Abstract
Prostate cancer (PCa) is the most common genitourinary malignancy in men, with a multifactorial etiology influenced by genetic, environmental, and microbial determinants. Although the prostate was traditionally considered sterile, advances in microbiome research have challenged this view, revealing potential links between microbial communities and PCa development, progression, and treatment response. This review synthesizes evidence on the gut, urinary, seminal fluid, and prostatic microbiomes, highlighting their potential contributions to PCa pathogenesis and therapeutic outcomes. Key studies utilizing next-generation sequencing (NGS), whole-genome sequencing (WGS), PCR, and metagenomic analyses have identified specific bacterial and fungal taxa associated with Pca; however, findings remain inconsistent across methodologies and cohorts. Microorganisms such as Propionibacterium acnes and Pseudomonas spp. may modulate inflammation, immune responses, and resistance to androgen-deprivation therapy. Further research is required to determine whether microbial signatures can serve as reliable biomarkers for early detection, prognosis, or novel therapeutic strategies in PCa management.
1. Introduction
Prostate cancer (PCa) represents the most frequently diagnosed malignancy of the male genitourinary tract and represents a major global public health burden. In 2020, approximately 1.41 million new cases were diagnosed, and PCa accounted for 375,304 deaths worldwide, corresponding to nearly 8.6 million disability-adjusted life years (DALYs) [1,2]. In the United States alone, 299,010 new PCa cases and 35,250 deaths were projected for 2024 [3]. Despite a relatively high five-year survival rate, PCa remains a significant contributor to global cancer-related mortality. The introduction of prostate-specific antigen (PSA) screening programs has facilitated the earlier detection of localized disease, but has also raised concerns about overtreatment, as many identified tumors may remain indolent and clinically insignificant if untreated [4]. Standard therapies, including radical prostatectomy and radiotherapy, are associated with substantial morbidity and impaired quality of life. Moreover, even with early-stage detection and treatment, many patients ultimately experience disease recurrence and progression to more aggressive phenotypes [5].
The prostate was historically considered a sterile organ. However, advances in sensitive microbiological techniques have challenged this paradigm, paralleling shifts in our understanding of microbial colonization in other sites once presumed sterile [6,7]. Increasing evidence now implicates microorganisms in the etiopathogenesis of diseases traditionally categorized as non-infectious, such as the associations of Helicobacter pylori with gastritis and peptic ulcer disease, Tropheryma whipplei with Whipple’s disease, and high-risk human papillomavirus (HPV) subtypes with cervical carcinoma [8,9,10]. The identification of both culturable and unculturable microorganisms in such environments suggests a potential role for microbes in the pathogenesis of other diseases historically regarded as non-infectious.
Emerging evidence supports the concept that microbe alterations can drive chronic inflammation (e.g., helicobacter pylori) by modulating local tissue immune microenvironments and triggering oncogenic cascades, such as metaplasia, dysplasia, and atypia, that facilitate tumorigenesis [10,11,12]. The impact of the tumor-associated microbiome on carcinogenesis, cancer progression, and responses to therapeutic interventions has become an area of significant research interest [13]. Furthermore, accumulating data suggest that the gut microbiome is not only linked to colorectal cancer tumorigenesis and prognosis but also across other malignancies as well [14,15]. Beyond statistical associations and prognostic implications, recent studies even suggest a potential causative role [16]. Importantly, they underscore the gut microbiome’s pivotal role in modulating host immune responses [17]. Some recent studies have also shown that the therapeutic manipulation of the gut microbiota via probiotic administration might be a promising strategy to enhance the efficacy of cancer immunotherapy [18]. Collectively, these findings support the hypothesis that both gut and tumor-localized microbial populations can substantially influence cancer development and progression.
Nevertheless, the contribution of microbiomes to PCa pathogenesis remains poorly defined. PCa is typically classified as an immunologically “cold” tumor, consistent with its limited responses to contemporary immunotherapeutic strategies [19]. While several mechanisms linking microbes to prostate carcinogenesis have been proposed, robust translational evidence is still lacking [20]. A key research priority is the identification of novel biomarkers that can reliably distinguish indolent from aggressive disease at diagnosis with high specificity and sensitivity, becoming the figurative holy grail of PCa research.
In this review, we systematically summarize current evidence on the associations between PCa and microbial communities within the gut (Section 2), urine, feces, seminal fluid, and prostatic secretions (Section 3), as well as prostate tissue microenvironments (Section 4). We also evaluate contemporary methodologies employed for microbiome detection. Our objective is to critically assess whether specific microbial taxa or microbiota signatures may serve as reliable biomarkers for PCa diagnosis or prognosis. The early detection and targeted modulation of relevant microbial populations could open new avenues for prevention and therapy.
2. Gut Microbiome and Prostate Cancer: Emerging Evidence and Therapeutic Implications
Investigation of the gut microbiome in PCa has become an essential research focus, supported by growing evidence of its potential role in cancer pathogenesis. In a prospective case-control study utilizing whole-genome next-generation sequencing (WGS), Colombos et al. analyzed the gut microbiota composition of 12 patients with intermediate or high-risk PCa compared with 8 individuals with benign prostate conditions. PCa patients exhibited a higher abundance of Bacteroides massiliensis, whereas controls showed an enrichment of Faecalibacterium prausnitzii and Eubacterium rectale [21]. Faecalibacterium prausnitzii plays a key role in maintaining gut homeostasis as an acetate-consuming species that produces butyrate and anti-inflammatory metabolites, including salicylic acid derivatives [22]. Similarly, Eubacterium rectale is a butyrate producer with established anti-inflammatory properties within the gut microenvironment [23].
Supporting these observations, Liu et al. characterized gut microbiota by sequencing theV3 and V4 hypervariable regions of the bacterial 16S rRNA gene in 21 PCa patients at matched hormone-sensitive and castration-resistant disease stages undergoing androgen deprivation therapy. Distinct microbial alterations were identified in castration-resistant PCa, including the enrichment of Phascolarctobacterium and Ruminococcus [24]. Pernigoni et al. provided complementary insights, proposing that androgen-producing gut microbiota significantly contribute to the progression toward castration-resistant prostate cancer (CRPC). Recently Wang et al. found that the gut microbiome member Clostridium scidens contains an enzyme that catalyzes the conversion of androstenedione to epitestosterone. The authors showed that the latter impacts the proliferation of androgen-dependent prostate cancer cell lines in vitro, while the responsible enzyme was elevated in patients who are not responsive to abiraterone therapy [25]. In murine models, gut microbiota depletion via antibiotic administration delayed the onset of CRPC, whereas fecal microbiota transplantation (FMT) from castration-resistant subjects conferred resistance to castration therapies in recipient mice. Conversely, the administration of Prevotella stercorea or FMT derived from hormone-sensitive PCa patients effectively suppressed tumor growth, highlighting potential microbiome-targeted interventions [26].
3. Beyond the Gut: Exploring Microbial Signatures in Urine, Feces, Seminal Fluid, and Prostatic Secretions in Prostate Cancer
Microbial compositions exhibit marked inter-individual variability, influencing metabolic processes, local and systemic inflammatory responses, and immune regulation. These characteristics suggest potential utility as non-invasive biomarkers for early cancer detection and risk stratification. Alanee et al. analyzed paired fecal and urine samples from 30 patients undergoing prostate biopsy for elevated PSA. Using 16S rRNA sequencing (V3–V5 region), distinct urinary microbial profiles were detected in 71.4% of PCa patients, clustering separately from controls. This clustering was associated with Gleason score in urine but not fecal samples. Urine from PCa was enriched in Veillonella, Streptococcus, and Bacteroides spp., while Faecalibacterium, Lactobacilli, and Acinetobacter spp. were reduced. By contrast, patients without PCa exhibited higher abundances of Clostridium XVIII & IV, Lachnospira, Acetanaerobacterium, and Faecalibacterium in urine samples. Fecal samples from PCa patients showed an increased abundance of Bacteroides species, although overall bacterial diversity was comparable [27].
Yu et al. further profiled expressed prostatic secretions (EPS), urine, and seminal fluid from PCa (n = 13) and benign prostatic hyperplasia (BPH; n = 34) patients by sequencing the 16S rRNA V3 region. PCa patients exhibited an enrichment of Bacteroidetes, Alphaproteobacteria, Firmicutes, Propionicimonas, Lachnospiraceae, and Ochrobactrum, whereas BPH samples were enriched in Eubacterium and Defluviicoccus. Alphaproteobacteria, often implicated in urinary tract infections, may contribute to inflammation in PCa. Firmicutes, associated with obesity and caloric metabolism, and Propionibacterineae, frequently linked to prostatitis, were also prominent. Ochrobactrum, an opportunistic pathogen, may indicate immune dysfunction. While Lachnospiraceae are generally beneficial butyrate producers, they have paradoxical associations with metabolic syndrome and diabetes. PCa patients showed reduced urinary Escherichia coli but an increased abundance of EPS and seminal fluid, along with elevated Enterococcus spp. in seminal fluid [28,29].
Shrestha et al. analyzed 129 urine samples (61 PCa, 63 benign biopsies, and 5 initially benign progressing to PCa) using a two-step PCR targeting the 16S rRNA V6 region. Cancer samples harbored slightly higher microbial diversity (67 vs. 60 genera), dominated frequently by Corynebacterium, Staphylococcus, or Streptococcus, with Anaerococcus, Lactobacillus, and Actinobaculum being prominent in selected samples. A cancer-associated cluster included urogenital pathogens such as Streptococcus anginosus, Anaerococcus obesiensis, Anaerococcus lactolyticus, Varibaculum cambriense, Actinobaculum schaalii, and Propionimicrobium lymphophilum, though Propionibacterium acnes (P. acnes) did not significantly differ between groups [30].
Hurst et al. employed NGS and qPCR in 215 patients, correlating urinary microbiota with D’Amico risk scores, clinical stage, PSA, and Gleason score. Total operational taxonomic unit (OTU) counts did not correlate with PCa risk groups. The bacterial characterization of urinary sediments revealed four novel bacteria with unassigned OTU sequences in the NCBI dataset, which were frequently found in the patient’s urine. These included Porphyromonas, Varibaculum, Peptoniphilus, and Fenollaria spp. All were detected in prostatic secretions, while Varibaculum and Peptoniphilus were detected in prostate tissue by qPCR. Anaerobic culture yielded 39 bacterial isolates from urine, as well as 8 isolates from PCa secretions. These mostly included Firmicutes, Fenollaria, and Anaerococcus species. Five anaerobic genera, including three of the novel isolates, were associated with PCa risk group in cancer tissue, urine sediment, and urine extracellular vesicles. Prostate secretions yielded microbes from Porphyromonas, Staphylococcus, Streptococcus, and Cutibacterium species [31]. Other studies confirmed distinct urinary microbiota in PCa compared with controls, even in the absence of tissue-level differences [32].
Liss et al. profiled rectal swabs from 105 patients (64 PCa, 41 non-PCa) using 16S rRNA V1–V2 sequencing. Distinct microbial signatures were found between cancer and non-cancer groups despite similar diversity. The enrichment of Bacteroides and Streptococcus in PCa patients suggested metabolic reprogramming favoring carbohydrate metabolism and reduced B-vitamin synthesis [33].
A comprehensive systematic review of 16 studies (n = 1486, including 9 PCa-focused studies) reported considerable heterogeneity in urinary microbiota across prostatic diseases. Despite this variability, certain bacterial taxa were consistently associated with PCa. For instance, increased abundances of specific phyla, genera, and species were observed in PCa patients compared to controls. Moreover, some bacterial species were linked to higher-grade disease, suggesting a potential role in PCa progression [34]. Wang et al. also showed that bacterial strains from the urine produced androgens and were able to promote prostate cancer cell growth via cortisol and prednisone metabolism [25]. Collectively, these findings underscore the intricate relationship between urinary microbiota and prostatic diseases, highlighting the need for further research to elucidate the potential of microbiome-related biomarkers in PCa diagnosis and prognosis.
4. Intratumoral and Intraprostatic Microbiome: Emerging Roles in Prostate Cancer Pathogenesis
Intratumoral microbial communities are increasingly recognized as key modulators of cancer initiation, progression, and therapeutic response. These microbes interact with host genomic stability, induce epigenetic alterations, and shape inflammation responses. Notably, intratumoral microbiota may exert dual effects: enhancing antitumor immunity or, conversely, promoting tumor progression through mechanisms such as reactive oxygen species (ROS) induction, T-cell exhaustion, and the creation of immunosuppressive microenvironments. Utilizing large-scale RNA-sequencing data integrated with clinical parameters, Ma et al. identified specific intratumoral bacteria correlating with immune pathways, PCa risk factors, and tumor aggressiveness. Bacteria such as Pediococcus pentosaceus, Listeria monocytogenes, Lactobacillus crispatus ST1, and Bacillus halodurans negatively correlated with Gleason scores, suggesting anti-tumor roles, whereas Nevskia ramosa positively correlated [35]. The former three have been associated with anti-tumor effects in various tumor models, while Listeria monocytogenes has been implicated in the activation of innate and adaptive immunity and with the release of pro-inflammatory cytokines [35,36]. Bacillus halodurans and Nevskia ramosa have not been previously associated with cancer. Rhodococcus erythropolis PR4, Delftia acidovorans SPH-1, Methylobacterium radiotolerans JCM 2831, Stenotrophomonas maltophilia K279a, and Meiothermus silvanus DSM 9946 demonstrated a negative correlation with TNM staging, while no bacteria correlated positively. Interestingly, Rhodococcus erythropolis PR4, Stenotrophomonas maltophilia K279a, and Meiothermus silvanus DSM 9946 are often observed in immunosuppressed patients with in-dwelling catheters. Overall, 234 microbes were associated with increased PSA levels, while the most strongly correlated bacteria were Campylobacter concisus UNSWCD, Thermus thermophilus HB27, and Streptococcus pneumoniae SPN032672. Thermus thermophilus produces L-asparaginase, which can pose anti-tumor effects in several cancer types. Streptococcus pneumoniae has been associated with an increased risk of lower esophageal adenocarcinoma risk, while Campylobacter concisus is pro-inflammatory in the esophagus. The most strongly negatively correlated microbes included Xanthomonas albilineans GPE PC73, Herminiimonas arsenicoxydans, and Pseudarthrobacter chlorophenolicus A6. Gardnerella vaginalis 409-05, Nitrobacter hamburgensis X14, and Delftia acidovorans SPH-1 were the most significant microbes positively correlated with the numbers of dysregulated immune-associated genes. Moreover, these bacteria were associated with downregulated genes that control immune system activation, suggesting that they are likely to promote PCa by suppressing immune cell expression, rather than by promoting inflammation. PCa tissue microbe abundance was associated with regulatory T-cell expression. Bradyrhizobium elkanii, Ochrobactrum anthropi ATCC 49188, and Bradyrhizobium japonicum demonstrated the strongest negative correlation with androgen receptor expression, while Escherichia coli ETEC H10407 and Escherichia coli str. K-12 substr. MG1655 showed the strongest positive correlation. The latter bacteria are frequently associated with prostatitis. Staphylococcus aureus subspecies MW2, Paraburkholderia phymatum STM815, Haemophilus parainfluenzae T3T1, and Pseudomonas putida F1 were the most strongly associated bacteria with stem cell gene expression in the PCa samples [35,36].
Feng et al. analyzed microbial content within PCa tissue from 22 men using host-derived whole-genome sequencing (WGS). Viral or other nonbacterial read counts were negligible. Dominant genera included Escherichia, Propionibacterium, and Pseudomonas. Escherichia spp. have been shown to promote PCa cell growth in vitro, while other studies have confirmed that Propionibacterium acne is more commonly found in the prostate tissue of patients with PCa compared to patients without PCa. No associations were found between microbial taxa and clinical presentation (low- vs. high-risk disease). Samples from African patients contained a higher abundance of bacteria, especially anaerobic, compared to Australian and Chinese cohorts. However, the authors noted that transrectal biopsy sampling in African patients cannot rule out the possibility of fecal contamination. While half of the core gut microbial genera were absent, Acidovorax and Escherichia species were significantly abundant. Total bacterial burden and Eubacterium species abundance were correlated with prostate tumor host hypermutation. The authors concluded that this correlation may potentially explain (at least partially) the aggressiveness of disease in African men [37].
Additional studies using genomic and 16S rDNA sequencing reported widespread microbial presence in 170 prostate tissue specimens from 30 patients undergoing radical prostatectomy. Validation with organism-specific PCR in 200 PCa patients confirmed microbial DNA in 87% of individuals, although only 37% of individual tissue cores were positive. On average, 4.5 microbial sequences were detected per patient (range 0–14). The most common sequences were identified as members of Gammaproteobacteria, Alphaproteobacteria, CFB group bacteria, Gram+ Low GC Content bacteria, Betaproteobacteria, and Gram+ High GC Content bacteria. The most frequently identified sequences in multiple patients were similar to Acinetobacter, Escherichia, Pseudomonas, Methylophilus, and Streptococcus spp. Overall, organism-specific PCR failed to detect several microorganisms previously considered to be common in the prostate, and no specific microbe sp. was associated with evidence of acute or chronic inflammation. These results suggest the presence of regional heterogeneity with respect to bacteria, and the absence of a ubiquitous “landmark” microbiome in the prostatic flora. The bacterial culture results from prostatic samples were not on par with the results from 16S rDNA PCR, suggesting that either 16S rDNA PCR samples were derived from non-viable bacteria or that most bacteria inside the prostate were unculturable [38]. Keay et al. analyzed 18 transperineal biopsy specimens from nine PCa patients using 16S rRNA PCR. Bacterial DNA was detected in 11/18 samples (8/9 patients). A single or dominant organism was identified in most cases, while some contained multiple taxa. Sequence comparisons with GenBank indicated that the predominant organisms were mainly Escherichia and Bacteroides [39].
Krieger et al. obtained prostate biopsies from 107 PCa patients and 170 patients with chronic prostatitis/pelvic pain syndrome, as well as numerous controls. Bacterial DNA sequences were detected in 19.6% of patients with PCa compared to 46.4% of those with chronic prostatitis [40]. Similarly, Hochreiter et al. analyzed 14 samples from 7 patients after radical prostatectomy and several samples from normal controls. The results ruled out the existence of a normal flora inside the prostate. They reported that the presence of bacteria and/or inflammation were localized and heterogeneous events in patients with PCa and other inflammatory conditions. The presence of inflammation was strongly associated with positive 16S rRNA-PCR results. Although methodological differences (e.g., primer selection) may partly explain discrepancies, these studies support the hypothesis that prostatic bacteria contribute to chronic inflammatory conditions and may play a role in carcinogenesis [41].
Alexeyev et al. assessed archival prostate samples from 325 patients with benign prostatic hyperplasia for bacterial 16S RNA by implementing bulk Sanger sequencing (implemented with BigDye™ Terminator Cycle Sequensing kit 1.1 (Applied biosystems, Foster City, CA, USA)) and evaluated whether it differed among patients who later developed PCa (n = 171) and those who did not (n = 181). Overall, they detected bacterial 16S RNA in 96/352 specimens. The most frequently identified microorganism was Propionibacterium acnes, which was detected in 23% of 16S RNA-positive patients. The presence of P. acnes was also associated with severe histological inflammation and the future development of PCa (OR 2.17, 95% CI 0.77–6.95). The second most common microorganism was Escherichia coli, which was found in 12 (12%) patients. Other isolates included Pseudomonas and Actinomyces species, Streptococcus mutans, as well as Corynebacterium, Nocardioides, Rhodococcus, and Veillonella species [42]. In another study, a microbiome profiling of tumor, peri-tumor, and non-tumor tissues from 16 radical prostatectomy specimens using ultra-deep pyrosequencing (V3–V5 region) revealed diverse bacterial communities. Several phyla, classes, and genera exceeded the 1% threshold, confirming a rich intraprostatic microbiota. Propionibacterium dominated overall, Staphylococcus was most abundant in tumor and peri-tumor tissues, and Enterococcus was almost exclusively found in non-tumor areas [43].
Davidsson et al. showed that P. acnes significantly increased PCa risk, supported by in vitro data demonstrating enhanced inflammation and cellular proliferation following bacterial co-culture [44]. In contrast, Yow et al. identified consistent microbial communities dominated by Enterobacteriaceae and Escherichia within high-grade tumors, though without evidence of active infection, highlighting a complex microbiota–host interplay [45].
The frequency in which P. acnes is isolated in prostate tissue samples from patients with PCa compared to patients without PCa was researched using both culture diagnostics and molecular techniques. A total of 100 cases and 50 controls were included in Davidsson’s S. et al. study [44]. P. acnes was cultured in roughly 60% of patients with PCa compared to 26% of the controls. Multivariate analysis revealed that the presence of P. acnes was associated with a four-fold increase in the odds of a diagnosis of PCa after adjusting for confounding factors such as age, smoking status, or calendar year of surgery. The researchers also conducted in vitro experiments in which they co-cultured P. acnes isolates with the PNT1A PCa cell line, and they reported increased cytokine/chemokine secretion and increased proliferation in infected cells [44]. Also, Chen et al. [46] used three RNA-seq sets in the Illumina (NGS) system to identify P. acnes in cancer samples. They underlined the fact that it did not detect any human–bacteria or human–virus fusion in any data set that may suggest that P. acnes species do not pose severe risks for the development of prostate cancer.
Yow et al. also applied 16S rRNA sequencing (V2–V3 and V4) to 20 high-grade tumor cores. The researchers identified a plethora of operational taxonomic units (OTUs) in every sample (mean number: 231.55, range: from 151 to 314) using 16S rRNA V4 hypervariable region. The family of Enterobacteriaceae was the most abundant taxa (70.1%), followed by the genus Escherichia (6.9%). A small proportion of the overall membership of the prostatic microbial community (18 OTUs) was present in 95% of samples and contributed to 84.6% of the relative abundance of the total communities. Enterobacteriacae and Escherichia abundance was consistent across samples. Analysis with 16S rRNA V2–V3 hypervariable region identified 117.95 OTUs (range: from 71 to 160) per sample. Enterobacteriaceae and Escherichia sequences were represented in every sample. Enterobacteriaceae, Streptococcaceae, Staphylococcus, Moraxella, Escherichia, P. acnes, and Streptococcus pseudopneumoniae were represented in 95% of the samples and constituted the core community within the samples. Total RNA sequencing detected human endogenous retroviral sequences, but there were no viral or bacterial transcripts; hence, there was no evidence for active infection [45].
A combined metagenomic–metatranscriptomic study of tumor and matched benign tissues from 65 patients identified Escherichia, Propionibacterium, Acinetobacter, and Pseudomonas as the core prostate microbiome. Microbial diversity did not differ between tumors and benign samples or across Gleason categories. Few viral sequences were detected, but Pseudomonas species were inversely associated with metastasis [47].
Alluri et al. [48] investigated periodontal pathogens in 90 prostate tissue samples from 30 men. Using real-time PCR, they concluded that Fusobacterium nucleatum was the only pathogen that showed a significant difference in the prostates that harbored cancer, chronic inflammation, and benign prostatic hyperplasia. A total of 50 prostate adenocarcinoma samples were also used in Banerjee’s et al. [49] research to define the microbiome (viral, bacterial, fungal, and parasitic) signatures associated with prostate cancer. Using PathoChip technology, a technology which combines both PCR (Poxviridae, Reoviridae, Papillomaviridae, Herpesviridaeand) and next-generation sequencing (NGS), they found a large number of bacteria (Rickettsia, Mycobacterium, Bordetella, Mycoplasma, Sphingomonas etc.), viruses, fungi (Alternaria, Malassezia, Candida, Cladosporium, Trichosporon, Cladophialophora etc.) and parasites (Plasmodium, Trichinella, Sarcocystis, Babesia, Entamoeba) that could be identified as diverse microbiome signatures associated with prostate cancer.
Finally, Miyake et al. screened 45 PCa tissue samples for sexually transmitted pathogens, detecting Mycoplasma genitalium predominantly in younger patients, though without correlation to inflammation severity [50]. Salachan et al. used whole-transcriptome sequencing in 106 PCa tissue samples, reporting associations of Shewanella, Vibrio parahaemolyticus, and Microbacterium spp. with tumor progression [51]. Similarly, Sarkar et al. compared PCa and benign prostatic hyperplasia (BPH) samples via 16S rRNA sequencing, finding Cupriavidus taiwanensis and Methylobacterium organophilum enriched in PCa, whereas Kocuria palustris and Cellvibrio mixtus were more abundant in BPH [52].
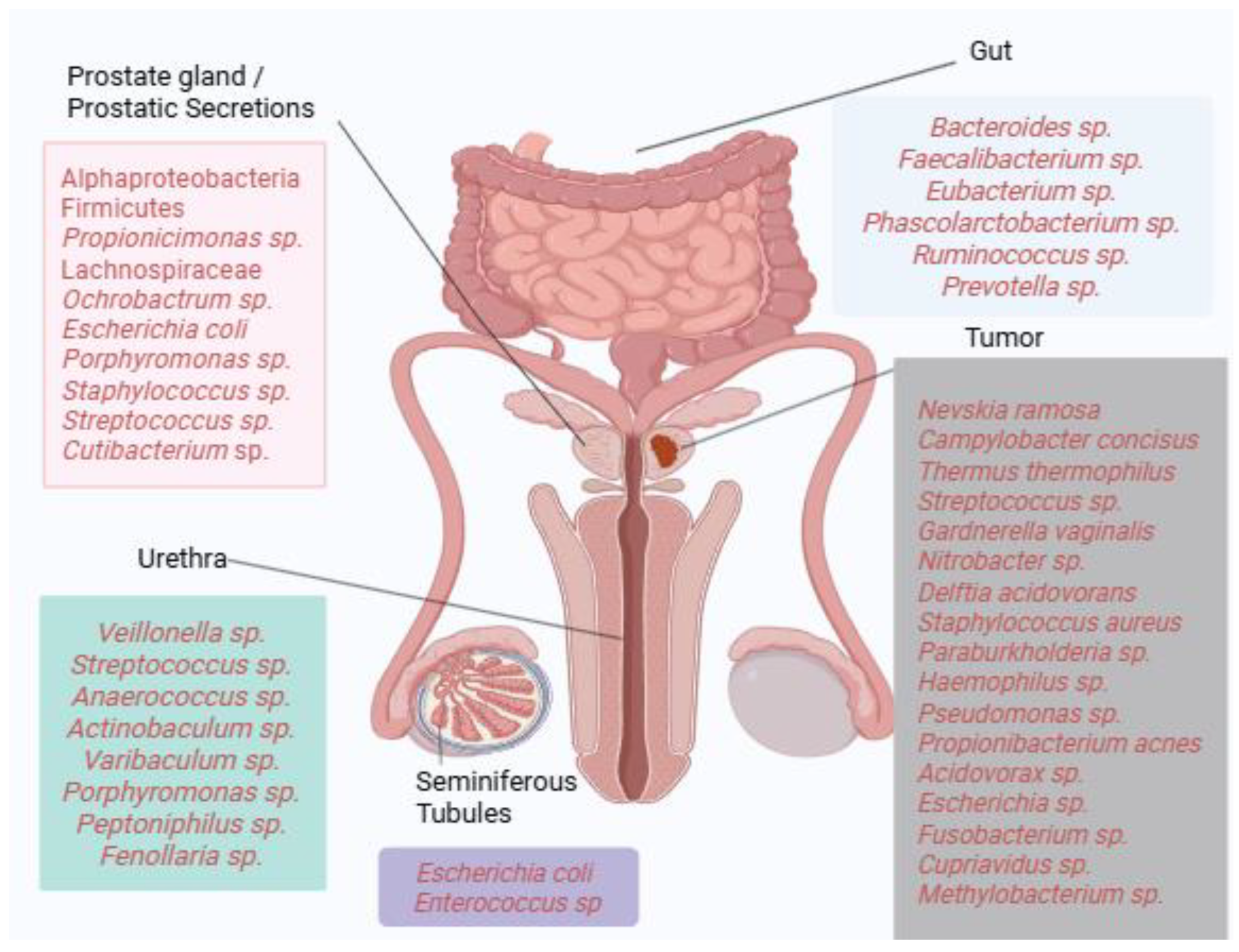
Collectively, these findings illustrate the substantial heterogeneity of microbial communities detected across prostate tissues, with recurrent enrichment of taxa such as Propionibacterium acnes, Escherichia, and Pseudomonas in multiple studies. However, results vary considerably across methodologies, patient cohorts, and disease stages, highlighting the absence of a universally defined “core” prostatic microbiome. Figure 1 provides a visual summary of dominant bacterial taxa identified across different anatomical compartments, including gut, urine, seminal fluid, prostatic secretions, and tumor tissue. Where available, reported associations with clinical parameters such as Gleason score, PSA levels, TNM staging, or castration resistance are indicated in the main text. This schematic underscores both the diversity and complexity of microbial signatures linked to prostate carcinogenesis.
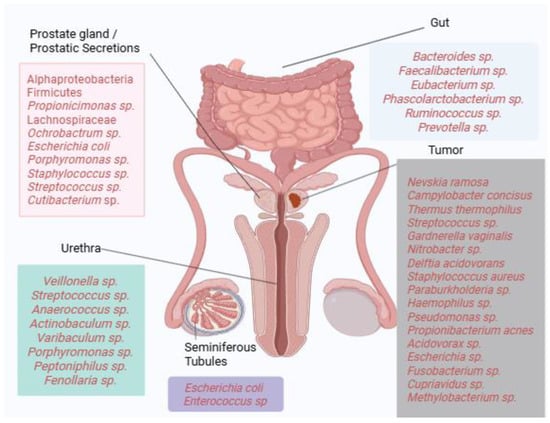
Figure 1.
Microbial landscape of prostate cancer. Dominant bacterial taxa identified across gut, urinary, seminal, and prostatic tissue compartments are depicted. Where available, associations with clinical parameters such as Gleason score, TNM staging, PSA levels, or castration resistance are indicated in the main text. The figure highlights taxa recurrently reported across multiple studies while acknowledging the heterogeneity of findings (Created in https://BioRender.com).
5. Mycobiome’s Role: Emerging Insights into Prostate Cancer
Emerging evidence indicates that fungal communities may also contribute to tumor biology. Certain fungal taxa are enriched in malignant tissues, suggesting potential roles in shaping the tumor microenvironment. The interaction between the mycobiome, bacterial microbiota, and host physiology opens novel avenues for cancer diagnostics and therapeutics [53]. In PCa, Wang et al. conducted a cross-sectional study of circulating plasma fungal microbiomes in PCa patients (n = 31) compared with age- and race-matched healthy controls (n = 34). Using the MR DNA (Shallowater, TX, USA) platform, the study revealed a pronounced enrichment of the fungal families Filobasidiales and Pyronemataceae, alongside the species Cryptococcus ater, exclusively in PCa patients. By contrast, diverse fungal classes and species were prominently elevated within the plasma microbiomes of the control group. Notably, a higher abundance of the genus Bipolaris was associated with lower PSA levels, while the increased representation of the class Sordariomycetes correlated with advanced pathological stages. These findings suggest that fungal signatures may hold diagnostic and prognostic value in Pca [53].
6. Virome
The potential role of the virome in prostate cancer has also been extensively investigated, although the overall conclusions so far remain inconclusive [54]. Several viruses have been detected in prostate tissue, including human papillomavirus (HPV), Epstein–Barr virus (EBV), herpes simplex virus type 2 (HSV-2), human herpesvirus 8 (HHV-8), cytomegalovirus (CMV), BK polyomavirus (BKV), and xenotropic murine leukemia virus-related virus (XMRV) [55,56,57,58,59,60,61]. Limitations related to detection methods, sample sizes, or study design variability lead to uncertainty regarding the consistency of these associations or the true frequency of infection. The HPV virus is of particular interest not only due to its proven tumorigenic role in other malignancies (cervix, vulva, anus, vagina, uterus, pelvis, head and neck) and its proximity to other urinary and anogenital sites, but also following preclinical evidence showing that viral proteins such as HPV E6/E7 have oncogenic potential by interfering with tumor suppressors like p53 and pRb [62,63]. Some studies have shown a statistical association of prostate cancer with sexually transmitted diseases, although a recent meta-analysis did not support this association [64]. High-risk HPV subtypes have been associated with prostate cancer in studies from Asian and some European populations (e.g., Greece, UK), but global results remain heterogeneous and non-confirmatory [11,54,55,56]. Moreover, XMRV has been isolated from prostate cancer tissue samples, but some evidence showed that the virus was formed in the laboratory and does not circulate in humans [65]. Although many viruses can interact with host proteins and result in genetic changes or immunological and inflammatory events that can favor the development or progression of tumors, it is believed that host genetic variations likely also play a role. For example, RNASEL R462Q polymorphism (that can lead to defective immunity and viral persistence) has been studied as a potential modulator of virome impact on prostate tissue [66,67]. Hence, the interplay between viral and host factors might exert pro-tumorigenic activity in prostate cancer, although more research needs to confirm this.
7. Discussion
Advances in sequencing technologies have fundamentally reshaped our understanding of microbial colonization, revealing diverse microbiome communities even in anatomical sites once considered sterile. These discoveries have spurred renewed interest in the role of gut, urinary, and tumor-associated microbiomes in PCa [7,10]. Although the studies reviewed here exhibit marked methodological and biological heterogeneity, along with some conflicting results, several consistent observations can be highlighted.
First, the increased abundance of Bacteroides spp. in the gut microbiome has been repeatedly associated with malignancy, while benign conditions were more often linked to beneficial genera such as Faecalibacterium and Eubacterium [21,22]. Although gut microbial composition has not generally correlated with Gleason score, some taxa have been associated with resistance to androgen-deprivation therapy. These findings, however, are limited by small sample sizes and should be interpreted cautiously [24].
Urinary microbiota have also been implicated in prostate carcinogenesis. Commonly reported genera include Bacteroides, Firmicutes, Varibaculum, and Streptococcus. The role of Propionibacterium acnes remains controversial, with inconsistent associations across different cohorts [28,30,34]. The frequent detection of identical bacterial species in both prostatic secretions and seminal fluid as well as urine samples suggests potential biological interplay among these compartments [28,29]. Despite variability between studies, Escherichia, Propionibacterium, Pseudomonas, and Acinetobacter are among the taxa most frequently identified in prostate tissue and peri-tumoral regions [35,36]. While Enterobacteriaceae are also commonly reported, some evidence suggests they may form part of the normal prostate microbiota and could exert protective or even antitumor effects [45].
The inconsistencies across studies highlight the complexity of microbial involvement in PCa and suggest that no single microorganism is solely responsible for tumor initiation or progression [68]. A “hit-and-run” model, in which pathogens contribute to early carcinogenesis but are no longer detectable in advanced stages, remains a plausible hypothesis [68,69,70].
Importantly, methodological heterogeneity likely explains many of the inconsistencies observed across studies. Differences in sequencing platforms, amplified regions, and bioinformatic pipelines can profoundly affect microbiome profiling and taxonomic resolution. As summarized in Table 1, most studies relied on amplicon-based 16S rRNA sequencing (e.g., Illumina MiSeq/HiSeq platforms targeting V3–V4, V3–V5, or V6 regions), which provides genus- or species-level resolution but limited functional insight. In contrast, a smaller number of studies employed whole-genome or whole-transcriptome sequencing, enabling a higher resolution of microbial taxa and functional pathways, albeit at greater cost and complexity. Table 2 further illustrates the variability in PCR-based methods, with diverse primer sets targeting different microbial taxa, including bacteria, viruses, and fungi. Such differences not only influence the detection of specific organisms (e.g., Propionibacterium acnes, Escherichia coli, or viral sequences) but also limit comparability across studies.

Table 1.
Overview of next-generation sequencing (NGS) platforms used in prostate cancer microbiome studies.

Table 2.
Overview of PCR-based methodologies applied in prostate cancer microbiome studies.
Taken together, these observations highlight the urgent need for standardized methodologies in prostate cancer microbiome research. The harmonization of sequencing approaches, target regions, and analytical pipelines will be essential to reduce variability and enable reproducibility. Only through methodological standardization and integration of multi-omic platforms (16S, shotgun metagenomics, metatranscriptomics, and targeted PCR) can reliable microbial biomarkers be identified and validated for clinical application in prostate cancer.
8. Conclusions
Microbiome research has considerably expanded our understanding of the potential role of microbiomes in prostate cancer (PCa) pathogenesis, progression, and treatment response. Across gut, urinary, seminal, and prostatic compartments, recurrent associations have been identified with taxa such as Bacteroides, Escherichia, and Propionibacterium acnes. However, significant heterogeneity persists, reflecting differences in patient cohorts, sampling methods, and analytical platforms. Importantly, no single microorganism has been established as a universal hallmark of PCa, and causality remains unproven.
Nevertheless, accumulating evidence suggests that microbial signatures may influence carcinogenesis through chronic inflammation, the modulation of local immune responses, and metabolic interactions, with implications for resistance to androgen-deprivation therapy and responsiveness to immunotherapy. This highlights the potential utility of microbiome profiles as biomarkers for diagnosis, risk stratification, and therapeutic guidance.
To translate these insights into clinical practice, future research should prioritize methodological standardization across sequencing platforms, genomic regions, and bioinformatic pipelines. Integration of multi-omic approaches—including 16S rRNA sequencing, shotgun metagenomics, metatranscriptomics, and targeted qPCR—will be essential to improve resolution, reproducibility, and functional interpretation. Large, prospective, and ethnically diverse cohorts will be required to validate candidate microbial biomarkers and clarify their mechanistic roles in PCa.
Ultimately, a standardized, systems-level approach to the prostate cancer microbiome could yield robust biomarkers for early detection and prognosis while paving the way for microbiome-targeted interventions as adjuncts to existing therapies. Such strategies hold promise for improving patient stratification, optimizing treatment response, and advancing the personalized management of prostate cancer.
Author Contributions
M.S., P.R. and A.B. conceptualized the study. M.S., P.R., E.P., K.A.P., I.B., K.G., F.A.-A., D.C. and A.B. made a substantial contribution to data interpretation and analysis and wrote, prepared, and revised the draft of the manuscript. M.S., P.R., I.B., F.A.-A., D.C. and A.B. analyzed the data and provided critical revisions. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable since no new data were created.
Acknowledgments
P.R. and K.A.P. were granted a PhD Fellowship by the Special Account for Research Grants (ELKE) of the Research Committee of the University of West Attica (UNIWA).
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| BPH | Benign prostatic hyperplasia |
| CI | Confidence interval |
| CRPC | Castration-resistant prostate cancer |
| EPS | Expressed prostatic secretion |
| FMT | Fecal microbiota transplantation |
| NGS | Next-generation sequencing |
| OTU | Operational taxonomic unit |
| PCR | Polymerase chain reaction |
| PSA | Prostate-specific antigen |
| TNM | Tumor–node–metastasis classification |
References
- Naghavi, M.; Ong, K.; Aali, A.; Ababneh, H.; Abate, Y.; Abbafati, C.; Abbasgholizadeh, R.; Abbasian, M.; Abbasi-Kangevari, M.; Abbastabar, H.; et al. Global burden of 288 causes of death and life expectancy decomposition in 204 countries and territories and 811 subnational locations, 1990–2021: A systematic analysis for the Global Burden of Disease Study 2021. Lancet 2024, 403, 2100–2132. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, A.J.; Santomauro, D.; Aali, A.; Abate, Y.; Abbafati, C.; Abbastabar, H.; Abd ElHafeez, S.; Abdelmasseh, M.; Abd-Elsalam, S.; Abdollahi, A.; et al. Global incidence, prevalence, years lived with disability (YLDs), disability-adjusted life-years (DALYs), and healthy life expectancy (HALE) for 371 diseases and injuries in 204 countries and territories and 811 subnational locations, 1990–2021: A systematic analysis for the Global Burden of Disease Study 2021. Lancet 2024, 403, 2133–2161. [Google Scholar] [CrossRef]
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Sakellakis, M.; Flores, L.; Ramachandran, S. Patterns of indolence in prostate cancer. Exp. Ther. Med. 2022, 23, 351. [Google Scholar] [CrossRef]
- Hamdy, F.C.; Donovan, J.; Lane, J.; Mason, M.; Metcalfe, C.; Holding, P.; Davis, M.; Peters, T.; Turner, E.; Martin, R.; et al. 10-Year Outcomes after Monitoring, Surgery, or Radiotherapy for Localized Prostate Cancer. N. Engl. J. Med. 2016, 375, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.J.; Brubaker, L. Sterile Urine and the Presence of Bacteria. Eur. Urol. 2015, 68, 173–174. [Google Scholar] [CrossRef]
- Meares, E.M., Jr. Bacterial prostatitis vs ‘prostatosis’. A clinical and bacteriological study. JAMA 1973, 224, 1372–1375. [Google Scholar] [CrossRef]
- Yan, L.; Chen, Y.; Chen, F.; Tao, T.; Hu, Z.; Wang, J.; You, J.; Wong, B.; Chen, J.; Ye, W. Effect of Helicobacter pylori Eradication on Gastric Cancer Prevention: Updated Report from a Randomized Controlled Trial With 26.5 Years of Follow-up. Gastroenterology 2022, 163, 154–162.e3. [Google Scholar] [CrossRef]
- Boumaza, A.; Azzouz, E.B.; Arrindell, J.; Lepidi, H.; Mezouar, S.; Desnues, B. Whipple’s disease and Tropheryma whipplei infections: From bench to bedside. Lancet Infect. Dis. 2022, 22, e280–e291. [Google Scholar] [CrossRef]
- Schiffman, M.; Doorbar, J.; Wentzensen, N.; De Sanjosé, S.; Fakhry, C.; Monk, B.; Stanley, M.; Franceschi, S. Carcinogenic human papillomavirus infection. Nat. Rev. Dis. Primers 2016, 2, 16086. [Google Scholar] [CrossRef]
- Lin, Y.; Mao, Q.; Zheng, X.; Yang, K.; Chen, H.; Zhou, C.; Xie, L. Human papillomavirus 16 or 18 infection and prostate cancer risk: A meta-analysis. Ir. J. Med. Sci. 2011, 180, 497–503. [Google Scholar] [CrossRef]
- Deng, R.; Zheng, H.; Cai, H.; Li, M.; Shi, Y.; Ding, S. Effects of helicobacter pylori on tumor microenvironment and immunotherapy responses. Front. Immunol. 2022, 13, 923477. [Google Scholar] [CrossRef] [PubMed]
- Sepich-Poore, G.D.; Zitvogel, L.; Straussman, R.; Hasty, J.; Wargo, J.A.; Knight, R. The microbiome and human cancer. Science 2021, 371, eabc4552. [Google Scholar] [CrossRef]
- Chen, G.; Ren, Q.; Zhong, Z.; Li, Q.; Huang, Z.; Zhang, C.; Yuan, H.; Feng, Z.; Chen, B.; Wang, N.; et al. Exploring the gut microbiome’s role in colorectal cancer: Diagnostic and prognostic implications. Front. Immunol. 2024, 15, 1431747. [Google Scholar] [CrossRef] [PubMed]
- Ağagündüz, D.; Cocozza, E.; Cemali, Ö.; Bayazıt, A.; Nanì, M.; Cerqua, I.; Morgillo, F.; Saygılı, S.; Berni, C.R.; Amero, P.; et al. Understanding the role of the gut microbiome in gastrointestinal cancer: A review. Front. Pharmacol. 2023, 14, 1130562. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.; Huang, G.; Wang, D.; Xu, Y.; Qiu, J.; Pei, B.; Qian, D.; Meng, X. Gut microbiome causal impacts on the prognosis of breast cancer: A Mendelian randomization study. BMC Genom. 2023, 24, 497. [Google Scholar] [CrossRef]
- Wiertsema, S.P.; van Bergenhenegouwen, J.; Garssen, J.; Knippels, L.M.J. The interplay between the gut microbiome and the immune system in the context of infectious diseases throughout life and the role of nutrition in optimizing treatment strategies. Nutrients 2021, 13, 886. [Google Scholar] [CrossRef]
- Dizman, N.; Meza, L.; Bergerot, P.; Alcantara, M.; Dorff, T.; Lyou, Y.; Frankel, P.; Cui, Y.; Mira, V.; Llamas, M.; et al. Nivolumab plus ipilimumab with or without live bacterial supplementation in metastatic renal cell carcinoma: A randomized phase 1 trial. Nat. Med. 2022, 28, 704–712. [Google Scholar] [CrossRef]
- Wang, I.; Song, L.; Wang, B.Y.; Kalebasty, A.R.; Uchio, E.; Zi, X. Prostate Cancer Immunotherapy: A Review of Recent Advancements with Novel Treatment Methods and Efficacy. Am. J. Clin. Exp. Urol. 2022, 10, 210–233. Available online: www.ajceu.us/ (accessed on 15 August 2022).
- Javier-DesLoges, J.; McKay, R.R.; Swafford, A.D.; Sepich-Poore, G.D.; Knight, R.; Parsons, J.K. The microbiome and prostate cancer. Prostate Cancer Prostatic Dis. 2022, 25, 159–164. [Google Scholar] [CrossRef]
- Golombos, D.A.; Ayangbesan, A.; O’Malley, P.; Lewicki, P.; Barlow, L.; Barbieri, C.; Chan, C.; DuLong, C.; Abu-Ali, G.; Huttenhower, C.; et al. The Role of Gut Microbiome in the Pathogenesis of Prostate Cancer: A Prospective, Pilot Study. Urology 2018, 111, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Parsaei, M.; Sarafraz, N.; Moaddab, S.Y.; Leylabadlo, H.E. The importance of Faecalibacterium prausnitzii in human health and diseases. New Microbes New Infect. 2021, 43, 100928. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Xu, X.; Fu, D.; Gu, Y.; Fan, R.; Yi, H.; He, X.; Wang, C.; Ouyang, B.; Zhao, P.; et al. Butyrate-producing Eubacterium rectale suppresses lymphomagenesis by alleviating the TNF-induced TLR4/MyD88/NF-κB axis. Cell Host Microbe 2022, 30, 1139–1150.e7. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jiang, H. Compositional differences of gut microbiome in matched hormone-sensitive and castration-resistant prostate cancer. Transl. Androl. Urol. 2020, 9, 1937–1944. [Google Scholar] [CrossRef]
- Wang, T.; Ahmad, S.; Cruz-Lebrón, A.; Ernst, S.E.; Olivos Caicedo, K.Y.; Jeong, Y.; Binion, B.; Mbuvi, P.; Dutta, D.; Francelys, V.; et al. An expanded metabolic pathway for androgen production by commensal bacteria. Nat. Res. 2025, 10, 1084–1098. [Google Scholar] [CrossRef]
- Pernigoni, N.; Zagato, E.; Calcinotto, A.; Troiani, M.; Mestre, R.P.; Calì, B.; Attanasio, G.; Troisi, J.; Minini, M.; Mosole, S.; et al. Commensal bacteria promote endocrine resistance in prostate cancer through androgen biosynthesis. Science 2021, 374, 216–224. [Google Scholar] [CrossRef]
- Alanee, S.; El-Zawahry, A.; Dynda, D.; Dabaja, A.; McVary, K.; Karr, M.; Braundmeier-Fleming, A. A prospective study to examine the association of the urinary and fecal microbiota with prostate cancer diagnosis after transrectal biopsy of the prostate using 16sRNA gene analysis. Prostate 2019, 79, 81–87. [Google Scholar] [CrossRef]
- Yu, H.; Meng, H.; Zhou, F.; Ni, X.; Shen, S.; Das, U.N. Urinary microbiota in patients with prostate cancer and benign prostatic hyperplasia. Arch. Med. Sci. 2015, 11, 385–394. [Google Scholar] [CrossRef]
- Vacca, M.; Celano, G.; Calabrese, F.M.; Portincasa, P.; Gobbetti, M.; De Angelis, M. The controversial role of human gut lachnospiraceae. Microorganisms 2020, 8, 573. [Google Scholar] [CrossRef]
- Shrestha, E.; White, J.; Yu, S.; Kulac, I.; Ertunc, O.; De Marzo, A.; Yegnasubramanian, S.; Mangold, L.; Partin, A.; Sfanos, K. Profiling the Urinary Microbiome in Men with Positive versus Negative Biopsies for Prostate Cancer. J. Urol. 2018, 199, 161–171. [Google Scholar] [CrossRef]
- Hurst, R.; Meader, E.; Gihawi, A.; Rallapalli, G.; Clark, J.; Kay, G.; Webb, M.; Manley, K.; Curley, H.; Walker, H.; et al. Microbiomes of Urine and the Prostate Are Linked to Human Prostate Cancer Risk Groups. Eur. Urol. Oncol. 2022, 5, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, M.; Pina-Vaz, T.; Fernandes, Â.; Miranda, I.; Silva, C.; Rodrigues, A.; Lisboa, C. Microbiota of Urine, Glans and Prostate Biopsies in Patients with Prostate Cancer Reveals a Dysbiosis in the Genitourinary System. Cancers 2023, 15, 1423. [Google Scholar] [CrossRef]
- Liss, M.; White, J.; Goros, M.; Gelfond, J.; Leach, R.; Johnson-Pais, T.; Lai, Z.; Rourke, E.; Basler, J.; Ankerst, D.; et al. Metabolic Biosynthesis Pathways Identified from Fecal Microbiome Associated with Prostate Cancer. Eur. Urol. Oncol. 2022, 5, 412–419. [Google Scholar] [CrossRef]
- Mjaess, G.; Karam, A.; Roumeguère, T.; Diamand, R.; Aoun, F.; McVary, K.; Moul, J.W.; De Nunzio, C.; Albisinni, S. Urinary microbiota and prostatic diseases: The key for the lock? A systematic review. Prostate Cancer Prostatic Dis. 2023, 26, 451–460. [Google Scholar] [CrossRef]
- Ma, J.; Gnanasekar, A.; Lee, A.; Li, W.; Haas, M.; Wang-Rodriguez, J.; Chang, E.; Rajasekaran, M.; Ongkeko, W. Influence of intratumor microbiome on clinical outcome and immune processes in prostate cancer. Cancers 2020, 12, 2524. [Google Scholar] [CrossRef]
- Wood, L.M.; Guirnalda, P.D.; Seavey, M.M.; Paterson, Y. Cancer immunotherapy using Listeria monocytogenes and listerial virulence factors. Immunol. Res. 2008, 42, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Jaratlerdsiri, W.; Patrick, S.; Lyons, R.; Haynes, A.; Collins, C.; Stricker, P.; Bornman, M.; Hayes, V. Metagenomic analysis reveals a rich bacterial content in high-risk prostate tumors from African men. Prostate 2019, 79, 1731–1738. [Google Scholar] [CrossRef] [PubMed]
- Sfanos, K.S.; Sauvageot, J.; Fedor, H.L.; Dick, J.D.; De Marzo, A.M.; Isaacs, W.B. A molecular analysis of prokaryotic and viral DNA sequences in prostate tissue from patients with prostate cancer indicates the presence of multiple and diverse microorganisms. Prostate 2008, 68, 306–320. [Google Scholar] [CrossRef] [PubMed]
- Keay, S.; Zhang, C.-O.; Baldwin, B.R.; Alexander, R.B. Polymerase Chain Reaction Amplification of Bacterial 16s rRNA Genes in Prostate Biopsies from Men Without Chronic Prostatitis. Urology 1999, 53, 487–491. [Google Scholar] [CrossRef]
- Krieger, J.N.; Riley, D.E.; Vesella, R.L.; Miner, D.C.; Ross, S.O.; Lange, P.H. Bacterial DNA Sequences in Prostate Tissue from Patients with Prostate Cancer and Chronic Prostatitis. J. Urol. 2000, 164, 1221–1228. [Google Scholar] [CrossRef]
- Hochreiter, W.W.; Duncan, J.L.; Schaeffer, A.J. Evaluation of the Bacterial Flora of the Prostate Using a 16S rRNA Gene Based Polymerase Chain Reaction. J. Urol. 2000, 163, 127–130. [Google Scholar] [CrossRef]
- Alexeyev, O.; Bergh, J.; Marklund, I.; Thellenberg-Karlsson, C.; Wiklund, F.; Grönberg, H.; Bergh, A.; Elgh, F. Association between the presence of bacterial 16S RNA in prostate specimens taken during transurethral resection of prostate and subsequent risk of prostate cancer (Sweden). Cancer Causes Control 2006, 17, 1127–1133. [Google Scholar] [CrossRef] [PubMed]
- Cavarretta, I.; Ferrarese, R.; Cazzaniga, W.; Saita, D.; Lucianò, R.; Ceresola, E.; Locatelli, I.; Visconti, L.; Lavorgna, G.; Briganti, A.; et al. The Microbiome of the Prostate Tumor Microenvironment. Eur. Urol. 2017, 72, 625–631. [Google Scholar] [CrossRef]
- Davidsson, S.; Mölling, P.; Rider, J.; Unemo, M.; Karlsson, M.; Carlsson, J.; Andersson, S.; Elgh, F.; Söderquis, B.; Andrén, O. Frequency and typing of Propionibacterium acnes in prostate tissue obtained from men with and without prostate cancer. Infect. Agents Cancer 2016, 11, 26. [Google Scholar] [CrossRef]
- Yow, M.; Tabrizi, S.; Severi, G.; Bolton, D.; Pedersen, J.; Giles, G.; Southey, M. Characterisation of microbial communities within aggressive prostate cancer tissues. Infect. Agents Cancer 2017, 12, 4. [Google Scholar] [CrossRef]
- Chen, Y.; Wei, J. Identification of pathogen signatures in prostate cancer using RNA-seq. PLoS ONE 2015, 10, e0128955. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Ramnarine, V.; Bell, R.; Volik, S.; Davicioni, E.; Hayes, V.; Ren, S.; Collins, C. Metagenomic and metatranscriptomic analysis of human prostate microbiota from patients with prostate cancer. BMC Genom. 2019, 20, 146. [Google Scholar] [CrossRef] [PubMed]
- Alluri, L.; Paes Batista da Silva, A.; Verma, S.; Fu, P.; Shen, D.; MacLennan, G.; Gupta, S.; Bissada, N. Presence of Specific Periodontal Pathogens in Prostate Gland Diagnosed with Chronic Inflammation and Adenocarcinoma. Cureus 2021, 13, e17742. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Alwine, J.C.; Wei, Z.; Tian, T.; Shih, N.; Sperling, C.; Guzzo, T.; Feldman, M.D.; Robertson, E.S. Microbiome signatures in prostate cancer. Carcinogenesis 2019, 40, 749–764. [Google Scholar] [CrossRef]
- Miyake, M.; Ohnishi, K.; Hori, S.; Nakano, A.; Nakano, R.; Yano, H.; Ohnishi, S.; Owari, T.; Morizawa, Y.; Itami, Y.; et al. Mycoplasma genitalium infection and chronic inflammation in human prostate cancer: Detection using prostatectomy and needle biopsy specimens. Cells 2019, 8, 212. [Google Scholar] [CrossRef]
- Salachan, P.V.; Rasmussen, M.; Fredsøe, J.; Ulhøi, B.; Borre, M.; Sørensen, K.D. Microbiota of the prostate tumor environment investigated by whole-transcriptome profiling. Genome Med. 2022, 14, 9. [Google Scholar] [CrossRef]
- Sarkar, P.; Malik, S.; Banerjee, A.; Datta, C.; Pal, D.; Ghosh, A.; Saha, A. Differential Microbial Signature Associated with Benign Prostatic Hyperplasia and Prostate Cancer. Front. Cell. Infect. Microbiol. 2022, 12, 894777. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, Z.; Turner, D.; Lilly, M.; Ou, T.; Jiang, W. Differential Circulating Fungal Microbiome in Prostate Cancer Patients Compared to Healthy Control Individuals. J. Immunol. Res. 2022, 2022, 2574964. [Google Scholar] [CrossRef]
- Abidi, S.H.; Bilwani, F.; Ghias, K.; Abbas, F. Viral etiology of prostate cancer: Genetic alterations and immune response. A literature review. Int. J. Surg. 2018, 52, 136–140. [Google Scholar] [CrossRef]
- Ahmed, M.S.; Salman, N.; Sandhu, S.; Cakir, M.; Seddon, A.; Kuehne, C.; Ashrafi, G. Detection of high-risk Human Papillomavirus in prostate cancer from a UK based population. Sci. Rep. 2023, 13, 7633. [Google Scholar] [CrossRef]
- Michopoulou, V.D.; Derdas, S.; Symvoulakis, E.; Mourmouras, N.; Nomikos, A.; Delakas, D.; Sourvinos, G.; Spandidos, D. Detection of human papillomavirus (HPV) DNA prevalence and p53 codon 72 (Arg72Pro) polymorphism in prostate cancer in a Greek group of patients. Tumor Biol. 2014, 35, 12765–12773. [Google Scholar] [CrossRef]
- Singh, N.; Hussain, S.; Kakkar, N.; Singh, S.K.; Sobti, R.C.; Bharadwaj, M. Implication of high risk Human papillomavirus HR-HPV infection in prostate cancer in Indian population-A pioneering case-control analysis. Sci. Rep. 2015, 5, 7822. [Google Scholar] [CrossRef]
- Ge, X.; Wang, X.; Shen, P. Herpes simplex virus type 2 or human herpesvirus 8 infection and prostate cancer risk: A meta-analysis. Biomed. Rep. 2013, 1, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Classon, J.; Stenudd, M.; Zamboni, M.; Alkass, K.; Eriksson, C.; Pedersen, L.; Schörling, A.; Thoss, A.; Bergh, A.; Wikström, P.; et al. Cytomegalovirus infection is common in prostate cancer and antiviral therapies inhibit progression in disease models. Mol. Oncol. 2025. [Google Scholar] [CrossRef] [PubMed]
- Keller, E.X.; Delbue, S.; Tognon, M.; Provenzano, M. Polyomavirus BK and prostate cancer: A complex interaction of potential clinical relevance. Rev. Med. Virol. 2015, 25, 366–378. [Google Scholar] [CrossRef] [PubMed]
- Silverman, R.H.; Nguyen, C.; Weight, C.J.; Klein, E.A. The human retrovirus XMRV in prostate cancer and chronic fatigue syndrome. Nat. Rev. Urol. 2010, 7, 392–402. [Google Scholar] [CrossRef] [PubMed]
- Leiros, G.J.; Galliano, S.R.; Sember, M.E.; Kahn, T.; Schwarz, E.; Eiguchi, K. Detection of human papillomavirus DNA and p53 codon 72 polymorphism in prostate carcinomas of patients from Argentina. BMC Urol. 2005, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.N.E.; Becker, G.L.; Jackson, J.B.; Rysavy, M.B. Human Papillomavirus and Associated Cancers: A Review. Viruses 2024, 16, 680. [Google Scholar] [CrossRef]
- Spence, A.R.; Rousseau, M.C.; Parent, M.É. Sexual partners, sexually transmitted infections, and prostate cancer risk. Cancer Epidemiol. 2014, 38, 700–707. [Google Scholar] [CrossRef]
- Sfanos, K.S.; Aloia, A.L.; De Marzo, A.M.; Rein, A. XMRV and prostate cancerg—A ‘final’ perspective. Nat. Rev. Urol. 2012, 9, 111–118. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Wang, J. Role of Tumor Microenvironment in Prostate Cancer Immunometabolism. Biomolecules 2025, 15, 826. [Google Scholar] [CrossRef]
- Martinez-Fierro, M.; Leach, R.; Gomez-Guerra, L.; Garza-Guajardo, R.; Johnson-Pais, T.; Beuten, J.; Morales-Rodriguez, I.; Hernandez-Ordoñez, M.; Calderon-Cardenas, G.; Ortiz-Lopez, R.; et al. Identification of Viral Infections in the Prostate and Evaluation of Their Association with Cancer. BMC Cancer 2010, 10, 326. [Google Scholar] [CrossRef]
- Thomas, R.M.; Jobin, C. The Microbiome and Cancer: Is the ‘Oncobiome’ Mirage Real? Trends Cancer 2015, 1, 24–35. [Google Scholar] [CrossRef]
- Hatakeyama, M. Helicobacter pylori CagA and gastric cancer: A paradigm for hit-and-run carcinogenesis. Cell Host Microbe 2014, 15, 306–316. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, Z.; Ma, A.; Li, Z.; Liu, B.; Ma, Q. Computational methods and challenges in analyzing intratumoral microbiome data. Trends Microbiol. 2023, 31, 707–722. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).