Abstract
Recently, medical image encryption has gained special attention due to the nature and sensitivity of medical data and the lack of effective image encryption using innovative encryption techniques. Several encryption schemes have been recommended and developed in an attempt to improve medical image encryption. The majority of these studies rely on conventional encryption techniques. However, such improvements have come with increased computational complexity and slower processing for encryption and decryption processes. Alternatively, the engagement of intelligent models such as deep learning along with encryption schemes exhibited more effective outcomes, especially when used with digital images. This paper aims to reduce and change the transferred data between interested parties and overcome the problem of building negative conclusions from encrypted medical images. In order to do so, the target was to transfer from the domain of encrypting an image to encrypting features of an image, which are extracted as float number values. Therefore, we propose a deep learning-based image encryption scheme using the autoencoder (AE) technique and the advanced encryption standard (AES). Specifically, the proposed encryption scheme is supposed to encrypt the digest of the medical image prepared by the encoder from the autoencoder model on the encryption side. On the decryption side, the analogous decoder from the auto-decoder is used after decrypting the carried data. The autoencoder was used to enhance the quality of corrupted medical images with different types of noise. In addition, we investigated the scores of structure similarity (SSIM) and mean square error (MSE) for the proposed model by applying four different types of noise: salt and pepper, speckle, Poisson, and Gaussian. It has been noticed that for all types of noise added, the decoder reduced this noise in the resulting images. Finally, the performance evaluation demonstrated that our proposed system improved the encryption/decryption overhead by 50–75% over other existing models.
1. Introduction
Technology and the Internet have become vital aspects of human lives in all scopes. Many institutions have converted their work to rely almost 100% on technology. All correspondence is exchanged by email. In some cases, data are being stored on the cloud, which has become more secure than personal vices or even institutions’ servers.
Health is one of the most important sectors that has been converted to technology in many aspects. With the development of scanning and imaging devices, such as MRI, X-ray, and others, medical images have been produced and stored in clinics, hospitals, and on physicians’ personal computers every day and in large amounts.
Medical images are considered the most sensitive data transferred or stored over the Internet [1]. Thus, the need to preserve their privacy has become a very hot research problem that has been tackled to propose proper solutions.
Encryption is one of the best solutions proposed for this problem. Several encryption algorithms have been created and used on data in general and medical images specifically.
Nevertheless, medical images’ sizes can vary from small to large, reaching over 4000 × 4000, which becomes even larger when dealing with colored images. Encrypting large-sized images may take time, especially with the additional steps aiming to sophisticate the encryption process to prevent possible malicious attacks related to medical images [2]. We should note that practical encryption techniques such as AES cannot solely provide authentication and integrity [3]; hence, they are usually combined with other techniques to be considered reliable.
Images, generally, have also been the subject of research in artificial intelligence (AI) systems. Various research studies have applied all types of AI models that perform classifications [4,5], clustering [6], segmentation [7,8], generation of fake images [9], denoising [10] and impainting [11].
Autoencoders are used with medical images to extract necessary features and reconstruct the images with remarkable accuracy [12]. Encoding medical images using autoencoders is a known deep learning method that reduces the dimensionality of the images into smaller, compact representations of the image as well [13]. The size of the generated data out of the autoencoder can be controlled according to the architecture of the used autoencoder. The encoded data, generated from autoencoders, can be used to regenerate the original images. However, the encoded data are entirely different from the original data and cannot be viewed as a representation of the original data. Hence, encrypting the encoded data of a medical image cannot be used to maliciously view the content of the medical image after encrypting the encoded data. On the other hand, encrypting the original image content can be used for malicious purposes [2].
Autoencoders are also used in much work as a powerful denoising tool. The work in [14,15,16] addressed the benefit of using autoencoders for medical image denoising. Medical images are prone to different types of noise and poor quality due to the technology used for taking the images [14,16]. In this work, we are interested in using the autoencoder to encrypt medical images to overcome the problem of malicious viewing, which is common with medical images. Encrypting the autoencoder’s extracted features can also reduce the required data to be encrypted and transferred, resulting in a faster encryption process.
The AES encryption algorithm has proven to be a robust and reliable encryption technique that can transfer data over the Internet [17]. AES is widely used in developing highly secure encryption techniques such as the one in [17,18,19] and many others.
The autoencoder, illustrated in Figure 1, is a deep learning model used to perform several tasks, such as denoising and impainting. It trains images by extracting important features and gathering them in a bottleneck layer in the encoder phase. Then the decoder uses these features to reconstruct the same image after removing noise, called denoising. Or, it can reconstruct the image by filling the empty spaces in it, called impainting.
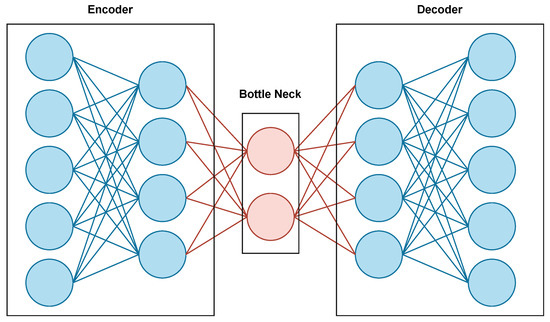
Figure 1.
Autoencoder architecture.
Both symmetric and asymmetric encryption techniques aim to protect data confidentiality, integrity, and authenticity over the Internet and other computer-based systems, such as computer clouds. Symmetric encryption uses the same key to encrypt and decrypt data. On the other hand, asymmetric encryption uses different keys on the encryption and decryption sides. Symmetric encryption is faster and requires fewer hardware and software resources. Transmitting large amounts of data via asymmetric encryption techniques can be considered impractical. Symmetric key algorithms alone cannot provide authentication and integrity. Hence, they should be embedded with other techniques to be considered practical [3].
This research proposes a medial image cryptosystem; the system uses an autoencoder to extract the important features from the image on the sender’s side. These features are the ones to be encrypted using the state-of-the-art advanced encryption standard (AES) [20], and they are then sent to the receiver. After decrypting the features, the receiver uses the decoder part to reconstruct the original image.
Consequently, in this study, we propose a robust medical image encryption algorithm that uses a deep learning model before encrypting the data using AES. The used deep learning model is an autoencoder, which is supposed to give us the ability to minimize the data being encrypted and transferred, as the data transmission is supposed to happen to the output of the encoder part of the autoencoder. This allows for transmitting medical images without sharing the real content of the image. On the decryption side, the decoder is supposed to regenerate the medical image from the encrypted transmitted data after applying decryption. Encrypting the encoder output, which is a part of the autoencoder, makes extracting information from the encrypted data over vulnerable transmission channels almost impossible. Even when malicious parties access the data, the image is not transferred. Even when the data is decrypted, no conclusions can be built over the data without the secret autoencoder model. The autoencoder is also used to enhance the quality of the encrypted images, as it is used as a denoising tool.
1.1. Summery of Contribution
The contributions of this research are listed in the following points:
- We present a new technique for image encryption where deep learning (autoencoder) has been used to generate the shared encrypted data.
- We present an encryption model that allows control of the size and structure of the data being encrypted and transmitted by using the autoencoders as a feature extraction instead of the actual images’ contents.
- We present an efficient encryption model that can denoise medical images during the decryption process.
Previous work that used deep learning techniques with cryptography applications used deep learning mainly as an obfuscation tool to enhance data hiding and prevent malicious views for the data carried in data ciphers. This work uses deep learning tools as an enhancing tool prior to the encryption process. During encryption, deep learning is used to minimize the size of the data to be encrypted and to take the original data into another scope where malicious views are almost impossible. During data transmission, even if the encryption process is broken, the transferred data are the extracted features from the auto-encoder; hence the attacker will get useless data. This use for deep learning tools such as auto-encoders can be considered a state-of-the-art technique that efficiently can improve the encryption process for medical images and many other forms of data.
1.2. Paper Organization
The paper is organized as follows. The next section reviews some significant work related to the current research. Then the proposed encryption model is presented. The fourth section presents the experiments and the results, and finally, the conclusion and future work are described in the last section.
2. Related Work
Data encryption has always been considered essential to protect digital data and information, especially during transmission over different channels—one form of information that has attracted special attention is medical images. Medical images usually require special encryption methods and techniques to hide the information in the image [21,22]. Medical images are also not tolerant of data loss during encryption and transmission due to the importance of the details carried in the images and their role in the diagnosis process.
Medical image encryption was recently discussed in [18,23,24]. In this work, we take a different path than the ones taken in the formerly noted work. We focus on using deep learning methods, namely, autoencoders, to safely and efficiently transfer medical images.
Special encryption methods for medical images were recently proposed, aiming to enhance the encryption process in many ways. For example, the work in [21] proposed an encryption technique using the SCAN technique and a chaotic tent map system to enhance the security measures of the encryption process.
Medical image homomorphic encryption was discussed in [25] to allow access to medical images in their encrypted form. Homomorphic encryption takes care of images being processed over clouds. The study showed that the encryption technique has a very high computational cost, for which they proposed a partition technique with a multi-agent technique to overcome this problem.
Machine learning and deep learning methods were used with medical images for multiple purposes, such as disease detection [26], dermatology health care services [27], and image segmentation [28,29]. Deep learning methods for improving medical image encryption techniques have also been proposed in the literature. The work presented in [30] aims to obfuscate medical images so human eyes cannot detect the important features. At the same time, they can be trained using deep learning models with an acceptable range of accuracy loss. The work used a variational autoencoder (VAE) and a random non-bijective pixel intensity mapping to protect the content of medical images. At the same time, the images could be used to train DL models and give good results.
At the same time, the authors of [2] proposed an image encryption algorithm based on a deep learning model. They proposed this model to encrypt medical images from the Internet of Medical Things systems. Their proposed model (DeepEDN) consists of a cycle generative adversarial network (GAN). This network is trained to transform the medical images into another form that works as a cipher image sent to the receiver. The original image is reconstructed (decrypted) from this cipher image on the receiver’s side. It has been proven to be secured against several types of attacks, such as ciphertext only and chosen ciphertext attacks, in addition to known plaintext and chosen plaintext attacks.
In some phases, the authors of [31] have depended on the work of [2]. They proposed an autoencoder network mainly used to scramble the image and generate a key. Then they used the Cycle GAN, presented by [2], to change the image into a different form. This should be done on the sender’s side. In contrast, at the receiver’s side, a reverse of the operation is applied by using the same structure to reconstruct the original, scrambled image and then descramble it to retrieve the original one. The parameters of the GAN network are used as public and private keys, creating an asymmetric encryption system.
As for [32], they also used GAN to change the linearity nature of an image encryption system. In their paper, they claim that using GAN combined with SHA-256 as a chaotic system could create a cryptosystem that is immune against known plaintext and chosen plaintext attacks that usually target linear image encryption systems. They start by creating and adding noise to the original image and then, by using the logistic maps, convert this image to a cipher image that can be sent to the receiver safely. Depending on the non-linear nature of the used GAN, they proved that their system could resist well-known attacks such as known or chosen plaintext attacks.
Similar techniques were applied in [33]. They started their encryption model by using logistic maps to scramble the image; then, an autoencoder is used to encrypt the image to create a cipher image.
On the other hand, ref. [14] studied the efficiency of using autoencoders in denoising medical images. The autoencoder was trained with a flattened dataset where each row representing an image was processed by adding Gaussian and Poisson noise with different parameter values. The testing results showed visual and measurable enhancement on corrupted images. The autoencoder enhanced the quality of noisy medical images, even with small datasets. Extremely corrupted images that almost did not show the original image content before adding the noise were clarified so that the image content was visible. The study presented in [14] showed that autoencoders could perform better than median filters commonly used in denoising medical images. Using autoencoders to denoise noisy bio-medical images was presented in [34]; the work showed that autoencoders can eliminate the added noise into the images even with a very high noising factor.
It has been noticed so far that GANs, autoencoders, or any neural network used in these systems, are not used alone to create encrypted images. A previous noising or scrambling phase is added to the image before inputting it into the network to create the cipher image. The main target in previous literature was to add randomness to the original image before encrypting it, either by the scrambling or the noising phase. Non-linearity has also been a target that has been added using the NN architecture. We dare to claim that our work is the first in the literature to use an autoencoder network as a pre-step to the encryption algorithm. In this encryption model, we propose that the encrypted data are not the image itself, but the extracted features are the data to be encrypted. We also present the efficiency of the autoencoder in denoising medical images through the encryption process.
Deep learning has found its way to many applications and succeeded in moving them into a more sophisticated intelligent scope. Deep learning is still new to cryptography applications, and few studies use deep learning methods in cryptography applications. This work is influenced by the exceptional cases rising when efficient encryption schemes are used with medical images and use the autoencoder model of deep learning to enhance the encryption process security and encryption process time efficiency. Other work that used deep learning to enhance the security of medical images mainly used it to produce ciphered data rather than produce suitable encrypted content. Table 1 summarizes other work in scope and compares it to our work.

Table 1.
Summary of selected related literature.
3. Proposed Model
The proposed model consists of many steps, from building and training the deep learning model to securely sending and receiving the images. Figure 2 shows the overall steps for the proposed model. At the same time, Figure 3 illustrates the full architecture for the proposed model. Following is a detailed description of each step.
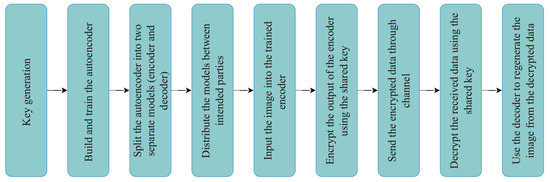
Figure 2.
Proposed model steps.
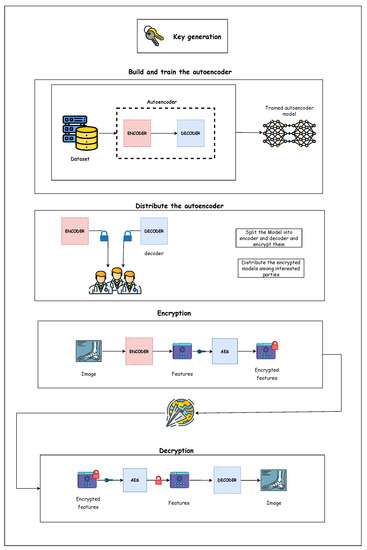
Figure 3.
Proposed model architecture.
3.1. Dataset Selection
Datasets play a critical part in deep learning, so choosing a suitable dataset is one of the essential steps. The proposed model uses the Messidor-2 (http:/www.adcis.net/en/third-party/messidor2/, accessed on 25 June 2022) [35], EyePac Balanced pre-processed dataset, which has 200 images of size (256X256) where each class contains 50 images. The dataset has five classes for diabetic retinopathy (DR) severity. The first class (0) represents the eye with no DR. The rest of the classes (1,2,3,4) represent mild non-proliferative DR, moderate non-proliferative DR, severe non-proliferative DR, and proliferative DR, respectively.
3.2. Deep Learning Model (Autoencoder)
Autoencoders consists of two main networks: the encoder and the decoder. The encoder encodes the images and extracts their features. On the other hand, the decoder decodes the features and reconstructs the image. The autoencoder is considered a semi-supervised learning algorithm, as no labels are involved in the training process. Nevertheless, the output is known, which is the image itself.
3.2.1. Encoder
As previously noted, the encoder is a part of an autoencoder, whose main goal is to learn how to encode the image to its features. The proposed encoder consists of four 2D-Conv layers, one max pooling layer, and a dense layer.
- The first 2D-Conv has 64 3 × 3 filters then a 2 × 2 max pooling layer.
- The second 2D-Conv has 64 3 × 3 filters.
- The third 2D-Conv has 32 filters.
- The last is the dense layer; this layer is added to the autoencoder to downsize the depth of the resulting vector to 3, so it can be visualized as an image.
3.2.2. Decoder
The output of the encoder is the input to the decoder network. Then, the decoder tries reconstructing the image using the feature map from the encoder.
- The first Conv2DTranspose has 32 3 × 3 filters.
- The third Conv2DTranspose has 64 3 × 3 filters then a 2 × 2 upsampling layer.
- The last Conv2DTranspose layer has 64 3 × 3 filters.
Finally, the image reconstruction process is done by a Conv2DTranspose layer with three 3 × 3 filters. Table 2 demonstrates the input and output of each layer in the proposed model.

Table 2.
Autoencoder structure.
3.3. Keys Generation
A symmetric key must be generated and exchanged to apply AES encryption between the involved parties. The scope of this paper concentrates on the usage of autoencoder in the proposed cryptosystem so that any key exchange approach can be used for this purpose.
3.4. Encryption and Decryption
Before describing the proposed cryptosystem, it is worth noting that the trained autoencoder is divided into two separate trained models; encoder and decoder. These models are distributed among the involved parties so that the encoder can be used whenever encryption is needed, and the decoder is used to decrypt any received encrypted images. It can be installed physically on the machines where the encryption and decryption operations will be conducted.
The proposed cryptosystem is illustrated in Figure 4 and can be described in the following steps:
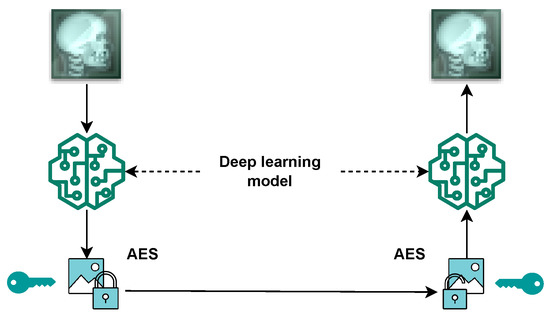
Figure 4.
Proposed encryption model.
- At Sender’s Side: The original image is inputted in the trained encoder to generate a matrix with extracted features.
- At Sender’s Side: The matrix is encrypted using the AES algorithm, using the exchanged symmetric key.
- Via the channel: The encrypted data are sent to the receiver.
- At Receiver’s Side: AES decryptor is used to decrypt the received data.
- At Receiver’s Side: The retrieved data from the decryption process are inputted into the trained decoder to reconstruct the original image.
4. Experiment and Results
The experiment was conducted on Colab Pro. The used dataset has been split into 80% training and 20% testing. The proposed model consists of three main steps.
- The first step was to train the autoencoder. In the proposed model, the autoencoder used ‘adamax’ as its optimizer function with 60 epochs and mean square error as the loss function. The model achieved an accuracy of 83%. Figure 5 shows the loss for training and testing. It has been noticed that the training and testing losses are almost the same, which indicates that the model performs very well.
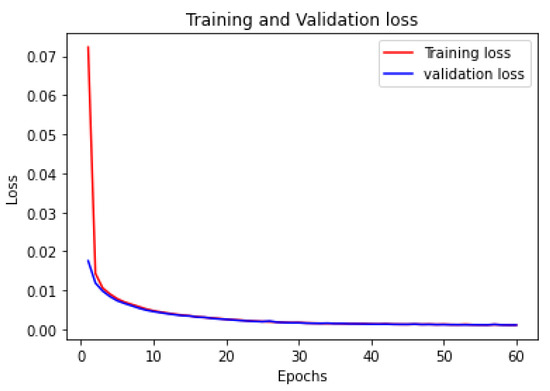 Figure 5. Training and validation loss of autoencoder.
Figure 5. Training and validation loss of autoencoder. - The second step was to use the trained encoder to extract the features from the image using the AES256 encryption algorithm and send the encrypted data to the intended party. Figure 6a shows the original image before any processing, and Figure 6b shows the feature extracted from the original image (the output of the encoder model). It is worth noting that the output of the encoder is not an image-like structure but has been represented in an image for visualization purposes only. In other words, the extracted features from the original image do not conform to an image; they are stored in an n-dimension matrix containing floating-point data. Because the output of the encoder has three dimensions, we were able to convert it into an image. The size of the feature matrix was (112 × 112 × 3). Figure 6c represents the encrypted data output; as previously noted, the extracted features were encrypted using 256 keys derived from the original shared key. The first 256 rows of the features were encrypted using the 256 keys, and the process was repeated until all features were encrypted. The size of the encrypted data was (112 × 112 × 3).
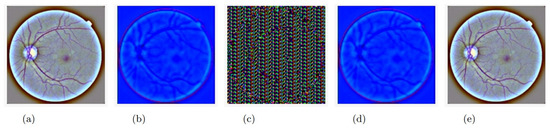 Figure 6. The image changes during its journey from receiver to sender: (a) original image, (b) image outputted from the encoder (sender’s side), (c) image after encryption (sender’s side), (d) image after decryption (receiver’s side), (e) image outputted from the decoder (receiver’s side).
Figure 6. The image changes during its journey from receiver to sender: (a) original image, (b) image outputted from the encoder (sender’s side), (c) image after encryption (sender’s side), (d) image after decryption (receiver’s side), (e) image outputted from the decoder (receiver’s side).
The encryption process takes place after the features are extracted using the autoencoder. AES’s encryption process is performed on the extracted features rather than the medical image data. Using the autoencoder on the decryption side to extract the original image from the decrypted feature data can give the system special robustness.
Table 3 shows the time analysis conducted on the proposed model. It can be noticed that the encryption and decryption times are reduced when using the proposed model.

Table 3.
Time analysis.
It is worth noting that the well-known evaluation metrics that have been used in the literature to evaluate the medical image encryption algorithms cannot be used in our case as the output of the encoder is not an image but rather a bulk of data that has positive and negative floating point values resulting from the deep learning model.
Thus, to compare our work with previous related work, ref. [2] has been chosen for this comparison. In their work, they used a deep learning model (GAN) but utilized it as a part of the encryption and decryption processes. In our case, the deep learning model (autoencoder) was trained to use the encoder as a preliminary step to the encryption process and the decoder as the next step of the decryption process. When comparing both models with the state-of-the-art AES, the model proposed by [2] reduced the original encryption time of AES by 50%; in our model, the reduction was 72%. Table 4 illustrates this comparison.

Table 4.
Comparison with related work.
As previously noted, one of the applications of an autoencoder is denoising images. Medical images may gain some noise during their capturing due to a device problem or even an unclear lens. Thus, in addition to reducing the size of the data to be encrypted, using an autoencoder has also helped reduce the percentage of noise in medical images produced by the decoder in the final step.
Certain experiments were applied to confirm this effect on some of the images in the testing dataset. Noise was added to certain images before inputting them into the model. These images passed through all the phases: encoding, encryption, decryption, and decoding. It was found that the noise amount was reduced from the resulting images.
Two metrics have been used to compare the noise amount on the inputted images and the outputted ones: structure similarity (SSIM) and mean square error (MSE) [36]. Structure similarity, illustrated in Equation (1), is a metric that gives a percentage of similarity between two images; thus, a higher value indicates better results. In the equation, and indicate the local means, and and represent the standard deviations. As for , it represents the cross-covariance of both images.
As for the mean square error metric, illustrated in Equation (2), it calculates the amount of error or difference between two images; thus, the lower the error value, the better. In the equation, O and N represent the original and the noisy images, respectively, and m and n represent number of pixels in each. For further illustration, Table 5 summarizes the used mathematical notations.

Table 5.
Mathematical notation.
Table 6 illustrates the resulting scores of SSIM and MSE and compares the amount of noise between the original and the noisy images on one side and the original and the decoder output images on the other. It has been noticed that for all types of noise added, the decoder reduced this noise in the resulting images.

Table 6.
Analysis of autoencoder effect on noisy images.
Analysis and Discussion
This sub-section is dedicated to discussing the results illustrated earlier. The encrypted data in our scheme is not the content of the original image but a compressed version of the image, meaning we have encrypted the features of the image. Thus, the features are presented as floating-point numbers whose values are not related to the image’s content, and AES is considered robust for encrypting floating-point data. In other words, the encrypted data are no longer a medical image.
Encrypting the extracted features has minimized the amount of data that are required to be encrypted. Accordingly, the time needed for encryption and decryption was reduced. Aside from changing the nature of the image, the encryption and decryption time were reduced by approximately 72%. It has also significantly aided in resisting malicious views on encrypted medical images.
The denoising effect of an autoencoder is caused by the usage of a max pooling layer in the model [37]. This layer reduces the size of an inputted image (matrix) by extracting the most important features. The noise, in this case, is not considered an important feature. Thus, it is discarded as a result of this layer.
In our model, one max pooling layer has been used. It has been noticed that if a second max pooling layer is added to the model, the resulting image will be even less noisy; it will be blurred. Because this model is intended to be used for medical images and the sensitivity of these images, it is more important to clarify the important features than remove a larger amount of noise. Thus, it was decided to settle for one max pooling layer for this purpose.
Regarding the quality of the output of the resulting image from the autoencoder when compared to the original image, the MSE was 0.0019% and the SSIM was 0.9528%, which indicates a loss of 0.0472, which can be neglected.
Returning to Figure 6b–d, they do not represent the actual outputs of the proposed model, which are indeed a set of floating-point values, not the values of the image content. However, the output was reframed in the form of the images noted earlier so that the reader can imagine the process that is taking place. In addition, we intentionally add dense layer (3) to the autoencoder so that we can present it to the reader as an image, but in real-life applications, this last layer will not be applied. Hence, the ability to present the transmitted data (features) as an image will not be an option.
5. Conclusions and Future Work
A hybrid cryptosystem for medical images has been proposed using an autoencoder and AES. The autoencoder is used mainly to convert the image into a matrix of features subsequently encrypted using AES. The decryption and reconstruction of the original image are done on the receiver side. The proposed model has been evaluated mainly by considering the enhanced encryption and decryption execution times when encrypting the features extracted from the original image. Another essential contribution of the proposed model is that the decoder preserves and enhances the quality of the encrypted image. The proposed model can denoise the image even if the image has been affected by unavoidable noise during the capturing phase. SSIM and MSE were calculated to show that the resulting image has small percentages of loss compared with the original image. It has been found that the error in the resulting image did not exceed 15%, but in most types of noises tested, 0% error is detected. Even for noiseless images, the autoencoder has preserved the image quality and resulted in a very small error value of 0.0019 and 0.0472 loss. The research has some limitations, however. First, the dataset used for training the autoencoder is considered small compared to datasets usually used for training tasks. It is intended, as future work, to re-train the model with a larger dataset to enhance the accuracy. The second important limitation, which resulted from the usage of medical images, is that for each type of image (e.g., eyes, chest, brain, etc.), a separate autoencoder model should be trained and used to encrypt and decrypt the specific type.
Author Contributions
Conceptualization, Y.A. (Yasmeen Asalaman) and E.A.; methodology, Y.A. (Yasmeen Asalaman) and E.A.; Formal Analysis, Y.A. (Yasmeen Asalaman) and A.A.; Software, Y.A. (Yasmeen Asalaman) and R.Y.; Resources, E.A. and Y.A. (Yousef AbuHour); Investigation, A.A.; Data curation, A.A. and Y.A. (Yousef AbuHour); Visualization, R.Y. and Q.A.A.-h.; Validation Q.A.A.-h.; Funding Acquisition, Y.A. (Yasmeen Asalaman), E.A., A.A., Y.A. (Yousef AbuHour), R.Y. and Q.A.A.-h.; writing—original draft, Y.A. (Yasmeen Asalaman), E.A., A.A., Y.A. (Yousef AbuHour), R.Y. and Q.A.A.-h.; writing—review and editing, Y.A. (Yasmeen Asalaman), E.A., A.A., Y.A. (Yousef AbuHour), R.Y. and Q.A.A.-h. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The data employed in this reseach is the Messidor-2 dataset. Can be retrived online: http:/www.adcis.net/en/thirdparty/messidor2/ (accessed on 25 June 2022).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Parameshachari, B.D.; Puranga, H.T.; Liberata Ullo, S. Analysis and computation of encryption technique to enhance security of medical images. IOP Conf. Ser. Mater. Sci. Eng. 2020, 925, 012028. [Google Scholar]
- Ding, Y.; Wu, G.; Chen, D.; Zhang, N.; Gong, L.; Cao, M.; Qin, Z. DeepEDN: A deep-learning-based image encryption and decryption network for internet of medical things. IEEE Internet Things J. 2020, 8, 1504–1518. [Google Scholar] [CrossRef]
- Cherniy, D. Securing Embedded Metadata with Symmetric and Asymmetric Encryption. Ph.D. Thesis, National College of Ireland, Dublin, Ireland, 2021. [Google Scholar]
- Abdar, M.; Fahami, M.A.; Chakrabarti, S.; Khosravi, A.; Pławiak, P.; Acharya, U.R.; Tadeusiewicz, R.; Nahavandi, S. BARF: A new direct and cross-based binary residual feature fusion with uncertainty-aware module for medical image classification. Inf. Sci. 2021, 577, 353–378. [Google Scholar] [CrossRef]
- Korot, E.; Guan, Z.; Ferraz, D.; Wagner, S.K.; Zhang, G.; Liu, X.; Faes, L.; Pontikos, N.; Finlayson, S.G.; Khalid, H.; et al. Code-free deep learning for multi-modality medical image classification. Nat. Mach. Intell. 2021, 3, 288–298. [Google Scholar] [CrossRef]
- Devunooru, S.; Alsadoon, A.; Chandana, P.; Beg, A. Deep learning neural networks for medical image segmentation of brain tumours for diagnosis: A recent review and taxonomy. J. Ambient. Intell. Humaniz. Comput. 2021, 12, 455–483. [Google Scholar] [CrossRef]
- Liu, X.; Song, L.; Liu, S.; Zhang, Y. A review of deep-learning-based medical image segmentation methods. Sustainability 2021, 13, 1224. [Google Scholar] [CrossRef]
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep learning techniques for medical image segmentation: Achievements and challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef]
- Shin, H.C.; Tenenholtz, N.A.; Rogers, J.K.; Schwarz, C.G.; Senjem, M.L.; Gunter, J.L.; Andriole, K.P.; Michalski, M. Medical image synthesis for data augmentation and anonymization using generative adversarial networks. In Proceedings of the International Workshop on Simulation and Synthesis in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2018; pp. 1–11. [Google Scholar]
- Rai, S.; Bhatt, J.S.; Patra, S.K. An unsupervised deep learning framework for medical image denoising. arXiv 2021, arXiv:2103.06575. [Google Scholar]
- Zhang, X.; Zhai, D.; Li, T.; Zhou, Y.; Lin, Y. Image inpainting based on deep learning: A review. Inf. Fusion 2022, 90, 74–94. [Google Scholar] [CrossRef]
- Chen, M.; Shi, X.; Zhang, Y.; Wu, D.; Guizani, M. Deep feature learning for medical image analysis with convolutional autoencoder neural network. IEEE Trans. Big Data 2017, 7, 750–758. [Google Scholar] [CrossRef]
- Sharma, S.; Umar, I.; Ospina, L.; Wong, D.; Tizhoosh, H.R. Stacked autoencoders for medical image search. In Proceedings of the International Symposium on Visual Computing; Springer: Berlin/Heidelberg, Germany, 2016; pp. 45–54. [Google Scholar]
- Gondara, L. Medical image denoising using convolutional denoising autoencoders. In Proceedings of the 2016 IEEE 16th International Conference on Data Mining Workshops (ICDMW), Barcelona, Spain, 12–15 December 2016; pp. 241–246. [Google Scholar]
- Ahmed, A.S.; El-Behaidy, W.H.; Youssif, A.A. Medical image denoising system based on stacked convolutional autoencoder for enhancing 2-dimensional gel electrophoresis noise reduction. Biomed. Signal Process. Control 2021, 69, 102842. [Google Scholar] [CrossRef]
- Mehta, J.; Majumdar, A. Rodeo: Robust de-aliasing autoencoder for real-time medical image reconstruction. Pattern Recognit. 2017, 63, 499–510. [Google Scholar] [CrossRef]
- Rachmat, N. Performance analysis of 256-bit AES encryption algorithm on android smartphone. J. Phys. Conf. Ser. 2019, 1196, 012049. [Google Scholar] [CrossRef]
- Ahmad, A.; AbuHour, Y.; Younisse, R.; Alslman, Y.; Alnagi, E.; Abu Al-Haija, Q. MID-Crypt: A Cryptographic Algorithm for Advanced Medical Images Protection. J. Sens. Actuator Netw. 2022, 11, 24. [Google Scholar] [CrossRef]
- Chhabra, S.; Lata, K. Obfuscated AES cryptosystem for secure medical imaging systems in IoMT edge devices. Health Technol. 2022, 12, 971–986. [Google Scholar] [CrossRef]
- Daemen, J.; Rijmen, V. Reijndael: The advanced encryption standard. Dr. Dobb’s J. Softw. Tools Prof. Program. 2001, 26, 137–139. [Google Scholar]
- Parameshachari, B.; Panduranga, H. Medical image encryption using SCAN technique and chaotic tent map system. In Recent Advances in Artificial Intelligence and Data Engineering; Springer: Berlin/Heidelberg, Germany, 2022; pp. 181–193. [Google Scholar]
- Al-Fayoumi, M.A.; Odeh, A.; Ismail Keshta, A.A. Techniques of medical image encryption taxonomy. Bull. Electr. Eng. Inform. 2022, 11, 1990–1997. [Google Scholar] [CrossRef]
- Li, M.; Pan, S.; Meng, W.; Guoyong, W.; Ji, Z.; Wang, L. Medical image encryption algorithm based on hyper-chaotic system and DNA coding. Cogn. Comput. Syst. 2022, 4, 378–390. [Google Scholar] [CrossRef]
- Vanitha, V.; Akila, D. Bio-medical Image Encryption Using the Modified Chaotic Image Encryption Method. In Artificial Intelligence on Medical Data; Springer: Berlin/Heidelberg, Germany, 2023; pp. 231–241. [Google Scholar]
- Vengadapurvaja, A.; Nisha, G.; Aarthy, R.; Sasikaladevi, N. An efficient homomorphic medical image encryption algorithm for cloud storage security. Procedia Comput. Sci. 2017, 115, 643–650. [Google Scholar] [CrossRef]
- Mim, T.A.; Rimi, T.A. A Review on Disease Detection from Medical Images using Machine Learning. In Proceedings of the 2022 6th International Conference on Trends in Electronics and Informatics (ICOEI), Tirunelveli, India, 28–30 April 2022; pp. 1437–1441. [Google Scholar]
- Islam, M.K.; Kaushal, C.; Amin, M.A.; Algarni, A.D.; Alturki, N.; Soliman, N.F.; Mansour, R.F. A secure framework toward IoMT-assisted data collection, modeling, and classification for intelligent dermatology healthcare services. Contrast Media Mol. Imaging 2022, 2022, 6805460. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Wang, G.; Liao, W.; Chen, J.; Song, T.; Chen, Y.; Zhang, S.; Metaxas, D.N.; Zhang, S. Semi-supervised medical image segmentation via uncertainty rectified pyramid consistency. Med. Image Anal. 2022, 80, 102517. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.; Yang, P.; Nie, D.; Wu, X.; Zhou, J.; Wang, Y. Unified medical image segmentation by learning from uncertainty in an end-to-end manner. Knowl.-Based Syst. 2022, 241, 108215. [Google Scholar] [CrossRef]
- Popescu, A.B.; Taca, I.A.; Vizitiu, A.; Nita, C.I.; Suciu, C.; Itu, L.M.; Scafa-Udriste, A. Obfuscation Algorithm for Privacy-Preserving Deep Learning-Based Medical Image Analysis. Appl. Sci. 2022, 12, 3997. [Google Scholar] [CrossRef]
- Bao, Z.; Xue, R.; Jin, Y. Image scrambling adversarial autoencoder based on the asymmetric encryption. Multimed. Tools Appl. 2021, 80, 28265–28301. [Google Scholar] [CrossRef]
- Wu, J.; Xia, W.; Zhu, G.; Liu, H.; Ma, L.; Xiong, J. Image encryption based on adversarial neural cryptography and SHA controlled chaos. J. Mod. Opt. 2021, 68, 409–418. [Google Scholar] [CrossRef]
- Sang, Y.; Sang, J.; Alam, M.S. Image encryption based on logistic chaotic systems and deep autoencoder. Pattern Recognit. Lett. 2022, 153, 59–66. [Google Scholar] [CrossRef]
- Nawarathne, T.; Withanage, T.; Gunarathne, S.; Delay, U.; Somathilake, E.; Senanayake, J.; Godaliyadda, R.; Ekanayake, P.; Rathnayake, C.; Wijayakulasooriya, J. Comprehensive Study on Denoising of Medical Images Utilizing Neural Network-Based Autoencoder. In Advanced Computational Paradigms and Hybrid Intelligent Computing; Springer: Berlin/Heidelberg, Germany, 2022; pp. 159–170. [Google Scholar]
- Yaqoob, M.K.; Ali, S.F.; Bilal, M.; Hanif, M.S.; Al-Saggaf, U.M. ResNet based deep features and random forest classifier for diabetic retinopathy detection. Sensors 2021, 21, 3883. [Google Scholar] [CrossRef] [PubMed]
- Alhayani, B.; Abbas, S.T.; Mohammed, H.J.; Mahajan, H.B. Intelligent secured two-way image transmission using corvus corone module over WSN. Wirel. Pers. Commun. 2021, 120, 665–700. [Google Scholar] [CrossRef]
- Fang, Z.; Jia, T.; Chen, Q.; Xu, M.; Yuan, X.; Wu, C. Laser stripe image denoising using convolutional autoencoder. Results Phys. 2018, 11, 96–104. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).