Exploring the Contribution of the AcrB Homolog MdtF to Drug Resistance and Dye Efflux in a Multidrug Resistant E. coli Isolate
Abstract
1. Introduction
2. Results and Discussion
2.1. Impact of mdtF Inactivation on Drug Susceptibility
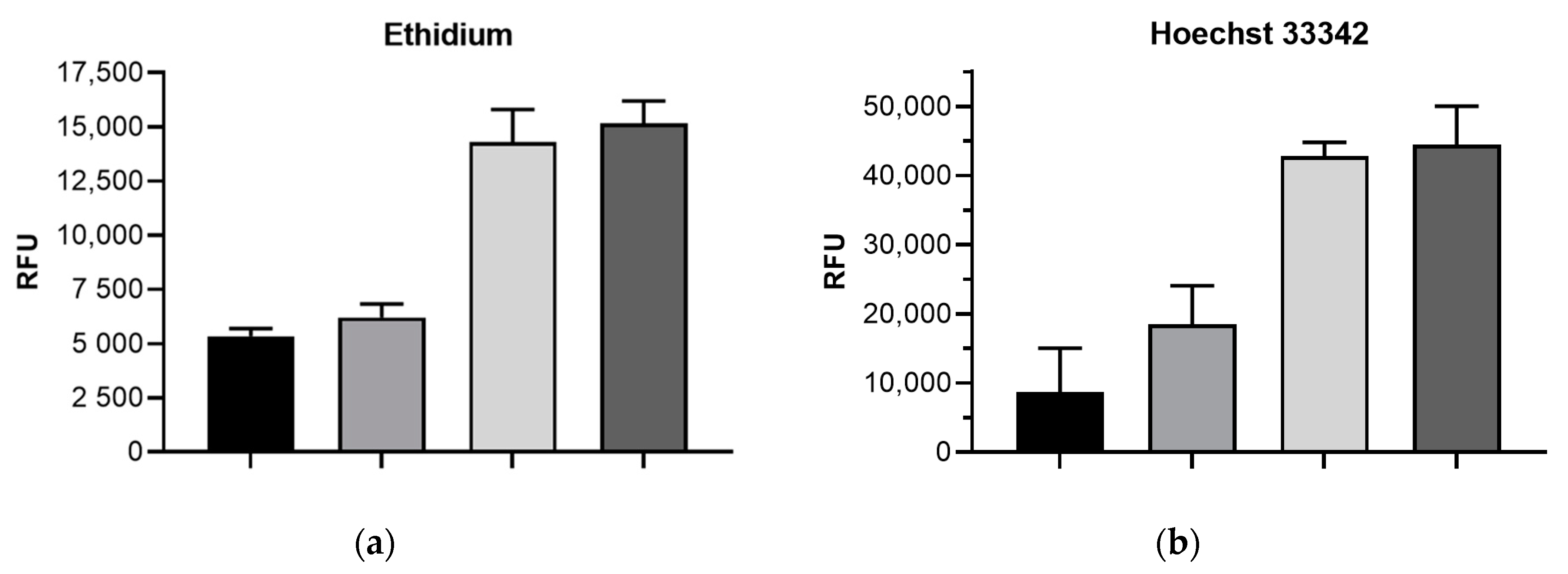
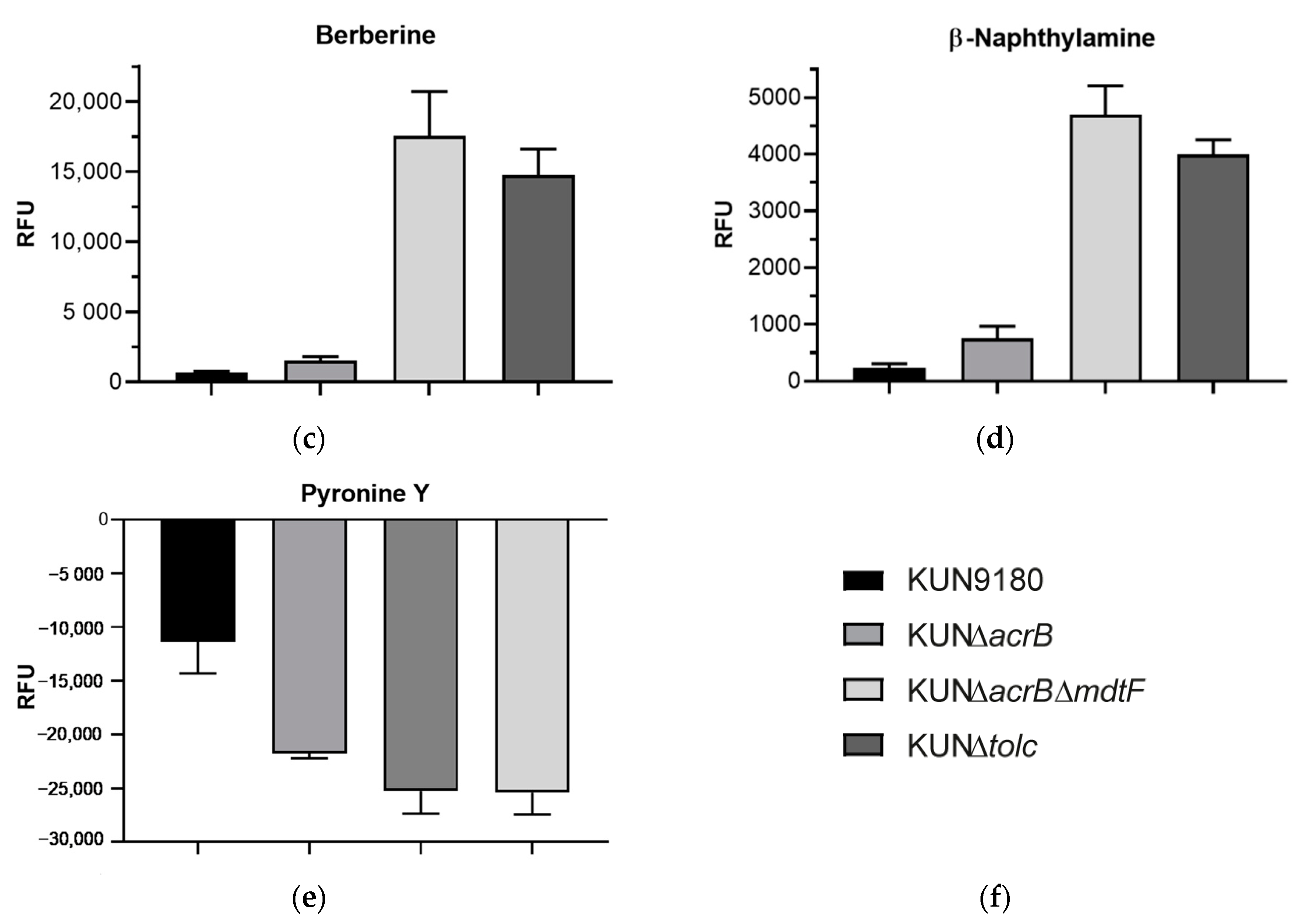
2.2. Impact of mdtF Inactivation on Intracellular Dye Accumulation
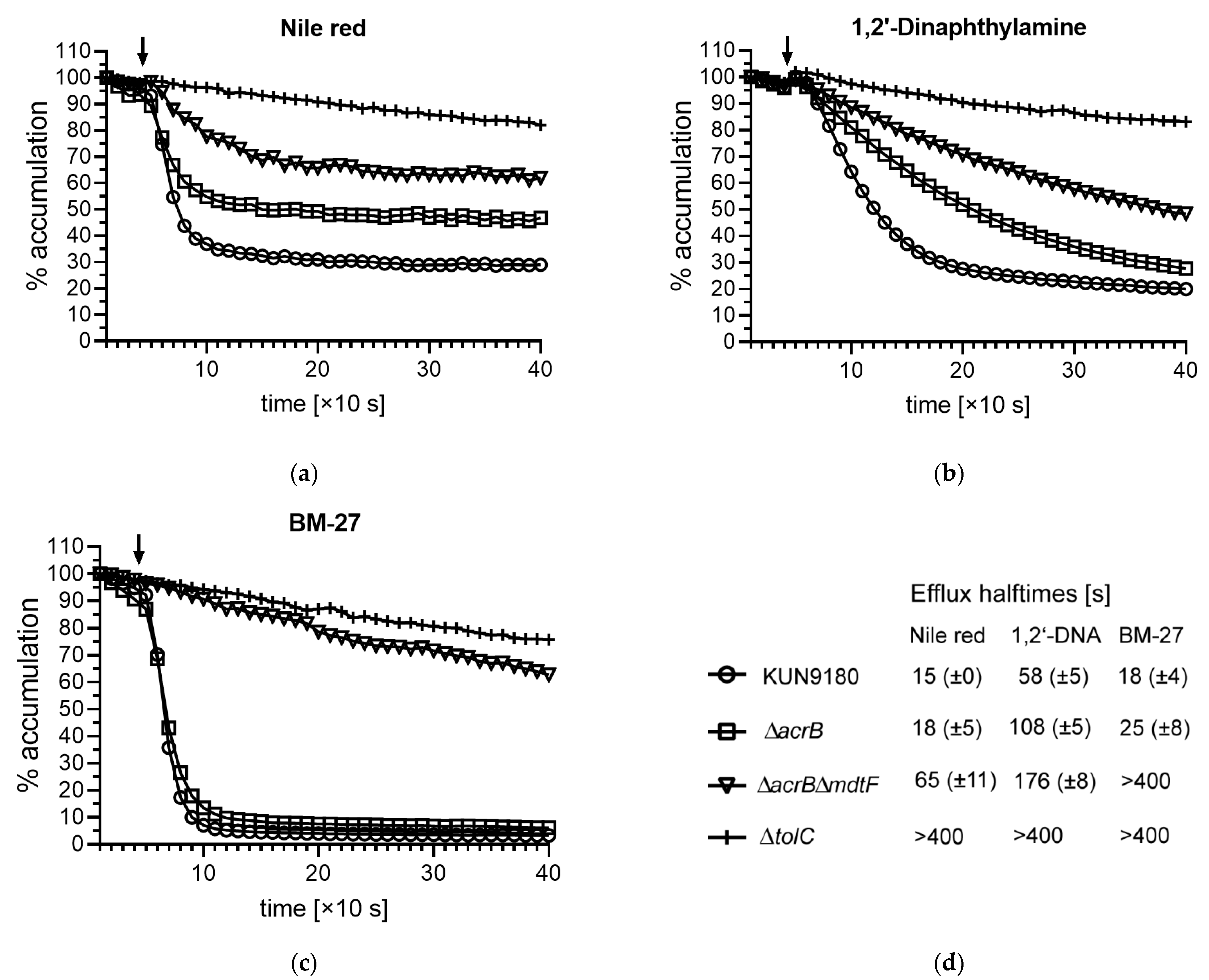
2.3. Impact of mdtF Inactivation on Real-Time Efflux
3. Materials and Methods
3.1. Strains, Growth Conditions, Chemicals
3.2. Susceptibility Testing
3.3. Engineering of Mutant KUN∆acrB∆mdtF
3.4. Dye Accumulation Assays
3.5. Real-Time Efflux Assays
3.6. Statistical Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Kern, W.V.; Oethinger, M.; Jellen-Ritter, A.S.; Levy, S.B. Non-Target Gene Mutations in the Development of Fluoroquinolone Resistance in Escherichia coli. Antimicrob. Agents Chemother. 2000, 44, 814–820. [Google Scholar] [CrossRef]
- Schuster, S.; Vavra, M.; Schweigger, T.M.; Rossen, J.W.; Matsumura, Y.; Kern, W.V. Contribution of AcrAB-TolC to multidrug resistance in an Escherichia coli sequence type 131 isolate. Int. J. Antimicrob. Agents 2017, 50, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Cunrath, O.; Meinel, D.M.; Maturana, P.; Fanous, J.; Buyck, J.M.; Auguste, P.S.; Seth-Smith, H.M.; Körner, J.; Dehio, C.; Trebosc, V.; et al. Quantitative contribution of efflux to multi-drug resistance of clinical Escherichia coli and Pseudomonas aeruginosa strains. EBioMedicine 2019, 41, 479–487. [Google Scholar] [CrossRef]
- Bialek-Davenet, S.; Lavigne, J.-P.; Guyot, K.; Mayer, N.; Tournebize, R.; Brisse, S.; Leflon-Guibout, V.; Nicolas-Chanoine, M.-H. Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2015, 70, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Gravey, F.; Cattoir, V.; Ethuin, F.; Fabre, L.; Beyrouthy, R.; Bonnet, R.; Le Hello, S.; Guérin, F. ramR deletion in an Enterobacter hormaechei isolate as a consequence of therapeutic failure to key antibiotics in a long-term hospitalized patient. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef]
- Ruzin, A.; Immermann, F.W.; Bradford, P.A. RT-PCR and Statistical Analyses of adeABC Expression in Clinical Isolates of Acinetobacter calcoaceticus–Acinetobacter baumannii Complex. Microb. Drug Resist. 2010, 16, 87–89. [Google Scholar] [CrossRef]
- Gajdács, M. Carbapenem-Resistant but Cephalosporin-Susceptible Pseudomonas aeruginosa in Urinary Tract Infections: Opportunity for Colistin Sparing. Antibiotics 2020, 9, 153. [Google Scholar] [CrossRef]
- Bohnert, J.A.; Schuster, S.; Fähnrich, E.; Trittler, R.; Kern, W.V. Altered spectrum of multidrug resistance associated with a single point mutation in the Escherichia coli RND-type MDR efflux pump YhiV (MdtF). J. Antimicrob. Chemother. 2006, 59, 1216–1222. [Google Scholar] [CrossRef]
- Camp, J.; Schuster, S.; Vavra, M.; Schweigger, T.; Rossen, J.W.A.; Reuter, S.; Kern, W.V. Limited Multidrug Resistance Efflux Pump Overexpression among Multidrug-Resistant Escherichia coli Strains of ST131. Antimicrob. Agents Chemother. 2021, 65, 01735-20. [Google Scholar] [CrossRef]
- Schuster, S.; Vavra, M.; Köser, R.; Rossen, J.W.A.; Kern, W.V. New Topoisomerase Inhibitors: Evaluating the Potency of Gepotidacin and Zoliflodacin in Fluoroquinolone-Resistant Escherichia coli upon tolC Inactivation and Differentiating Their Efflux Pump Substrate Nature. Antimicrob. Agents Chemother. 2020, 65. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Yamaguchi, A. Analysis of a Complete Library of Putative Drug Transporter Genes in Escherichia coli. J. Bacteriol. 2001, 183, 5803–5812. [Google Scholar] [CrossRef] [PubMed]
- Zgurskaya, H.I.; Krishnamoorthy, G.; Ntreh, A.; Lu, S. Mechanism and Function of the Outer Membrane Channel TolC in Multidrug Resistance and Physiology of Enterobacteria. Front. Microbiol. 2011, 2, 189. [Google Scholar] [CrossRef] [PubMed]
- Bleuel, C.; Grosse, C.; Taudte, N.; Scherer, J.; Wesenberg, D.; Krauß, G.J.; Nies, D.H.; Grass, G. TolC Is Involved in Enterobactin Efflux across the Outer Membrane of Escherichia coli. J. Bacteriol. 2005, 187, 6701–6707. [Google Scholar] [CrossRef]
- Vega, D.E.; Young, K.D. Accumulation of periplasmic enterobactin impairs the growth and morphology of Escherichia coli tolC mutants. Mol. Microbiol. 2013, 91, 508–521. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, M.; Horiyama, T.; Zhang, Y.; Li, X.; Nishino, K.; Yan, A. The Multidrug Efflux Pump MdtEF Protects against Nitrosative Damage during the Anaerobic Respiration in Escherichia coli. J. Biol. Chem. 2011, 286, 26576–26584. [Google Scholar] [CrossRef]
- Sjuts, H.; Vargiu, A.V.; Kwasny, S.M.; Nguyen, S.T.; Kim, H.-S.; Ding, X.; Ornik, A.R.; Ruggerone, P.; Bowlin, T.L.; Nikaido, H.; et al. Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc. Natl. Acad. Sci. USA 2016, 113, 3509–3514. [Google Scholar] [CrossRef]
- Lomovskaya, O.; Warren, M.S.; Lee, A.; Galazzo, J.; Fronko, R.; Lee, M.; Blais, J.; Cho, D.; Chamberland, S.; Renau, T.; et al. Identification and Characterization of Inhibitors of Multidrug Resistance Efflux Pumps in Pseudomonas aeruginosa: Novel Agents for Combination Therapy. Antimicrob. Agents Chemother. 2001, 45, 105–116. [Google Scholar] [CrossRef]
- Wang-Kan, X.; Rodríguez-Blanco, G.; Southam, A.D.; Winder, C.L.; Dunn, W.B.; Ivens, A.; Piddock, L.J.V. Metabolomics Reveal Potential Natural Substrates of AcrB in Escherichia coli and Salmonella enterica Serovar Typhimurium. mBio 2021, 12. [Google Scholar] [CrossRef]
- Bohnert, J.A.; Karamian, B.; Nikaido, H. Optimized Nile Red Efflux Assay of AcrAB-TolC Multidrug Efflux System Shows Competition between Substrates. Antimicrob. Agents Chemother. 2010, 54, 3770–3775. [Google Scholar] [CrossRef]
- Bohnert, J.A.; Schuster, S.; Szymaniak-Vits, M.; Kern, W.V. Determination of Real-Time Efflux Phenotypes in Escherichia coli AcrB Binding Pocket Phenylalanine Mutants Using a 1,2′-Dinaphthylamine Efflux Assay. PLoS ONE 2011, 6, e21196. [Google Scholar] [CrossRef]
- Bohnert, J.A.; Schuster, S.; Kern, W.V.; Karcz, T.; Olejarz, A.; Kaczor, A.; Handzlik, J.; Kieć-Kononowicz, K. Novel Piperazine Arylideneimidazolones Inhibit the AcrAB-TolC Pump in Escherichia coli and Simultaneously Act as Fluorescent Membrane Probes in a Combined Real-Time Influx and Efflux Assay. Antimicrob. Agents Chemother. 2016, 60, 1974–1983. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; A Shoemaker, B.; A Thiessen, P.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Schumacher, A.; Steinke, P.; Bohnert, J.A.; Akova, M.; Jonas, D.; Kern, W.V. Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Enterobacteriaceae other than Escherichia coli. J. Antimicrob. Chemother. 2005, 57, 344–348. [Google Scholar] [CrossRef]
- Schuster, S.; Kohler, S.; Buck, A.; Dambacher, C.; König, A.; Bohnert, J.A.; Kern, W.V. Random Mutagenesis of the Multidrug Transporter AcrB from Escherichia coli for Identification of Putative Target Residues of Efflux Pump Inhibitors. Antimicrob. Agents Chemother. 2014, 58, 6870–6878. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.; Bohnert, J.A.; Vavra, M.; Rossen, J.W.; Kern, W.V. Proof of an Outer Membrane Target of the Efflux Inhibitor Phe-Arg-β-Naphthylamide from Random Mutagenesis. Molecules 2019, 24, 470. [Google Scholar] [CrossRef]
| MIC in µg/mL 1 | |||
|---|---|---|---|
| E. coli Strain/Mutant | Nadifloxacin | Zoliflodacin | Novobiocin |
| KUN9180 | >512 (0) | 4 (0) | 128 (64) |
| KUN∆acrB | 32 (16) | 0.25 (0) | 4 (2) |
| KUN∆acrB∆mdtF | 16 (0) | 0.125 (0) | 2 (0) |
| KUN∆tolC | 4 (2) | 0.045 (0.02) | 1 (0) |
| Oligonucleotide | Sequence (5′-3′) 1 | Application |
|---|---|---|
| upOl-mdtF-FRT-PGKgb2neo | gtcactcaggtgattgagcaaaatatgaatgggcttgatggcctgatgtaaattaaccctcactaaagggcg | FRT-PGK-gb2neo-FRTcassette amplification |
| lowOl-mdtF-FRT-PGKgb2neo | gcggtgccatcgtgccagaggcgttgcgtacataccattggttgatgttataatacgactcactatagggctc | FRT-PGK-gb2neo-FRTcassette amplification |
| Check-mdtF-fw | ggcgatcatgaacttaccgg | Check-primer for mdtF insertions |
| Check-mdtF-rv | ggatgccgttgtagcgttc | Check-primer for mdtF insertions |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schuster, S.; Vavra, M.; Greim, L.; Kern, W.V. Exploring the Contribution of the AcrB Homolog MdtF to Drug Resistance and Dye Efflux in a Multidrug Resistant E. coli Isolate. Antibiotics 2021, 10, 503. https://doi.org/10.3390/antibiotics10050503
Schuster S, Vavra M, Greim L, Kern WV. Exploring the Contribution of the AcrB Homolog MdtF to Drug Resistance and Dye Efflux in a Multidrug Resistant E. coli Isolate. Antibiotics. 2021; 10(5):503. https://doi.org/10.3390/antibiotics10050503
Chicago/Turabian StyleSchuster, Sabine, Martina Vavra, Ludwig Greim, and Winfried V. Kern. 2021. "Exploring the Contribution of the AcrB Homolog MdtF to Drug Resistance and Dye Efflux in a Multidrug Resistant E. coli Isolate" Antibiotics 10, no. 5: 503. https://doi.org/10.3390/antibiotics10050503
APA StyleSchuster, S., Vavra, M., Greim, L., & Kern, W. V. (2021). Exploring the Contribution of the AcrB Homolog MdtF to Drug Resistance and Dye Efflux in a Multidrug Resistant E. coli Isolate. Antibiotics, 10(5), 503. https://doi.org/10.3390/antibiotics10050503






