Abstract
Infectious diseases contribute significantly to the global disease burden. Sensitive and accurate screening methods are some of the most effective means of identifying sources of infection and controlling infectivity. Conventional detecting strategies such as quantitative polymerase chain reaction (qPCR), DNA sequencing, and mass spectrometry typically require bulky equipment and well-trained personnel. Therefore, mass screening of a large population using conventional strategies during pandemic periods often requires additional manpower, resources, and time, which cannot be guaranteed in resource-limited settings. Recently, emerging microfluidic technologies have shown the potential to replace conventional methods in performing point-of-care detection because they are automated, miniaturized, and integrated. By exploiting the spatial separation of detection sites, microfluidic platforms can enable the multiplex detection of infectious diseases to reduce the possibility of misdiagnosis and incomplete diagnosis of infectious diseases with similar symptoms. This review presents the recent advances in microfluidic platforms used for multiplex detection of infectious diseases, including microfluidic immunosensors and microfluidic nucleic acid sensors. As representative microfluidic platforms, lateral flow immunoassay (LFIA) platforms, polymer-based chips, paper-based devices, and droplet-based devices will be discussed in detail. In addition, the current challenges, commercialization, and prospects are proposed to promote the application of microfluidic platforms in infectious disease detection.
1. Introduction
Infectious diseases are caused by pathogens, including viruses, bacteria, and parasites [1]. As one of the greatest threats to human health and global security, infectious diseases contribute the most to the global disease burden [2,3]. For example, according to the World Health Organization, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) had infected 6 billion people and caused 6 million deaths by December, 2022 [4]. Compared to other diseases, infectious diseases are characterized by their high infectivity [1]. Managing their infectivity is crucial for preventing and controlling the pandemics of infectious diseases [5,6,7]. It is reported that timely diagnosis can help control the source of infection by isolating and treating infected individuals. A rapid, sensitive, and accurate diagnostic assay is urgently required to effectively investigate and locate the source of infection and control the spread of infectious diseases [7].
Laboratory tests usually detect infectious diseases with methods such as enzyme-linked immunosorbent assays (ELISA), DNA sequencing, and qPCR [8]. These techniques usually require well-trained operators, professional procedures, and expensive testing equipment [9]. For example, ELISA technology requires a complex labeling procedure and a bulky optical reader [10,11]. As the other gold standard technology for nucleic acid detection [12], qPCR is limited by its many manual steps and precise equipment such as thermocyclers [13]. Therefore, current laboratory detection technologies are struggling to meet the need for rapid mass screening and surveillance of infectious diseases, especially in resource-limited settings. In addition, in many cases (e.g., to distinguish between the different types of sepsis), clinical evidence based on a single biomarker is insufficient to adequately diagnose a disease or monitor its treatment [14]. Moreover, simultaneous detection of infectious diseases with similar symptoms (e.g., respiratory viruses) can rapidly identify the source of infection [15]. In this context, there is a great need for multiplex detection platforms, that can ensure the performance and quality requirements of diagnostics of infectious diseases that can be performed in a short time by laypersons [14,16].
Recently, microfluidics has been highlighted and extensively researched due to its outstanding advantages, such as fast operation (less than 1 h), low reagent volume (microliter or even nanoliter), and high integration capability (integration of sample preparation, detection, and analysis into the platform) [17]. In addition, the characteristics of microfluidic platforms are well-suited for multiplex detection [14,18]. The general strategies of multiplex detection include spatial separation of detection sites, discrete regions of a channel network or array, or the detection of different markers in integrated microfluidic chips which can effectively reduce the interference between different reaction systems in the simultaneous detection of a large number of targets [14]. Moreover, because of the relatively independent reaction space on the microfluidic platforms, the sensitivity and specificity of each assay can be ensured in multiplex detection. Therefore, multiplex detection technology based on microfluidic platforms shows great potential for the diagnosis and mass screening of infectious diseases [19,20]. Although there are several reviews that comprehensively discuss microfluidic detection technologies for infectious disease diagnosis [21,22,23], few of them focus on microfluidic platforms for multiplex detection [21]. Therefore, this review summarizes recent advances in various microfluidic platforms for multiplex detection of infectious diseases (Figure 1). In particular, immunosensors and nucleic acid sensors that are based on microfluidics for multiplex detection of pathogens are discussed. The current challenges, commercialization, and prospects are also proposed to improve the development of more efficient multiplex microfluidic platforms, especially in the rapid and accurate diagnosis of infectious diseases.
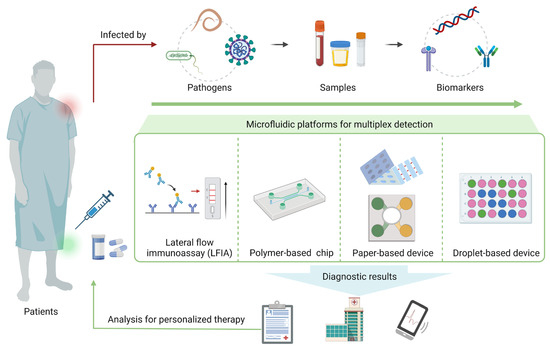
Figure 1.
Overview of microfluidic platforms for multiplex detection of pathogens.
2. Multiplex Immunosensors on Microfluidic Platforms
The microfluidic immunosensor is a well-developed diagnostic tool for detecting analytes at low concentrations, which use antibodies as the biological recognition element to convert an antibody-antigen binding event into a measurable physical signal [24]. In the microfluidic multiplex immunosensor, a series of discriminatory biomarkers are simultaneously recognized by the antibodies with high specificity and sensitivity, generating signals in proportion to the antigen concentration in the samples [16,25,26,27,28,29,30,31,32,33,34,35]. For infectious disease diagnosis, multiplex immunosensors based on microfluidic platforms mainly use spatial multiplexing and barcode multiplexing strategies. According to the principle of fluid propulsion, they can be classified into capillary, pressure-driven, centrifugal, electrokinetic, and acoustic systems [36,37]. Among them, capillary force-driven microfluidic platforms are widely used because they do not require external energy to enable continuous fluid automation, with lateral flow immunoassay (LFIA) being the most typical representative [36].
LFIA has become one of the most successful analytical techniques because it meets the ASSURED (affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and delivered) criteria of the WHO [38]. The rapidity, simplicity, relative cost-effectiveness, and the ability to be used by unskilled personnel have contributed to the widespread acceptance of LFIA [38,39]. In fact, the global lateral flow assay market was estimated to be approximately $5.98 billion in 2019 and is expected to reach $10.36 billion by 2027 [39]. Simultaneous detection of multiple analytes is mainly realized by arranging multiple test lines (TL) in a single strip, which enables discrimination of different targets by spatial resolution [39,40]. There are three typical signal readout strategies for LFIA platforms, including colorimetric signal, surface-enhanced Raman scattering (SERS) signal, and fluorescent signal, which are discussed below.
Colorimetric LFIA is the most commonly used for simple and rapid detection of pathogens [41,42]. The analytes (pathogens or their antigens) are first captured by antibodies on the strip, followed by the recognition of detection antibody which is usually conjugated with colorimetric readout elements such as gold nanoparticles (AuNPs) to produce a visible signal on the test line [42]. A number of LFIA platforms based on different nanoparticles have been developed for multiplex detection of infectious diseases [26,27,28]. For example, a LFIA platform using a color-mixing encoding and readout strategy was established (Figure 2A). After binding nanoparticles and IgM/IgG antibodies against dengue virus (DENV) and chikungunya virus (CHIKV), labeled antibody-nanoparticle complexes bind to the immobilized capture reagents on the different test lines in sandwich format to show different colors. By processing RGB data (from photographs on the LFIA) captured by the reader, the test results could be read semi-quantitatively. Furthermore, this platform has been validated in 50 human plasma samples, demonstrating that it can be used for infectious disease management by providing accessible, evidence-based laboratory diagnosis [27]. However, nanoparticle-based colorimetric assays have two inherent shortcomings: limited sensitivity and poor quantitative capability [43]. To improve the sensitivity and quantitative detection capability of LFIA in the diagnosis of infectious diseases, a SERS signal-based system [29,30,31] and a fluorescence signal-based strategy [32,33,44] have been recently reported. SERS-LFIA platforms integrate functional SERS-encoded nanoparticles (NPs), also known as SERS nanotags, into the LFIA system instead of the commonly used AuNPs as signal reporters [45,46]. This platform can provide specific (fingerprint feature), strong (high sensitivity), and stable (no photobleaching) SERS signals [46]. For example, Wang’s group developed a sensitive and quantitative SERS-LFIA platform using Fe3O4@Ag nanoparticles as magnetic SERS nanotags (Figure 2B), which could simultaneously detect human adenovirus (HAdV) and influenza A H1N1 viruses from human whole blood, serum, and sputum samples. The limits of detection (LOD) for HAdV and H1N1 were 10 and 50 pfu/mL respectively, which is 2000 times higher sensitivity than the standard colloidal gold strip method [29]. In general, fluorescent LFIA in which quantum dots (QDs) are considered as one of the most commonly used signal labels, show higher sensitivity than the conventional AuNPs-based LFIA due to their excellent luminescence properties [47]. A LFIA platform based on a dual-channel fluorescent immunochromatographic assay (ICA) was developed for ultrasensitive and simultaneous qualification of SARS-CoV-2 and influenza A virus. A high-performance quantum dot nanobead (QB) was fabricated by adsorbing multiple layers of dense quantum dots (QDs) on the SiO2 surface and used as a highly luminescent label of the ICA system to ensure the high-sensitivity and stability of the assay. The LOD was 5 pg/mL for SARS-CoV-2 antigen and 50 pfu/mL for H1N1 within 15 min, respectively. In addition, this platform showed high accuracy and specificity in throat swab samples with two orders of magnitude improvement in sensitivity compared to a conventional AuNP-based platform [32].
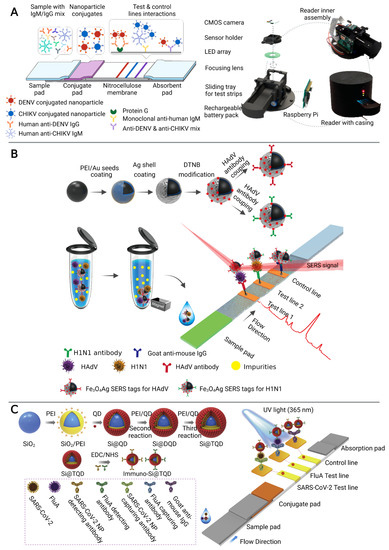
Figure 2.
The multiplex detection of infectious diseases using microfluidic immunosensors. (A) Rapid diagnostic platform for DENV and CHIKV IgM/IgG antibodies, consisting of a multiplex color encoded lateral flow test strip and optical reader. Adapted with permission from Ref. [27]. Copyright 2019, American Chemical Society. (B) The multiplex detection of H1N1 and HAdV of the magnetic SERS-LFIA. Adapted with permission from Ref. [29]. Copyright 2019, American Chemical Society. (C) The multiplex detection of H1N1 and SARS-CoV-2 based on a dual-channel fluorescent immunochromatographic assay. Adapted with permission from Ref. [32]. Copyright 2021, Elsevier.
There are various commercial LFIA products. Such as, ActiveXpress (Edinburgh Genetics Ltd, Edinburgh, UK), Roche (SD Biosensor Inc./Roche Diagnostics, Basel, Switzerland), and Standard-Q (SD Biosensor Inc, Gyeonggi-do, Republic of Korea) [48]. Despite the extensive LFIAs research and the availability of several commercial products for detection, some challenges remain. First, the sensitivity should be enhanced by fabrication of high-resolution instrumentation and label materials with high response signal [33]. Second, the false positives may be reduced by multi-signal synchronous detection based on multiplexed type and multi-signal type [49]. Third, the false negatives caused by mutations are required to decrease through selecting new immune targets in a timely manner [50,51].
3. Multiplex Nucleic Acid Sensors on Microfluidic Platforms
Nucleic acid is another biomarker that can be used for the diagnosis of infectious diseases, due to its unique and outstanding characteristics (e.g., molecular recognition, biocompatibility, functionalization, and programmability), which endow nucleic acids with potential application as powerful sensing elements and provide key information of specific genes and species [52]. PCR is typically considered as the gold standard detection method of nucleic acid-based diagnosis, but involves costly/advanced equipment and skilled personnel, so it cannot be easily combined with microfluidic technology [53]. Therefore, isothermal amplification and clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (CRISPR/Cas) system, which have the advantage of low-cost, reliability, and do not require for bulky equipment, can be compatible with the microfluidic platforms for nucleic acid detection [20,54]. Based on the spatial separation of detection sites on microfluidic platforms, the multiplex detection of different nucleic acids can be realized [14]. Multiplex nucleic acid sensors on microfluidic platforms mainly include polymer-based microfluidic chips, paper-based microfluidic devices, and droplet-based microfluidic devices [55]. They are characterized by cost-efficiency, portability, low sample consumption (µL-fL), miniaturization (with dimensions of tens to hundreds of micrometers chambers), simplicity (no training is required), and multiplex detection (providing multiple spatially separated detection channels) [56]. In this section, we will discuss nucleic acid sensors on microfluidic platforms for multiplex diagnosis of infectious diseases [57,58,59].
3.1. Polymer-Based Microfluidics
Fabrication of microfluidic devices is an important step in integrated automated nucleic acid sensing. In this regard, polymers (e.g., polydimethylsiloxane, PDMS) are one of the most common materials, due to their advantages such as cost-effectiveness, good biocompatibility, and simple fabrication protocol [20]. Polymer-based microfluidic chip is highly automated (multistep continuous reactions can be realized via sophisticated microstructures) and integrated, which has been widely used for infectious disease detections [60].
Depending on the presence or absence of moving mechanical parts, fluid flow support techniques for polymer-based microfluidic chips are generally divided into mechanical drive (e.g., centrifugal force drive and micropump drive) and nonmechanical drive (e.g., electric drive and capillary force drive) [61,62]. The mechanical drive chips have the advantages of easy large-scale integration, high drive pressure, wide range of flow rates, and high adaptability. For example, Nguyen et al. designed a centrifugal disc equipped with a glass filter extraction column and multiple reaction chambers. Such a portable analyzer could simultaneously detect four pathogens of upper respiratory diseases within 90 min (Figure 3A) [63]. The non-mechanical drive is also suitable for integration, operation, and accurate control [61,63,64]. A pump-free microfluidic chip with capillary force drive was developed by Ciftci et al., which could directly collect the nucleic acid amplification products through the extraction solution without loading or washing procedures, enabling accurate diagnosis of the Ebola, Zika, and dengue viruses simultaneously [64].
The other key element for multiplex detection of infectious diseases is the design of microfluidic channels. The general processing technology of the polymer-based microfluidic channel includes the hot pressing, injection molding, photolithography, and laser etching [65]. For example, Huang et al. used laser cauterization to fabricate a basement layer consisting of two sides: side A contained microstructures for the recombinase polymerase amplification (RPA) reaction, and side B for the loop-mediated isothermal amplification (LAMP) reaction. Thus, the two-step of isothermal amplification could be completed on the chip to achieve a higher detection sensitivity (10 copies/μL) [66]. Similarly, Choi et al. developed a CRISPR/Cas strategy nucleic acid amplification-free on a polymer-based microfluidic chip. The activated CRISPR-Cas 12a in the presence of viral DNA was combined with a Raman-sensitive system consisting of ssDNA-immobilized Raman probe-functionalized AuNPs on the graphene oxide/triangle Au nanoflower array. Using this platform, simultaneous detection of hepatitis B virus (HBV), human papillomavirus 16 (HPV-16), and HPV-18 could be achieved with high sensitivity range from 1 aM to 100 pM without any amplification steps [67]. However, these methods require sophisticated instrumentation and high cost, and are difficult to achieve rapid prototyping [68]. To address this problem, three dimensional (3D) printing technology has been gradually adapted for the fabrication of polymer-based microfluidic chip manufacturing [69]. For example, our group used 3D printing technology to develop a sensitive, multiplex colorimetric detection (SMCD) method for the detection of pathogens in wastewater samples (Figure 3B). This SMCD method integrated nucleic acid extraction, RPA, LAMP, and colorimetric detection into microfluidic chips, and detected multi-gene targets of SARS-CoV-2 and multiple human enteric pathogens from the wastewater. The detection time was about 60 min, which was half of the time required for the qPCR method. Moreover, a smart, connected, and on-site detection was achieved with a reporting framework embedded in a smartphone-based detection platform, which exhibited the rapid spatiotemporal epidemiological data collection potential regarding the transmission and persistence of infectious diseases [70].
Biological manufacturers have developed several polymer-based microfluidic commercial devices [e.g., Revogene (Meridian Bioscience, Cincinnati, OH, USA) and GenPlex® (BOHUI, Beijing, China)] for the diagnosis of infectious diseases [1,71,72,73]. Nevertheless, weakness of polymer materials includes thermal conductivity and low thermal resistance. The surface modification of polymers also needs to be further explored [74,75]. In addition, a practical solution for the processing and integration of the whole microfluidic system should be presented and optimized [74]. Finally, microfluidic devices need to be tested in large-scale clinical trials before being commercialized [75].
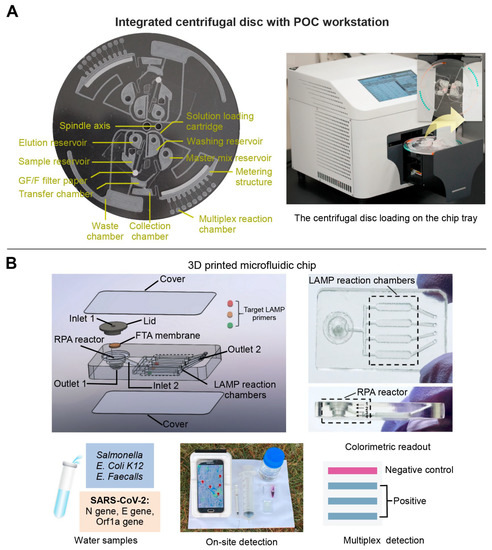
Figure 3.
The multiplex detection of infectious diseases of polymer-based microfluidic chip. (A) A centrifugal microfluidic chip equipped with a glass-filter extraction column for purifying nucleic acid and multiple reaction chambers for multiplex detection. Adapted with permission from Ref. [63]. Copyright 2021, Elsevier. (B) A 3D printed microfluidic chip integrated on-chip nucleic acid extraction, two-stage isothermal amplification, and colorimetric detection. Adapted with permission from Ref. [70]. Copyright 2021, Elsevier.
3.2. Paper-Based Microfluidics
The paper-based microfluidic device is a miniature laboratory analysis system that utilizes a paper substrate to replace the conventional substrates (e.g., quartz, silicon, and glass) [76]. There are several advantages of paper-based microfluidic devices: (1) paper is cheaper than conventional substrates; (2) the paper itself has a capillary effect, which can guide reagent flow without external forces; and (3) the flexibility and elasticity of paper facilitate customization [77,78]. Therefore, paper-based microfluidic devices, including lateral flow assays (LFA) and microfluidic paper-based analytical devices (μPAD) are widely applied to the multiplex detection of infectious diseases [79].
Generally, the LFA consists of a sample pad, a conjugate pad, a nitrocellulose filter membrane, an absorbent pad, and a back card. Each part has varied degrees of overlapping to ensure the continuity of sample flow [80]. Simultaneous detection of different targets can be achieved by fixing and modifying multiple identification elements and signal transduction elements on the LFA test strip [58]. Zhang’s group reported that SHERLOCKv2 can simultaneously detect dengue and Zika virus single-stranded RNA using a fantastic CRISPR/Cas detection system on the LFA platform [81,82]. Similarly, a CRISPR/Cas9-mediated triple-line LFA (TL-LFA) was combined with a multiplex RPA to achieve rapid and simultaneous detection of the E and ORF1ab genes of SARS-CoV-2 in a single strip test (Figure 4A). The TL-LFA showed high sensitivity with an LOD of four copies/mL. The total detection time, including sample pretreatment and analysis, was within 60 min [58].
In 2007, Whiteside’s group at Harvard University first proposed the concept of µPAD [83]. Generally, the fabrication technologies of μPADs are categorized as photolithography, PDMS stamping and printing, laser cutting, engraving, plasma treatment, screen printing, and vapor phase deposition [84]. The µPAD can simultaneously quantify multi-component objects, which is hardly achieved by conventional test papers [84,85]. Therefore, μPAD has gained significant interest as a promising analytical platform for point-of-care testing in the last ten years [85]. Nae Yoon Lee’s group developed a series of μPADs for the detection of several infectious diseases [86,87]. For example, a μPAD based on LAMP was fabricated to detect Escherichia coli O157:H7, Salmonella spp., Staphylococcus aureus, and Cochlodinium polykrikoides, followed by on-chip fluorescent readouts (Figure 4B). This platform can detect as low as 0.12 ng/μL and 0.13 ng/μL for S. aureus and E. coli O157:H7 DNA, respectively [86]. Moreover, a slidable μPAD contained three layers to accomplish DNA extraction, LAMP reaction, and multiplex colorimetric signal output sequentially (Figure 4C). A novel colorimetric fuchsin-based method was used to detect LAMP amplicons. The limit of detection of Salmonella spp., Staphylococcus aureus, and Escherichia coli O157:H7 were 3.0 × 101, 3.0 × 102, and 3.0 × 101 CFU/sample, respectively [87].
There are several successful commercialized LFAs (e.g., urine dipstick and pregnancy test kit) [88,89] and μPADs (e.g., Diagnostics For All and INSiGHT) [90,91]. However, problems remain. Firstly, the LFAs often allows only qualitative/semi-quantitative results, and thus researchers have tried to solve this by introducing smart phone attachments to achieve more accurate result analysis [92]. In addition, the insufficient validation on their compatibility with real sample matrices may cause discrepancy between the detection results obtained from real samples and standard samples and can eventually require major modification of the device design [93]. Thus, the examination of μPADs using clinical samples in a real environment is deemed to be essential [90]. Finally, the integration degree of the µPADs can be improved by adding sample processing modules which can simplify the operational steps and improve the integrated capability of the devices [94].
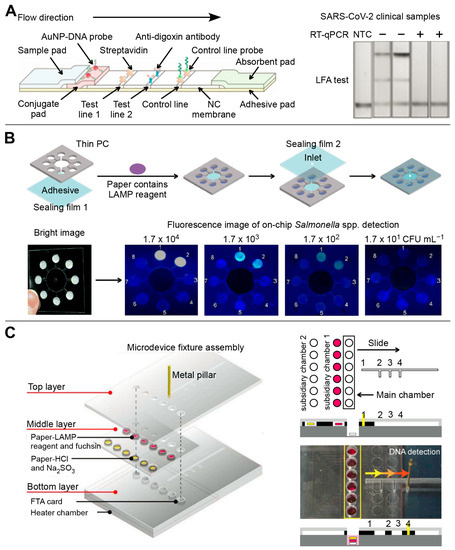
Figure 4.
The multiplex detection of infectious diseases based on paper-based microfluidic devices. (A) The multiplex detection for E and ORF1ab genes of SARS-CoV-2 based on CRISPR/Cas9-mediated LFA. Adapted with permission from Ref. [58]. Copyright 2021, WILEY. (B) Fluorescent μPAD-based LAMP was prepared by simple craft-cutting for the simultaneous detection of four pathogens. Adapted with permission from Ref. [86]. Copyright 2018, Royal Society of Chemistry. (C) The colorimetric slidable μPAD was prepared by etching method for simultaneous detection of Salmonella spp., Staphylococcus aureus, and Escherichia coli O157:H7. Adapted with permission from Ref. [87]. Copyright 2019, Elsevier.
3.3. Droplet-Based Microfluidics
A droplet-based microfluidic device is an alternative strategy for large-scale and parallel biological and chemical reactions [95]. The key technology is to generate small and mono-dispersed droplets (picoliter to nanoliter level) under high frequency (∼kHz) and precise control. Generally, microdroplets are generated from two incompatible liquids as a continuous phase and discrete phase, respectively, and different size distributions can be formed by controlling the microsphere structure and flow ratio of the two phases (the volume of microdroplets varies greatly from microliters to femtoliters) [96]. Meanwhile, the spatial separation of detection sites on the chips can be used as independent bioreactors to effectively distinguish different reactions [95,96]. Therefore, high-throughput droplet-based microfluidic devices enable the large-scale screening and multiplex detection of infectious diseases [97].
Depending on the mode of droplet generation, droplet-based microfluidic devices can be divided into active and passive modes. The passive mode requires additional energy input to generate droplets, while the active mode generates droplets without external propulsion [98]. The passive modes, which do not require programmable syringe pumps or other automated instrumentation to control fluid flow, have been designed to construct droplet-based microfluidic devices [99]. For example, the first combination of droplet digital PCR technique was developed to simultaneously detect ORF1ab and N genes of SARS-CoV-2 (Figure 5A). Compared with the standard qPCR method, the droplet digital PCR system showed similar accuracy but with a lower turnaround time and a lower false-negative results [100]. In another study, a microdroplet platform integrating multiple LAMP, scorpion-shaped probes, and fluorescence microscopic counting, was developed using the flow-focusing method. The platform successfully detected Hepatitis C virus (HCV) and HIV from clinical plasma samples, with an LOD of four copies/reaction [101].
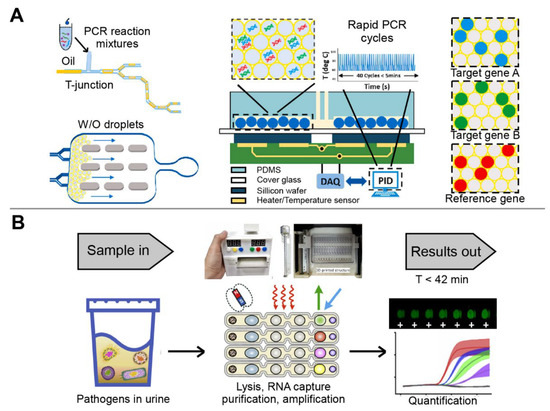
Figure 5.
The multiplex detection of infectious diseases using droplet-based microfluidic devices. (A) Droplet PCR detection system fabricated by T-type structure method. Adapted with permission from Ref. [100]. Copyright 2021, Elsevier. (B) Automatic droplet-based microfluidic platform controlled by magnetic force. Adapted with permission from Ref. [59]. Copyright 2021, Elsevier.
In contrast, active designs enable on-demand generation of droplets with a short response time and a better control of droplet size, content, and motion [102]. Specifically, compared with a few seconds or even minutes in a passive approach, the response time can be reduced to a few milliseconds in an active method. In addition, active methods control droplet size and production rate with higher flexibility and additional handles, and allow on-demand droplet generation, thus greatly promoting practical applications of microfluidic droplets [103]. However, in the passive approach, it is almost impossible to control droplet size and generation frequency independently because they are interrelated through mass conservation [99,102]. To date, the cutting-edge active techniques mainly apply magnetic, electrical, thermal, optical, mechanical, and centrifugal methods, by which magnetic, electrical, and centrifugal forces are introduced, and the viscosity, flow, interfacial tension, fluid density, and channel wettability are varied [104]. A cost-effective and automated multiplex micro-droplet detection platform was created using 3D-printed structural parts incorporated with off-the-shelf mechanic/electronic components (Figure 5B). The platform used magnetic force to control the linear displacement of the multichannel array chip, and seamlessly integrated multiple steps including bacterial lysis, RNA extraction, and amplification through droplet combination. The sample–answer assay could be completed within 42 min, with 100% concordance with qPCR [59]. In addition, the combinatorial arrayed reactions for multiplex evaluation of nucleic acids (CARMEN) were developed for scalable and multiplex pathogen detection. As a result, the platform simultaneously detected 169 human-associated viruses with at least 10 published genome sequences. In addition, SARS-CoV-2 was also detected by incorporating CARMEN-Cas 13a and an additional crRNA. The multiplex and throughput abilities of CARMEN made it practical to scale-up, as miniaturization reduced reagent cost per test by more than 300-fold [2]. Therefore, CARMEN enables large-scale CRISPR-based diagnosis, which is an important step toward routine, comprehensive infectious disease surveillance to improve patient care and public health [105,106].
Significant progress has been made in droplet-based microfluidics and some commercial products (e.g., 10× g and Drop-seq) have been developed [107,108]. To largely facilitate commercialization, several challenges remain to be addressed. For passive mode, it occurs at quite low flow rate ratio, tip-streaming is stable typically in less than few minutes, and then is destabilized by the variation in flow rate of syringe pumps [103]. Thus, it is indispensable to design a system that facilitates a stable tip-streaming over a long period of time [103,109]. In active design, the system should be parallelized and miniaturized. Furthermore, the piezoelectric dispenser and pulse laser-driven droplet generation show great potential to develop the fast-responding actuation and smart design of microfluidic junctions [108].
4. Conclusions and Future Prospects
Recent research on multiplex microfluidic detection platforms (Table 1) have demonstrated their potential for developing accurate, convenient, and rapid diagnoses of infectious diseases. LFIA is known as an ideal diagnostic assay characterized by fast, easy operation, durable stability, and low cost. Polymer-based microfluidic chips are a highly automated and integrated microfluidic platform. Paper-based microfluidic devices offer advantages such as ease of processing, control of reagent flow without external forces, and ease of customization. Droplet-based microfluidic devices are an alternative strategy for large-scale and parallel biological and chemical reactions, providing advantages of high throughput, low-cost, and multiplex detection. The industrialization of microfluidic platforms is also still in its infancy, and challenges such as liquid leakage and difficulties in reusability need to be improved by enhancing functional modularity of sample processing and target detection, as well as by automating platform fabrication and finding cost-effective substrate substitutes. Moreover, to improve the sensitivity and specificity of detection, the combination of two or more technologies may enhance the signal output or minimize interference [22]. Finally, cross-contamination and sensitivity attenuation of multiplex detections, and biosafety, data security, and privacy issues of microfluidic platforms should also be examined. A cohesive collaboration of industry and academic institutes will be increasingly desired to put microfluidic platforms into mass production and market distribution. In summary, microfluidic platforms have the outstanding advantages of integration, miniaturization, automation, and high-throughput, and the detection assays own the capabilities of portable signal readouts, simplicity, sensitivity, and specificity. Therefore, it is expected that microfluidic platforms integrated with various detection methods will provide a conceptually novel tool for infectious disease diagnoses.

Table 1.
Summary of multiplex microfluidic platforms based on sensor types.
Author Contributions
Conceptualization, K.Y. and X.G.; writing—original draft preparation, F.C.; visualization, F.C., Q.H., H.L., Y.X. and L.X.; writing—polishing language, Y.Z.; writing—review and editing, K.Y., Q.H. and X.G.; supervision, K.Y. and X.G.; funding acquisition, K.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (22104090) and the Natural Science Foundation of Shanghai (22ZR1436200).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (22104090) and the Natural Science Foundation of Shanghai (22ZR1436200). Unless otherwise stated in the figure captions, all images in the figures are adapted with permission from Biorender.com with a paid academic subscription.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, X.; Hong, X.-Z.; Li, Y.-W.; Li, Y.; Wang, J.; Chen, P.; Liu, B.-F. Microfluidics-based strategies for molecular diagnostics of infectious diseases. Mil. Med. Res. 2022, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.-S.F.; Kehe, J.; et al. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-X.; Liu, J.-S.; Han, L.-F.; Xia, S.; Li, S.-Z.; Li, O.Y.; Kassegne, K.; Li, M.; Yin, K.; Hu, Q.-Q.; et al. Towards a global One Health index: A potential assessment tool for One Health performance. Infect. Dis. Poverty 2022, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- The Overview of COVID-19. Available online: https://covid19.who.int/ (accessed on 16 December 2022).
- Teixeira, R.; Doetsch, J. The multifaceted role of mobile technologies as a strategy to combat COVID-19 pandemic. Epidemiol. Infect. 2020, 148, e244. [Google Scholar] [CrossRef]
- Piret, J.; Boivin, G. Pandemics Throughout History. Front. Microbiol. 2020, 11, 631736. [Google Scholar] [CrossRef]
- Heesterbeek, H.; Anderson, R.M.; Andreasen, V.; Bansal, S.; De Angelis, D.; Dye, C.; Eames, K.T.D.; Edmunds, W.J.; Frost, S.D.W.; Funk, S.; et al. Modeling infectious disease dynamics in the complex landscape of global health. Science 2015, 347, aaa4339. [Google Scholar] [CrossRef]
- Liu, L.; Moore, M.D. A Survey of Analytical Techniques for Noroviruses. Foods 2020, 9, 318. [Google Scholar] [CrossRef]
- Foddai, A.C.G.; Grant, I.R. Methods for detection of viable foodborne pathogens: Current state-of-art and future prospects. Appl. Microbiol. Biotechnol. 2020, 104, 4281–4288. [Google Scholar] [CrossRef]
- Li, J.; Macdonald, J. Multiplexed lateral flow biosensors: Technological advances for radically improving point-of-care diagnoses. Biosens. Bioelectron. 2016, 83, 177–192. [Google Scholar] [CrossRef]
- Li, H.; Xie, Y.; Chen, F.; Bai, H.; Xiu, L.; Zhou, X.; Guo, X.; Hu, Q.; Yin, K. Amplification-free CRISPR/Cas detection technology: Challenges, strategies, and perspectives. Chem. Soc. Rev. 2023, 52, 361–382. [Google Scholar] [CrossRef]
- Suther, C.; Stoufer, S.; Zhou, Y.; Moore, M.D. Recent Developments in Isothermal Amplification Methods for the Detection of Foodborne Viruses. Front. Microbiol. 2022, 13, 841875. [Google Scholar] [CrossRef]
- Qian, W.; Huang, J.; Wang, X.; Wang, T.; Li, Y. CRISPR-Cas12a combined with reverse transcription recombinase polymerase amplification for sensitive and specific detection of human norovirus genotype GII.4. Virology 2021, 564, 26–32. [Google Scholar] [CrossRef]
- Dincer, C.; Bruch, R.; Kling, A.; Dittrich, P.S.; Urban, G.A. Multiplexed Point-of-Care Testing–xPOCT. Trends Biotechnol. 2017, 35, 728–742. [Google Scholar] [CrossRef]
- Teixeira, W.; Pallás-Tamarit, Y.; Juste-Dolz, A.; Sena-Torralba, A.; Gozalbo-Rovira, R.; Rodríguez-Díaz, J.; Navarro, D.; Carrascosa, J.; Gimenez-Romero, D.; Maquieira, A.; et al. An all-in-one point-of-care testing device for multiplexed detection of respiratory infections. Biosens. Bioelectron. 2022, 213, 114454. [Google Scholar] [CrossRef]
- Gil Rosa, B.; Akingbade, O.E.; Guo, X.; Gonzalez-Macia, L.; Crone, M.A.; Cameron, L.P.; Freemont, P.; Choy, K.-L.; Güder, F.; Yeatman, E.; et al. Multiplexed immunosensors for point-of-care diagnostic applications. Biosens. Bioelectron. 2022, 203, 114050. [Google Scholar] [CrossRef]
- Manessis, G.; Gelasakis, A.I.; Bossis, I. Point-of-Care Diagnostics for Farm Animal Diseases: From Biosensors to Integrated Lab-on-Chip Devices. Biosensors 2022, 12, 455. [Google Scholar] [CrossRef]
- Kim, H.; Huh, H.J.; Park, E.; Chung, D.-R.; Kang, M. Multiplex Molecular Point-of-Care Test for Syndromic Infectious Diseases. BioChip J. 2021, 15, 14–22. [Google Scholar] [CrossRef]
- Mitchell, K.R.; Esene, J.E.; Woolley, A.T. Advances in multiplex electrical and optical detection of biomarkers using microfluidic devices. Anal. Bioanal. Chem. 2022, 414, 167–180. [Google Scholar] [CrossRef]
- Xie, Y.; Li, H.; Chen, F.; Udayakumar, S.; Arora, K.; Chen, H.; Lan, Y.; Hu, Q.; Zhou, X.; Guo, X.; et al. Clustered Regularly Interspaced short palindromic repeats-Based Microfluidic System in Infectious Diseases Diagnosis: Current Status, Challenges, and Perspectives. Adv. Sci. 2022, 9, e2204172. [Google Scholar] [CrossRef]
- Yadav, S.; Sharma, N.N.; Akhtar, J. Nucleic acid analysis on paper substrates (NAAPs): An innovative tool for Point of Care (POC) infectious disease diagnosis. Analyst 2021, 146, 3422–3439. [Google Scholar] [CrossRef]
- Wang, C.; Liu, M.; Wang, Z.; Li, S.; Deng, Y.; He, N. Point-of-care diagnostics for infectious diseases: From methods to devices. Nano Today 2021, 37, 101092. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xu, Y.; Fohlerova, Z.; Chang, H.; Iliescu, C.; Neuzil, P. LAMP-on-a-chip: Revising microfluidic platforms for loop-mediated DNA amplification. TrAC Trends Anal. Chem. 2019, 113, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Mani, V.; Durmus, C.; Khushaim, W.; Ferreira, D.C.; Timur, S.; Arduini, F.; Salama, K.N. Multiplexed sensing techniques for cardiovascular disease biomarkers—A review. Biosens. Bioelectron. 2022, 216, 114680. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-R.; Bong, J.-H.; Jung, J.; Sung, J.S.; Kang, M.-J.; Park, J.-G.; Pyun, J.-C. An On-chip Chemiluminescent Immunoassay for Bacterial Detection using in Situ-synthesized Cadmium Sulfide Nanowires with Passivation Layers. BioChip J. 2020, 14, 268–278. [Google Scholar] [CrossRef]
- Yen, C.-W.; de Puig, H.; Tam, J.O.; Gómez-Márquez, J.; Bosch, I.; Hamad-Schifferli, K.; Gehrke, L. Multicolored silver nanoparticles for multiplexed disease diagnostics: Distinguishing dengue, yellow fever, and Ebola viruses. Lab Chip 2015, 15, 1638–1641. [Google Scholar] [CrossRef]
- Wang, R.; Ongagna-Yhombi, S.Y.; Lu, Z.; Centeno-Tablante, E.; Colt, S.; Cao, X.; Ren, Y.; Cárdenas, W.B.; Mehta, S.; Erickson, D. Rapid Diagnostic Platform for Colorimetric Differential Detection of Dengue and Chikungunya Viral Infections. Anal. Chem. 2019, 91, 5415–5423. [Google Scholar] [CrossRef]
- Cavalera, S.; Colitti, B.; Rosati, S.; Ferrara, G.; Bertolotti, L.; Nogarol, C.; Guiotto, C.; Cagnazzo, C.; Denina, M.; Fagioli, F.; et al. A multi-target lateral flow immunoassay enabling the specific and sensitive detection of total antibodies to SARS COV-2. Talanta 2021, 223 Pt 1, 121737. [Google Scholar] [CrossRef]
- Wang, C.; Wang, C.; Wang, X.; Wang, K.; Zhu, Y.; Rong, Z.; Wang, W.; Xiao, R.; Wang, S. Magnetic SERS Strip for Sensitive and Simultaneous Detection of Respiratory Viruses. ACS Appl. Mater. Interfaces 2019, 11, 19495–19505. [Google Scholar] [CrossRef]
- Chen, S.; Meng, L.; Wang, L.; Huang, X.; Ali, S.; Chen, X.; Yu, M.; Yi, M.; Li, L.; Chen, X.; et al. SERS-based lateral flow immunoassay for sensitive and simultaneous detection of anti-SARS-CoV-2 IgM and IgG antibodies by using gap-enhanced Raman nanotags. Sens. Actuators B Chem. 2021, 348, 130706. [Google Scholar] [CrossRef]
- Liu, H.; Dai, E.; Xiao, R.; Zhou, Z.; Zhang, M.; Bai, Z.; Shao, Y.; Qi, K.; Tu, J.; Wang, C.; et al. Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sens. Actuators B Chem. 2021, 329, 129196. [Google Scholar] [CrossRef]
- Wang, C.; Yang, X.; Zheng, S.; Cheng, X.; Xiao, R.; Li, Q.; Wang, W.; Liu, X.; Wang, S. Development of an ultrasensitive fluorescent immunochromatographic assay based on multilayer quantum dot nanobead for simultaneous detection of SARS-CoV-2 antigen and influenza A virus. Sens. Actuators B Chem. 2021, 345, 130372. [Google Scholar] [CrossRef]
- Wang, C.; Cheng, X.; Liu, L.; Zhang, X.; Yang, X.; Zheng, S.; Rong, Z.; Wang, S. Ultrasensitive and Simultaneous Detection of Two Specific SARS-CoV-2 Antigens in Human Specimens Using Direct/Enrichment Dual-Mode Fluorescence Lateral Flow Immunoassay. ACS Appl. Mater. Interfaces 2021, 13, 40342–40353. [Google Scholar] [CrossRef]
- Guo, Y.; Zhou, Y.; Fu, J.; Fang, H.; Li, Y.; Huang, X.; Xiong, Y. A self-luminous bifunctional bacteria directed fluorescent immunosensor for the simultaneous detection and quantification of three pathogens in milk. Sens. Actuators B Chem. 2021, 338, 129757. [Google Scholar] [CrossRef]
- Layqah, L.A.; Eissa, S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim. Acta 2019, 186, 224. [Google Scholar] [CrossRef]
- Li, P.; Zhang, Z.; Zhang, Q.; Zhang, N.; Zhang, W.; Ding, X.; Li, R. Current development of microfluidic immunosensing approaches for mycotoxin detection via capillary electromigration and lateral flow technology. Electrophoresis 2012, 33, 2253–2265. [Google Scholar] [CrossRef]
- Xing, K.-Y.; Shan, S.; Liu, D.-F.; Lai, W.-H. Recent advances of lateral flow immunoassay for mycotoxins detection. TrAC Trends Anal. Chem. 2020, 133, 116087. [Google Scholar] [CrossRef]
- Chen, X.; Ding, L.; Huang, X.; Xiong, Y. Tailoring noble metal nanoparticle designs to enable sensitive lateral flow immunoassay. Theranostics 2022, 12, 574–602. [Google Scholar] [CrossRef]
- Di Nardo, F.; Chiarello, M.; Cavalera, S.; Baggiani, C.; Anfossi, L. Ten Years of Lateral Flow Immunoassay Technique Applications: Trends, Challenges and Future Perspectives. Sensors 2021, 21, 5185. [Google Scholar] [CrossRef]
- Jauset-Rubio, M.; El-Shahawi, M.S.; Bashammakh, A.S.; Alyoubi, A.O.; O′sullivan, C.K. Advances in aptamers-based lateral flow assays. TrAC Trends Anal. Chem. 2017, 97, 385–398. [Google Scholar] [CrossRef]
- Bishop, J.D.; Hsieh, H.V.; Gasperino, D.J.; Weigl, B.H. Sensitivity enhancement in lateral flow assays: A systems perspective. Lab Chip 2019, 19, 2486–2499. [Google Scholar] [CrossRef]
- Huang, Z.; Hu, S.; Xiong, Y.; Wei, H.; Xu, H.; Duan, H.; Lai, W. Application and development of superparamagnetic nanoparticles in sample pretreatment and immunochromatographic assay. TrAC Trends Anal. Chem. 2019, 114, 151–170. [Google Scholar] [CrossRef]
- Liu, S.; Dou, L.; Yao, X.; Zhang, W.; Zhao, M.; Yin, X.; Sun, J.; Zhang, D.; Wang, J. Nanozyme amplification mediated on-demand multiplex lateral flow immunoassay with dual-readout and broadened detection range. Biosens. Bioelectron. 2020, 169, 112610. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Shi, D.; Wan, N.; Yang, X.; Liu, H.; Gao, H.; Zhang, M.; Bai, Z.; Li, D.; Dai, E.; et al. Development of spike protein-based fluorescence lateral flow assay for the simultaneous detection of SARS-CoV-2 specific IgM and IgG. Analyst 2021, 146, 3908–3917. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Covián, L.; Montes-García, V.; Girard, A.; Fernández-Abedul, M.T.; Pérez-Juste, J.; Pastoriza-Santos, I.; Faulds, K.; Graham, D.; Blanco-López, M.C. Au@Ag SERRS tags coupled to a lateral flow immunoassay for the sensitive detection of pneumolysin. Nanoscale 2017, 9, 2051–2058. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Hwang, J.; Kim, K.; Jeon, J.; Lee, S.; Ko, J.; Lee, J.; Kang, M.; Chung, D.R.; Choo, J. Quantitative Serodiagnosis of Scrub Typhus Using Surface-Enhanced Raman Scattering-Based Lateral Flow Assay Platforms. Anal. Chem. 2019, 91, 12275–12282. [Google Scholar] [CrossRef]
- Fang, B.; Xiong, Q.; Duan, H.; Xiong, Y.; Lai, W. Tailored quantum dots for enhancing sensing performance of lateral flow immunoassay. TrAC Trends Anal. Chem. 2022, 157, 116754. [Google Scholar] [CrossRef]
- Cubas-Atienzar, A.I.; Kontogianni, K.; Edwards, T.; Wooding, D.; Buist, K.; Thompson, C.R.; Williams, C.T.; Patterson, E.I.; Hughes, G.L.; Baldwin, L.; et al. Limit of detection in different matrices of 19 commercially available rapid antigen tests for the detection of SARS-CoV-2. Sci. Rep. 2021, 11, 18313. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Fang, L.; Jia, B.; Long, N.; Shi, L.; Zhou, L.; Zhao, H.; Kong, W. Optical lateral flow test strip biosensors for pesticides: Recent advances and future trends. TrAC Trends Anal. Chem. 2021, 144, 116427. [Google Scholar] [CrossRef]
- Pei, F.; Feng, S.; Hu, W.; Liu, B.; Mu, X.; Hao, Q.; Cao, Y.; Lei, W.; Tong, Z. Sandwich mode lateral flow assay for point-of-care detecting SARS-CoV-2. Talanta 2023, 253, 124051. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, Y.; Leng, Y.; Lai, W.; Huang, X.; Xiong, Y. Emerging design strategies for constructing multiplex lateral flow test strip sensors. Biosens. Bioelectron. 2020, 157, 112168. [Google Scholar] [CrossRef]
- Pu, F.; Ren, J.; Qu, X. Recent progress in sensor arrays using nucleic acid as sensing elements. Coord. Chem. Rev. 2022, 456, 214379. [Google Scholar] [CrossRef]
- Aman, R.; Mahas, A.; Mahfouz, M. Nucleic Acid Detection Using CRISPR/Cas Biosensing Technologies. ACS Synth. Biol. 2020, 9, 1226–1233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X. Development of CRISPR-Mediated Nucleic Acid Detection Technologies and Their Applications in the Livestock Industry. Genes 2022, 13, 2007. [Google Scholar] [CrossRef] [PubMed]
- Trinh, T.N.D.; Lee, N.Y. Advances in Nucleic Acid Amplification-Based Microfluidic Devices for Clinical Microbial Detection. Chemosensors 2022, 10, 123. [Google Scholar] [CrossRef]
- Yang, S.-M.; Lv, S.; Zhang, W.; Cui, Y. Microfluidic Point-of-Care (POC) Devices in Early Diagnosis: A Review of Opportunities and Challenges. Sensors 2022, 22, 1620. [Google Scholar] [CrossRef]
- Murdock, R.C.; Gallegos, K.M.; Hagen, J.A.; Kelley-Loughnane, N.; Weiss, A.A.; Papautsky, I. Development of a point-of-care diagnostic for influenza detection with antiviral treatment effectiveness indication. Lab Chip 2017, 17, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Xiong, E.; Jiang, L.; Tian, T.; Hu, M.; Yue, H.; Huang, M.; Lin, W.; Jiang, Y.; Zhu, D.; Zhou, X. Simultaneous Dual-Gene Diagnosis of SARS-CoV-2 Based on CRISPR/Cas9-Mediated Lateral Flow Assay. Angew. Chem. Int. Ed. 2021, 60, 5307–5315. [Google Scholar] [CrossRef]
- Shu, B.; Lin, L.; Wu, B.; Huang, E.; Wang, Y.; Li, Z.; He, H.; Lei, X.; Xu, B.; Liu, D. A pocket-sized device automates multiplexed point-of-care RNA testing for rapid screening of infectious pathogens. Biosens. Bioelectron. 2021, 181, 113145. [Google Scholar] [CrossRef] [PubMed]
- Nasseri, B.; Soleimani, N.; Rabiee, N.; Kalbasi, A.; Karimi, M.; Hamblin, M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018, 117, 112–128. [Google Scholar] [CrossRef]
- Narayanamurthy, V.; Jeroish, Z.E.; Bhuvaneshwari, K.S.; Bayat, P.; Premkumar, R.; Samsuri, F.; Yusoff, M.M. Advances in passively driven microfluidics and lab-on-chip devices: A comprehensive literature review and patent analysis. RSC Adv. 2020, 10, 11652–11680. [Google Scholar] [CrossRef]
- Xu, B.; Guo, J.; Fu, Y.; Chen, X.; Guo, J. A review on microfluidics in the detection of food pesticide residues. Electrophoresis 2020, 41, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Van Nguyen, H.; Phan, V.M.; Seo, T.S. A portable centrifugal genetic analyzer for multiplex detection of feline upper respiratory tract disease pathogens. Biosens. Bioelectron. 2021, 193, 113546. [Google Scholar] [CrossRef] [PubMed]
- Ciftci, S.; Neumann, F.; Abdurahman, S.; Appelberg, K.S.; Mirazimi, A.; Nilsson, M.; Madaboosi, N. Digital Rolling Circle Amplification–Based Detection of Ebola and Other Tropical Viruses. J. Mol. Diagn. 2020, 22, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Yan, C.; Wu, W.; Li, J. Application of Microfluidic Chip Technology in Food Safety Sensing. Sensors 2020, 20, 1792. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Shan, X.; Cao, R.; Jin, X.; Lin, X.; He, Q.; Zhu, Y.; Fu, R.; Du, W.; Lv, W.; et al. Microfluidic Chip with Two-Stage Isothermal Amplification Method for Highly Sensitive Parallel Detection of SARS-CoV-2 and Measles Virus. Micromachines 2021, 12, 1582. [Google Scholar] [CrossRef]
- Choi, J.-H.; Shin, M.; Yang, L.; Conley, B.; Yoon, J.; Lee, S.-N.; Lee, K.-B.; Choi, J.-W. Clustered Regularly Interspaced Short Palindromic Repeats-Mediated Amplification-Free Detection of Viral DNAs Using Surface-Enhanced Raman Spectroscopy-Active Nanoarray. ACS Nano 2021, 15, 13475–13485. [Google Scholar] [CrossRef]
- Arshavsky-Graham, S.; Enders, A.; Ackerman, S.; Bahnemann, J.; Segal, E. 3D-printed microfluidics integrated with optical nanostructured porous aptasensors for protein detection. Microchim. Acta 2021, 188, 67. [Google Scholar] [CrossRef]
- Chang, N.; Zhai, J.; Liu, B.; Zhou, J.; Zeng, Z.; Zhao, X. Low cost 3D microfluidic chips for multiplex protein detection based on photonic crystal beads. Lab Chip 2018, 18, 3638–3644. [Google Scholar] [CrossRef]
- Yin, K.; Ding, X.; Xu, Z.; Li, Z.; Wang, X.; Zhao, H.; Otis, C.; Li, B.; Liu, C. Multiplexed colorimetric detection of SARS-CoV-2 and other pathogens in wastewater on a 3D printed integrated microfluidic chip. Sens. Actuators B Chem. 2021, 344, 130242. [Google Scholar] [CrossRef]
- Harding-Esch, E.; Cousins, E.; Chow, S.-L.; Phillips, L.; Hall, C.; Cooper, N.; Fuller, S.; Nori, A.; Patel, R.; Thomas-William, S.; et al. A 30-Min Nucleic Acid Amplification Point-of-Care Test for Genital Chlamydia trachomatis Infection in Women: A Prospective, Multi-center Study of Diagnostic Accuracy. Ebiomedicine 2018, 28, 120–127. [Google Scholar] [CrossRef]
- The Binx, Io. Available online: https://mybinxhealth.com/point-of-care (accessed on 12 March 2023).
- Sadek, M.; Demord, A.; Poirel, L.; Nordmann, P. Fast and reliable detection of carbapenemase genes in various Gram negatives using a new commercially available fluorescence-based real-time polymerase chain reaction platform. Diagn. Microbiol. Infect. Dis. 2020, 98, 115127. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Zhang, Y.; Tian, J.; Wu, T.; Li, Z.; Xing, F.; Fu, S. Polymer-based microfluidic devices: A comprehensive review on preparation and applications. Polym. Eng. Sci. 2021, 62, 3–24. [Google Scholar] [CrossRef]
- Iyer, V.; Yang, Z.; Ko, J.; Weissleder, R.; Issadore, D. Advancing microfluidic diagnostic chips into clinical use: A review of current challenges and opportunities. Lab Chip 2022, 22, 3110–3121. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Liu, J.; Zhang, Z.; Li, J.; Yuan, L.; Zhang, Z.; Chen, L. Microfluidic paper-based chips in rapid detection: Current status, challenges, and perspectives. TrAC Trends Anal. Chem. 2021, 143, 116371. [Google Scholar] [CrossRef]
- Wang, X.; Li, B.; Guo, Y.; Shen, S.; Zhao, L.; Zhang, P.; Sun, Y.; Sui, S.-F.; Deng, F.; Lou, Z. Molecular basis for the formation of ribonucleoprotein complex of Crimean-Congo hemorrhagic fever virus. J. Struct. Biol. 2016, 196, 455–465. [Google Scholar] [CrossRef]
- Liu, P.; Li, B.; Fu, L.; Huang, Y.; Man, M.; Qi, J.; Sun, X.; Kang, Q.; Shen, D.; Chen, L. Hybrid Three Dimensionally Printed Paper-Based Microfluidic Platform for Investigating a Cell’s Apoptosis and Intracellular Cross-Talk. ACS Sens. 2020, 5, 464–473. [Google Scholar] [CrossRef]
- Christodouleas, D.C.; Kaur, B.; Chorti, P. From Point-of-Care Testing to eHealth Diagnostic Devices (eDiagnostics). ACS Cent. Sci. 2018, 4, 1600–1616. [Google Scholar] [CrossRef]
- Huang, L.; Tian, S.; Zhao, W.; Liu, K.; Ma, X.; Guo, J. Aptamer-based lateral flow assay on-site biosensors. Biosens. Bioelectron. 2021, 186, 113279. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- Martinez, A.W.; Phillips, S.T.; Butte, M.; Whitesides, G.M. Patterned Paper as a Platform for Inexpensive, Low-Volume, Portable Bioassays. Angew. Chem. Int. Ed. 2007, 46, 1318–1320. [Google Scholar] [CrossRef]
- Selvakumar, B.; Kathiravan, A. Sensory materials for microfluidic paper based analytical devices—A review. Talanta 2021, 235, 122733. [Google Scholar] [CrossRef]
- Noviana, E.; Ozer, T.; Carrell, C.S.; Link, J.S.; McMahon, C.; Jang, I.; Henry, C.S. Microfluidic Paper-Based Analytical Devices: From Design to Applications. Chem. Rev. 2021, 121, 11835–11885. [Google Scholar] [CrossRef]
- Trinh, T.N.D.; Lee, N.Y. A rapid and eco-friendly isothermal amplification microdevice for multiplex detection of foodborne pathogens. Lab Chip 2018, 18, 2369–2377. [Google Scholar] [CrossRef]
- Trinh, K.T.L.; Trinh, T.N.D.; Lee, N.Y. Fully integrated and slidable paper-embedded plastic microdevice for point-of-care testing of multiple foodborne pathogens. Biosens. Bioelectron. 2019, 135, 120–128. [Google Scholar] [CrossRef]
- Harpaldas, H.; Arumugam, S.; Rodriguez, C.C.; Kumar, B.A.; Shi, V.; Sia, S.K. Point-of-care diagnostics: Recent developments in a pandemic age. Lab Chip 2021, 21, 4517–4548. [Google Scholar] [CrossRef]
- Sachdeva, S.; Davis, R.W.; Saha, A.K. Microfluidic Point-of-Care Testing: Commercial Landscape and Future Directions. Front. Bioeng. Biotechnol. 2020, 8, 602659. [Google Scholar] [CrossRef]
- Yamada, K.; Shibata, H.; Suzuki, K.; Citterio, D. Toward practical application of paper-based microfluidics for medical diagnostics: State-of-the-art and challenges. Lab Chip 2017, 17, 1206–1249. [Google Scholar] [CrossRef]
- Lee, S.; Mehta, S.; Erickson, D. Two-Color Lateral Flow Assay for Multiplex Detection of Causative Agents Behind Acute Febrile Illnesses. Anal. Chem. 2016, 88, 8359–8363. [Google Scholar] [CrossRef]
- Luo, K.; Kim, H.-Y.; Oh, M.-H.; Kim, Y.-R. Paper-based lateral flow strip assay for the detection of foodborne pathogens: Principles, applications, technological challenges and opportunities. Crit. Rev. Food Sci. Nutr. 2020, 60, 157–170. [Google Scholar] [CrossRef]
- Yamada, K.; Henares, T.G.; Suzuki, K.; Citterio, D. Distance-Based Tear Lactoferrin Assay on Microfluidic Paper Device Using Interfacial Interactions on Surface-Modified Cellulose. ACS Appl. Mater. Interfaces 2015, 7, 24864–24875. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Ding, F.; Yang, Y.; Zhao, G.; Zhang, C.; Wang, R.; Huang, X. Research Progress and Future Trends of Microfluidic Paper-Based Analytical Devices in In-Vitro Diagnosis. Biosensors 2022, 12, 485. [Google Scholar] [CrossRef] [PubMed]
- Ning, R.; Fan, J.; Kong, L.; Jiang, X.; Qian, Y.; Du, T.; Zhang, G.; Wu, W. Recent developments of droplets-based microfluidics for bacterial analysis. Chin. Chem. Lett. 2021, 33, 2243–2252. [Google Scholar] [CrossRef]
- Kalantarifard, A.; Saateh, A.; Elbuken, C. Label-Free Sensing in Microdroplet-Based Microfluidic Systems. Chemosensors 2018, 6, 23. [Google Scholar] [CrossRef]
- Liu, W.-W.; Zhu, Y. “Development and application of analytical detection techniques for droplet-based microfluidics”—A review. Anal. Chim. Acta 2020, 1113, 66–84. [Google Scholar] [CrossRef]
- Kaushik, A.M.; Hsieh, K.; Wang, T. Droplet microfluidics for high-sensitivity and high-throughput detection and screening of disease biomarkers. WIREs Nanomed. Nanobiotechnol. 2018, 10, e1522. [Google Scholar] [CrossRef]
- Kaminski, T.S.; Scheler, O.; Garstecki, P. Droplet microfluidics for microbiology: Techniques, applications and challenges. Lab Chip 2016, 16, 2168–2187. [Google Scholar] [CrossRef]
- Yin, H.; Wu, Z.; Shi, N.; Qi, Y.; Jian, X.; Zhou, L.; Tong, Y.; Cheng, Z.; Zhao, J.; Mao, H. Ultrafast multiplexed detection of SARS-CoV-2 RNA using a rapid droplet digital PCR system. Biosens. Bioelectron. 2021, 188, 113282. [Google Scholar] [CrossRef]
- Tan, Y.-L.; Huang, A.-Q.; Tang, L.-J.; Jiang, J.-H. Multiplexed droplet loop-mediated isothermal amplification with scorpion-shaped probes and fluorescence microscopic counting for digital quantification of virus RNAs. Chem. Sci. 2021, 12, 8445–8451. [Google Scholar] [CrossRef]
- Ding, Y.; Howes, P.D.; Demello, A.J. Recent Advances in Droplet Microfluidics. Anal. Chem. 2020, 92, 132–149. [Google Scholar] [CrossRef]
- Zhu, P.; Wang, L. Passive and active droplet generation with microfluidics: A review. Lab Chip 2016, 17, 34–75. [Google Scholar] [CrossRef]
- Lee, C.-P.; Lan, T.-S.; Lai, M.-F. Fabrication of two-dimensional ferrofluid microdroplet lattices in a microfluidic channel. J. Appl. Phys. 2014, 115, 17B527. [Google Scholar] [CrossRef]
- Kocak, D.D.; Gersbach, C.A. From CRISPR scissors to virus sensors. Nature 2018, 557, 168–169. [Google Scholar] [CrossRef]
- Chertow, D.S. Next-generation diagnostics with CRISPR. Science 2018, 360, 381–382. [Google Scholar] [CrossRef]
- Lignos, I.; Morad, V.; Shynkarenko, Y.; Bernasconi, C.; Maceiczyk, R.M.; Protesescu, L.; Bertolotti, F.; Kumar, S.; Ochsenbein, S.T.; Masciocchi, N.; et al. Exploration of Near-Infrared-Emissive Colloidal Multinary Lead Halide Perovskite Nanocrystals Using an Automated Microfluidic Platform. ACS Nano 2018, 12, 5504–5517. [Google Scholar] [CrossRef]
- Sohrabi, S.; Kassir, N.; Moraveji, M.K. Droplet microfluidics: Fundamentals and its advanced applications. RSC Adv. 2020, 10, 27560–27574. [Google Scholar] [CrossRef]
- Amirifar, L.; Besanjideh, M.; Nasiri, R.; Shamloo, A.; Nasrollahi, F.; de Barros, N.R.; Davoodi, E.; Erdem, A.; Mahmoodi, M.; Hosseini, V.; et al. Droplet-based microfluidics in biomedical applications. Biofabrication 2022, 14, 022001. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).