Simple Summary
RSAD2, an interferon-inducible gene and an emerging biomarker in cancer, was found to drive tumor progression in preclinical studies. A clinical study showed supportive evidence that, in resected hepatocellular carcinoma (HCC), RSAD2 gene upregulation had an association with blood vessel invasion, which is a proven risk factor for developing metastasis. However, clinical research on RSAD2 in HCC is lacking, and it is of clinical interest whether RSAD2 has an association with metastatic disease. Furthermore, RSAD2 gene upregulation has been reported to be associated with poor survival in patients with breast and stomach cancer, but its prognostic value in HCC has not been formally evaluated. This study involved 309 HCC patients and investigated the clinical implications of RSAD2 gene upregulation in peripheral blood, in terms of its associations with survival, the presence of metastasis, and other clinical manifestations. Higher RSAD2 expression in the blood was significantly associated with poorer survival and was associated with the presence of metastasis. Moreover, HCC patients with increased RSAD2 expression were more likely to have nutritional disturbance, appetite loss, fatigue, and functional impairment.
Abstract
Background: JAK/STAT interferon signaling interacts with the PI3K/AKT/mTOR pathway to drive hepatocellular carcinoma (HCC) progression and metastasis. RSAD2, an interferon-inducible gene, is upregulated by the PI3K/AKT/mTOR pathway and serves as a key factor for metabolic reprogramming to promote stem-like properties of cancer stem cells and tumor proliferation. In patients with resected HCC, RSAD2 upregulation showed an association with microvascular invasion, which is a proven risk factor for developing HCC metastasis. This clinical observation was compatible with preclinical findings. On the other hand, RSAD2 upregulation has been reported to confer poor prognosis in breast and gastric cancers. However, further clinical study of RSAD2 in HCC is lacking. As a result, we investigated the clinical implications of RSAD2 gene expression in HCC patients, in terms of its associations with survival, the presence of extra-hepatic metastasis, and other clinical manifestations. Methods: We studied 309 treatment-naïve HCC patients, as well as data from the TCGA and GTEx databases. Results: RSAD2 gene expression was differentially upregulated in HCC tumors when compared to normal liver tissues (p < 0.01). Elevated RSAD2 mRNA levels in the blood and the presence of extra-hepatic metastasis were independent prognostic factors for poor overall survival (OS) (p < 0.01). The median OS of patients with high RSAD2 expression vs. low expression were 5.4 vs. 14.2 months, respectively (p < 0.01). A high RSAD2 mRNA level was significantly correlated with the presence of extra-hepatic metastasis, nutritional disturbance, and functional impairment after controlling for confounding clinical factors (p < 0.05). Conclusions: High RSAD2 gene expression is associated with poorer OS, the presence of extra-hepatic metastasis, and quality-of-life disturbances in HCC patients.
1. Introduction
Hepatocellular carcinoma (HCC) accounts for the sixth most common cancer and the third leading cause of cancer death in the world [1]. The majority of the world’s HCC cases are diagnosed in China [2].
Inflammation, immune system activation, and immune evasion play key roles in HCC development and progression [3]. In particular, the interferon response in HCC has been shown to induce the metabolic reprogramming of tumor cells, the tumor micro-environment, and immune cells to facilitate HCC progression and metastasis [4]. The interferon response has also been shown to interact with PI3K/AKT/mTOR signaling to drive HCC proliferation and tumor cell migration [5]. The interferon response gene RSAD2, upregulated mechanistically by the JAK/STAT interferon response and PI3K/AKT/mTOR pathways, is a key factor for metabolic reprogramming to enhance lipogenesis and glycolysis, thereby promoting stem-like properties of cancer stem cells and tumor proliferation [6]. RSAD2 knockdown in cancer cell lines was shown to suppress tumor progression and reverse metabolic reprogramming, implicating it as a driver of tumor proliferation [7].
However, the current literature indicates a lack of clinical studies on RSAD2 in HCC patients to validate these pre-clinical mechanistic findings. In a report on patients with resected HCC, RSAD2 upregulation in tumors was linked to microvascular invasion, which is a proven risk factor for developing HCC metastasis [8,9]. Microvascular invasion in resected HCC has been used to identify high-risk patients who will develop extra-hepatic metastases [9]. Moreover, Bertuzzo et al. reported that microvascular invasion and a cell-count-based inflammatory marker were the strongest risk factors for HCC recurrence [10]. This calls for an investigation on the clinical implications of RSAD2 gene expression in HCC patients—whether RSAD2 upregulation is associated with the presence of metastasis and poorer survival.
Studies evaluating RSAD2 in other cancers commonly reported its prognostic value with mixed findings. In particular, RSAD2 was reported to be an adverse prognostic factor in breast and gastric cancers but a favorable prognostic factor in oral cancer and melanoma [7,11,12,13,14]. Prognostic studies of RSAD2 in cancer were mainly genomic analyses using public databases. A caveat concerning interpreting online database studies is that clinical factors provided by these databases may not be adequate to support post hoc prognostic analyses, thereby compromising the accuracy of prognostic models. Dedicated prognostic studies of RSAD2 with relevant and comprehensive clinical data from patients with cancer are lacking.
As a result, we set out to investigate the clinical significance of RSAD2 gene expression in an HCC patient cohort. In the first part of the study, we evaluated whether RSAD2 was differentially expressed in HCC tumor samples compared to normal tissues using genomic repositories. The second part of this study involved a prospective cohort of treatment-naïve HCC patients. We determined their peripheral blood leucocytic RSAD2 mRNA transcript levels and then evaluated their clinical implications in terms of prognostic value and associations with clinical factors (including extra-hepatic metastasis) and patients’ symptoms as captured by patient-reported outcome measures.
2. Materials and Methods
2.1. RSAD2 Gene Expression in HCC
To explore whether RSAD2 is differentially expressed in HCC compared to normal liver tissues, we analyzed the combined database of The Cancer Genome Analysis (TCGA) (https://www.cancer.gov/tcga, accessed on 26 April 2025) and Genotype-Tissue Expression (GTEx) (https://gtexportal.org, accessed on 26 April 2025), in which the batch effect was minimized after sample re-analyses using a uniform RNA sequencing (RNA-seq) pipeline. Data were analyzed using the Xena multi-omic exploration tool, University of California, Santa Cruz, CA, USA [15]. RSAD2 gene expressions in HCC samples (the database of Genotypes and Phenotypes [dbGaP] accession number phs000178.v11.p8) were compared to normal liver tissue samples (dbGaP accession number phs000424.v10.p2) using Welch’s t-test.
2.2. Local HCC Patient Cohort
This study was approved by the Joint Chinese University of Hong Kong—New Territories East Cluster Clinical Research Ethics Committee of Prince of Wales Hospital, Hong Kong on 13 October 2006 (reference number CRE-2006.340). Newly diagnosed HCC patients were prospectively recruited. Diagnosis was based on either tumor biopsy or a combination of typical dynamic imaging findings and elevated α-fetoprotein (AFP). Patients were eligible for the study if they were treatment-naïve. Major exclusion criteria included cognitive impairment or a history of another malignancy. Written informed consent was obtained from every patient.
2.3. Demographic, Clinical, and Laboratory Data
Baseline demographic and clinical data were collected. On the day of consent, blood tests were taken for cell counts, liver and renal biochemistries, coagulation profile, AFP, hepatitis serology, and RSAD2 mRNA level.
2.4. Peripheral Blood RSAD2 Gene Expression Quantification
2.4.1. Isolation of Peripheral Blood Leucocytes and Extraction of Total RNA
Peripheral blood in ethylenediaminetetraacetic acid (EDTA) was used for RSAD2 expression analysis. Peripheral blood mononuclear cells (PBMCs) and granulocytes were isolated by density gradient centrifugation using Lymphoprep™ (Serumwerk Bernburg AG, Bernburg, Germany). Total RNA from leucocytes and whole blood were extracted via a modified acid guanidinium thiocyanate–phenol–chloroform (AGPC) protocol suggested by the manufacturer (Molecular Research Center, Inc., OH, USA) [16]. Then, 250 μL of the TRI Reagent® LS (TB126) was added to the PBMCs and granulocytes, while 300 µL of the TRI Reagent® BD (TB120) was added to the whole blood. 1-Bromo-3-chloropropane (B9673, Sigma-Aldrich, MA, USA) was used for phase separation. Subsequent washing and elution were performed by isopropanol (I9516, Sigma-Aldrich, MA, USA) and ethanol (4116-4104, Daejung, Siheung-si, South Korea).
2.4.2. cDNA Synthesis
First-strand cDNA was reverse transcribed from the total RNA using the PrimeScript RT Reagent Kit (#RR037A, Takara Bio, Shiga, Japan). The 10 µL reverse transcription system was used, including 2 µL of 5X PrimeScript Buffer (for real-time), 0.5 µL PrimeScript RT Enzyme Mix I, 0.5 µL Oligo dT Primer (50 µM), 2 µL Random 6mers (100 µM), and 5 µL of RNA (up to 1 µg/µL) from the samples. Reverse transcription was performed in the C1000 Touch Thermal Cycler (Bio-Rad, Hercules, CA, USA) under the following conditions: sample incubation at 37 °C for 15 min, followed by enzyme denaturation at 85 °C for 5 s, and then storage at 4 °C for short-term preservation before refrigeration storage.
2.4.3. RSAD2 Gene Expression Quantification
The synthesized cDNA was diluted and duplicated to optimize the real-time qPCR performance. In each batch of the qPCR experiment, the same reference sample was used, and a serial 2-fold dilution was carried out to establish PCR efficiency. The TB Green® Premix Ex Taq™ II (Tli RNase H Plus) kit (#RR820A, Takara Bio, Shiga, Japan) and RSAD2 gene primers (see Table 1) were used in the reaction, performed in an LC480 thermal cycler (Roche, Basel, Switzerland). The qPCR conditions included a pre-incubation step at 95 °C for 30 s, followed by 45 cycles of amplification (95 °C for 5 s, 55 °C for 30 s, and 72 °C for 20 s), a melting phase (95 °C for 1 min, 40 °C for 1 min and 65 °C for 20 s), and a cooling step at 40 °C for 30 s. Patients’ RSAD2 mRNA transcript levels were expressed as fold change (FC) against the same normal healthy donor as the calibrator, using the amplification efficiency-corrected delta delta cycle threshold (ddCT) method [17].
FC = eff RSAD2 ^ (CtRSAD2_calibrator − CtRSAD2_mean) ÷ eff UBC ^ (CtUBC_calibrator − CtUBC_mean)]

Table 1.
Primer sequences used for quantification of RSAD2 mRNA transcript levels in peripheral blood.
2.5. Health-Related Quality of Life (HRQoL) Assessment
Patients filled in the European Organization for Research and Treatment of Cancer (EORTC) QLQ-C30 and QLQ-HCC18 questionnaires at study entry [18,19]. C30 and HCC18 index scores were calculated [20].
2.6. Follow-Up
Patients were followed up until death.
2.7. Sample Size Estimation and Missing Data Handling
The median overall survival (OS) was estimated to be around 6 months from prior local data [21]. Assuming a hazard ratio (HR) of 2.0 for high RSAD2 expression with two-sided alpha at 0.05 and 80% power, we aimed to recruit 298 patients. There was no prior observation for the novel variable of peripheral blood RSAD2 mRNA levels to guide reasonable imputation for missing data. Furthermore, in order to calculate C30 and HCC18 index scores, all scores from QLQ-C30 and QLQ-HCC18, respectively, had to be available. As a result, we adopted the complete-case analysis approach.
2.8. Statistical Analyses
Baseline RSAD2 mRNA transcript levels and clinical and HRQoL factors were analyzed by standard descriptive tests. OS was defined as the time between the date of recruitment and the date of death. Patients alive or lost to follow-up were censored at the date of last follow-up. Kaplan–Meier analysis was used for survival estimates. A log-rank test was used to compare survival distributions. Cox proportional hazards models were used to identify non-overlapping independent prognostic factors among clinical and HRQoL data for OS. Two-tailed p-values of less than 0.05 were considered statistically significant. Correlations between RSAD2 mRNA levels and HRQoL factors were analyzed using Student’s t-test, Wilcoxon rank-sum test (parametric and non-parametric tests were conducted as sensitivity analyses to account for different distributional assumptions of the novel RSAD2 variable), and univariate logistic regressions. Multivariate logistic regressions were used to control for demographic and clinical factors. Data were analyzed using SAS version 9.4 (SAS institute, Cary, NC, USA). Study methods and results are reported in accordance with the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement [22].
3. Results
3.1. RSAD2 Gene Expression in HCC Tumors
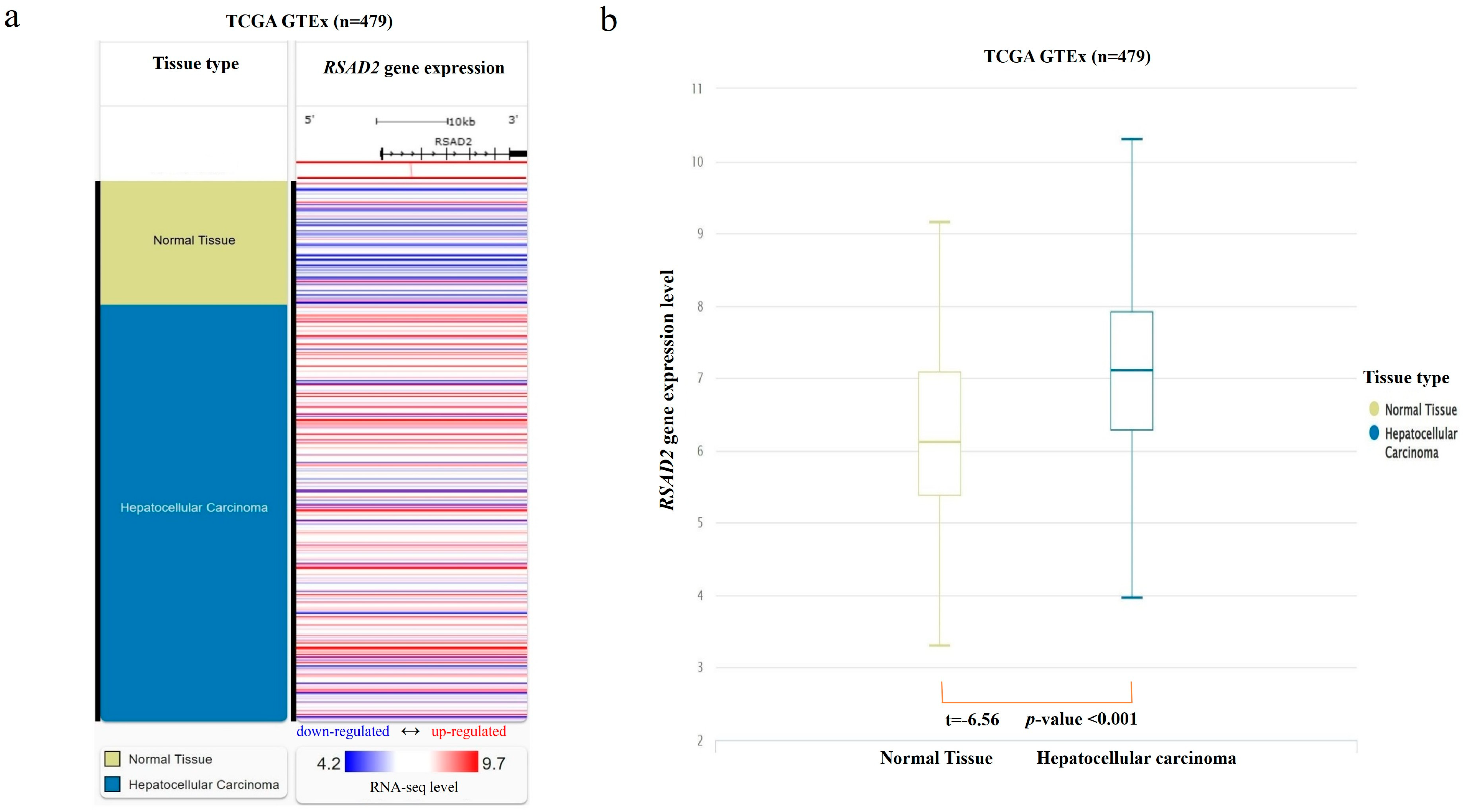
From the combined TCGA and GTEx database, RNA-seq data were found in 369 HCC and 110 normal liver tissue samples. RSAD2 mRNA transcript abundance in HCC tumors was compared against matching controls. The results showed that RSAD2 gene expression was significantly higher in HCC tumors when compared to normal liver tissues (p < 0.01) (shown in Figure 1).
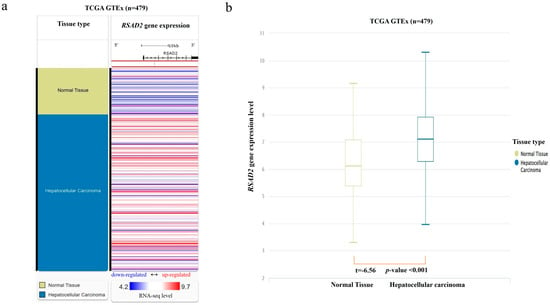
Figure 1.
RSAD2 mRNA expression in the combined TCGA and GTEx databases. (a) Heatmap of RSAD2 mRNA expression in HCC tumors vs. normal liver tissue samples. Each horizontal bar represents an individual patient’s RSAD2 mRNA gene expression level, and a color gradient is applied according to the RNA-seq level (red representing upregulated mRNA expression; white representing average expression; and blue representing downregulated expression). (b) Boxplot of RSAD2 mRNA expression in HCC compared to normal liver. mRNA expression level was quantified according to the RNA-Seq by Expectation–Maximization algorithm.
3.2. Characteristics of the HCC Patient Cohort
From 2009 to 2017, 309 HCC patients had complete data for analysis (among the 340 recruited patients, 11 patients’ RSAD2 mRNA levels could not be determined, and 20 patients had incomplete HRQoL data). Table 2 shows their baseline clinical characteristics. The median age at diagnosis was 59 (range 27–86) years. Eighty-seven percent were male. Most patients (80%) had evidence of hepatitis B infection. Focal, multifocal, diffuse liver tumors were found in 98 (32%), 90 (29%), and 121 (39%) patients, respectively. Seventy-one (23%) patients had extra-hepatic metastasis (AJCC 8th edition stage IV) [23]. The median follow-up duration was 116 (95% confidence interval [CI] 87–120) months. The median OS was 9.5 (95% CI 6.1–14.3) months. Supplementary Table S1 shows their baseline HRQoL scores in QLQ-C30 and QLQ-HCC18. The mean ‘global health status’ score was 53.4 (standard deviation [SD] 25.1).

Table 2.
Demographic and clinical characteristics of the patients.
3.3. Peripheral Blood RSAD2 mRNA Transcript Level Analyses
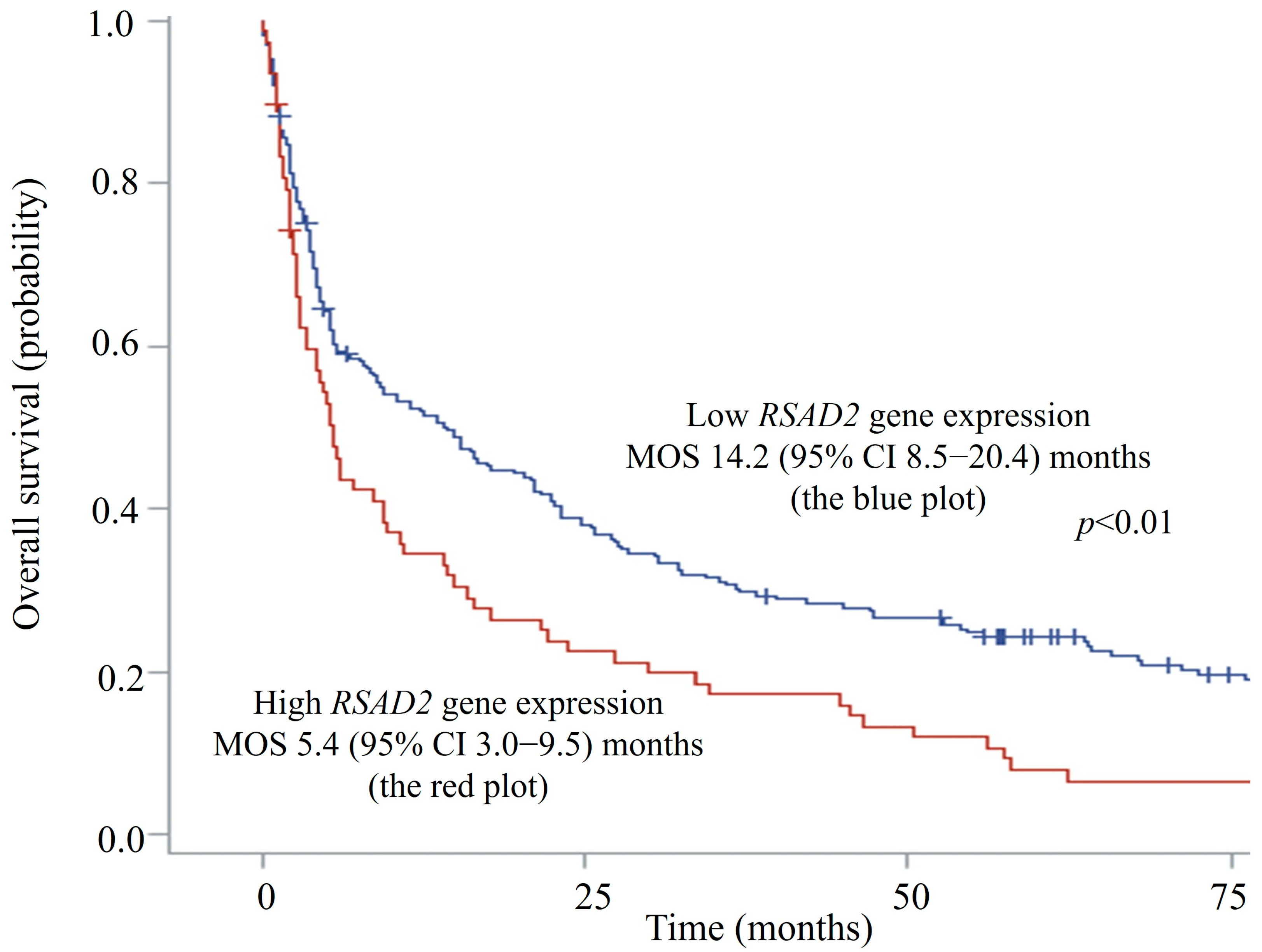
The median RSAD2 mRNA transcript level in patients’ circulating leucocytes was 1.16 (interquartile range 0.33–2.89) FC. Since RSAD2 gene expression demonstrated an asymmetrical skewed distribution, expression levels of more than the 75th percentile (2.89 FC) were ranked as high to facilitate risk of death analyses using Cox models. Seventy-six patients had high RSAD2 gene expression in peripheral blood, whereas 233 patients had low expression. The median OS was significantly shorter in patients with high RSAD2 gene expression when compared to those with low RSAD2 expression, 5.4 (95% CI, 3.0–9.5) vs. 14.2 (95% CI, 8.5–20.4) months, respectively (log-rank p < 0.01). Figure 2 shows the Kaplan–Meier survival curves of patients with respect to RSAD2 gene expressions.
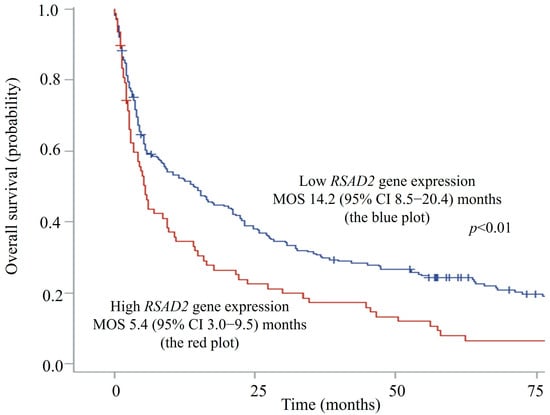
Figure 2.
Overall survival plots of hepatocellular carcinoma patients with high vs. low RSAD2 mRNA expression. CI—confidence interval; MOS—median overall survival.
3.4. Univariate Prognostic Analyses of RSAD2 mRNA Level and Clinical Factors
Table 2 shows the results of univariate Cox proportional hazards analyses of clinical factors. Higher RSAD2 mRNA levels, the presence of extra-hepatic metastasis, a higher white cell count (WCC), lower hemoglobin levels, and higher alanine transferase were significantly associated with poorer OS (p < 0.01). Patients with poor liver function (Child–Pugh classes B or C) had significantly poorer OS than patients with preserved liver function (class A) (p < 0.01). Patients with more advanced HCC (Barcelona Clinic Liver Cancer stage B, C or D) had significantly poorer OS than patients with earlier disease (stage A) (p < 0.001) [24].
3.5. Multivariate Analyses for Independent Prognostic Factors
The results of multivariate Cox proportional hazards models with RSAD2 gene expression and non-overlapping clinical and HRQoL factors are shown in Table 3. The presence of extra-hepatic metastasis, high RSAD2 mRNA gene expression, higher WCC, and anemia were independent poor prognostic factors for OS (p < 0.01). When AFP was included in the multivariate Cox model, the RSAD2 mRNA level was no longer identified as a significant factor. In other words, AFP was a stronger prognostic factor than RSAD2.

Table 3.
Multivariate Cox proportional hazards analyses of prognostic factors for overall survival.
3.6. Correlates of RSAD2 mRNA Levels and Extra-Hepatic Metastases, as Well as Other Clinical Manifestations
Higher RSAD2 gene expression was significantly correlated with the presence of extra-hepatic metastasis (p < 0.03). However, RSAD2 levels had no significant association with age, sex, cell counts, hepatitis B, hepatitis C, or liver or renal function. Regarding other clinical manifestations, higher RSAD2 gene expression in circulating leucocytes were associated with worse symptoms in ‘appetite loss’, ‘fatigue’, ‘nutritional concern’, and ‘body image’ (p < 0.03) (see Table 4).

Table 4.
Clinical symptoms showing significant correlations with RSAD2 mRNA transcript level.
3.7. Multivariate Correlation Models Between RSAD2 mRNA Transcript Levels and Extra-Hepatic Metastases, as Well as Other Clinical Manifestations
After controlling for potentially confounding clinical factors in multivariate logistic regressions, extra-hepatic metastasis remained significantly correlated with upregulated RSAD2 gene expression (OR, 2.94; 95% CI, 1.29–6.70; p < 0.03) (Table 5). The presence of extra-hepatic metastasis was among the factors that best predicted a higher RSAD2 transcript level in peripheral blood.

Table 5.
Multivariate logistic regression analyses between RSAD2 mRNA transcript levels and extra-hepatic metastasis and other clinical manifestations for clinical factors.
4. Discussion
From the current study, RSAD2 gene expression was found to be heightened in HCC patients’ tumor tissues and/or peripheral blood leucocytes. RSAD2 gene expression level and extra-hepatic metastasis were both identified as independent prognostic factors for OS in HCC. Patients with a high RSAD2 mRNA transcript level had significantly shorter median OS than those with a low RSAD2 level (5 vs. 14 months, respectively). Although its prognostic value was not superior to AFP, further research is needed to evaluate whether integrating RSAD2 can improve HCC staging systems. For instance, various staging systems have been incorporated with non-anatomical prognostic biomarkers [25,26].
A high RSAD2 mRNA level was significantly correlated with the presence of extra-hepatic metastasis. Multivariate correlation analyses were performed after controlling for potentially confounding factors, including age, sex, cell counts, hepatitis B/C, and liver or renal function. Since treatment-naïve patients were recruited, therapy-related alteration in RSAD2 levels could be avoided.
The association between RSAD2 and extra-metastasis is compatible with the current mechanistic knowledge of RSAD2 in cancers. Clinical reports have also shown an association between inflammatory status and the development of metastasis in melanoma and breast cancer [27,28]. Microvascular invasion, a known risk factor for developing metastasis, was more frequently observed in HCC patients with RSAD2 upregulation [8,9]. These findings are compatible with the results of our current study.
To date, AFP has remained the most widely used tumor marker for HCC detection. However, 30–40% of HCC patients had negative AFP [29]. Even among patients with elevated AFP, the level cannot differentiate between the presence or absence of extra-hepatic spread. Therefore, there has been a clinical need for a biomarker to indicate the presence of HCC metastasis. From the multivariate regressions between RSAD2 gene expression, adjusting for clinical factors, the presence of extrahepatic metastasis was among the factors that best predicted a higher RSAD2 gene expression level. Based on the findings of our study, RSAD2 upregulation in peripheral blood would suggest a need for systemic imaging to search for extra-hepatic metastasis.
This study used HRQoL instruments to measure and quantify patients’ symptoms into analyzable data, in order to examine the relationships between RSAD2 gene upregulation and patients’ clinical manifestations. Remarkably, peripheral blood RSAD2 gene upregulation (signifying inflammatory cascade activation) demonstrated significant correlations with appetite loss, nutritional concern, fatigue, and social functional impairment. These results are logical since such clinical manifestations are typically found in patients in an inflammatory state and with constitutional symptoms [30]. Chiba et al. demonstrated a correlation between pro-inflammatory state and cancer cachexia in an animal model [31]. This observation also substantiated our current clinical findings.
We acknowledge certain limitations in this study. Firstly, it is unclear whether the current results are generalizable to patients with other cancers. Further studies are warranted. Secondly, this study provided valuable observations in Chinese patients, but it is unknown if the findings are applicable to HCC patients of other ethnicities. Future validation in a multi-ethnic cohort is indicated.
5. Conclusions
RSAD2 gene expression was differentially upregulated in HCC tumors when compared to normal liver tissues. Increased RSAD2 gene expression in peripheral blood and extra-hepatic metastasis are significant independent prognostic factors for OS in HCC. An elevated RSAD2 mRNA transcript level in circulating leucocytes was significantly correlated with the presence of extra-hepatic metastasis, and it was associated with clinical features that were suggestive of constitutional symptoms (nutritional concern, appetite loss, fatigue, and functional impairment). An elevated peripheral blood RSAD2 level could suggest a search for extra-hepatic metastasis. Further research is needed to examine the clinical significance of RSAD2 gene expression in other cancer types.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/diseases13120395/s1. Table S1. Health-related quality of life characteristics of the patients.
Author Contributions
Conceptualization, L.L., N.L.S.T., and W.Y.; methodology, L.L., N.L.S.T., T.-K.K., F.M., and W.Y.; data acquisition, interpretation or curation, L.L., N.L.S.T., F.M., J.K., E.P.H., S.L.C., D.R.J., T.-K.K., L.L.C., K.F.L., S.C.H.Y., and W.Y.; formal analysis, F.M., L.L., N.L.S.T., and W.Y.; writing—original draft, L.L., N.L.S.T., F.M., and W.Y.; writing—review and editing, all authors; supervision, L.L., N.L.S.T., and W.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Research Grant Council (Hong Kong) through the Co-funding Mechanism on Joint Laboratories with the CAS—KIZ-CUHK Joint Laboratory of Bioresources and Molecular Research in Common Diseases under the funding number JLFS/M-403/24.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Joint Chinese University of Hong Kong—New Territories East Cluster Clinical Research Ethics Committee of Prince of Wales Hospital, Hong Kong on 13 Oct 2006 (reference number CRE-2006.340). This article does not contain any studies with experimentation on human participants or animals performed by any of the authors.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets generated and analyzed during the current study are not publicly available due to embedded individual privacy but are available on request from the corresponding author.
Conflicts of Interest
N.L.S.T. is the inventor of the patent ‘Determination of gene expression levels of a cell type’, which has been assigned to The Chinese University of Hong Kong. N.L.S.T. is a share-holder of Cytomics Ltd. Cytomics Ltd. holds a license to use a patent related to the DIRECT LS-TA assay, patent application pending (CN116334204A). T.-K.K. is an employee of Cytomics Ltd. Other authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| AFP | α-fetoprotein |
| AJCC | the American Joint Committee on Cancer staging system |
| AGPC | acid guanidinium thiocyanate–phenol–chloroform |
| cDNA | complementary deoxyribonucleic acid |
| CI | confidence interval |
| CRP | C-reactive protein |
| dbGaP | the database of Genotypes and Phenotypes |
| ddCT | delta delta cycle threshold |
| EDTA | ethylenediaminetetraacetic acid |
| EORTC | the European Organization for Research and Treatment of Cancer |
| FC | fold change |
| GTEx | genotype–tissue expression |
| HRQoL | health-related quality of life |
| IL | interleukin |
| mRNA | messenger ribonucleic acid |
| OS | overall survival |
| PBMC | peripheral blood mononuclear cell |
| qPCR | quantitative polymerase chain reaction |
| RNA-seq | RNA sequencing |
| ROC | receiver operator characteristic |
| RSAD2 | radical S-adenosyl methionine domain containing 2 |
| SD | standard deviation |
| TCGA | The Cancer Genome Analysis |
| Viperin | virus inhibitory protein, endoplasmic reticulum-associated, interferon-inducible |
| WCC | white cell count |
References
- Konyn, P.; Ahmed, A.; Kim, D. Current epidemiology in hepatocellular carcinoma. Expert Rev. Gastroenterol. Hepatol. 2021, 15, 1295–1307. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B.; Rudolph, K.L. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef]
- Hernandez-Gea, V.; Toffanin, S.; Friedman, S.L.; Llovet, J.M. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology 2013, 144, 512–527. [Google Scholar] [CrossRef]
- Xia, Y.; Brown, Z.J.; Huang, H.; Tsung, A. Metabolic reprogramming of immune cells: Shaping the tumor microenvironment in hepatocellular carcinoma. Cancer Med. 2021, 10, 6374–6383. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, S.; Gao, M.; Chang, J.; Sun, J.; Qin, L.; Li, A.; Lv, F.; Lou, J.; Zhang, Y.; et al. Interferon-Induced Transmembrane Protein 3 Expression Upregulation Is Involved in Progression of Hepatocellular Carcinoma. BioMed Res. Int. 2021, 2021, 5612138. [Google Scholar] [CrossRef]
- Weinstein, A.G.; Godet, I.; Gilkes, D.M. The rise of viperin: The emerging role of viperin in cancer progression. J. Clin. Investig. 2022, 132, e165907. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.M.; Kim, J.J.; Yoo, J.; Kim, K.S.; Gu, Y.; Eom, J.; Jeong, H.; Kim, K.; Nam, K.T.; Park, Y.S.; et al. The interferon-inducible protein viperin controls cancer metabolic reprogramming to enhance cancer progression. J. Clin. Investig. 2022, 132, e157302. [Google Scholar] [CrossRef]
- Xin, Z.; Chen, H.; Xu, J.; Zhang, H.; Peng, Y.; Ren, J.; Guo, Q.; Song, J.; Jiao, L.; You, L.; et al. Exosomal mRNA in plasma serves as a predictive marker for microvascular invasion in hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2024, 39, 2228–2238. [Google Scholar] [CrossRef]
- Zhang, X.; Li, J.; Shen, F.; Lau, W.Y. Significance of presence of microvascular invasion in specimens obtained after surgical treatment of hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2018, 33, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Bertuzzo, V.R.; Cescon, M.; Ravaioli, M.; Grazi, G.L.; Ercolani, G.; Del Gaudio, M.; Cucchetti, A.; D’Errico-Grigioni, A.; Golfieri, R.; Pinna, A.D. Analysis of factors affecting recurrence of hepatocellular carcinoma after liver transplantation with a special focus on inflammation markers. Transplantation 2011, 91, 1279–1285. [Google Scholar] [CrossRef]
- Reyimu, A.; Chen, Y.; Song, X.; Zhou, W.; Dai, J.; Jiang, F. Identification of latent biomarkers in connection with progression and prognosis in oral cancer by comprehensive bioinformatics analysis. World J. Surg. Oncol. 2021, 19, 240. [Google Scholar] [CrossRef]
- Sun, S.; Zhi, Z.; Su, Y.; Sun, J.; Li, Q. A CD8+ T cell-associated immune gene panel for prediction of the prognosis and immunotherapeutic effect of melanoma. Front. Immunol. 2022, 13, 1039565. [Google Scholar] [CrossRef]
- Lewis, M.W.; Wisniewska, K.; King, C.M.; Li, S.; Coffey, A.; Kelly, M.R.; Regner, M.J.; Franco, H.L. Enhancer RNA Transcription Is Essential for a Novel CSF1 Enhancer in Triple-Negative Breast Cancer. Cancers 2022, 14, 1852. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yang, Q.; Cui, Q.; Zhang, D.; Kong, D.; Liao, X.; Ren, J.; Gong, Y.; Wu, G. Weighted gene correlation network analysis identifies RSAD2, HERC5, and CCL8 as prognostic candidates for breast cancer. J. Cell. Physiol. 2020, 235, 394–407. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. Quantification strategies in real-time PCR. In A–Z of Quantitative PCR; Bustin, S.A., Ed.; International University Line (IUL): La Jolla, CA, USA, 2004; pp. 87–112. [Google Scholar]
- Aaronson, N.K.; Ahmedzai, S.; Bergman, B.; Bullinger, M.; Cull, A.; Duez, N.J.; Filiberti, A.; Flechtner, H.; Fleishman, S.B.; de Haes, J.C.; et al. The European Organization for Research and Treatment of Cancer QLQ-C30: A quality-of-life instrument for use in international clinical trials in oncology. J. Natl. Cancer Inst. 1993, 85, 365–376. [Google Scholar] [CrossRef]
- Blazeby, J.M.; Currie, E.; Zee, B.C.; Chie, W.C.; Poon, R.T.; Garden, O.J. Development of a questionnaire module to supplement the EORTC QLQ-C30 to assess quality of life in patients with hepatocellular carcinoma, the EORTC QLQ-HCC18. Eur. J. Cancer 2004, 40, 2439–2444. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Mo, F.K.; Chan, S.L.; Hui, E.P.; Tang, N.S.; Koh, J.; Leung, L.K.; Poon, A.N.; Hui, J.; Chu, C.M.; et al. Prognostic values of EORTC QLQ-C30 and QLQ-HCC18 index-scores in patients with hepatocellular carcinoma—Clinical application of health-related quality-of-life data. BMC Cancer 2017, 17, 8. [Google Scholar] [CrossRef]
- Yeo, W.; Mo, F.K.; Koh, J.; Chan, A.T.; Leung, T.; Hui, P.; Chan, L.; Tang, A.; Lee, J.J.; Mok, T.S.; et al. Quality of life is predictive of survival in patients with unresectable hepatocellular carcinoma. Ann. Oncol. 2006, 17, 1083–1089. [Google Scholar] [CrossRef]
- von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gøtzsche, P.C.; Vandenbroucke, J.P. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: Guidelines for reporting observational studies. BMJ (Clin. Res. Ed.) 2007, 335, 806–808. [Google Scholar] [CrossRef]
- Kamarajah, S.K.; Frankel, T.L.; Sonnenday, C.; Cho, C.S.; Nathan, H. Critical evaluation of the American Joint Commission on Cancer (AJCC) 8th edition staging system for patients with Hepatocellular Carcinoma (HCC): A Surveillance, Epidemiology, End Results (SEER) analysis. J. Surg. Oncol. 2018, 117, 644–650. [Google Scholar] [CrossRef]
- Llovet, J.M.; Bru, C.; Bruix, J. Prognosis of hepatocellular carcinoma: The BCLC staging classification. Semin. Liver Dis. 1999, 19, 329–338. [Google Scholar] [CrossRef]
- Cserni, G.; Chmielik, E.; Cserni, B.; Tot, T. The new TNM-based staging of breast cancer. Virchows Arch. 2018, 472, 697–703. [Google Scholar] [CrossRef]
- Paner, G.P.; Stadler, W.M.; Hansel, D.E.; Montironi, R.; Lin, D.W.; Amin, M.B. Updates in the Eighth Edition of the Tumor-Node-Metastasis Staging Classification for Urologic Cancers. Eur. Urol. 2018, 73, 560–569. [Google Scholar] [CrossRef]
- Lollini, P.L.; De Giovanni, C.; Del Re, B.; Nicoletti, G.; Prodi, G.; Nanni, P. Interferon-mediated enhancement of metastasis. Are MHC Antigens Involved? Clin. Exp. Metastasis 1987, 5, 277–287. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, S.; Srivastava, R.K.; Nandi, A.; Thacker, G.; Murali, H.; Kim, S.; Baldeon, M.; Tobias, J.; Blanco, M.A.; et al. Loss of ELF5-FBXW7 stabilizes IFNGR1 to promote the growth and metastasis of triple-negative breast cancer through interferon-γ signalling. Nat. Cell Biol. 2020, 22, 591–602. [Google Scholar] [CrossRef]
- Galle, P.R.; Foerster, F.; Kudo, M.; Chan, S.L.; Llovet, J.M.; Qin, S.; Schelman, W.R.; Chintharlapalli, S.; Abada, P.B.; Sherman, M.; et al. Biology and significance of alpha-fetoprotein in hepatocellular carcinoma. Liver Int. 2019, 39, 2214–2229. [Google Scholar] [CrossRef]
- Bodewes, I.L.A.; van der Spek, P.J.; Leon, L.G.; Wijkhuijs, A.J.M.; van Helden-Meeuwsen, C.G.; Tas, L.; Schreurs, M.W.J.; van Daele, P.L.A.; Katsikis, P.D.; Versnel, M.A. Fatigue in Sjögren’s Syndrome: A Search for Biomarkers and Treatment Targets. Front. Immunol. 2019, 10, 312. [Google Scholar] [CrossRef]
- Chiba, F.; Soda, K.; Yamada, S.; Tokutake, Y.; Chohnan, S.; Konishi, F.; Rikiyama, T. The importance of tissue environment surrounding the tumor on the development of cancer cachexia. Int. J. Oncol. 2014, 44, 177–186. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).