Surfactant-Assisted Label-Free Fluorescent Aptamer Biosensors and Binding Assays
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Effect of Time-Dependent Stabilization
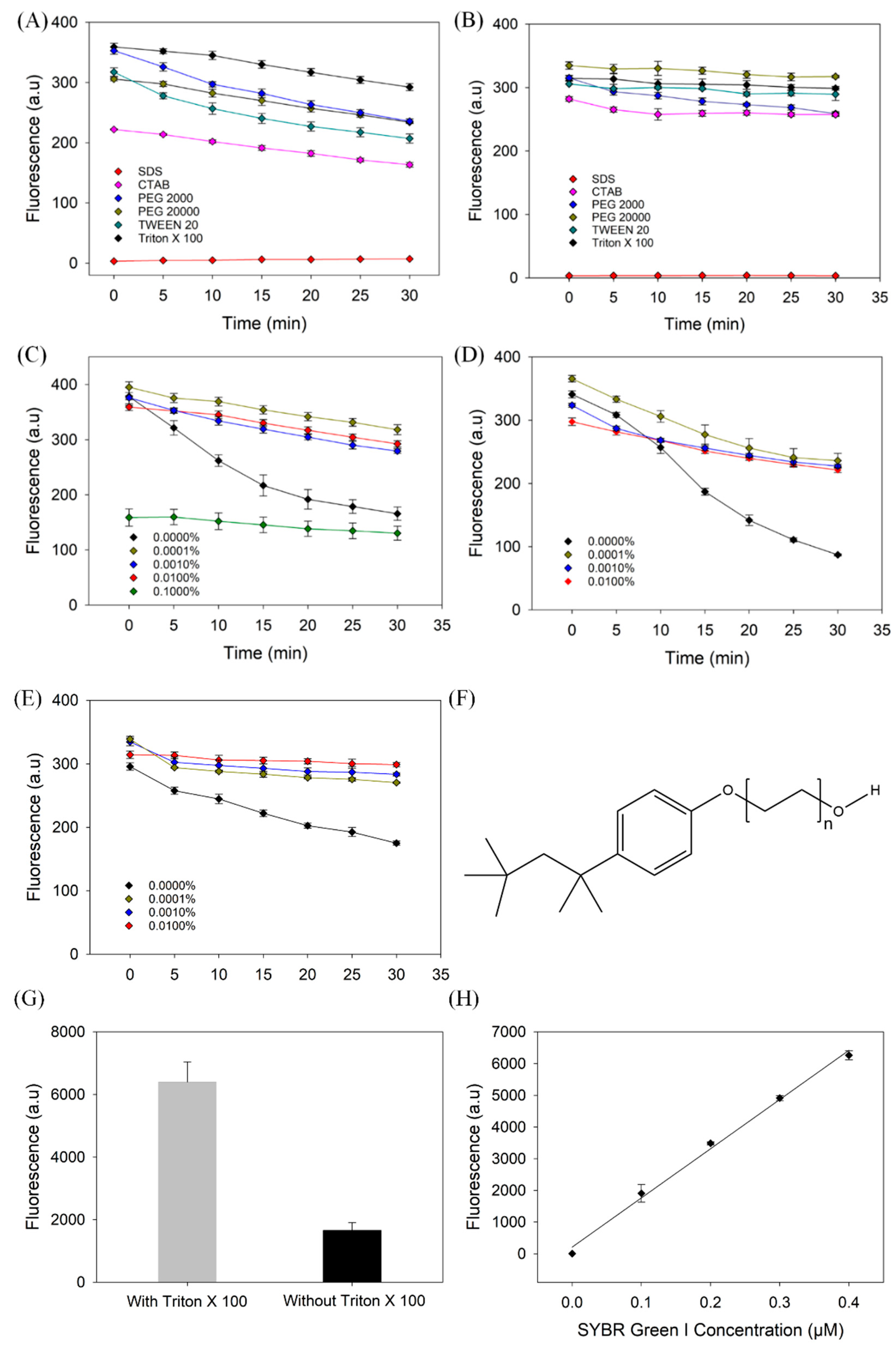
2.3. Test of Different Surfactants and Assay Materials
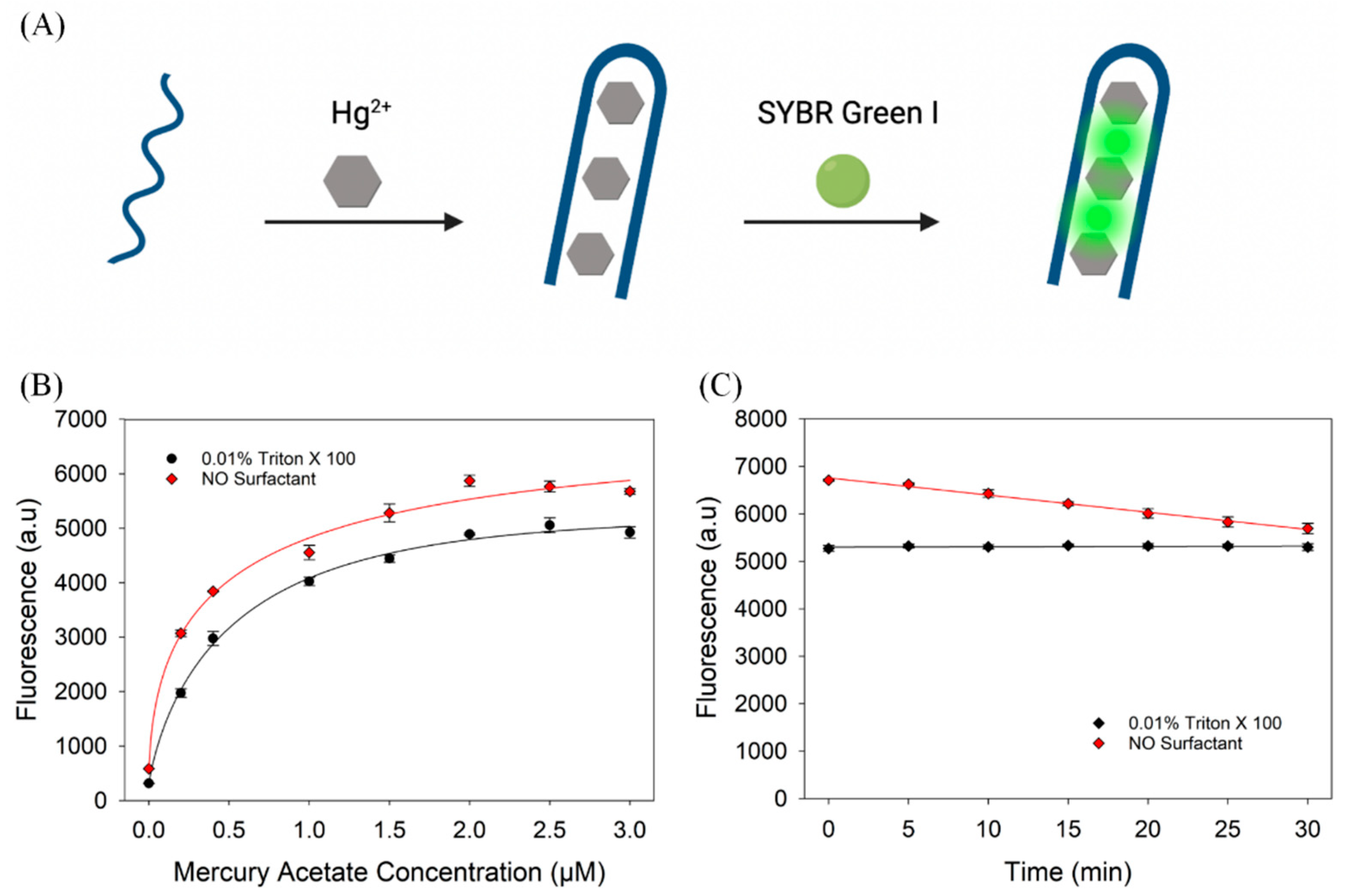
2.4. Hg2+ Sensing Assay
2.5. Adenosine Sensing Assay
2.6. Cortisol Sensing Assay
3. Results and Discussion
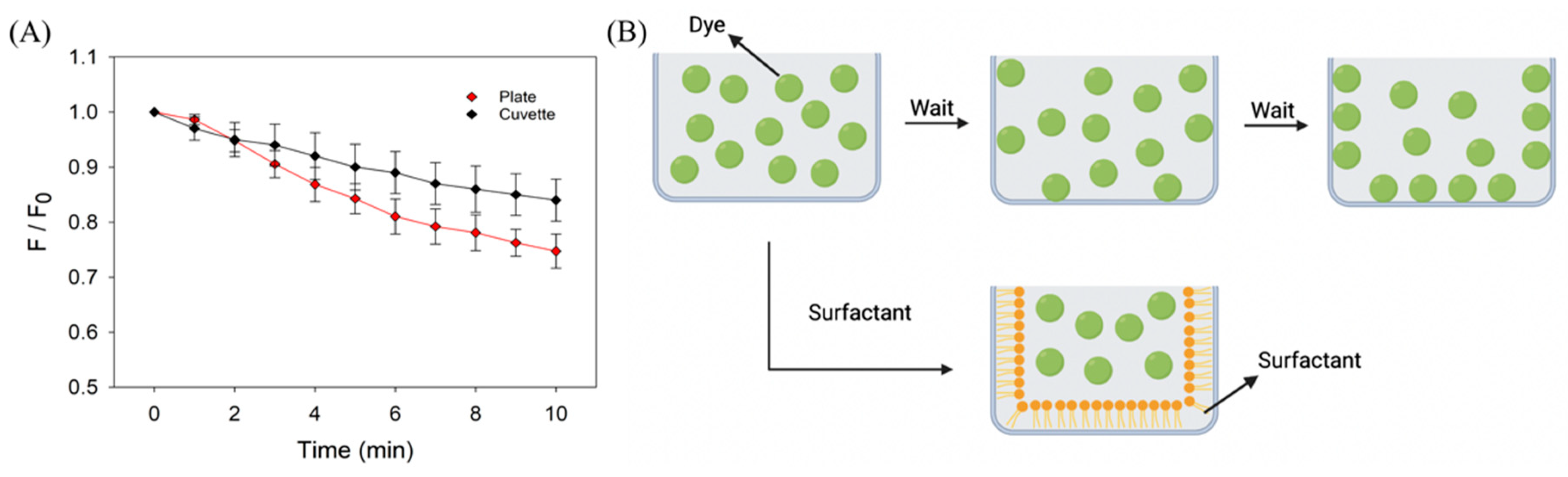
3.1. Effect of Stabilization in Microplates and Cuvettes
3.2. Time-Dependent Signal Stability
3.3. The Protective Effect of Surfactants
3.4. Sensing Mercury Using a Thymine-Rich DNA
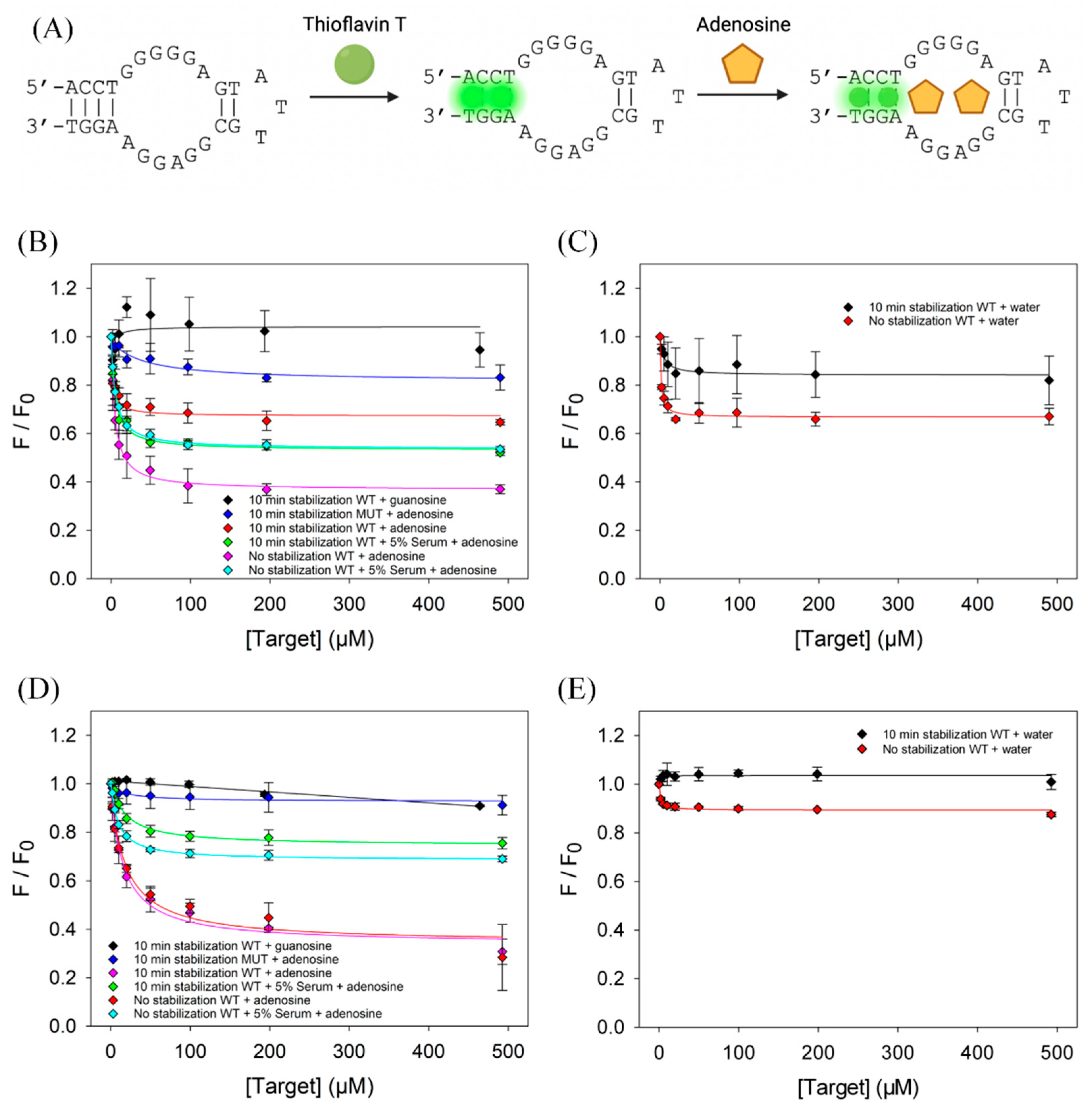
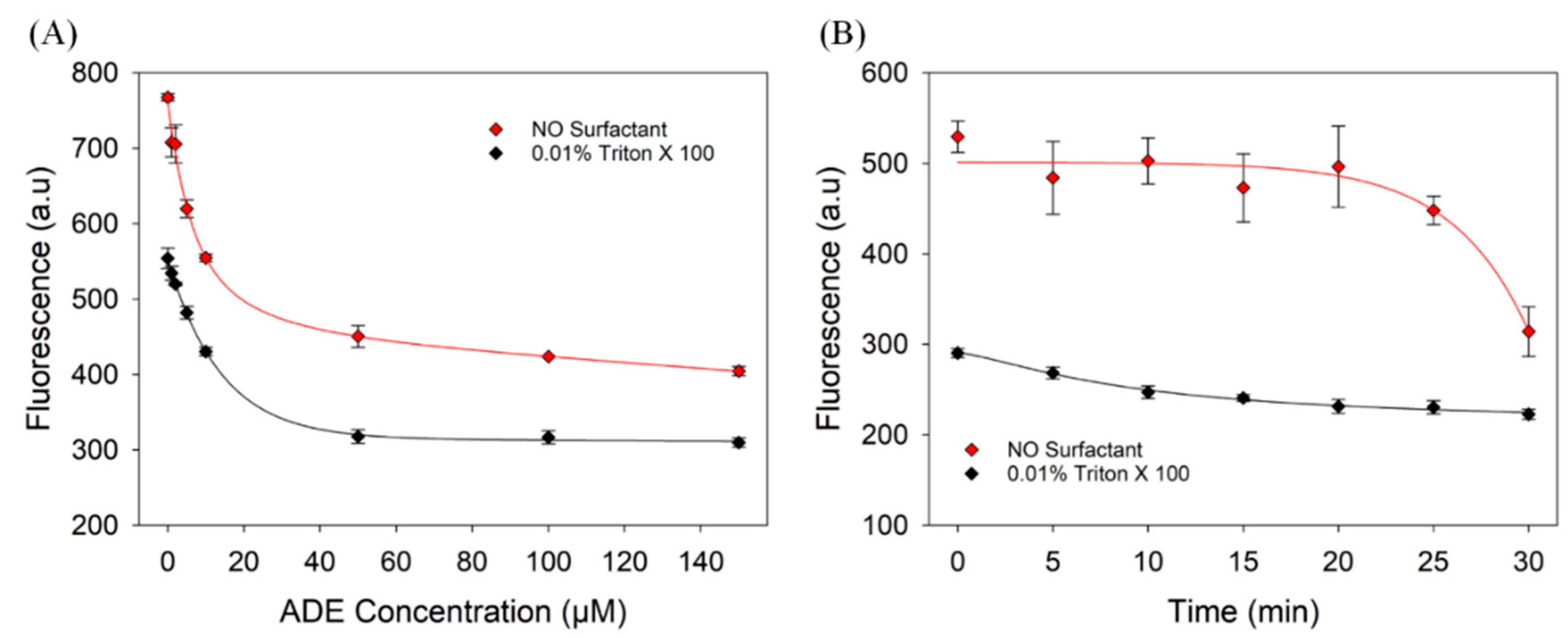
3.5. Evaluation of the Adenosine Aptamer
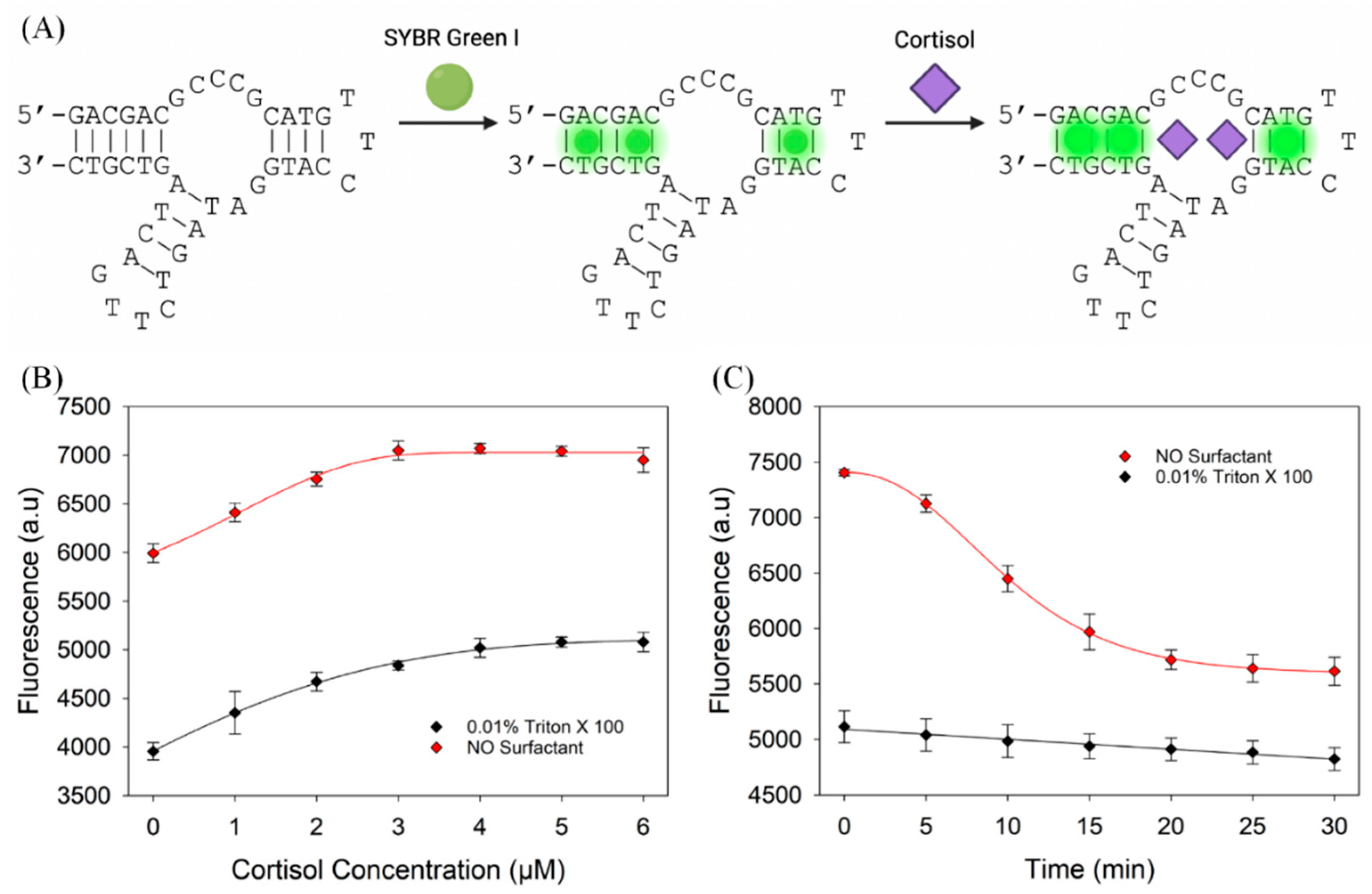
3.6. Evaluation of the Cortisol Aptamer
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Daems, E.; Moro, G.; Campos, R.; De Wael, K. Mapping the gaps in chemical analysis for the characterisation of aptamer-target interactions. TrAC Trends Anal. Chem. 2021, 142, 116311. [Google Scholar] [CrossRef]
- McKeague, M.; De Girolamo, A.; Valenzano, S.; Pascale, M.; Ruscito, A.; Velu, R.; Frost, N.R.; Hill, K.; Smith, M.; McConnell, E.M.; et al. Comprehensive Analytical Comparison of Strategies Used for Small Molecule Aptamer Evaluation. Anal. Chem. 2015, 87, 8608–8612. [Google Scholar] [CrossRef]
- Yu, H.; Alkhamis, O.; Canoura, J.; Liu, Y.; Xiao, Y. Advances and Challenges in Small-Molecule DNA Aptamer Isolation, Characterization, and Sensor Development. Angew. Chem. Int. Ed. 2021, 60, 16800–16823. [Google Scholar] [CrossRef]
- He, L.; Huang, R.; Xiao, P.; Liu, Y.; Jin, L.; Liu, H.; Li, S.; Deng, Y.; Chen, Z.; Li, Z.; et al. Current signal amplification strategies in aptamer-based electrochemical biosensor: A review. Chin. Chem. Lett. 2021, 32, 1593–1602. [Google Scholar] [CrossRef]
- Liu, J.; Cao, Z.; Lu, Y. Functional Nucleic Acid Sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Qin, K.; Lopez, A.; Li, Z.; Liu, J. General Label-Free Fluorescent Aptamer Binding Assay Using Cationic Conjugated Polymers. Anal. Chem. 2022, 94, 15456–15463. [Google Scholar] [CrossRef]
- Zhao, Y.; Gao, B.; Sun, P.; Liu, J.; Liu, J. Metal and pH-Dependent Aptamer Binding of Tetracyclines Enabling Highly Sensitive Fluorescence Sensing. Biosensors 2022, 12, 717. [Google Scholar] [CrossRef]
- Li, L.-L.; Ge, P.; Selvin, P.R.; Lu, Y. Direct Detection of Adenosine in Undiluted Serum Using a Luminescent Aptamer Sensor Attached to a Terbium Complex. Anal. Chem. 2012, 84, 7852–7856. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Lopez, A.; Zheng, J.; Liu, J. Using the Intrinsic Fluorescence of DNA to Characterize Aptamer Binding. Molecules 2022, 27, 7809. [Google Scholar] [CrossRef]
- Yeasmin Khusbu, F.; Zhou, X.; Chen, H.; Ma, C.; Wang, K. Thioflavin T as a fluorescence probe for biosensing applications. TrAC Trends Anal. Chem. 2018, 109, 1–18. [Google Scholar] [CrossRef]
- NK, R.; Kamali, R.V.; Gorthi, S.S. A rapid aptamer-based fluorescence assay for the detection of lipopolysaccharides using SYBR Green I. Luminescence 2021, 36, 1632–1637. [Google Scholar]
- Xiao, S.; Sun, L.; Lu, J.; Dong, Z. A label-free aptasensor for rapid detection of clenbuterol based on SYBR Green I. New J. Chem. 2022, 46, 16177–16182. [Google Scholar] [CrossRef]
- Li, B.; Dong, S.; Wang, E. Homogeneous Analysis: Label-Free and Substrate-Free Aptasensors. Chem. Asian J. 2010, 5, 1262–1272. [Google Scholar] [CrossRef] [PubMed]
- Suss, O.; Motiei, L.; Margulies, D. Broad Applications of Thiazole Orange in Fluorescent Sensing of Biomolecules and Ions. Molecules 2021, 26, 2828. [Google Scholar] [CrossRef]
- Zhang, P.; Zandieh, M.; Ding, Y.; Wu, L.; Wang, X.; Liu, J.; Li, Z. A Label-Free, Mix-and-Detect ssDNA-Binding Assay Based on Cationic Conjugated Polymers. Biosensors 2023, 13, 122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Liu, J. Label-Free Colorimetric Biosensors Based on Aptamers and Gold Nanoparticles: A Critical Review. Anal. Sens. 2021, 1, 30–43. [Google Scholar] [CrossRef]
- Liu, X.; He, F.; Zhang, F.; Zhang, Z.; Huang, Z.; Liu, J. Dopamine and Melamine Binding to Gold Nanoparticles Dominates Their Aptamer-based Label-free Colorimetric Sensing. Anal. Chem. 2020, 92, 9370–9378. [Google Scholar] [CrossRef]
- Zhang, F.; Huang, P.-J.J.; Liu, J. Sensing Adenosine and ATP by Aptamers and Gold Nanoparticles: Opposite Trends of Color Change from Domination of Target Adsorption Instead of Aptamer Binding. ACS Sens. 2020, 5, 2885–2893. [Google Scholar] [CrossRef]
- Zahraee, H.; Khoshbin, Z.; Ramezani, M.; Alibolandi, M.; Abnous, K.; Taghdisi, S.M. A tag-free fluorescent aptasensor for tobramycin detection using a hybridization of three aptamer strands and SYBR Green I dye. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 290, 122305. [Google Scholar] [CrossRef]
- Kong, L.; Xu, J.; Xu, Y.; Xiang, Y.; Yuan, R.; Chai, Y. A universal and label-free aptasensor for fluorescent detection of ATP and thrombin based on SYBR Green Idye. Biosens. Bioelectron. 2013, 42, 193–197. [Google Scholar] [CrossRef]
- Sarpong, K.; Datta, B. Nucleic-Acid-Binding Chromophores as Efficient Indicators of Aptamer-Target Interactions. J. Nucleic Acids 2012, 2012, 247280. [Google Scholar] [CrossRef] [PubMed]
- Zipper, H.; Brunner, H.; Bernhagen, J.; Vitzthum, F. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. Nucleic Acids Res. 2004, 32, e103. [Google Scholar] [CrossRef] [PubMed]
- Kolbeck, P.J.; Vanderlinden, W.; Gemmecker, G.; Gebhardt, C.; Lehmann, M.; Lak, A.; Nicolaus, T.; Cordes, T.; Lipfert, J. Molecular structure, DNA binding mode, photophysical properties and recommendations for use of SYBR Gold. Nucleic Acids Res. 2021, 49, 5143–5158. [Google Scholar] [CrossRef]
- Huizenga, D.E.; Szostak, J.W. A DNA Aptamer That Binds Adenosine and ATP. Biochemistry 1995, 34, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, J. Aptamer-based strategies for recognizing adenine, adenosine, ATP and related compounds. Analyst 2020, 145, 6753–6768. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Oni, O.; Liu, J. New insights into a classic aptamer: Binding sites, cooperativity and more sensitive adenosine detection. Nucleic Acids Res. 2017, 45, 7593–7601. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.M.; Jahnke, F.M.; Heemstra, J.M. Modulating the Substrate Selectivity of DNA Aptamers Using Surfactants. Langmuir 2015, 31, 11769–11773. [Google Scholar] [CrossRef]
- Liu, B.; Huang, P.-J.J.; Zhang, X.; Wang, F.; Pautler, R.; Ip, A.C.F.; Liu, J. Parts-per-Million of Polyethylene Glycol as a Non-Interfering Blocking Agent for Homogeneous Biosensor Development. Anal. Chem. 2013, 85, 10045–10050. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, N.; Giridhar, V.; Vasudevan, D. Surfactant effects on methanol oxidation at Pt–Ru/C coated glassy carbon electrode. J. Solid State Electrochem. 2010, 14, 877–881. [Google Scholar] [CrossRef]
- Lavkush Bhaisare, M.; Pandey, S.; Shahnawaz Khan, M.; Talib, A.; Wu, H.-F. Fluorophotometric determination of critical micelle concentration (CMC) of ionic and non-ionic surfactants with carbon dots via Stokes shift. Talanta 2015, 132, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Bak, A.; Podgorska, W. Investigation of drop breakage and coalescence in the liquid-liquid system with nonionic surfactants Tween 20 and Tween 80. Chem. Eng. Sci. 2012, 74, 181–191. [Google Scholar] [CrossRef]
- Wang, J.; Liu, B. Highly sensitive and selective detection of Hg2+ in aqueous solution with mercury-specific DNA and Sybr Green I. Chem. Commun. 2008, 39, 4759–4761. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.-A.; Chun, H.; Zhang, Y.; Pecic, S.; Nakatsuka, N.; Andrews, A.M.; Worgall, T.S.; Stojanovic, M.N. High-Affinity Nucleic-Acid-Based Receptors for Steroids. ACS Chem. Biol. 2017, 12, 3103–3112. [Google Scholar] [CrossRef]
- Niu, C.; Ding, Y.; Zhang, C.; Liu, J. Comparing two cortisol aptamers for label-free fluorescent and colorimetric biosensors. Sens. Diagn. 2022, 1, 541–549. [Google Scholar] [CrossRef]
- Yu, H.; Luo, Y.; Alkhamis, O.; Canoura, J.; Yu, B.; Xiao, Y. Isolation of Natural DNA Aptamers for Challenging Small-Molecule Targets, Cannabinoids. Anal. Chem. 2021, 93, 3172–3180. [Google Scholar] [CrossRef]
- Huang, P.-J.J.; Liu, J. Simultaneous Detection of L-Lactate and D-Glucose Using DNA Aptamers in Human Blood Serum. Angew. Chem. Int. Ed. 2023, 62, e20221287. [Google Scholar] [CrossRef]
- Niu, C.; Zhang, C.; Liu, J. Capture-SELEX of DNA Aptamers for Estradiol Specifically and Estrogenic Compounds Collectively. Environ. Sci. Technol. 2022, 56, 17702–17711. [Google Scholar] [CrossRef]
- Zhao, Y.; Yavari, K.; Liu, J. Critical evaluation of aptamer binding for biosensor designs. TrAC Trends Anal. Chem. 2022, 146, 116480. [Google Scholar] [CrossRef]
| Name | Sequences (5′-3′) |
|---|---|
| DNA1 | ACGACACGGAGGCTTAGTTTGCTAAATGGTCATGTCGT |
| CSS.1-42 | GACGACGCCCGCATGTTCCATGGATAGTCTTGACTAGTCGTC |
| Ade Apt | ACCTGGGGGAGTATTGCGGAGGAAGGT |
| Ade Apt M2 | ACCTGGGGTAGTATTGCGGAGTAAGGT |
| T30 | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Li, A.Z.; Liu, J. Surfactant-Assisted Label-Free Fluorescent Aptamer Biosensors and Binding Assays. Biosensors 2023, 13, 434. https://doi.org/10.3390/bios13040434
Zhang H, Li AZ, Liu J. Surfactant-Assisted Label-Free Fluorescent Aptamer Biosensors and Binding Assays. Biosensors. 2023; 13(4):434. https://doi.org/10.3390/bios13040434
Chicago/Turabian StyleZhang, Hanxiao, Albert Zehan Li, and Juewen Liu. 2023. "Surfactant-Assisted Label-Free Fluorescent Aptamer Biosensors and Binding Assays" Biosensors 13, no. 4: 434. https://doi.org/10.3390/bios13040434
APA StyleZhang, H., Li, A. Z., & Liu, J. (2023). Surfactant-Assisted Label-Free Fluorescent Aptamer Biosensors and Binding Assays. Biosensors, 13(4), 434. https://doi.org/10.3390/bios13040434





