. Although the gut microbiota is a potential biomarker for IBS, there is no firm conclusion on the characteristics of IBS-related gut microbiota, and no biomarkers have been identified to date.
5. Conclusions
In conclusion, having reviewed a range of data types and reported pathways that were identified across the studies, there was significant heterogeneity among the studies and no uniform characteristics of IBS-related gut microbiota. There is still a paucity of human studies and a need to ensure that these putative mechanisms can be translated to therapeutic benefits for patients with IBS. Despite the advances in metabolomics and microbiome understanding, studies have not uncovered distinct changes that underlie the symptoms in IBS patients.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/metabo13040484/s1, Table S1: Full search strategy for the various databases.
Author Contributions
Conceptualization, Q.X.N.; Data curation, Q.X.N., C.E.Y., C.Y.L.Y., R.I.H.C., N.Z.-Y.C., S.E.T., Y.L.L., A.Y.S.S., W.K.N. and J.T.; Formal analysis, Q.X.N., C.E.Y., C.Y.L.Y., R.I.H.C., N.Z.-Y.C., S.E.T. and Y.L.L.; Investigation, Q.X.N., C.E.Y., C.Y.L.Y., R.I.H.C., N.Z.-Y.C. and Y.L.L.; Methodology, Q.X.N., C.E.Y., C.Y.L.Y., R.I.H.C., N.Z.-Y.C., S.E.T., Y.L.L., A.Y.S.S., W.K.N. and J.T.; Supervision, A.Y.S.S., W.K.N. and J.T.; Writing—original draft, Q.X.N., C.E.Y., C.Y.L.Y., R.I.H.C., N.Z.-Y.C., S.E.T. and Y.L.L.; Writing—review & editing, Q.X.N., C.E.Y., C.Y.L.Y., R.I.H.C., N.Z.-Y.C., S.E.T., Y.L.L., A.Y.S.S., W.K.N. and J.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lovell, R.M.; Ford, A.C. Global prevalence of and risk factors for irritable bowel syndrome: A meta-analysis. Clin. Gastroenterol. Hepatol. 2012, 10, 712–721.e4. [Google Scholar] [CrossRef] [PubMed]
- Vork, L.; Weerts, Z.Z.R.M.; Mujagic, Z.; Kruimel, J.W.; Hesselink, M.A.M.; Muris, J.W.M.; Keszthelyi, D.; Jonkers, D.M.A.E.; Masclee, A.A.M. Rome III vs Rome IV criteria for irritable bowel syndrome: A comparison of clinical characteristics in a large cohort study. Neurogastroenterol. Motil. 2018, 30, e13189. [Google Scholar] [CrossRef] [PubMed]
- Cassar, G.E.; Youssef, G.J.; Knowles, S.; Moulding, R.; Austin, D.W. Health-Related Quality of Life in Irritable Bowel Syndrome: A Systematic Review and Meta-analysis. Gastroenterol. Nurs. 2020, 43, E102–E122. [Google Scholar] [CrossRef] [PubMed]
- Buono, J.L.; Mathur, K.; Averitt, A.J.; Andrae, D.A. Economic Burden of Irritable Bowel Syndrome with Diarrhea: Retrospective Analysis of a U.S. Commercially Insured Population. J. Manag. Care Spec. Pharm. 2017, 23, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Ng, Q.X.; Soh, A.Y.S.; Loke, W.; Lim, D.Y.; Yeo, W.S. The role of inflammation in irritable bowel syndrome (IBS). J. Inflamm. Res. 2018, 11, 345–349. [Google Scholar] [CrossRef]
- Enck, P.; Aziz, Q.; Barbara, G.; Farmer, A.D.; Fukudo, S.; Mayer, E.A.; Niesler, B.; Quigley, E.M.; Rajilić-Stojanović, M.; Schemann, M.; et al. Irritable bowel syndrome. Nat. Rev. Dis. Prim. 2016, 2, 16014. [Google Scholar] [CrossRef]
- Martin, C.R.; Osadchiy, V.; Kalani, A.; Mayer, E.A. The Brain-Gut-Microbiome Axis. Cell. Mol. Gastroenterol. Hepatol. 2018, 6, 133–148. [Google Scholar] [CrossRef]
- Holschneider, D.P.; Bradesi, S.; Mayer, E.A. The role of experimental models in developing new treatments for irritable bowel syndrome. Expert Rev. Gastroenterol. Hepatol. 2011, 5, 43–57. [Google Scholar] [CrossRef]
- Miller, L.E. Study design considerations for irritable bowel syndrome clinical trials. Ann. Gastroenterol. 2014, 27, 338–345. [Google Scholar]
- Karczewski, K.J.; Snyder, M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef]
- Mihindukulasuriya, K.A.; Mars, R.A.T.; Johnson, A.J.; Ward, T.; Priya, S.; Lekatz, H.R.; Kalari, K.R.; Droit, L.; Zheng, T.; Blekhman, R.; et al. Multi-Omics Analyses Show Disease, Diet, and Transcriptome Interactions with the Virome. Gastroenterology 2021, 161, 1194–1207.e8. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.S.; Rodriguez-Saona, L.; Hackshaw, K.V. Metabolomics in Central Sensitivity Syndromes. Metabolites 2020, 10, 164. [Google Scholar] [CrossRef]
- Healey, G.R.; Murphy, R.; Brough, L.; Butts, C.A.; Coad, J. Interindividual variability in gut microbiota and host response to dietary interventions. Nutr. Rev. 2017, 75, 1059–1080. [Google Scholar] [CrossRef]
- Duan, R.; Zhu, S.; Wang, B.; Duan, L. Alterations of Gut Microbiota in Patients with Irritable Bowel Syndrome Based on 16S rRNA-Targeted Sequencing: A Systematic Review. Clin. Transl. Gastroenterol. 2019, 10, e00012. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Mars, R.A.T.; Yang, Y.; Ward, T.; Houtti, M.; Priya, S.; Lekatz, H.R.; Tang, X.; Sun, Z.; Kalari, K.R.; Korem, T.; et al. Longitudinal Multi-omics Reveals Subset-Specific Mechanisms Underlying Irritable Bowel Syndrome. Cell 2020, 182, 1460–1473.e17. [Google Scholar] [CrossRef]
- Aerssens, J.; Camilleri, M.; Talloen, W.; Thielemans, L.; Göhlmann, H.W.; Van Den Wyngaert, I.; Thielemans, T.; De Hoogt, R.; Andrews, C.N.; Bharucha, A.E.; et al. Alterations in mucosal immunity identified in the colon of patients with irritable bowel syndrome. Clin. Gastroenterol. Hepatol. 2008, 6, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Ankersen, D.V.; Weimers, P.; Bennedsen, M.; Haaber, A.B.; Fjordside, E.L.; Beber, M.E.; Lieven, C.; Saboori, S.; Vad, N.; Rannem, T.; et al. Long-Term Effects of a Web-Based Low-FODMAP Diet versus Probiotic Treatment for Irritable Bowel Syndrome, Including Shotgun Analyses of Microbiota: Randomized, Double-Crossover Clinical Trial. J. Med. Internet Res. 2021, 23, e30291. [Google Scholar] [CrossRef]
- Bonfrate, L.; Di Palo, D.M.; Celano, G.; Albert, A.; Vitellio, P.; De Angelis, M.; Gobbetti, M.; Portincasa, P. Effects of Bifidobacterium longum BB536 and Lactobacillus rhamnosus HN001 in IBS patients. Eur. J. Clin. Investig. 2020, 50, e13201. [Google Scholar] [CrossRef]
- Ek, W.E.; Reznichenko, A.; Ripke, S.; Niesler, B.; Zucchelli, M.; Rivera, N.V.; Schmidt, P.T.; Pedersen, N.L.; Magnusson, P.; Talley, N.J.; et al. Exploring the genetics of irritable bowel syndrome: A GWA study in the general population and replication in multinational case-control cohorts. Gut 2015, 64, 1774–1782. [Google Scholar] [CrossRef]
- Kim, J.; Cho, K.; Kim, J.S.; Jung, H.C.; Kim, B.; Park, M.S.; Ji, G.E.; Cho, J.Y.; Hong, K.S. Probiotic treatment induced change of inflammation related metabolites in IBS-D patients/double-blind, randomized, placebo-controlled trial. Food Sci. Biotechnol. 2019, 29, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Kuo, B.; Bhasin, M.; Jacquart, J.; Scult, M.A.; Slipp, L.; Riklin, E.I.; Lepoutre, V.; Comosa, N.; Norton, B.A.; Dassatti, A.; et al. Genomic and clinical effects associated with a relaxation response mind-body intervention in patients with irritable bowel syndrome and inflammatory bowel disease. PLoS ONE 2015, 10, e0123861. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, G.; Noor, S.O.; Ridgway, K.; Scovell, L.; Jamieson, C.; Johnson, I.T.; Colquhoun, I.J.; Kemsley, E.K.; Narbad, A. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J. Proteome Res. 2011, 10, 4208–4218. [Google Scholar] [CrossRef] [PubMed]
- Mack, A.; Bobardt, J.S.; Haß, A.; Nichols, K.B.; Schmid, R.M.; Stein-Thoeringer, C.K. Changes in gut microbial metagenomic pathways associated with clinical outcomes after the elimination of malabsorbed sugars in an IBS cohort. Gut Microbes 2020, 11, 620–631. [Google Scholar] [CrossRef]
- Moser, A.M.; Spindelboeck, W.; Halwachs, B.; Strohmaier, H.; Kump, P.; Gorkiewicz, G.; Högenauer, C. Effects of an oral synbiotic on the gastrointestinal immune system and microbiota in patients with diarrhea-predominant irritable bowel syndrome. Eur. J. Nutr. 2019, 58, 2767–2778. [Google Scholar] [CrossRef]
- Ng, S.C.; Lam, E.F.; Lam, T.T.; Chan, Y.; Law, W.; Tse, P.C.; Kamm, M.A.; Sung, J.J.; Chan, F.K.; Wu, J.C. Effect of probiotic bacteria on the intestinal microbiota in irritable bowel syndrome. J. Gastroenterol. Hepatol. 2013, 28, 1624–1631. [Google Scholar] [CrossRef] [PubMed]
- Noorbakhsh, H.; Yavarmanesh, M.; Mortazavi, S.A.; Adibi, P.; Moazzami, A.A. Metabolomics analysis revealed metabolic changes in patients with diarrhea-predominant irritable bowel syndrome and metabolic responses to a synbiotic yogurt intervention. Eur. J. Nutr. 2019, 58, 3109–3119. [Google Scholar] [CrossRef] [PubMed]
- Nybacka, S.; Simrén, M.; Störsrud, S.; Törnblom, H.; Winkvist, A.; Lindqvist, H.M. Changes in serum and urinary metabolomic profile after a dietary intervention in patients with irritable bowel syndrome. PLoS ONE 2021, 16, e0257331. [Google Scholar] [CrossRef]
- Stenlund, H.; Nilholm, C.; Chorell, E.; Roth, B.; D’Amato, M.; Ohlsson, B. Metabolic Profiling of Plasma in Patients with Irritable Bowel Syndrome after a 4-Week Starch- and Sucrose-Reduced Diet. Metabolites 2021, 11, 440. [Google Scholar] [CrossRef]
- Wang, R.S.; Lembo, A.J.; Kaptchuk, T.J.; Cheng, V.; Nee, J.; Iturrino, J.; Rao, M.; Loscalzo, J.; Silvester, J.A.; Hall, K.T. Genomic Effects Associated with Response to Placebo Treatment in a Randomized Trial of Irritable Bowel Syndrome. Front. Pain Res. 2022, 2, 775386. [Google Scholar] [CrossRef]
- Yamamoto, M.; Pinto-Sanchez, M.I.; Bercik, P.; Britz-McKibbin, P. Metabolomics reveals elevated urinary excretion of collagen degradation and epithelial cell turnover products in irritable bowel syndrome patients. Metabolomics 2019, 15, 82. [Google Scholar] [CrossRef]
- Kelly, C.J.; Zheng, L.; Campbell, E.L.; Saeedi, B.; Scholz, C.C.; Bayless, A.J.; Wilson, K.E.; Glover, L.E.; Kominsky, D.J.; Magnuson, A.; et al. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe 2015, 17, 662–671. [Google Scholar] [CrossRef]
- Sun, Q.; Jia, Q.; Song, L.; Duan, L. Alterations in fecal short-chain fatty acids in patients with irritable bowel syndrome: A systematic review and meta-analysis. Medicine 2019, 98, e14513. [Google Scholar] [CrossRef]
- Camilleri, M.; Lasch, K.; Zhou, W. Irritable bowel syndrome: Methods, mechanisms, and pathophysiology. The confluence of increased permeability, inflammation, and pain in irritable bowel syndrome. Am. J. Physiol. Gastrointest Liver Physiol. 2012, 303, G775–G785. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Chumpitazi, B.P.; Cope, J.L.; Hollister, E.B.; Tsai, C.M.; McMeans, A.R.; Luna, R.A.; Versalovic, J.; Shulman, R.J. Randomised clinical trial: Gut microbiome biomarkers are associated with clinical response to a low FODMAP diet in children with the irritable bowel syndrome. Aliment. Pharmacol. Ther. 2015, 42, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Bennet, S.M.P.; Böhn, L.; Störsrud, S.; Liljebo, T.; Collin, L.; Lindfors, P.; Törnblom, H.; Öhman, L.; Simrén, M. Multivariate modelling of faecal bacterial profiles of patients with IBS predicts responsiveness to a diet low in FODMAPs. Gut 2018, 67, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Zilberman-Schapira, G.; Suez, J.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Kotler, E.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.; et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell 2018, 174, 1388–1405.e21. [Google Scholar] [CrossRef]
- Ng, Q.X.; Lim, Y.L.; Yaow, C.Y.; Ng, W.K.; Thumboo, J.; Liew, T.M. Effect of Probiotic Supplementation on Gut Microbiota in Patients with Major Depressive Disorders: A Systematic Review. Nutrients 2023, 15, 1351. [Google Scholar] [CrossRef]
- Kalyuzhnaya, M.G.; Lapidus, A.; Ivanova, N.; Copeland, A.C.; McHardy, A.C.; Szeto, E.; Salamov, A.; Grigoriev, I.V.; Suciu, D.; Levine, S.R.; et al. High-resolution metagenomics targets specific functional types in complex microbial communities. Nat. Biotechnol. 2008, 26, 1029–1034. [Google Scholar] [CrossRef]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).