Abstract
Daptomycin, produced by Streptomyces roseosporus, is a clinically important cyclic lipopeptide antibiotic used for the treatment of human infections caused by drug-resistant Gram-positive pathogens. In contrast to most Streptomyces antibiotic biosynthetic gene clusters (BGCs), daptomycin BGC has no cluster-situated regulator (CSR) genes. DasR, a GntR-family transcriptional regulator (TR) widely present in the genus, was shown to regulate antibiotic production in model species S. coelicolor by binding to promoter regions of CSR genes. New findings reported here reveal that DasR pleiotropically regulates production of daptomycin and reddish pigment, and morphological development in S. roseosporus. dasR deletion enhanced daptomycin production and morphological development, but reduced pigment production. DasR inhibited daptomycin production by directly repressing dpt structural genes and global regulatory gene adpA (whose product AdpA protein activates daptomycin production and morphological development). DasR-protected regions on dptEp and adpAp contained a 16 nt sequence similar to the consensus DasR-binding site dre in S. coelicolor. AdpA was shown to target dpt structural genes and dptR2 (which encodes a DeoR-family TR required for daptomycin production). A 10 nt sequence similar to the consensus AdpA-binding site was found on target promoter regions dptAp and dptR2p. This is the first demonstration that DasR regulates antibiotic production both directly and through a cascade mechanism. The findings expand our limited knowledge of the regulatory network underlying daptomycin production, and will facilitate methods for construction of daptomycin overproducers.
1. Introduction
Gram-positive Streptomyces species are abundant in soil and decaying vegetation. Their morphological development (involving formation of substrate hyphae, aerial hyphae, and chains of spores) is typically associated with the synthesis of secondary metabolites, including antibiotics that display well-documented antibacterial, antiviral, antifungal, anthelmintic, anticancer, and immunosuppressive activities. Antibiotic biosynthetic genes are usually clustered, with one or more cluster-situated regulator (CSR) genes located within the cluster. Antibiotic production is tightly controlled by complex regulatory networks based on CSRs and various higher-level pleiotropic/global regulators that respond to environmental and physiological changes [1,2,3].
Streptomyces roseosporus produces daptomycin, a clinically important cyclic lipopeptide antibiotic that strongly inhibits multidrug-resistant Gram-positive pathogens, including methicillin-resistant Staphylococcus aureus (MRSA), through a distinctive action mechanism [4]. Daptomycin is used as a “last resort” treatment for right-sided infective endocarditis and complex skin and soft tissue infections [5]. It contains 13 amino acids to form a 10-membered cyclic peptide and a 3-amino-acid tail with a straight-chain decanoic acid moiety [6]. Daptomycin is biosynthesized through a nonribosomal peptide synthase (NRPS) pathway [7], and its biosynthetic cluster dpt consists of 12 structural genes. Among these, dptA, dptBC, and dptD encode three subunits of NRPS; dptE and dptF are involved in activation of decanoic acid; and dptG, -H, -I, -J, -M, -N, and -P are responsible for precursor supply, resistance, or transport [8,9]. Three regulatory genes (dptR1, dptR2, dptR3) are located adjacent to the dpt structural genes. In view of the clinical and commercial importance of daptomycin, many studies have addressed the regulatory mechanism of its biosynthesis. Our group and YQ Li’s group observed that DptR1 did not affect daptomycin production [10,11], whereas GH Yu’s group reported that daptomycin production was repressed by both deletion and overexpression of dptR1 [12]. The reason for these seemingly contradictory findings is unclear. DptR2 is required for daptomycin production, but does not regulate the expression of the dpt cluster [13]. We found that DptR3 promotes daptomycin production by activating transcription of dpt structural genes in an indirect manner [11]. These findings suggest that the dpt cluster does not contain CSR genes, in contrast to other typical antibiotic biosynthetic gene clusters (BGCs) of Streptomyces. YQ Li’s group demonstrated that transcriptional regulators (TRs) AtrA [14], DepR1 [10], DepR2 [15], and PhaR [16] regulate daptomycin production by binding directly to dptEp, the promoter for core operon dptE-dptF-dptA-dptBC-dptD-dptG-dptH of the dpt cluster. They also reported that two global regulators, AdpA and PhoP, bind competitively to atrAp, activate atrA expression, and thereby promote daptomycin production [17]. WblA (WhiB-family TR) inhibits daptomycin production [18], and the cyclic AMP receptor protein Crp has a positive effect on daptomycin production [19], but whether their regulatory effect is direct or indirect is unclear. We recently demonstrated [20] that BldD, the master repressor of Streptomyces development, also activates daptomycin production directly and through a cascade mechanism based on binding to dptEp, dptR3p, adpAp, and afsRp. Our knowledge of the regulatory network underlying daptomycin production remains quite limited and fragmentary despite the above findings, presenting an obstacle to the rational design of daptomycin high-yielding strains through genetic manipulation.
DasR, a GntR-family TR, was initially identified in model species S. coelicolor as a global regulator involved in control of GlcNAc/chitin catabolism [21,22], antibiotic production [23], and morphological development [22]. Consensus DasR-binding site dre (DasR-responsive element) is a 16 nt imperfect palindromic sequence 5′-DSWGGWSTVVDCMHBN-3′ (D = A/G/T; S = C/G; W = A/T; V = A/C/G; M = A/C; H = A/C/T; B = G/C/T; N = A/G/C/T) [3,21]. DasR represses actinorhodin (Act) and undecylprodigiosin (Red) production by binding to dre sites upstream of cluster-situated activator genes actII-ORF4 and redZ, respectively [23]. DasR is also essential for S. coelicolor development; dasR deletion results in “bald” phenotype [22]. In S. cinnamonensis, DasR acts as an activator of monensin production by binding to promoter regions of CSR and structural genes of mon cluster [24]. DasR homologs are present in actinomycetes other than Streptomyces, including the erythromycin producer Saccharopolyspora erythraea. DasR in this species directly activates reddish pigment production by binding to dre site upstream of rpp cluster, and indirectly promotes erythromycin production. dasR deletion in S. erythraea causes delayed development, but does not abolish formation of aerial hyphae and spores as it does in S. coelicolor [25]. The above findings reflect the diverse roles of DasR in secondary metabolism and development, and the complex regulatory mechanisms involved. To date, DasR functions have not been investigated in S. roseosporus.
Here, we describe the characterization of DasR in S. roseosporus as a dual repressor/activator, i.e., repressor of daptomycin production and morphological development, and activator of pigment production. Its inhibitory effect on daptomycin production is mediated by both direct and cascade mechanisms, based on binding to dptEp and adpAp. Our findings demonstrate the essential role of DasR in control of Streptomyces antibiotic production.
2. Results
2.1. DasR Inhibits Daptomycin Production and Morphological Development, but Promotes Pigment Production
S. roseosporus dasR gene contains 765 nucleotides (nt) and encodes a 254-amino-acid protein, DasR. DasR is a highly conserved protein, as revealed by sequence alignment analysis; it has 89.8, 92.1, 98.8, and 93.7% amino acid identities with its homologs in S. coelicolor, S. avermitilis, S. griseus, and S. venezuelae, respectively, reflecting its important functional role in the genus.
To investigate the role of DasR in S. roseosporus, the dasR-deletion mutant DdasR was constructed by homologous recombination (Figure S1A). Daptomycin production by DdasR grown in fermentation medium for 10 days was 26% higher than that of the wild-type (WT) strain NRRL11379 [26] (Figure 1A). Complementation of DdasR (strain CdasR) restored daptomycin production to WT level. Enhancement of dasR expression in WT (strain OdasR) reduced daptomycin production by 25%. For plasmid control strains WT/pKC1139 and WT/pSET152, daptomycin production was close to that of WT (Figure 1A). Time course measurements of growth and daptomycin production showed that biomass (dry cell weight) values of DdasR and OdasR were similar to that of WT (Figure 1B), whereas daptomycin production was upregulated by dasR deletion and downregulated by dasR overexpression (Figure 1C). These findings indicate that DasR acts as a repressor of daptomycin production and has no effect on cell growth.
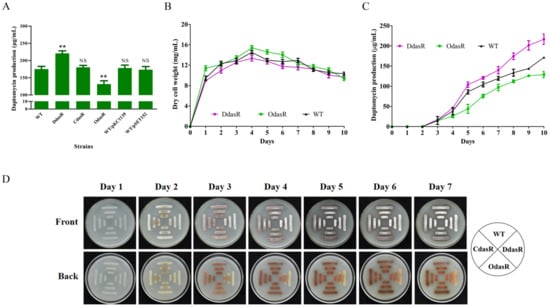
Figure 1.
Effects of DasR on daptomycin and pigment production, cell growth, and morphological development in S. roseosporus. (A) Daptomycin production in WT, dasR-deletion mutant DdasR, complemented strain CdasR, overexpression strain OdasR, and plasmid control strains WT/pKC1139 and WT/pSET152 cultured in fermentation medium for 10 days. Statistical notations: NS, not significant; **, p < 0.01 for comparison with WT (t-test). (B) Growth curves for WT, DdasR, and OdasR. Biomass is presented as dry cell weight. (C) Daptomycin production curves for WT, DdasR, and OdasR. Error bars in panels (A–C): SD for three replicates. (D) Phenotypes of WT, DdasR, CdasR, and OdasR grown on DA1 plates at 28 °C.
WT, DdasR, CdasR, and OdasR were grown on DA1 plates for sporulation in order to assess the effect of DasR on morphological development. Spore formation occurred earlier for DdasR, whereas development of CdasR and OdasR did not differ notably from that of WT (Figure 1D), indicating that DasR is a repressor of S. roseosporus development.
S. roseosporus produces a reddish pigment (a secondary metabolite). Pigment production was much lower for DdasR than for WT, but did not differ notably for CdasR or OdasR (Figure 1D), indicating that DasR is an activator of pigment production.
2.2. DasR Regulates the Transcription of dpt Genes
To investigate the causes of daptomycin overproduction in DdasR, we performed RT-qPCR analysis of RNAs prepared from WT and DdasR cultured in fermentation medium at days 2 (exponential phase), 4 (early stationary phase), and 6 (middle stationary phase). Transcription levels of structural genes dptA, dptBC, dptD, dptE, dptF, dptG, dptH, dptI, dptM, and dptP in the dpt cluster were higher for DdasR than for WT at one, two, or all three of these time points (Figure 2). These data are consistent with the daptomycin production data. In DdasR, all dpt structural genes were upregulated on day 2, suggesting that DasR represses transcription of these genes mainly during the early fermentation stage.
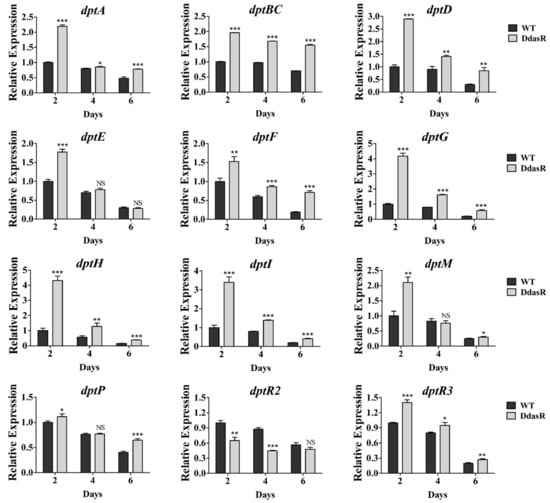
Figure 2.
RT-qPCR analysis of dpt genes in WT and DdasR grown in fermentation medium for 2, 4, or 6 days. WT transcription level for each gene on day 2 was defined as 1. NS, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001 (t-test). Error bars: SD for three replicates.
There are three regulatory genes (dptR1, dptR2, dptR3) adjacent to the dpt cluster. Because we found that DptR1 was not involved in control of daptomycin production [11], we only assessed transcription levels of dptR2 and dptR3 in WT and DdasR using the same RNA preparations. RT-qPCR results revealed that dptR2 was downregulated, whereas dptR3 was upregulated in DdasR (Figure 2). DptR2 and DptR3 were previously shown to promote daptomycin production [11,13]. Therefore, the observed change of daptomycin production in DdasR reflects a combined effect of altered transcription levels of dpt structural and regulatory genes.
2.3. DasR Binds Specifically to the dptE-Promoter Region
To determine whether the dpt genes described above are regulated directly by DasR, we performed electrophoretic mobility shift assays (EMSAs) using soluble His6-DasR purified from E. coli and respective promoter probes. The dpt cluster consists of four transcriptional units: dptP, dptM-dptN, dptE-dptF-dptA-dptBC-dptD-dptG-dptH (hereafter termed “dptE operon”), and dptI-dptJ [20,27] (Figure 3A). dptA, dptBC, and dptD are large, contiguous genes that encode subunits of NRPS for biosynthesis of daptomycin skeleton. We therefore performed 5′ RACE analysis to test the possibility that dptA has its own transcriptional start site (TSS). dptA TSS was mapped to G, 453 nt upstream of dptA translational start codon (TSC) (Figure S2), indicating that dptA has its own promoter for dptA-dptBC-dptD-dptG-dptH (hereafter termed “dptA operon”). Promoter probes dptP-M (containing bidirectional promoters), dptEp, dptAp, dptIp, dptR2p, and dptR3p were designed and used for EMSAs (Figure 3A). TRs generally do not bind to open reading frame (ORF) of genes. We therefore used nonspecific probe hrdB within hrdB ORF as negative control. EMSA results indicated that His6-DasR formed complexes with probe dptEp, but not with dptP-M, dptAp, dptIp, dptR2p, dptR3p, or hrdB (Figure 3B). Binding specificity was evaluated by competition assays using ~100-fold excess unlabeled specific probe dptEp (lane S) (which abolished delayed bands) and hrdB (lane N) (which did not). Our findings indicate that DasR directly represses dptE operon, whereas it represses dptI, dptM, dptP, and dptR3, and activates dptR2, in an indirect manner.
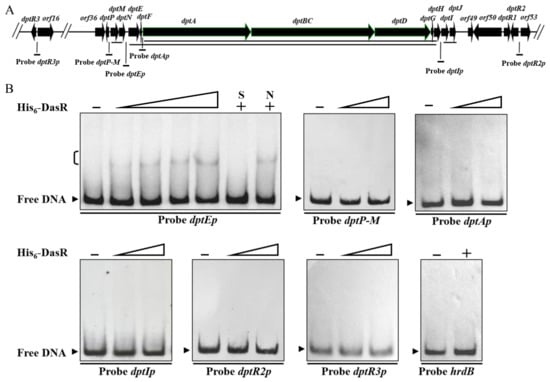
Figure 3.
Binding of DasR to dptE-promoter region. (A) Promoter probes used for EMSAs (schematic). Long solid lines: transcriptional units. (B) EMSAs of interactions of His6-DasR with promoter probes of dpt genes. Negative control probe: hrdB. Concentrations of His6-DasR for probe dptEp: 1, 1.5, 3, and 4.5 μM; for other dpt promoter probes: 1 and 4.5 μM. Lanes –: EMSAs without His6-DasR. A measure of 4.5 μM His6-DasR was used for competition assays and control probe hrdB (lanes +). Lanes N and S: competition assays with ~100-fold excess of unlabeled nonspecific probe hrdB (N) and specific probe dptEp (S). Bracket (upper left): DasR-DNA complex. Arrowheads: free probe.
DNase I footprinting assays were performed to determine the precise DasR-binding site on dptEp, and to clarify the mechanism whereby DasR regulates target dptE operon. DasR protected a 25 nt region containing a 16 nt sequence (5’-AGTGGTTTGGTCCGCC-3’) (Figure 4A), similar to the conserved DasR-binding site dre (5’-DSWGGWSTVVDCMHBN-3’) (D = A/G/T; S = C/G; W = A/T; V = A/C/G; M = A/C; H = A/C/T; B = G/C/T; N = A/G/C/T) in S. coelicolor [3,21], suggesting that the DNA-binding property of DasR is conserved in the genus.
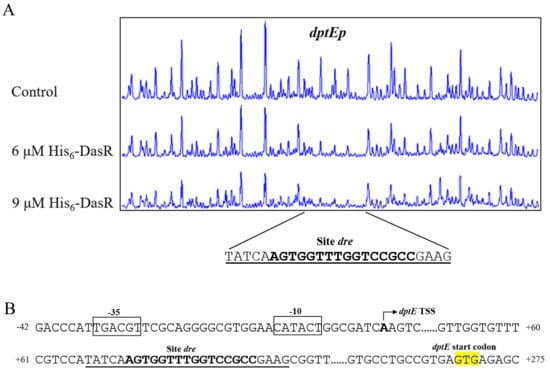
Figure 4.
Determination of DasR-binding site on dptEp. (A) DNase I footprinting assay of DasR on dptEp. Protection patterns were acquired with increasing His6-DasR concentrations. Control: reaction without His6-DasR. (B) Nucleotide sequences of dptE-promoter region and DasR-binding site. Numbers: distance (nt) from dptE TSS. Bent arrow: dptE TSS. Boxes: putative −10 and −35 regions. Yellow highlight: dptE TSC. Solid line: DasR-binding site. Bold font: dre-like sequence.
We previously mapped the TSS of dptE to A, 267 nt upstream of dptE TSC [20], and accordingly predicted −10 and −35 promoter elements (Figure 4B). The 16 nt DasR-binding site was found to extend from positions +72 to +87, relative to dptE TSS (Figure 4B). DasR presumably represses dptE operon transcription by blocking transcriptional extension of RNA polymerase.
2.4. DasR Directly Represses adpA Involved in Daptomycin Production and Morphological Development
Additional DasR target genes involved in daptomycin production were investigated by using dre sequence with MAST/MEME program (http://meme-suite.org, accessed on 29 June 2022) to scan the S. roseosporus genome. Global regulatory gene adpA was identified as a putative DasR target. AdpA protein distributes widely among Streptomyces species, and controls antibiotic production and morphological development [1]. The accuracy of bioinformatic prediction was tested by performing EMSAs using His6-DasR and promoter probe adpAp. DasR bound specifically to adpAp (Figure 5A), indicating that it targets adpA. adpA transcription level was enhanced at three time points during growth of DdasR in fermentation medium (Figure 5B), indicating that DasR acts as a repressor of adpA.
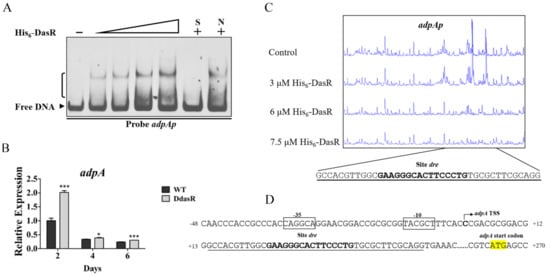
Figure 5.
Identification of DasR target adpA. (A) EMSAs of His6-DasR with probe adpAp. Lanes 2–5: 1, 1.5, 3, and 4.5 μM His6-DasR. Notations are as in Figure 3B. (B) RT-qPCR analysis of adpA in WT and DdasR grown in fermentation medium. *, p < 0.05; ***, p < 0.001 (t-test). Error bars: SD for three replicates. (C) DNase I footprinting assay of DasR on adpAp. (D) Nucleotide sequences of adpA promoter region and DasR-binding site. Formatting conventions as in Figure 4B.
We reported previously that adpA TSS is localized at C, 263 nt upstream of adpA TSC [20] DNase I footprinting assays revealed that the 40 nt protected site of DasR on adpAp extends from positions +14 to +53 nt relative to adpA TSS, and contains a 16 nt dre-like sequence 5′-GAAGGGCACTTCCCTG-3′ (Figure 5C,D). This sequence is located downstream of adpA TSS (Figure 5D), suggesting that DasR represses adpA transcription through a mechanism similar to that of dptE operon repression.
The 2015 study by YQ Li’s group indicated that AdpA promotes daptomycin production and morphological development in S. roseosporus [14]. That study involved construction of an adpA-deletion mutant, based on WT strain SW0702, that abolished daptomycin production, but did not involve construction of an adpA overexpression strain. We further investigated the role of AdpA in S. roseosporus by constructing adpA-deletion mutant DadpA (Figure S1B), complemented strain CadpA, and overexpression strain OadpA based on WT strain NRRL11379. Quantitative analysis of cultures following 10-day fermentation revealed that, relative to WT, daptomycin production was reduced by 33% in DadpA, and increased by 49% in OadpA (Figure 6A). Daptomycin production by CadpA was close to that of WT, demonstrating that reduced production in DadpA was due solely to adpA deletion. Examination of WT, DadpA, and OadpA growth and fermentation curves revealed that adpA deletion and overexpression did not notably affect cell growth (Figure 6B), but reduced and increased daptomycin production, respectively (Figure 6C). The above findings clearly indicate that AdpA positively regulates daptomycin production, regardless of the reduced production observed for DadpA. Enhanced adpA expression in dasR-deletion mutant DdasR thus contributes to increased daptomycin production.
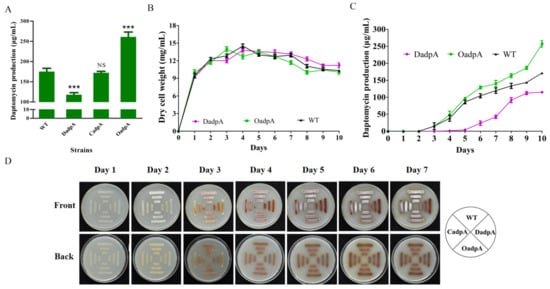
Figure 6.
Effects of AdpA on daptomycin and pigment production, cell growth, and morphological development in S. roseosporus. (A) Daptomycin production in WT, DadpA, CadpA, and OadpA cultured in fermentation medium for 10 days. NS, not significant; ***, p < 0.001 (t-test). (B) Growth curves for WT, DadpA, and OadpA. (C) Daptomycin production curves for WT, DadpA, and OadpA. Error bars in panels (A–C): SD for three replicates. (D) Phenotypes of WT, DadpA, CadpA, and OadpA grown on DA1 plates at 28 °C.
The role of AdpA in S. roseosporus morphological phenotype was investigated by streaking WT, DadpA, CadpA, and OadpA strains on DA1 plates. DadpA displayed “bald” phenotype, i.e., grew in substrate mycelia (Figure 6D), consistent with the 2015 study by YQ Li’s group [14]. CadpA and OadpA were phenotypically similar to WT (Figure 6D). These findings indicate that normal development requires the presence of AdpA. Pigment production showed no notable changes in any of the four strains (Figure 6D), suggesting that AdpA is not involved in this process.
2.5. AdpA Directly Regulates Expression of dptA Operon and dtpR2
The 2015 study by YQ Li’s group identified atrA, which encodes TetR-family TR AtrA that directly activates expression of dptE operon, as an AdpA target [14]. To determine whether AdpA also directly regulates dpt genes, we performed a series of EMSAs using soluble His6-AdpA and dpt promoter probes. His6-AdpA clearly retarded probes dptAp and dptR2p, but did not bind to dptP-M, dptEp, dptIp, dptR3p, or control probe hrdB (Figure 7A), indicating that dptA operon and dptR2 are novel AdpA targets. Effects of AdpA on expression of target dpt genes were assessed by RT-qPCR. In DadpA grown in fermentation medium, transcription levels of dptA, dptBC, dptD, dptG, and dptH within dptA operon were reduced at two (days 2 and 4) or three time points (days 2, 4, and 6), and that of dptR2 was increased at days 2 and 4 (Figure 7B), indicating that AdpA functions as an activator of dptA operon but as a repressor of dptR2. AdpA was shown to be an activator of atrA, and adpA deletion resulted in decreased atrA expression [14]. Therefore, reduced daptomycin production in DadpA is a collective result of altered expression of AdpA-targeted atrA and dpt genes.
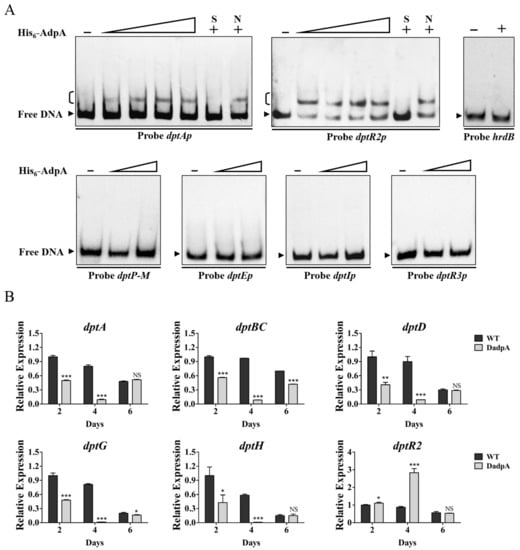
Figure 7.
Identification of AdpA targets dptA and dptR2. (A) EMSAs of His6-AdpA with indicated promoter probes. Concentrations of His6-AdpA for probes dptAp and dptR2p: 50, 100, 150, and 200 nM; for control probe hrdB: 200 nM; for other probes: 50 and 200 nM. A measure of 200 nM His6-AdpA was used for competition assays. Notations are same as in Figure 3B. (B) RT-qPCR analysis of dptR2 and dpt structural genes within dptA operon in WT and DadpA grown in fermentation medium. NS, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001 (t-test). Error bars: SD for three replicates.
We determined precise AdpA-binding sites on target promoters dptAp and dptR2p by DNase I footprinting assays. AdpA protected a 35 nt region extending from +26 to +60 nt relative to dptA TSS and containing a 10 nt sequence (5′-TGGCACGCCA-3′) (Figure 8A), similar to the consensus AdpA-binding site 5′-TGGCSNGWWY-3′ (S = C/G; N = A/G/C/T; W = A/T; Y = C/T) [28]. Binding of a transcriptional activator to a site downstream of the TSS is uncommon; however, BldD [20] and DepR1 [10] were shown to activate dptE by binding to sites downstream of dptE TSS. The regulatory mechanism underlying such transcriptional activation remains to be elucidated.
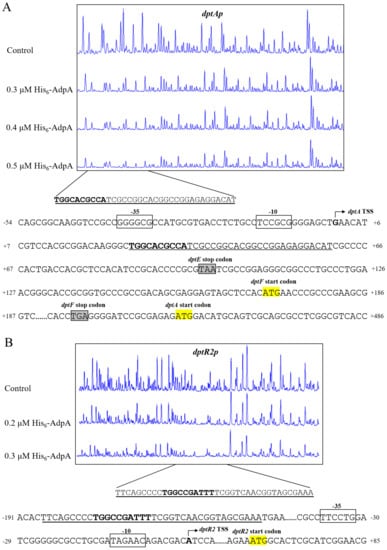
Figure 8.
DNase I footprinting assays and nucleotide sequences of AdpA-binding sites on dptAp (A) and dptR2p (B). Numbers: distance (nt) from respective TSSs. Bent arrows: TSSs. Boxes: putative −10 and −35 regions. Yellow highlight: TSCs. Solid lines: AdpA protected regions. Bold font: sequence similar to the consensus AdpA-binding site.
The dptR2 TSS was identified by 5′ RACE and mapped to A, 65 nt upstream of dptR2 TSC (Figure S3). A 38 nt AdpA protected region was detected on dptR2p, located −187 to −150 nt relative to dptR2 TSS (Figure 8B). A 10 nt sequence (5′-TGGCCGATTT-3′) similar to the consensus AdpA-binding site was also found in the AdpA-protected region (Figure 8B). This is analogous to our previous finding that binding sites of AvaR2 (TetR-family TR) on aveRp (promoter of cluster-situated activator gene aveR) are far upstream of aveR TSS, and that AvaR2 represses aveR [29]. The mechanism underlying transcriptional repression in this case also remains to be elucidated.
3. Discussion
The role of DasR in control of antibiotic production has been well characterized in S. coelicolor and S. cinnamonensis. DasR inhibits Act and Red production through CSR genes [23], and promotes monensin production through both CSR and biosynthetic genes [24]. The present study was focused on the molecular mechanism whereby DasR regulates production of daptomycin, a clinically important antibiotic whose BGC lacks CSRs, and revealed that DasR acts as a repressor in this process. DasR directly represses expression of dpt biosynthetic genes and of regulatory gene adpA, whose product AdpA controls daptomycin production through activation of atrA [14] and dptA operon, but repression of dptR2. Thus, DasR regulates daptomycin production both directly and in a cascade manner. This is the first report of such cascade regulatory mechanism of DasR for antibiotic production control in Streptomyces. DasR and AdpA are widely present in Streptomyces species. Therefore, DasR presumably regulates AdpA in other species besides S. roseosporus.
adpA, a target of DasR, is a global regulatory gene involved in secondary metabolism and morphological development in Streptomyces. It was first identified in S. griseus and shown to be directly repressed by A-factor receptor ArpA [30]. Derepression of adpA causes activation of secondary metabolic and developmental processes. In S. coelicolor, adpA is directly repressed by its encoding protein AdpA [31], BldD [32], and pseudo γ-butyrolactone receptor ScbR2 [33], and plays essential roles in Act and Red production and morphological development. In S. ansochromogenes, AdpA is required for nikkomycin production and morphological development [34], but inhibits oviedomycin production [35]. In S. xiamenensis, AdpA represses development and differentially regulates production of polycyclic tetramate macrolactams and xiamenmycin [36]. In S. roseosporus, adpA has been demonstrated to be repressed by ArpA [14] and activated by BldD [20]. We found in the present study that adpA is also repressed by DasR. Thus, AdpA plays differing roles in secondary metabolism and morphological development in various Streptomyces species, and regulation of its expression is complex. The adpA-deletion mutant constructed based on S. roseosporus SW0702 in the 2015 study by YQ Li’s group abolished daptomycin production [14], whereas the mutant constructed based on strain NRRL11379 [26] in the present study reduced—but did not abolish—daptomycin production. This seeming discrepancy may be due to differences in the parental strains and the growth media used.
dptEp drives the expression of the core dptE operon, which contains seven daptomycin biosynthetic genes and is the major promoter within the dpt cluster. Transcriptional regulation of dptEp is highly complex; to date, six TRs have been shown to target this promoter: AtrA [14], DepR1 [10], DepR2 [15], PhaR [16], BldD [20], and DasR (present study). The signaling molecules that DepR1, DepR2, and PhaR sense and respond to are unclear. atrA is targeted by AdpA, a key TR in the A-factor signaling pathway; therefore, it is likely that extracellular A-factor-like signals affect dptE operon expression via AtrA [14]. BldD’s control of its targets is based on its response to signaling molecule c-di-GMP [37]. The DNA-binding activity of DasR is modulated by multiple signaling molecules that act as allosteric effectors. Binding of DasR to its target DNAs is inhibited by glucosamine-6-phosphate (GlcN-6P) and N-acetylglucosamine-6-phosphate (GlcNAc-6P) catabolized from N-acetylglucosamine (GlcNAc), but enhanced by organic phosphate metabolites from glucose catabolism (e.g., glucose-6-phosphate, glucose-1-phosphate, glycerol-3-phosphate, fructose-1,6-bisphosphate, fructose-6-phosphate) and by inorganic phosphate [22,38]; The metabolic status of a particular cell thus determines the effect of DasR on expression of its targets. In a well-established S. coelicolor model, GlcN-6P and GlcNAc-6P (metabolic intermediates of GlcNAc released by cell wall autolysis of substrate mycelium during development) act as DasR allosteric effectors, impair repressive effect of DasR on target CSR genes, and thereby induce antibiotic production [22,23,38]. In view of our findings that DasR represses daptomycin production, and that its target promoters dptEp and adpAp both contain consensus dre-like site, it seems likely that DasR responds to GlcN-6P and GlcNAc-6P in control of daptomycin production in S. roseosporus, just as its role in antibiotic production in S. coelicolor. The possibility cannot be ruled out, however, that other metabolites (e.g., phosphorylated sugars) act as signaling molecules in DasR-mediated regulation of daptomycin production, depending on culture conditions. Identification of additional environmental or physiological signals that affect daptomycin production will help clarify the complex regulatory mechanisms of this process.
We propose a model of DasR-mediated regulation of daptomycin production in S. roseosporus (Figure 9), based on present and previous findings. According to this model, DasR exerts its regulatory effect on daptomycin production through several mechanisms: (i) direct repression of dptE operon (which contains dptE, dptF, dptA, dptBC, dptD, dptG, and dptH); (ii) indirect repression of other dpt structural genes (dptP, dptM, dptI); (iii) direct repression of regulatory gene adpA, which activates daptomycin production; (iv) indirect activation of dptR2 through global regulator AdpA (and perhaps other regulators); (v) indirect repression of dptR3 through yet-unknown mechanism(s). DasR also regulates morphological development and pigment production, besides daptomycin production. Future studies will identify additional DasR targets involved in these biological processes.
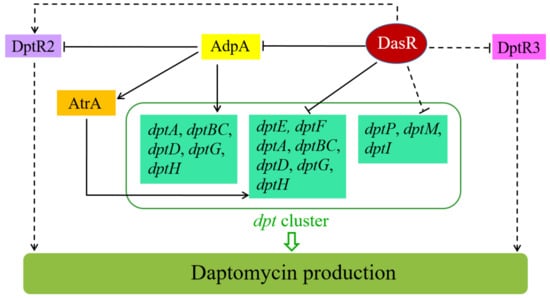
Figure 9.
Proposed model of regulatory role of DasR in control of daptomycin production in S. roseosporus. Solid arrows: activation. Bars: repression. Solid lines: direct control. Dashed lines: indirect control.
4. Materials and Methods
4.1. Strains, Plasmids, Primer Pair, and Growth Conditions
Strains and plasmids used are listed in Table S1, and primers are listed in Table S2. Growth conditions for S. roseosporus and Escherichia coli strains were described previously [11,20]. Solid DA1 [11] was used for S. roseosporus sporulation and phenotype observation. Seed medium and fermentation medium [11] were used for daptomycin production.
4.2. Construction of S. roseosporus Mutant Strains
For in-frame gene deletion of dasR, a 427 bp 5′ flanking region (positions from −391 to +36 relative to dasR start codon) and a 605 bp 3′ flanking region (positions from −30 to +575 relative to dasR stop codon) were amplified, respectively, with primer pairs CQ9/CQ10 and CQ11/CQ12 from WT genomic DNA. The two fragments were connected by fusion PCR with primer pair CQ9/CQ12 and ligated into XbaI/HindⅢ-digested pKC1139 [39] to generate dasR-deletion plasmid pDdasR, which was then transformed into WT protoplasts. dasR-deletion mutant DdasR was isolated as described previously [11], confirmed by PCR with primer pairs CQ13/CQ14 (flanking exchange regions) and CQ15/CQ16 (located within deletion region of dasR ORF) (Figure S1A), and DNA-sequenced. Use of primer pair CQ13/CQ14 generated a 1389 bp band in the mutant and a 2085 bp band in WT. When primer pair CQ15/CQ16 was used, only WT produced a 418 bp band (data not shown).
For complementation of DdasR, a 1118 bp DNA fragment carrying dasR ORF and its promoter was amplified with primer pair CQ75/CQ76. The PCR product was excised with EcoRI/XbaI and ligated into pSET152 [39] to generate dasR-complemented plasmid pCdasR, which was then transformed into DdasR to obtain complemented strain CdasR. For overexpression of dasR, an 897 bp DNA fragment carrying dasR ORF was amplified with primer pair CQ21/CQ22 and excised with HindIII/XbaI. A 188 bp ermE*p fragment was excised from pJL117 [40] with EcoRI/HindIII. The two fragments were ligated simultaneously into EcoRI/XbaI-digested pKC1139 to generate dasR overexpression plasmid pOdasR, which was transformed into WT to construct dasR overexpression strain OdasR.
To construct an adpA-gene-deletion mutant, a 423 bp 5′ flanking region (positions from −335 to +88 relative to adpA start codon) was amplified with primer pair CQ1/CQ2, and a 560 bp 3′ flanking region (positions from −55 to +505 relative to adpA stop codon) was amplified with primer pair CQ3/CQ4. The two fragments were fused by PCR with primer pair CQ1/CQ4 and ligated into EcoRI/XbaI-digested pKC1139 to generate adpA-deletion plasmid pDadpA, which was then transformed into WT. The mutant, termed DadpA, was isolated by selection of DdasR and confirmed by PCR with primer pairs CQ5/CQ6 (flanking exchange regions) and CQ7/CQ8 (located within deletion region of adpA ORF) (Figure S1B). Use of primer pair CQ5/CQ6 generated a 1414 bp band in the mutant and a 2450 bp band in WT. When primer pair CQ7/CQ8 were used, only WT produced a 298 bp band (data not shown).
For complementation of DadpA, a 1705 bp DNA fragment carrying adpA ORF and its promoter was amplified with primer pair CQ19/CQ20, excised with EcoRI/XbaI, and ligated into pSET152 to generate adpA-complemented plasmid pCadpA, which was transformed into DadpA to obtain complemented strain CadpA. For adpA overexpression, a 1391 bp DNA fragment carrying adpA ORF was amplified using primer pair CQ17/CQ18, excised with HindIII/XbaI, and ligated simultaneously with the 188 bp EcoRI/HindIII-ermE*p fragment into EcoRI/XbaI-digested pKC1139 to generate adpA overexpression plasmid pOadpA, which was transformed into WT to construct adpA overexpression strain OadpA.
4.3. Production and Analysis of Daptomycin
The fermentation process of S. roseosporus strains and the quantitative analysis of daptomycin production by HPLC were performed as described previously [11]. Briefly, spores of S. roseosporus prepared from DA1 plates were added to 50 mL primary seed medium in flasks and incubated at 28 °C, 250 rpm for 48 h. The culture in primary seed medium was inoculated at 6% (v/v) into 50 mL secondary seed medium and incubated at same condition for 30 h. Then, a 6% (v/v) inoculation volume of the culture in secondary seed medium was inoculated into 50 mL fermentation medium and cultured for 10 days. Sodium decanoate (final concentration 0.02%, w/v) was added every 12 h after 48 h of fermentation until the end of fermentation.
After fermentation, the broth was centrifuged and the supernatant was analyzed by HPLC on a C18 reverse-phase column (5 μm, 4.6 mm × 100 mm; Waters, Milford, MA, USA) with UV detection at 218 nm at a flow rate of 1.0 mL/min. The mobile phase contained 0.1% (v/v) trifluoroacetic acid in water and acetonitrile (55:45, v/v). Authentic daptomycin sample was used as a standard.
4.4. Reverse Transcription and Quantitative Real-Time PCR (RT-qPCR) Analysis
Samples of S. roseosporus strains grown in fermentation medium were collected at various time points and frozen in liquid nitrogen. Total RNAs were prepared using TRIzol reagent (Tiangen; Beijing, China), and samples were treated with DNase I (TaKaRa; Kusatsu, Japan) to eliminate genomic DNA contamination. Reverse transcription for cDNA synthesis and quantitative PCR assay of transcription levels of tested genes using respective primer pair (Table S2) were performed as described previously [11]. Relative transcription value of each gene was normalized relative to internal control gene hrdB value using comparative Ct method. Experiments were performed in triplicate.
4.5. Overexpression and Purification of His6-DasR and His6-AdpA
To prepare His6-DasR protein, an 855 bp fragment containing dasR ORF was amplified with primer pair CQ25/CQ26. To prepare His6-AdpA protein, primer pair CQ23/CQ24 was used to amplify a 1259 bp fragment containing adpA ORF. PCR products were excised with EcoRI/XhoI and ligated into pET-28a (+) to generate pET28-dasR and pET28-adpA, which were transformed separately into E. coli BL21 (DE3) for protein overexpression. Cells were induced by 0.4 mM IPTG for 10 h at 16 °C, and those containing recombinant protein were collected, washed, disrupted in lysis buffer [41] by sonication on ice, and centrifuged. Recombinant His6-tagged protein from supernatant was purified using Ni2+-NTA resin (Qiagen; Hilden, Germany), eluted with lysis buffer plus 250 mM imidazole, and dialyzed against binding buffer for electrophoretic mobility shift assays (EMSAs) to eliminate imidazole. Purified His6-DasR and His6-AdpA were stored at −80 °C until use.
4.6. EMSAs
Probes carrying promoter regions of tested genes were amplified using corresponding primer pair (Table S2), labeled at the 3′ end with digoxigenin (DIG), and EMSAs were performed using a DIG gel shift kit (2nd generation; Roche, Mannheim, Germany) as described previously [41]. Each binding reaction system (20 μL) contained 1 μg poly [d(I-C)], 0.15 nM labeled probe, and various amounts of His6-DasR or His6-AdpA as indicated. Specificity of DasR (or AdpA)-probe interaction was tested by adding ~100-fold excess of each specific or nonspecific hrdB unlabeled probe to the reaction system.
4.7. DNase I Footprinting Assay
To identify binding sites of DasR and AdpA, 5′-FAM fluorescence-labeled DNA fragments corresponding to upstream regions of tested genes were PCR-synthesized using primers listed in Table S2, and gel-purified. DNase I footprinting assays were performed using a nonradiochemical capillary electrophoresis method as described previously [11,42], and electropherogram data were analyzed using software program GeneMarker V. 2.2.0.
4.8. 5′ Rapid Amplification of cDNA Ends (5′ RACE)
TSSs of dptA and dptR2 were identified by 5′ RACE using a 5′/3′ RACE Kit (Roche; Mannheim, Germany). A measure of 2 µg total RNA extracted from 48 h culture of S. roseosporus WT grown in fermentation medium was used for cDNA synthesis with 20 pmol gene-specific primer dptA-SP1 or dptR2-SP1. Obtained cDNAs were purified, and oligo-dA tails were added to 3′ ends by terminal transferase (TaKaRa). First, a PCR round was performed using tailed cDNA as template, and oligo dT-anchor primer and second inner gene-specific primer dptA-SP2 or dptR2-SP2. Second PCR round was performed to yield a specific single band, using 10-fold diluted original PCR product as template, and anchor primer and nested primer dptA-SP3 or dptR2-SP3. The final PCR product was sent for sequencing, and TSS was determined as the first nucleotide following oligo-dA sequence.
5. Conclusions
In summary, we characterized S. roseosporus DasR as a key repressor during daptomycin production through both direct and cascade mechanisms, based on binding to promoter regions of dptE operon and global regulatory gene adpA. DasR was also demonstrated to negatively regulate morphological development, but positively regulate pigment production. Furthermore, we verified AdpA function as an activator of daptomycin production and development, and identified dptA operon and dptR2 as novel AdpA targets. Our work contributes to the clarification of the complex regulatory mechanisms of daptomycin production and rationally construct daptomycin overproducers.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics11081065/s1. Figure S1: Strategy for deletion of dasR (A) and adpA (B) in WT (schematic). Large arrows: genes and their directions. Small arrows: positions of primers. Rectangles: homologous exchange regions used for gene deletion. Figure S2: Determination of dptA TSS. (A) 5′ RACE analysis of dptA TSS. Box: complementary sequence of oligo dT-anchor primer. Bent arrow: complementary base of TSS. (B) Promoter structure of dptA. Boxes: putative −10 and −35 regions. Bent arrow: dptA TSS. Yellow highlight: TSCs. Shading: translational stop codons. Figure S3: Determination of dptR2 TSS. (A) 5′ RACE analysis of dptR2 TSS. Box: complementary sequence of oligo dT-anchor primer. Bent arrow: complementary base of TSS. (B) Promoter structure of dptR2. Boxes: putative −10 and −35 regions. Bent arrow: dptR2 TSS. Yellow highlight: dptR2 TSC. Table S1: Strains and plasmids used in this study. Table S2: Primers used in this study. References [39,40,43] are cited in the Supplementary Materials.
Author Contributions
Conceptualization, Y.W.; methodology, Q.C., J.Z., and X.L.; validation, Q.C. and J.Z.; formal analysis, Y.W.; investigation, Q.C.; data curation, Y.W.; writing—original draft preparation, J.Z.; writing—review and editing, Y.W.; supervision, Y.W.; project administration, Y.W.; funding acquisition, Y.W. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 32170081).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are grateful to S. Anderson for English editing of the manuscript and to Yinghua Lu (Xiamen University, China) for providing S. roseosporus NRRL11379.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef] [PubMed]
- Urem, M.; Swiatek-Polatynska, M.A.; Rigali, S.; van Wezel, G.P. Intertwining nutrient-sensory networks and the control of antibiotic production in Streptomyces. Mol. Microbiol. 2016, 102, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Van Wezel, G.P.; McDowall, K.J. The regulation of the secondary metabolism of Streptomyces: New links and experimental advances. Nat. Prod. Rep. 2011, 28, 1311–1333. [Google Scholar] [CrossRef] [PubMed]
- Zuttion, F.; Colom, A.; Matile, S.; Farago, D.; Pompeo, F.; Kokavecz, J.; Galinier, A.; Sturgis, J.; Casuso, I. High-speed atomic force microscopy highlights new molecular mechanism of daptomycin action. Nat. Commun. 2020, 11, 6312. [Google Scholar] [CrossRef]
- Gonzalez-Ruiz, A.; Seaton, R.A.; Hamed, K. Daptomycin: An evidence-based review of its role in the treatment of Gram-positive infections. Infect. Drug Resist. 2016, 9, 47–58. [Google Scholar]
- Debono, M.; Abbott, B.J.; Molloy, R.M.; Fukuda, D.S.; Hunt, A.H.; Daupert, V.M.; Counter, F.T.; Ott, J.L.; Carrell, C.B.; Howard, L.C.; et al. Enzymatic and chemical modifications of lipopeptide antibiotic A21978C: The synthesis and evaluation of daptomycin (LY146032). J. Antibiot. 1988, 41, 1093–1105. [Google Scholar] [CrossRef]
- Robbel, L.; Marahiel, M.A. Daptomycin, a bacterial lipopeptide synthesized by a nonribosomal machinery. J. Biol. Chem. 2010, 285, 27501–27508. [Google Scholar] [CrossRef]
- Miao, V.; Coeffet-Legal, M.F.; Brian, P.; Brost, R.; Penn, J.; Whiting, A.; Martin, S.; Ford, R.; Parr, I.; Bouchard, M.; et al. Daptomycin biosynthesis in Streptomyces roseosporus: Cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology 2005, 151, 1507–1523. [Google Scholar] [CrossRef]
- Wittmann, M.; Linne, U.; Pohlmann, V.; Marahiel, M.A. Role of DptE and DptF in the lipidation reaction of daptomycin. FEBS J. 2008, 275, 5343–5354. [Google Scholar] [CrossRef]
- Yuan, P.; Zhou, R.; Chen, X.; Luo, S.; Wang, F.; Mao, X.; Li, Y. DepR1, a TetR family transcriptional regulator, positively regulates daptomycin production in an industrial producer, Streptomyces roseosporus SW0702. Appl. Environ. Microbiol. 2016, 82, 1898–1905. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, Q.; Zhuang, S.; Chen, Z.; Wen, Y.; Li, J. A MarR family transcriptional regulator, DptR3, activates daptomycin biosynthesis and morphological differentiation in Streptomyces roseosporus. Appl. Environ. Microbiol. 2015, 81, 3753–3765. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Hui, M.; Li, R.; Zhang, S. Pleiotropic regulation of daptomycin synthesis by DptR1, a LuxR family transcriptional regulator. World J. Microbiol. Biotechnol. 2020, 36, 173. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Ren, N.; Luo, S.; Chen, X.; Mao, X.; Li, Y. DptR2, a DeoR type auto-regulator, is required for daptomycin production in Streptomyces roseosporus. Gene 2014, 544, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Luo, S.; Zhou, R.; Wang, F.; Yu, P.; Sun, N.; Chen, X.; Tang, Y.; Li, Y. Transcriptional regulation of the daptomycin gene cluster in Streptomyces roseosporus by an autoregulator, AtrA. J. Biol. Chem. 2015, 290, 7992–8001. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Luo, S.; Li, Y. Negative regulation of daptomycin production by DepR2, an ArsR-family transcriptional factor. J. Ind. Microbiol. Biotechnol. 2017, 44, 1653–1658. [Google Scholar] [CrossRef]
- Luo, S.; Chen, X.; Mao, X.; Li, Y. Transposon-based identification of a negative regulator for the antibiotic hyper-production in Streptomyces. Appl. Microbiol. Biotechnol. 2018, 102, 6581–6592. [Google Scholar] [CrossRef]
- Zheng, Y.; Sun, C.; Fu, Y.; Chen, X.; Li, Y.; Mao, X. Dual regulation between the two-component system PhoRP and AdpA regulates antibiotic production in Streptomyces. J. Ind. Microbiol. Biotechnol. 2019, 46, 725–737. [Google Scholar] [CrossRef]
- Huang, X.; Ma, T.; Tian, J.; Shen, L.; Zuo, H.; Hu, C.; Liao, G. wblA, a pleiotropic regulatory gene modulating morphogenesis and daptomycin production in Streptomyces roseosporus. J. Appl. Microbiol. 2017, 123, 669–677. [Google Scholar] [CrossRef]
- Wu, J.; Chen, D.; Wu, J.; Chu, X.; Yang, Y.; Fang, L.; Zhang, W. Comparative transcriptome analysis demonstrates the positive effect of the cyclic AMP receptor protein Crp on daptomycin biosynthesis in Streptomyces roseosporus. Front. Bioeng. Biotechnol. 2021, 9, 618029. [Google Scholar] [CrossRef]
- Yan, H.; Lu, X.; Sun, D.; Zhuang, S.; Chen, Q.; Chen, Z.; Li, J.; Wen, Y. BldD, a master developmental repressor, activates antibiotic production in two Streptomyces species. Mol. Microbiol. 2020, 113, 123–142. [Google Scholar] [CrossRef]
- Colson, S.; Stephan, J.; Hertrich, T.; Saito, A.; van Wezel, G.P.; Titgemeyer, F.; Rigali, S. Conserved cis-acting elements upstream of genes composing the chitinolytic system of streptomycetes are DasR-responsive elements. J. Mol. Microbiol. Biotechnol. 2007, 12, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Rigali, S.; Nothaft, H.; Noens, E.E.; Schlicht, M.; Colson, S.; Muller, M.; Joris, B.; Koerten, H.K.; Hopwood, D.A.; Titgemeyer, F.; et al. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol. Microbiol. 2006, 61, 1237–1251. [Google Scholar] [CrossRef]
- Rigali, S.; Titgemeyer, F.; Barends, S.; Mulder, S.; Thomae, A.W.; Hopwood, D.A.; van Wezel, G.P. Feast or famine: The global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008, 9, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lin, C.; Li, X.; Tang, Z.; Qiao, J.; Zhao, G. DasR positively controls monensin production at two-level regulation in Streptomyces cinnamonensis. J. Ind. Microbiol. Biotechnol. 2016, 43, 1681–1692. [Google Scholar] [CrossRef]
- Liao, C.; Xu, Y.; Rigali, S.; Ye, B. DasR is a pleiotropic regulator required for antibiotic production, pigment biosynthesis, and morphological development in Saccharopolyspora erythraea. Appl. Microbiol. Biotechnol. 2015, 99, 10215–10224. [Google Scholar] [CrossRef]
- Ng, I.S.; Ye, C.; Zhang, Z.; Lu, Y.; Jing, K. Daptomycin antibiotic production processes in fed-batch fermentation by Streptomyc-es roseosporus NRRL11379 with precursor effect and medium optimization. Bioprocess Biosyst. Eng. 2014, 37, 415–423. [Google Scholar] [CrossRef]
- Coeffet-Le Gal, M.F.; Thurston, L.; Rich, P.; Miao, V.; Baltz, R.H. Complementation of daptomycin dptA and dptD deletion mutations in trans and production of hybrid lipopeptide antibiotics. Microbiology 2006, 152, 2993–3001. [Google Scholar] [PubMed][Green Version]
- Higo, A.; Hara, H.; Horinouchi, S.; Ohnishi, Y. Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res. 2012, 19, 259–273. [Google Scholar] [CrossRef]
- Zhu, J.; Sun, D.; Liu, W.; Chen, Z.; Li, J.; Wen, Y. AvaR2, a pseudo gamma-butyrolactone receptor homologue from Streptomyces avermitilis, is a pleiotropic repressor of avermectin and avenolide biosynthesis and cell growth. Mol. Microbiol. 2016, 102, 562–578. [Google Scholar] [CrossRef]
- Ohnishi, Y.; Yamazaki, H.; Kato, J.Y.; Tomono, A.; Horinouchi, S. AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 2005, 69, 431–439. [Google Scholar] [CrossRef]
- Wolanski, M.; Donczew, R.; Kois-Ostrowska, A.; Masiewicz, P.; Jakimowicz, D.; Zakrzewska-Czerwinska, J. The level of AdpA directly affects expression of developmental genes in Streptomyces coelicolor. J. Bacteriol. 2011, 193, 6358–6365. [Google Scholar] [CrossRef] [PubMed]
- Den Hengst, C.D.; Tran, N.T.; Bibb, M.J.; Chandra, G.; Leskiw, B.K.; Buttner, M.J. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 2010, 78, 361–379. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ji, J.; Li, X.; Wang, J.; Li, S.; Pan, G.; Fan, K.; Yang, K. Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA 2014, 111, 5688–5693. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Liu, G.; Yang, H.; Tian, Y.; Tan, H. The pleiotropic regulator AdpA-L directly controls the pathway-specific activator of nikkomycin biosynthesis in Streptomyces ansochromogenes. Mol. Microbiol. 2009, 72, 710–723. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, J.; Zhuo, J.; Li, Y.; Tian, Y.; Tan, H. Activation and mechanism of a cryptic oviedomycin gene cluster via the disruption of a global regulatory gene, adpA, in Streptomyces ansochromogenes. J. Biol. Chem. 2017, 292, 19708–19720. [Google Scholar] [CrossRef]
- Bu, X.; Weng, J.; He, B.; Xu, M.; Xu, J. A novel AdpA homologue negatively regulates morphological differentiation in Streptomyces xiamenensis 318. Appl. Environ. Microbiol. 2019, 85, e03107–e03118. [Google Scholar] [CrossRef]
- Tschowri, N.; Schumacher, M.A.; Schlimpert, S.; Chinnam, N.B.; Findlay, K.C.; Brennan, R.G.; Buttner, M.J. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 2014, 158, 1136–1147. [Google Scholar] [CrossRef]
- Tenconi, E.; Urem, M.; Swiatek-Połatynska, M.A.; Titgemeyer, F.; Muller, Y.A.; van Wezel, G.P.; Rigali, S. Multiple allosteric effectors control the affinity of DasR for its target sites. Biochem. Biophys. Res. Commun. 2015, 464, 324–329. [Google Scholar] [CrossRef]
- Bierman, M.; Logan, R.; O’Brien, K.; Seno, E.T.; Rao, R.N.; Schoner, B.E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 1992, 116, 43–49. [Google Scholar] [CrossRef]
- Li, L.; Guo, J.; Wen, Y.; Chen, Z.; Song, Y.; Li, J. Overexpression of ribosome recycling factor causes increased production of avermectin in Streptomyces avermitilis strains. J. Ind. Microbiol. Biotechnol. 2010, 37, 673–679. [Google Scholar] [CrossRef]
- Luo, S.; Sun, D.; Zhu, J.; Chen, Z.; Wen, Y.; Li, J. An extracytoplasmic function sigma factor, σ(25), differentially regulates avermectin and oligomycin biosynthesis in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 2014, 98, 7097–7112. [Google Scholar] [CrossRef] [PubMed]
- Zianni, M.; Tessanne, K.; Merighi, M.; Laguna, R.; Tabita, F.R. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J. Biomol. Tech. 2006, 17, 103–113. [Google Scholar] [PubMed]
- Macneil, D.J.; Klapko, L.M. Transformation of Streptomyces avermitilis by plasmid DNA. J. Ind. Microbiol. 1987, 2, 209–218. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).