Abstract
Cervical cancer is the fourth most diagnosed cancer and the fourth leading cause of cancer death in women globally. Its onset and progression have been attributed to high-risk human papillomavirus (HPV) types, especially 16 and 18, while the Epstein–Barr virus (EBV) is believed to also significantly contribute to cervical cancer growth. The E6 protein associated with high-risk HPV strains, such as HPV16 and HPV18, is known for its role in promoting cervical cancer and other anogenital cancers. E6 proteins contribute to the malignant transformation of infected cells by targeting and degrading tumor suppressor proteins, especially p53. On the other hand, EBV nuclear antigen 1 (EBNA1) plays a crucial role in the maintenance and replication of the EBV genome in infected cells. EBNA1 is believed to increase HPV E6 and E7 levels, as well as c-MYC, and BIRC5 cellular genes in the HeLa cell line, implying that HPV/EBV co-infection accelerates cervical cancer onset and growth. Thus, the E6 and EBNA1 antigens of HPV and EBV, respectively, are attractive targets for cervical cancer immunotherapy. This study, therefore, virtually screened for potential drug candidates with good binding affinity to all three oncoviral proteins, HPV16 E6, HPV18 E6, and EBNA1. The compounds were further subjected to ADMET profiling, biological activity predictions, molecular dynamics (MD) simulations, and molecular mechanics Poisson–Boltzmann surface area (MM/PBSA) calculations. A total of six compounds comprising ZINC000013380012, ZINC000070454124, ZINC000014588133, ZINC000085568136, ZINC000095909247, and ZINC000085597263 demonstrated very strong affinity (≤−60 kJ/mol) to the three oncoviral proteins (EBNA1, HPV16 E6, and HPV18 E6) after being subjected to docking, MD, and MM/PBSA. These compounds demonstrated relatively stronger binding than the controls used, inhibitors of EBNA1 (VK-1727) and HPV E6 (baicalein and gossypetin). Biological activity predictions also corroborated their antineoplastic, p53-enhancing, Pin1 inhibitory, and JAK2 inhibitory activities. Further experimental testing is required to validate the ability of the shortlisted compounds to silence the insidious effects of HPV E6 and EBNA1 proteins in cervical cancers.
1. Introduction
Several cancers have been linked to various viruses, including human papillomaviruses (HPVs), hepatitis viruses (HBV and HCV), Epstein–Barr virus (EBV), Kaposi’s sarcoma herpesvirus (KSHV), human T cell leukemia virus (HTLV-I), and Merkel cell polyomavirus (MCPyV) [1]. Virus-associated cancers account for about 10 to 15% of cancer cases globally [2,3], representing over 1.4 million cases annually [1]. These two human oncoviruses, HPV and EBV, are responsible for approximately 38% of all tumor-associated cancers [3]. Cervical cancer is one such cancer and is primarily associated with persistent infection by high-risk HPV types, emphasizing the pivotal role of viral oncogenesis in its development [4,5,6]. Although the disease is preventable and treatable when detected early, challenges persist in ensuring widespread access to preventive measures, which include HPV vaccination and regular screening programs [7,8,9,10]. Globally, approximately 604,000 women are diagnosed with cervical cancer, while over 342,000 die annually [11]. The death rate is estimated to reach over 440,000 by 2030 [12]. Cervical cancer ranks as the fourth most commonly diagnosed cancer and the fourth primary contributor to cancer-related fatalities among women [11,12].
HPVs typically target layered epithelial tissues and are directly linked to various cancers affecting the anogenital region, such as cervical, anal, vulvar, and vaginal carcinomas [1]. Additionally, they are associated with carcinomas in other mucosal epithelial areas, particularly in the oropharyngeal region [1]. HPV is diagnosed in more than 90% of cervical cancer cases [12], with the most common HPV types being 16 and 18 (approximately 70%) [13]. HPV is a relatively small virus (55 nm in diameter) that possesses a non-enveloped circular double-stranded genomic DNA of approximately 8 kb in size [14,15,16]. Another oncovirus, EBV, which is an oncogenic herpesvirus, is associated with a spectrum of diseases, ranging from infectious mononucleosis to various neoplastic conditions [17,18,19]. These neoplastic diseases include lymphoproliferative disorders in immunocompromised individuals, Burkitt’s lymphoma, Hodgkin’s lymphoma, and nasopharyngeal carcinoma [20,21,22]. More than 95% of healthy adults globally experience asymptomatic infection with EBV [23,24]. EBV possesses a linear double-stranded DNA genome of around 175 kb, which undergoes circularization within latently infected cells [23]. Within the EBV genome, there are over 80 open reading frames (ORFs), multiple non-coding RNAs, and 44 mature microRNAs (miRNA) [23,25,26]. Latent membrane protein 1 (LMP1) and four EBNA proteins comprising EBNA1, EBNA2, EBNA3A, and EBNA3C are crucial EBV gene products required for B cell transformation [25]. There are currently no approved EBV drugs, as the drugs that demonstrated efficacy in replicating EBV had limited clinical successes [24,27]. A meta-analysis combining data from 25 articles revealed that women with EBV infection had a fourfold higher occurrence of cervical carcinoma compared to women without EBV [28]. Moreover, the study indicated that EBV/HPV co-infection is associated with an elevated risk of cervical cancer [28].
Oncoproteins of EBV (LMP1, LMP2A, and EBNA1) and high-risk HPVs (E5, E6, and E7) have been documented to contribute to the onset and growth of various human cancers, including head and neck, cervical, and colorectal [29]. In HPV-positive cervical, oral, and lung cancer cells, the upregulation of Foxhead box M1 (FOXM1) expression occurs through E6-mediated NKX2-1 [30,31]. The induction of FOXM1 by E6 is orchestrated through the MZF1/NKX2-1 axis, and this regulatory mechanism is responsible for HPV-mediated traits such as soft agar growth, invasiveness, and stemness [30]. The activation of the Wnt/β-catenin signaling pathway is implicated in mediating these effects, underscoring the intricate molecular interactions contributing to the oncogenic potential of HPV in these cancer types [30]. Also, the interaction between E6 and the cellular ubiquitin ligase E6AP facilitates the degradation of p53, a tumor suppressor protein [32,33]. Consequently, a crucial strategy for hindering the survival and proliferation of infected cells involves suppressing the formation of the E6–E6AP complex [34,35].
EBNA1, a nuclear antigen, is crucial for maintaining, replicating, and segregating the EBV genome and is required for preserving the episomal DNA of EBV within infected cells [27,36]. It is the sole protein necessary for the replication of the latent virus [36]. However, the replication and persistence of the EBV episomal genome rely significantly on EBNA1 binding to the EBV origin of plasmid replication (oriP) element [37,38]. EBNA1 is believed to elevate HPV18 E6 and E7 levels along with the cellular genes c-MYC and BIRC5 in the HeLa cell line [39]. This implies that co-infection of cervical cells with HPV and EBV might accelerate cervical cancer growth and progression [39]. Considering the crucial roles of HPV E6 and EBNA1 in the tumorigenesis, proliferation, and progression of cervical cancer cells, it is imperative to identify drug candidates to augment the therapeutic options for combating this cancer, which is characterized by a high mortality rate.
Polypharmacology, which describes the capacity of a single drug to engage with multiple targets or biological pathways within an organism, is increasingly prominent in drug discovery research. The complexity of certain diseases, including cancers, often renders targeting a single pathway inadequate for achieving the desired therapeutic effect [40,41,42]. Hence, exploring compounds with polypharmacological properties has emerged as a promising approach to addressing the multifaceted mechanisms underlying such diseases and enhancing therapeutic efficacy [42]. A recent study conducted at the oncology center of the University of Gondar Comprehensive Specialized Hospital (Ethiopia) analyzed data from 124 cervical cancer patients to identify common medication-related problems (MRPs) [43]. The study included all patients diagnosed with cervical cancer between 1 January 2016 and 31 December 2020. The study revealed that the most common MRPs included suboptimal dosage, requirement for supplementary drug therapy, and adverse drug reactions (ADRs), accounting for 24.4%, 22.6%, and 22% of cases, respectively [43]. Furthermore, 16.1% (27 patients) experienced minor to moderate drug–drug interactions (DDIs), with 48% of these classified as moderate, necessitating close monitoring of the outcome, and 8% categorized as serious reactions, prompting the use of alternative medications in the treatment regimen [43]. Interestingly, the study also found that patients with polypharmacy (≥5 drugs) were seven times more likely to experience MRPs compared to their counterparts [43]. Polypharmacology presents an opportunity to mitigate drug–drug interactions and enhance patient compliance through simplified dosing regimens [44] while potentially reducing the risk of drug resistance [45,46].
Natural products provide a large and diverse source for finding these treatment options [47]. Harnessing the potential of natural products may lead to the discovery of novel compounds with novel mechanisms of action against cervical cancer, contributing to the development of more effective and targeted therapeutic interventions [48,49]. Additionally, the exploration of natural products aligns with the growing interest in sustainable and environmentally friendly approaches to drug discovery. Computational-based drug discovery approaches offer an efficient and cost-effective avenue to expeditiously identify potential drug candidates [50,51]. Using a robust computational drug discovery pipeline, the enrichment factor for shortlisting promising drug candidates can be significantly enhanced. The integration of computational techniques, such as molecular docking, molecular dynamics simulations, biological activity prediction, and toxicity predictions enables the screening of large compound libraries, shortlisting compounds with the highest likelihood of success.
This study therefore employed computational techniques to sift through a vast traditional Chinese medicine (TCM) chemical landscape [52], seeking compounds that can potentially inhibit the E6 proteins of both HPV16 and HPV18. These compounds, displaying commendable binding affinity to both E6 proteins (docking scores ≤ −8.5 kcal/mol) were re-docked against the EBNA1 protein, unveiling potential polypharmacologic drugs that extend beyond a single viral or cancer target. The shortlisted candidates underwent further rigorous scrutiny through ADMET screening and profiling and biological activity predictions prior to molecular dynamics (MD) simulations and molecular mechanics Poisson–Boltzmann surface area (MM/PBSA) calculations.
2. Materials and Methods
This study employed a molecular docking technique to virtually screen natural products from the TCM database against three oncoproteins, EBNA1, HPV16 E6, and HPV18 E6 (Figure 1). The top compounds were subjected to safety and toxicity profiling using SwissADME and DataWarrior 5.0. Compounds that were predicted to be drug-like, non-tumorigenic, non-mutagenic, and non-irritants and had no reproductive effects were shortlisted, and their biological activities were predicted. Ten top candidates in complex with each of the proteins were further subjected to 100 ns MD simulations and MM/PBSA calculations (Figure 1). Similarity searches were then performed for the topmost compounds that were predicted to have strong binding affinity to all three oncoproteins.
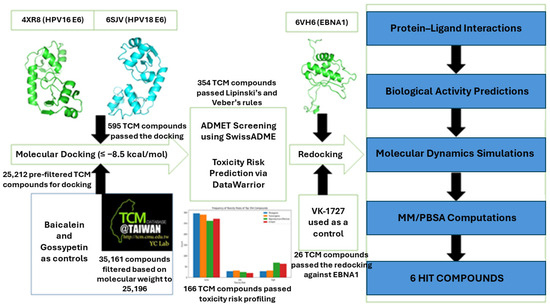
Figure 1.
Step-by-step procedure employed in this study to identify potential small molecule inhibitors of HPV E6 and EBNA1. Compounds from traditional Chinese medicine were screened against EBNA1 and E6 proteins of HPV16 and HPV18. Top compounds were subjected to ADMET profiling and biological activity predictions. The shortlisted compounds were further subjected to molecular dynamics simulations and MM/PBSA computations to evaluate their binding affinities.
2.1. Obtaining Protein Structures
Structures of HPV16 E6, HPV18 E6, and EBNA1 were searched and retrieved from the Research Collaboratory for Structural Bioinformatics (RCSB) protein databank (PDB) [53,54]. The RCSB PDB is a repository that houses experimentally determined protein structures [53,54]. In selecting the protein structures to be used in this study, factors such as resolution, the count of missing residues, and the year of structure determination were considered. The lower the resolution value of the protein structure, the higher the level of detail (or resolution) and accuracy in depicting the atomic arrangement within the protein [55].
2.2. Protein and Ligand Preparation
The 3D structure data file (sdf) format of two E6 inhibitors, gossypetin and baicalein, were obtained from PubChem [56,57,58] with corresponding compound identifiers (CIDs) of 5280647 and 5281605. Baicalein and gossypetin have been shown to inhibit E6 (of both HPV16 and HPV18), limit E6 binding to E6AP, and prevent E6-mediated p53 degradation [59]. The 2D sdf of EBNA1 inhibitor VK-1727 [60] was also obtained from PubChem and then converted to 3D using OpenBabel [61] due to the unavailability of a 3D sdf format. The screening library consisted of 35,161 compounds from the traditional Chinese Medicine (TCM) catalogue found on ZINC15 database [62,63]. The TCM compounds were pre-filtered using a molecular weight criterion of 150 to 600 g/mol, as previously performed [64]. Also, duplicates were removed using OSIRIS DataWarrior (Idorsia Pharmaceuticals Ltd., version 5.0.0, Allschwil, Switzerland) [65]. After filtering, 25,212 TCM compounds were obtained and used for the molecular docking study. All the ligands were then subjected to energy minimization using the Universal force field (UFF) [66,67] and were converted to protein data bank, partial charge (Q), and atom type (T) (“pdbqt”) formats via PyRx 0.9 [68,69]. The three protein structures (HPV16 E6, HPV18 E6, and EBNA1) were also subjected to energy minimization via GROMACS version 5.1.5 using the optimized potentials for liquid simulations (OPLS) force field prior to docking. After energy minimization, each protein structure was converted to pdbqt format via PyRx using the “Make Macromolecule” option.
2.3. Molecular Docking Studies (E6 Proteins of HPV16 and HPV18)
Molecular docking, a computational structure-based approach in drug discovery, aids in the identification of potential therapeutic compounds by predicting interactions between proteins and ligands [70,71,72]. A major advantage of molecular docking is its capacity to perform these tasks without the need for prior knowledge of the chemical structure of other target modulators [71]. This study employed a molecular docking tool, AutoDock Vina [73] to virtually screen the TCM ligands and the two known E6 inhibitors against both E6 proteins. Compounds that demonstrated docking scores less than −8.4 kcal/mol for both proteins were shortlisted for analysis. For the HPV16 E6 protein, a grid box dimension of 51.88 × 42.19 × 43.44 Å3 was used centering the protein at x = 39.12 Å, y = 38.85 Å, and z = 38.90 Å. Using a center of x = 38.89 Å, y = 39.18 Å, and z = 39 Å, a grid box of size of 45.59 × 46.03 × 44.56 Å3 was also set up for the HPV18 E6 protein. For all the docking runs, the exhaustiveness was set to 8.
2.4. Predictive Evaluation of ADMET Properties
The shortlisted compounds that demonstrated good binding affinities to both E6 proteins were subjected to ADMET profiling using SwissADME [74]. The compounds were assessed based on Lipinski’s rule of 5 [75] and Veber’s rule [76]. Toxicity risks, including mutagenicity, tumorigenicity, irritancy, and reproductive effects of the shortlisted compounds, were also determined using OSIRIS DataWarrior 5.0 [65]. These analyses aimed to provide a more thorough understanding of the compounds’ pharmacokinetic and safety profiles, contributing valuable insights into their potential as therapeutic candidates. These analyses were also performed to identify and eliminate potentially toxic candidates or compounds with low safety profiles.
2.5. Re-Docking Top Compounds against EBNA1
The shortlisted compounds with insignificant toxicities and good binding affinities to both E6 proteins were further re-docked against the EBNA1 protein using AutoDock Vina. A literature search revealed two pockets of the EBNA1 that are close and may overlap. The first is the DNA binding pocket, which comprises residues Arg469, Gly472, Lys514, Tyr518, Arg521, Pro535, and Leu536 [77,78]. The other pocket, which is a deep hydrophobic pocket [79] and is also found in the DNA binding domain (DBP), includes residues Lys477, Asn480, Ile481, Gly484, Ser516, Asn519, Leu520, Leu582, Val583, Thr585, Lys586, Thr590, and Ile593 [77,80]. EBNA1 inhibitor H20 was shown to bind in this pocket [77]. This region is crucial for DNA binding to EBNA1 [79]. The grid box with dimensions of 19.68 × 19.41 × 20.85 Å3 was set up to cover both pockets, and the protein was centered at x = 35.33 Å, y = 40.31 Å, and z = 47.11 Å prior to docking.
2.6. Further Re-Docking of Top Compounds against All 3 Oncoproteins Using DockThor-VS
The top TCM compounds and the known inhibitors were further re-docked against all three proteins using DockThor-VS [81,82] to provide an extra layer of confidence and reliability in the docking scores obtained via AutoDock Vina. For all 3 proteins, the grid size was set at x = 40 Å, y = 40 Å, and z = 40 Å to cover the proteins, while the discretization was 0.42. Also, “virtual screening” was selected as the algorithm precision option. Default parameters were used for number of evaluations (500,000), population size (750), initial seed (−1985), and number of runs (12). The soft docking option was also selected. For EBNA1, the grid center was set at x = 36.1225 Å, y = 34.7735 Å, and z = 35.9885 Å. For HPV16 E6, a grid center of x = 35.7925 Å, y = 36.861 Å, and z = 38.2585 Å was used, while x = 40.9985 Å, y = 39.821 Å, and z = 42.948 Å was specified for HPV18 E6 docking.
2.7. Visualizing Protein–Ligand Interaction Profiles
Shortlisted compounds and the known inhibitors in complex with the various proteins were subjected to protein–ligand interaction profiling using LigPlot+ (version v.2.2.8) [83]. LigPlot+ generates interaction maps of the query complex and displays the hydrogen bonds and hydrophobic interactions that exist between the protein and the ligand.
2.8. Prediction of Biological Activity of Lead Compounds
Prediction of activity spectra for substances (PASS), hosted on the Way2Drug platform, was used to determine the biological activities of the shortlisted compounds [84,85,86]. PASS employs a predictive algorithm to predict diverse biological activities associated with compounds based on their chemical structures. The SMILES format of the query is submitted as the input, and the PASS algorithm evaluates the query structure by comparing it to structures in its database. PASS operates on the principle that the biological activity of a compound corresponds to its structure. The predictive accuracy of PASS averages around 95% as per leave-one-out cross-validation estimates [85].
2.9. Molecular Dynamics Simulation
The three unbound proteins (EBNA1, HPV16 E6, and HPV18 E6 proteins), as well as the protein–ligand complexes, were subjected to molecular dynamics simulations using OPLS force field via GROMACS version 5.1.5 [87,88]. The MD simulations were performed to analyze and compare the dynamic behavior, structural stability, and intermolecular interactions of the proteins in their unbound states and when bound to the shortlisted ligands. Trajectory analysis, root mean square deviation (RMSD), root mean square fluctuation (RMSF), the radius of gyration (Rg), and hydrogen bond analyses were performed to gain insights into the conformational changes associated with the protein–ligand interactions. These simulations aimed to provide a comprehensive understanding of the molecular mechanisms governing the binding affinity and stability of the investigated protein–ligand complexes.
For the protein–ligand complexes, prior to the MD simulations, hydrogens were added to the ligands, and their respective charges were computed using UCSF Chimera [89]. The ligands were then submitted to LigParGen [90] to generate their respective topology files (“gro” and “itp” files) that are compatible with GROMACS. The 1.14*CM1A charge model was used to generate the topology files [91]. Protein–ligand complexes in “gro” format were then prepared using the Sublime Text editor. The complexes were then inspected using PyMOL [92]. The unbound proteins and the complexes were then centered in cubic boxes 1 nm away from the edges of the box. Each system was solvated using the TIP4P water model, and chlorine ions were then added to neutralize the charges. Each system was energy-minimized using the steepest descent algorithm through a maximum of 5000 steps or until the maximum force reached <10.0 kJ/mol, whichever occurred first. The systems were then subjected to 100 ps isothermal–isobaric ensemble (NPT) and 100 ps canonical ensemble (NVT) equilibrations before running the 100 ns MD simulations. For the MD runs, the velocity rescaling (V-rescale) thermostat and the Parrinello–Rahman barostat were used. After the simulations, Xmgrace was used to plot the RMSD, RMSF, Rg, and hydrogen bond graphs.
2.10. Molecular Mechanics Poisson–Boltzmann Surface Area Calculations
Using g_mmpbsa [93], MMPBSA decomposition was carried out to analyze the energetic contributions of the complexes. The energy terms analyzed include van der Waals (vdW), electrostatic, polar solvation, solvent-accessible surface area (SASA), and enthalpy-based estimates of binding energies. The binding free energy calculations help to understand the energetics associated with the protein–ligand interactions. The per-residue energy contributions of each system were also determined to identify hotspot residues that contribute significantly to ligand binding.
2.11. Structural Similarity Search of Top Compounds
DrugBank [94,95] was employed to perform a structural similarity search of the shortlisted compounds using their SMILES and chemical structure as input. A similarity threshold of ≥0.7 was used, as this threshold is often considered indicative of substantial structural similarity [96,97]. A literature search was conducted to establish the connection between the analogs and their relevance to EBV, HPV, and cervical cancer.
3. Results and Discussion
3.1. Retrieving Protein Structures
A search via the RCSB PDB [53,54] revealed several EBNA1 structures and fragments; however, PDB ID 6VH6 was selected due to its superior resolution (1.3 Å) and year published (2020) [79]. Aligning another EBNA1 structure (PDB ID: 6NPP, solved at a resolution of 1.3 Å) to the 6VH6 yielded RMSD of 0.09 Å, implying minimal disparity between the two EBNA1 structures. The 6NPP structure is bound to 3-(phenylethynyl)-2-(1H-pyrrol-1-yl)benzoic acid [60], while the 6VH6 is bound to 4-hydroxy-6-methyl-2H-1-benzopyran-2-one [79]. For the HPV16 E6 protein, PDB ID 4XR8 was selected. 4XR8 represents a ternary complex of HPV16 E6, E6AP, and p53 solved at a resolution of 2.25 Å [33]. PDB ID 6SJV, which was solved in 2019 using X-ray diffraction at a resolution of 2.03 Å, was selected as the best HPV18 E6 structure for this study. Further aligning only the E6 structures of both 4XR8 and 6SJV in PyMOL produced an RMSD of 0.830 Å.
3.2. Molecular Docking Analysis
A total of 25,131 TCM compounds were successfully screened against both HPV16 and HPV18 E6 proteins. A total of 595 compounds had docking scores of less than −8.4 kcal/mol for both E6 proteins and were shortlisted for further analysis.
3.2.1. Docking against HPV16 E6
The known E6 inhibitors, baicalein and gossypetin, had docking scores of −7.9 and −6.8 kcal/mol, respectively. After virtual screening, a total of 801 natural product compounds had docking scores of −8.5 kcal/mol and lower with the HPV16 E6 and were thus shortlisted for further analysis. Compound ZINC000070454124 demonstrated the lowest docking score of −10.8 kcal/mol (Table 1), implying the strongest binding affinity with E6, followed by ZINC000103580868 and ZINC000085543515, both with a docking score of −10.4 kcal/mol. Compounds ZINC000070451100 and ZINC000103579839 both had a docking score of −10.3 kcal/mol, while ZINC000103526876 also had a good docking score of −10.2 kcal/mol. ZINC000004097766 had a docking score of −10.1, while ZINC000095912674, ZINC000085631224, and ZINC000103527856 had a docking score of −10 kcal/mol. A previous study docked 20 natural compounds with known antiviral and anticancer properties against the HPV16 E6 protein using Autodock 4.2.6 [98]. The top 20 compounds had docking scores ranging from −4.33 to −8.46 kcal/mol, with ginkgetin demonstrating the strongest affinity [98]. Ginkgetin had docking scores of −8.45, −8.46, and −7.22 kcal/mol when docked against the EA6P, p53, and Myc binding sites [98]. These binding sites have been shown to overlap [99]. Another study also docked four compounds from marine sources against HPV16 E6 using Autodock 4.2 [100]. The four compounds, salicylihalamide B, salicylihalamide A, frigocyclinone, and ascorbic acid, had docking scores of −8.92, −8.76, −8.01, and −3.82 kcal/mol, respectively [100]. Herein, the shortlisted compounds demonstrated better docking scores than those previously obtained, suggesting improved or better affinity (Table 1).

Table 1.
Docking scores of the known inhibitors and shortlisted compounds after virtual screening against the three oncoproteins, HPV16 E6, HPV18 E6, and EBNA1. “E6 Average” represents the average docking score for each compound after docking with both HPV16 and HPV18 E6.
3.2.2. Docking against HPV18 E6
Baicalein and gossypetin, which are known inhibitors of E6, had docking scores of −6.8 and −6.5 kcal/mol when screened against the E6 protein of HPV18. Compound ZINC000103584225 from the TCM library demonstrated the highest binding affinity to the HPV18 E6 protein with a low docking score of −11.4 kcal/mol, followed by ZINC000085593983 with −11.2 kcal/mol and ZINC000095909496 and ZINC000095911538, both with a docking score of −11.1 kcal/mol. ZINC000095913861 also had a docking score of −10.9 kcal/mol, while ZINC000103574367, ZINC000103554058, ZINC000095911347, and ZINC000103526876 had a docking score of −10.8 kcal/mol. Also, compounds ZINC000085530466, ZINC000070454124, ZINC000067902771, and ZINC000085541858 all had a docking score of −10.7 kcal/mol. In all, a total of 3063 compounds had docking scores of −8.5 kcal/mol and lower when screened against the HPV18 E6 protein. A previous study docked 12 natural products with known anti-HPV activity against the HPV18 E6 protein using Autodock 4.2, and their docking scores ranged from −2.86 to −5.85 kcal/mol [101]. Another study also docked known antivirals (natural products) against the HPV18 E6 protein using Autodock 4.2, and the docking scores of the top 19 compounds ranged from −7.48 to −14.77 kcal/mol [102]. Also, doxorubicin and pinoresinol monomethyl ether-β-D-glucoside (PMG), isolated from the dichloromethane extract of Jurinea macrocephala subsp. elbursensis, had docking scores of −3.08 and −4.75 when docked against the HPV18 E6 protein [103]. Doxorubicin and PMG demonstrated cytotoxicity against HeLa cells with average IC50 values of 1.33 and 10.13 μg/mL (after 24 h exposure) and 0.136 and 3.54 μg/mL (after 48 h exposure), respectively [103]. However, doxorubicin and PMG are not known E6 inhibitors. The docking scores of the compounds obtained in this study imply better binding affinity to the E6, warranting further experimental testing.
3.2.3. Average Docking Scores for Both HPV16 and HPV18 E6 Proteins
A total of 595 compounds were observed to have docking scores of −8.5 kcal/mol and lower for both E6 proteins and were shortlisted for further analysis. A multi-target docking score was determined by averaging the docking scores for both HPV16 and HPV18 E6 proteins. Averaging the docking scores resulted in ZINC000070454124 having the lowest average docking score of −10.75 kcal/mol, implying the best binding affinity (Table 1). ZINC000070454124 was also observed to have the highest binding affinity to the HPV16 E6 protein (−10.8 kcal/mol). For both E6 proteins, compounds ZINC000103526876, ZINC000103584225, ZINC000085593983, and ZINC000103580868 had average docking scores of −10.5, −10.35, −10.3, and −10.3 kcal/mol, respectively. Furthermore, compounds ZINC000095910492, ZINC000095912055, ZINC000031155872, ZINC000095909496, ZINC000103584057, ZINC000103527856, and ZINC000103543220 had average docking scores of −10.2, −10.2, −10.1, −10.1, −10.0, −10.0, and −10.0 kcal/mol, respectively.
3.3. Predictive Evaluation of Drug-Like Properties: ADMET Profiling
The top compounds were subjected to ADMET prediction using SwissADME (Table 2) [74]. All ADME properties were predicted for all compounds except for ZINC000085571466. A total of 418 and 176 compounds passed and failed Lipinski’s rule, respectively. A total of 436 and 158 compounds passed and failed Veber’s rule, respectively. Veber’s rule requires a drug-like or orally bioavailable compound to have a topological surface area (TPSA) less than 140 Å2 and fewer than 10 rotatable bonds [76]. Lipinski’s rule, on the other hand, requires a drug-like molecule to not violate more than one of the following criteria: molecular weight should be less than 500 Daltons; number of hydrogen bond acceptor groups (usually nitrogen or oxygen atoms) should be less than or equal to 10; number of hydrogen bond donor groups (usually OH or NH groups) should be less than or equal to five; and the octanol–water partition coefficient (LogP) should be less than five [75]. A total of 82 compounds failed Lipinski’s rule only, 64 failed Veber’s rule only, and 94 compounds failed both rules simultaneously. In all, a total of 354 compounds passed both Lipinski’s and Veber’s rules and were further subjected to toxicity risk prediction using OSIRIS DataWarrior 5.0 [65].

Table 2.
Predicted physicochemical and biological characteristics of some selected compounds using SwissADME. TPSA: Topological Polar Surface Area; ESOL: Estimated Solubility; GI: Gastrointestinal; BBB: Blood-brain barrier; and Pgp: P-glycoprotein.
A total of 29, 29, and 296 compounds were predicted to have low, high, and no mutagenic properties, respectively. For tumorigenicity, 32 compounds were predicted to be low, 32 were predicted to be high, and 290 had no tumorigenic properties. A total of 25 compounds were predicted to possess low reproductive effects, 68 had high reproductive effects, and 261 were predicted to have no reproductive effects (Figure 2). For the irritant toxicity risk prediction, 20, 63, and 271 compounds had low, high, and no irritancy, respectively (Figure 2). Altogether, 166 compounds were predicted to exhibit characteristics of non-mutagenicity, non-tumorigenicity, absence of irritant properties, and no adverse reproductive effects. These compounds were consequently chosen as reasonably safe and used for further analysis.
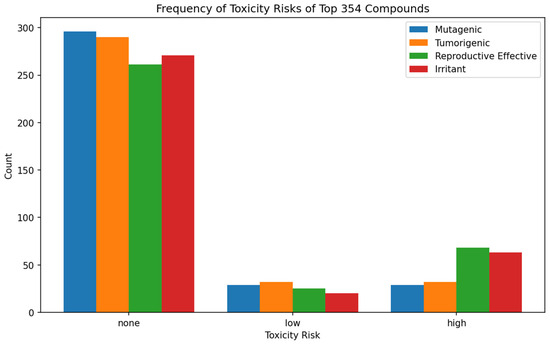
Figure 2.
Bar plot illustrating the count of compounds categorized as “high”, “low”, or “none” for each of the four toxicity risks, including mutagenicity, tumorigenicity, reproductive effect, and irritant.
3.4. Re-Docking Top Compounds against EBNA1
The top 166 TCM compounds and the known inhibitors were re-docked against the EBNA1 protein. Only compounds with docking scores ≤−7.0 kcal/mol were considered for further studies, as this AutoDock Vina threshold has been shown to effectively separate good binders from weak binders of viral proteins [104]. A total of 26 compounds were obtained as potentially good binders of E6 (both HPV16 and HPV18) and EBNA1 proteins. Compound ZINC000033831887 demonstrated the highest binding affinity, with a docking score of −7.8 kcal/mol, followed by ZINC000085597263 and ZINC000085543579, both with a docking score of −7.7 kcal/mol (Table 1). Compounds ZINC000000899994, ZINC000085628525, ZINC000070454501, ZINC000085950180, and ZINC000095914132 all had a docking score of −7.6 kcal/mol. A previous study virtually screened known sphingosine-1-phosphate receptor (S1PR) modulators (fingolimod, ozanimod, and siponimod), teriflunomide, dimethyl fumarate (tecfidera), monomethyl fumarate, diroximel fumarate (vumerity), cladribine, andrographolide, angelicin, EGCG, emodin, lignans, moronic acid, and protoapigenone against EBNA1 [80]. The docking scores ranged from −4.1 kcal/mol to −8.6 kcal/mol, with ozanimod demonstrating the highest affinity [80]. The docking scores obtained herein are comparable to those from the previous study [80].
3.5. Further Re-Docking of Top Compounds against All Three Oncoproteins Using DockThor
The top 26 TCM compounds, along with the three known inhibitors, were re-docked against all three proteins using a different docking tool, DockThor-VS [81,82]. This step aimed to bolster confidence and reliability in the shortlisted compounds and docking outcomes. All three known inhibitors were successfully screened against the proteins; however, for the TCM compounds, all but ZINC000095909247, ZINC000095913339, ZINC000095914132, and ZINC000070454552 were screened successfully (Table S1). A previous study that docked drugs against various SARS-CoV-2 targets reported that DockThor-VS docking scores higher than −6.8 kcal/mol indicate binding affinities above approximately 10 µM, which were considered as low binding affinities [82]. Compounds with docking scores between −6.8 and −8.2 kcal/mol were considered to have moderate binding affinities, while those with docking scores below −8.2 kcal/mol demonstrated high binding with submicromolar affinities [82]. EBNA1 inhibitor, VK-1727 demonstrated high binding affinity with all three proteins with docking scores of −8.702, −8.301, and −9.363 against EBNA1, HPV16 E6, and HPV18 E6, respectively (Table S1). Further studies are required to explore the potential repurposing of VK-1727 as an effective HPV E6 inhibitor. Similarly, compounds ZINC000014588133, ZINC000085568150, ZINC000070454501, ZINC000085597263, ZINC000085568136, ZINC000085543579, and ZINC000014768164 demonstrated high binding affinities with all three proteins (Table S1). Baicalein demonstrated moderate binding affinities with all three proteins, having docking scores of −7.178, −7.463, and −7.43 kcal/mol against EBNA1, HPV16 E6, and HPV18 E6 proteins, respectively. Gossypetin, on the other hand, demonstrated moderate docking scores against EBNA1 (−6.984 kcal/mol) and HPV18 E6 (−7.28 kcal/mol) while exhibiting a low binding affinity for HPV16 E6 (−6.751 kcal/mol) [Table S1]. For both HPV E6 proteins, all the TCM compounds demonstrated higher binding affinities than the two known E6 inhibitors, baicalein and gossypetin. The results obtained with DockThor-VS are consistent with those from AutoDock Vina.
3.6. Exploring Molecular Interactions
The protein–ligand interaction profiles of the various complexes were evaluated (Figure 3a,b and Figures S1–S3, and Table 3). For the EBNA1–ligand complexes, all the selected compounds were observed to interact with Lys477 and Lys586 (Figure 3a,b and Figure S1a–i). For the EBNA1–ligand complexes, residues Lys477, Asn480, Ile481, Gly484, Leu582, Thr585, Lys586, Pro587, and Thr590 were involved in most ligand interactions and are suggested to be hotspot residues (Table 3). EBNA1 inhibitor, VK-0497 was previously shown to interact with Lys477, Asn519, Lys586, and Thr590 [60]. Previous molecular docking studies also identified EBNA1 residues Lys477, Ile481, Leu582, and Lys586 as hotspot residues crucial for ligand binding [80]. Another study that screened malaria box compounds against the EBNA1 reported that Asn480, Ile481, Asn519, Leu582, Val583, Lys586, and Thr590 were involved in most ligand interactions [105].
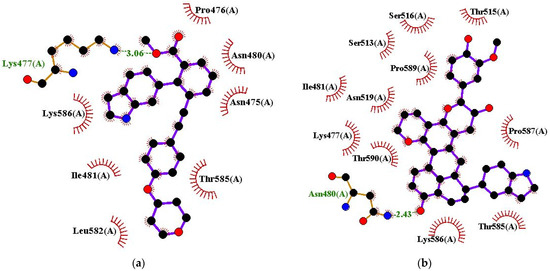
Figure 3.
Protein–ligand interaction profile for EBNA1 in complex with (a) VK-1727 and (b) ZINC000085568136. Ligand is represented as purple, hydrophobic bonds are represented as red arc spokes, and hydrogen bonds are represented as green dashed lines.

Table 3.
Protein–ligand interactions between three oncoproteins (HPV16 E6, HPV18 E6, and EBNA1) and the shortlisted compounds after molecular docking.
For the HPV16 E6–ligand complexes, residues Tyr32, Leu50, Cys51, Val53, Val62, Leu67, Tyr70, Ser71, Ser74, Arg102, Gln107, and Arg131 interacted with most of the ligands and are potential hotspot residues crucial for ligand binding (Figure 4a,b and Figure S2a–j, and Table 3). Residues Leu50 and Cys51 were identified previously as key E6 residues for high affinity ligand binding, while residues Cys51, Leu50, Arg102, Arg131, Leu67, Val62, and Gln107 were suggested to be important in E6 inhibition [106]. Previous docking studies also showed that Arg10, Lys11, Val31, Tyr32, Leu50, Cys51, Val53, Arg55, Val62, Leu67, Tyr70, Ser74, Arg77, His78, Leu100, Arg102, Gln107, Arg129, and Arg131 are involved in E6–E6AP binding [98]. For E6 and p53 interaction, E6 residues Gln6, Glu7, Arg8, Arg10, Gln14, Glu18, Tyr43, Asp44, Phe47, Asp49, Leu100, and Pro112 are involved, while Phe2, Pro5, Arg8, and Tyr54 are involved in E6 interaction with Myc [98]. Herein, the shortlisted compounds were also observed to interact with these same residues (Table 3) and could inhibit E6 interaction with p53, EA6P, and Myc.
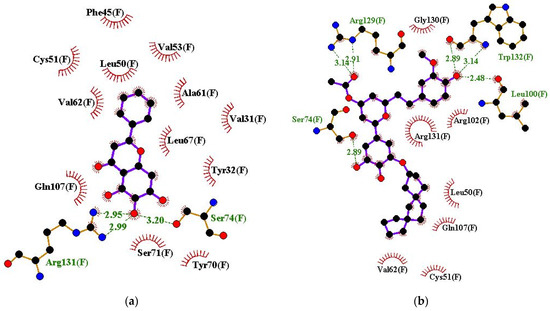
Figure 4.
Protein–ligand interaction profile for HPV16 E6 in complex with (a) baicalein and (b) ZINC000085597263.
For the HPV18 E6–ligand complexes, residues Tyr1034 (Tyr34), Leu1052 (Leu52), Phe1053 (Phe53), Val1055 (Val55), Ala1064 (Ala64), Ile1069 (Ile69), Tyr1072 (Tyr72), Arg1076 (Arg76), Arg1104 (Arg104), Gln1109 (Gln109), and Arg1135 (Arg135) were observed to interact with ≥6 compounds (out of 12) and can be considered as potential hotspot residues (Figure 5a,b and Figure S3a–j, and Table 3). The amino acid numbering in the 6SJV structure corresponds to an offset of 1000 from the canonical HPV18 E6 protein sequence (Uniprot ID: P06463). The canonical numbering is enclosed in brackets, whereas the 6SJV numbering is presented without brackets. A previous study investigated the interactions between HPV18 E6 with doxorubicin and PMG [103]. Doxorubicin formed hydrogen bonds with Tyr99 and Asn113 and hydrophobic contacts with Phe49, Leu112, Pro114, Ala115, and Leu118 [103]. PMG also interacted with residues Tyr99, Leu112, and Arg119 via hydrogen bonds and formed hydrophobic bonds with Pro111, Asn113, Pro114, Ala115, Glu116, and His139 [103]. This binding site is in proximity to the binding cavity identified for the shortlisted candidates in this study; however, they are different.
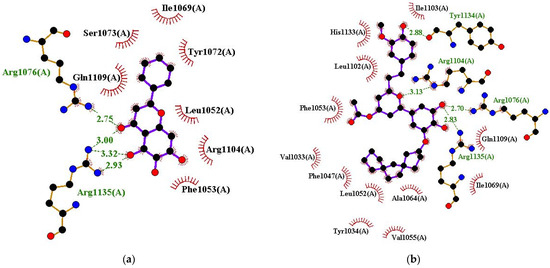
Figure 5.
Protein–ligand interaction profile for HPV18 E6 in complex with (a) baicalein and (b) ZINC000085597263.
3.7. Biological Activity Prediction of Selected Lead Compounds
The biological activities of the top 26 compounds obtained were predicted using the prediction of activity spectra for substances (PASS) [85,86]. Biological activity predictions add depth to the previously obtained results, corroborating the potential inhibitory activities of the most promising candidates. PASS computes a compound’s probability to be active (Pa) and probability to be inactive (Pi) using the activity data of structurally similar molecules [84,85,107]. Only activities with Pa > 0.3 and Pa > Pi were considered for each compound. The biological activities of three compounds (ZINC000085568150, ZINC000085568136, and ZINC000103533241) could not be predicted using PASS. Most of the compounds were predicted to be antivirals. Compounds ZINC000003588336, ZINC000014768164, and ZINC000013380012 were predicted to possess antineoplastic activity specifically against cervical cancer, with Pa values of 0.311, 0.306, and 0.302 and Pi values of 0.017, 0.018, and 0.018, respectively.
Compounds ZINC000095909247, ZINC000013380012, ZINC000070454124, ZINC000003588336, ZINC000014768164, ZINC000085628525, ZINC000014588133, ZINC000085597263, ZINC000085597267, ZINC000085950180, ZINC000033831887, ZINC000085631200, ZINC000013378520, ZINC000013378519, ZINC000014697269, ZINC000070454552, and ZINC000000899994, were predicted to be antineoplastics with Pa values of 0.93, 0.911, 0.827, 0.787, 0.711, 0.697, 0.655, 0.623, 0.615, 0.563, 0.536, 0.528, 0.516, 0.487, 0.485, 0.426, and 0.34 and corresponding Pi values of 0.005, 0.005, 0.009, 0.013, 0.024, 0.027, 0.034, 0.04, 0.042, 0.053, 0.061, 0.063, 0.067, 0.076, 0.076, 0.095, and 0.13, respectively. The compounds were predicted to exhibit antineoplastic activities against various types of cancers, including breast, colorectal, colon, brain, renal, bladder, endocrine, lung, small cell lung, and non-small cell lung cancers. Additionally, the PASS predictions suggested efficacy against glioma, melanoma, lymphoma, sarcoma, squamous cell carcinoma, glioblastoma multiforme, solid tumors, and carcinoma. A total of eight compounds, including ZINC000070454124, ZINC000013380012, ZINC000070454501, ZINC000013378519, ZINC000013378520, ZINC000014697269, ZINC000085950180, and ZINC000014649947, were also predicted to be antimutagenic.
A total of 16 of the 23 compounds, including ZINC000013380012 (Pa: 0.869 and Pi: 0.007), ZINC000013378520 (Pa: 0.737 and Pi: 0.019), ZINC000013378519 (Pa: 0.723 and Pi: 0.022), ZINC000095909247 (Pa: 0.694 and Pi: 0.027), ZINC000085950180 (Pa: 0.678 and Pi: 0.03), ZINC000014697269 (Pa: 0.671 and Pi: 0.032), ZINC000070454124 (Pa: 0.653 and Pi: 0.036), ZINC000033831887 (Pa: 0.648 and Pi: 0.037), ZINC000070454501 (Pa: 0.604 and Pi: 0.048), ZINC000014649947 (Pa: 0.592 and Pi: 0.052), ZINC000014768164 (Pa: 0.54 and Pi: 0.072), ZINC000000899994 (Pa: 0.512 and Pi: 0.085), ZINC000014588133 (Pa: 0.44 and Pi: 0.12), ZINC000003588336 (Pa: 0.439 and Pi: 0.12), ZINC000085597267 (Pa: 0.393 and Pi: 0.145), and ZINC000085597263 (Pa: 0.34 and Pi: 0.179), were predicted to be TP53 expression enhancers. E6 proteins degrade tumor suppressor protein p53, thereby promoting cervical cancer progression [32,108].
Ten compounds, including ZINC000070454124, ZINC000085628525, ZINC000085597263, ZINC000095909247, ZINC000003588336, ZINC000070454552, ZINC000085597267, ZINC000033831887, ZINC000085950180, and ZINC000014649947, were also predicted to be antineoplastics for treating non-Hodgkin’s lymphoma (NHL) (with Pa > Pi and Pa > 0.3). EBV is known to cause mononucleosis and is implicated in certain types of NHL, particularly in immunocompromised individuals [109]. EBV is most commonly linked to the development of Burkitt lymphoma and post-transplant lymphoproliferative disorder (PTLD) [110]. Burkitt lymphoma, in particular, has a well-established association with EBV, especially in endemic regions [22,109]. In immunocompromised individuals, EBV can lead to the development of lymphomas, including diffuse large B cell lymphoma (DLBCL) [109,111].
Furthermore, nine compounds, including ZINC000070454124 (Pa: 0.343 and Pi: 0.126), ZINC000070454501 (Pa: 0.739 and Pi: 0.005), ZINC000003588336 (Pa: 0.392 and Pi: 0.091), ZINC000085543579 (Pa: 0.312 and Pi: 0.153), ZINC000013378519 (Pa: 0.575 and Pi: 0.023), ZINC000013378520 (Pa: 0.622 and Pi: 0.015), ZINC000014697269 (Pa: 0.621 and Pi: 0.015), ZINC000085950180 (Pa: 0.702 and Pi: 0.007), and ZINC000014649947 (Pa: 0.633 and Pi: 0.014), were predicted to be Pin1 inhibitors. The overexpression of Pin1 has been shown to play a role in both the tumorigenesis and proliferation of cervical cancer [112,113]. Pin1 also contributes to chemo-resistance in cervical cancer by upregulating FOXM1 [114]. Notably, the use of thiostrepton, a FOXM1 inhibitor, has been reported to significantly decrease metastatic lung tumor nodules in HPV-positive cancer cells [30].
Seventeen compounds were predicted to be apoptosis agonists, while thirteen were predicted to be Janus kinase 2 (JAK2) expression inhibitors. The activation of the JAK/STAT pathway has been proposed to induce the expression of antiapoptotic genes, potentially contributing significantly to the progression of cervical cancer tumorigenesis [115,116,117,118]. Four compounds (ZINC000085597263, ZINC000095909247, ZINC000014768164, and ZINC000085597267) were predicted to be transcription factor NF kappa B (NF-κB) inhibitors. The activation of the NF-κB signaling pathway has been implicated in cervical cancer cell proliferation, invasion, and metastasis [119,120]. The HPV E6 and E7 proteins have been shown to be involved in the activation of NF-κB [119,120,121,122,123,124].
3.8. MD Simulation Analysis
Molecular dynamics (MD) simulations have been extensively used in computational drug discovery projects to investigate the conformational transitions of proteins resulting from the binding and unbinding of ligands [125]. Using MD simulations, the dynamic behaviors of biomolecules at the atomic level can be explored, providing insights into the structural changes associated with ligand interactions. Herein, 100 ns were performed for the apo and holo forms of all three proteins. Average RMSD, RMSF, Rg, and number of hydrogen bonds formed were calculated for each system throughout the 100 ns MD runs (Table 4). A significant constraint is the absence of simulations conducted under the typical physiological salt concentration of ~0.15 M. Future studies could explore conducting simulations under both physiological salt concentrations and those found in cancer cells, which often tend to be higher than physiological levels [126].

Table 4.
Average RMSD, Rg, RMSF, and number of hydrogen bonds computed using data from the 100 ns MD simulations of the unbound protein and protein–ligand complexes.
3.8.1. Test for Convergence
To evaluate convergence, a clustering analysis (gmx cluster) was employed to generate clusters from the first 80 ns of each simulation and the last 20 ns using a similarity cutoff of 0.2 nm and a time interval of 1 ns (Table S2). A small cutoff value was used to obtain well-defined clusters with structurally similar members. For each system, a total of 81 (0–80 ns) and 20 (81–100 ns) frames were generated from the first 80 ns and the last 20 ns segments, respectively. For the first 80 ns of each simulation, several distinct clusters were identified and denoted as first1, first2, first3, etc. Similarly, for the last 20 ns, clusters labeled last1, last2, last3, etc., were generated. The clusters are labeled according to decreasing cluster population size. In other words, cluster1 (first1 or last1) has the highest number of members followed by cluster2 (first2 or last2), and subsequent clusters follow suit in decreasing order. To assess convergence, the cluster representative structures obtained from the first 80 ns of the simulation were compared to those from the last 20 ns (Table S3). Aligning the C-alpha atoms of each conformation from the final 20 ns to each conformation from the first 80 ns provides a measure of how similar the structures are between the two segments of the simulation. This alignment can reveal significant structural changes or consistency between conformations from the two segments. The structural similarity was quantified using the RMSD metric (Table S3), which measures the average distance between atoms of aligned structures [127,128]. By comparing the cluster populations between the first 80 ns and the last 20 ns, any significant shifts in the distribution of conformations can be observed. A simulation with a relatively converged state will exhibit stability or similarity between the populations of the two segments. However, if there are major differences in cluster populations, it may indicate ongoing dynamics or lack of convergence. Lower RMSD values indicate greater similarity between the conformations, while higher RMSD values suggest more significant structural changes. Relatively low and consistent RMSD values (0–3 Å) indicate a stable or converged simulation, whereas high RMSD values (>3 Å) suggest ongoing dynamics or a lack of convergence [129]. Lower RMSD values indicate greater structural similarity between the clusters.
For the EBNA1 systems, all the RMSD values were below 1.5 Å (most were below 1 Å) (Table S3), implying no significant differences between the first 80 ns and the last 20 ns. It is also worth mentioning that for all the EBNA1 systems, the RMSD values obtained from aligning the representative structures of the largest clusters (first1 and last1) were below 1 Å except for the EBNA1–ZINC000085631200 complex, which had an RMSD of 1.148 Å (Table S3). These results suggest that all the EBNA1 systems reached a state of convergence. For all the HPV16 and HPV18 E6 systems, varying degrees of similarity were observed between clusters from the last 20 ns and those from the first 80 ns (Table S3). Most aligned clusters exhibited relatively low RMSD values, suggesting a high level of structural conservation throughout the simulation (Table S3). For the HPV16 E6 systems, the RMSD values obtained from the alignment of the representative structures from the largest clusters (first1 and last1) were below 3 Å, indicative of converged states during the simulation. A few RMSDs above 3 Å were observed when representative structures from clusters with smaller population sizes were aligned. This is not surprising as clusters with smaller populations tend to deviate (or are structurally different). For the HPV18 E6 systems, alignments of the representative structures from the largest clusters (first1 and last1) yielded RMSDs less than 2 Å, except for the unbound HPV18 E6 protein, and HPV18 E6–ZINC000085631200, HPV18 E6–ZINC000095913339, and HPV18 E6–ZINC000085543530 complexes (Table S3). Overall, the clustering analyses showed that the systems converged, suggesting the reproducibility of the simulations and that 100 ns is sufficient to study the dynamic behavior of EBNA1, HPV16 E6, and HPV18 E6 systems.
3.8.2. RMSD Analysis
To assess the stability of the protein–ligand complexes, the RMSDs were analyzed over the course of a 100 ns MD simulation. For the EBNA1–ligand systems, the apo-EBNA1 had an average RMSD of 0.36 nm (Figure 6a,b, and Table 4). From the plot, complexes EBNA1–VK1727, EBNA1–ZINC000070454124, EBNA1–ZINC000085568136, EBNA1–ZINC000085631200, EBNA1–ZINC000095913339, and EBNA1–ZINC000085543530 demonstrated lower RMSDs than the apo-EBNA1 (Table 4 and Figure 6a,b), implying they achieved better stability after ligand binding. For the HPV16 E6 systems, E6 complexed with baicalein, ZINC000013380012, ZINC000070454124, ZINC000085631200, ZINC000095913339, ZINC000085628525, and ZINC000085543530 had lower average RMSD values than the apo protein, implying better stability after binding to the E6 protein (Figure S4a,b). A previous MD study on HPV16 E6–salicylihalamide B complex revealed a stable RMSD with an average of 0.35 nm [100]. All complexes except ZINC000014588133, ZINC000085568136, ZINC000095909247, and ZINC000085597263 demonstrated lower RMSDs than salicylihalamide B [100]. For the HPV18 E6 systems, all the complexes except E6–baicalein achieved better stability (lower RMSDs) than the unbound E6 protein (Table 4 and Figure S5a,b). The average RMSD of the E6–baicalein complex was observed to be 0.46 nm, which was the same as that of the apo-HPV18 E6 protein (Table 4).
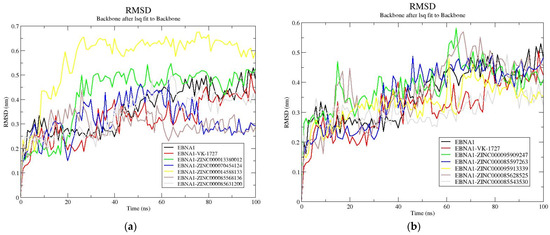
Figure 6.
RMSD plots for the unbound EBNA1 protein and EBNA1–ligand complexes. The RMSD plots are shown for apo-EBNA1 and VK-1727 as well as (a) ZINC000013380012, ZINC000070454124, ZINC000014588133, ZINC000085568136, and ZINC000085631200 and (b) ZINC000095909247, ZINC000085597263, ZINC000095913339, ZINC000085628525, and ZINC000085543530.
3.8.3. Rg Analysis
The radius of gyration (Rg) measures the level of compactness in a protein’s structure [130]. A higher Rg value indicates reduced compactness, while a low Rg value corresponds to a high level of compactness [130]. Rg is also used to assess the stability of an unbound protein or protein–ligand complexes during MD simulations, serving as an indicator of whether the structure remains stably folded or unfolds [130,131]. Consistency in the Rg values throughout the MD simulation suggests a stable, folded structure, while fluctuations may indicate an unfolded structure or a poorly folded structure [130].
For the MD simulations involving the apo-EBNA1 and holo-EBNA1, the average Rg of all the complexes were comparable to or lower than that of the unbound protein, with the exception of EBNA1–VK1727, EBNA1–ZINC000070454124, EBNA1–ZINC000085631200, and EBNA1–ZINC000085628525 complexes. The Rg of the unbound EBNA1 remained stable (average of 1.6 nm) until about 60 ns, at which a rise to an average of 1.65 nm was observed (Table 4 and Figure 7a,b). From the Rg plots, EBNA1 complexed with ZINC000013380012, ZINC000014588133, ZINC000085597263, ZINC000095913339, ZINC000085628525, and ZINC000085543530 demonstrated lower Rg than the unbound EBNA1 toward the end of the 100 ns simulation period. For the HPV16 E6 systems, the apo E6 had an average Rg of 1.72 nm (Table 4 and Figure S6a,b). All the protein–ligand complexes demonstrated comparable Rg to that of the apo-E6. Previous MD simulation study on HPV16 E6–salicylihalamide B complex revealed a stable Rg ranging between 1.65 and 1.75 nm (average of 1.7 nm) [100], consistent with the results obtained in this study. For the HPV18 E6 systems, the unbound protein had an average Rg of 1.84 nm, while the E6–ligand complexes had lower average Rg values throughout the 100 ns simulation period (Table 4 and Figure S7a,b). This implies that upon binding, the known inhibitors and the shortlisted compounds caused the HPV18 E6 to be more compact.
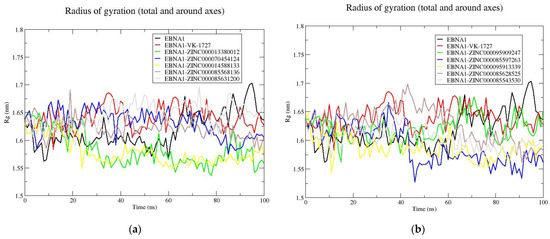
Figure 7.
Rg plots for the unbound EBNA1 protein and EBNA1–ligand complexes. The Rg plots are shown for apo-EBNA1 and VK-1727 as well as (a) ZINC000013380012, ZINC000070454124, ZINC000014588133, ZINC000085568136, and ZINC000085631200 and (b) ZINC000095909247, ZINC000085597263, ZINC000095913339, ZINC000085628525, and ZINC000085543530.
3.8.4. RMSF Analysis
Consistent trends in RMSF were observed across all EBNA1 systems, encompassing both the apo and holo forms (Figure 8). Regions exhibiting substantial fluctuations were identified around residues 485–500, 540–560, and 583–595 within the EBNA1 systems (Figure 8). Conversely, regions spanning residue indexes 475–487, 503–510, 520–535, 562–582, and 592–597 displayed comparatively lower fluctuations, suggesting their potential involvement in ligand binding (Figure 8). This observation aligns with the protein–ligand interaction analysis, highlighting residues Lys477, Asn480, Ile481, Gly484, Leu582, Thr585, Lys586, Pro587, and Thr590 as interacting with multiple ligands. For HPV16 E6, similar fluctuations were observed across all systems, with elevated RMSFs noted at positions 30–36, 55–58, 64–100, 113–124, and 125–134 (Figure S8). In contrast, regions spanning residue indexes 10–30, 37–54, 59–63, and 100–110 exhibited reduced fluctuations in all HPV16 E6 systems (Figure S8). For HPV18, the unbound E6 displayed the highest fluctuations, averaging 0.4 nm (Figure S9 and Table 4). However, the holo forms of HPV18 E6 demonstrated decreased RMSF values. Residue indexes 1055–1060, 1074–1085, and 1087–1105 exhibited notable fluctuations across all HPV18 E6–ligand complexes, while indexes 1010–1030, 1040–1055, 1061–1074, and 1105–1120 generally exhibited lower fluctuations (Figure S9), suggesting their potential involvement in ligand binding.
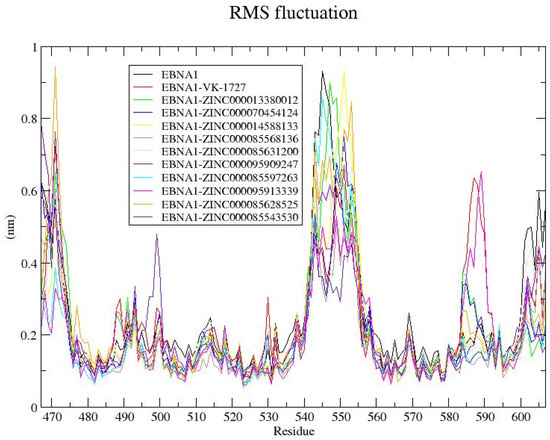
Figure 8.
RMSF plot for the unbound EBNA1 and EBNA1–ligand complexes after 100 ns MD simulation. The unbound EBNA1 protein, EBNA1 complexed with VK-1727, ZINC000013380012, ZINC000070454124, ZINC000014588133, ZINC000085568136, ZINC000085631200, ZINC000095909247, ZINC000085597263, ZINC000095913339, ZINC000085628525, and ZINC000085543530 are colored black, red, green, blue, yellow, brown, grey, violet, cyan, magenta, orange, and indigo, respectively.
3.9. Post-MD MM/PBSA Analysis
Aside from the dynamical structural information obtained through MD simulations, energetic information about protein–ligand interactions can also be explored using MD simulations [132]. The wealth of data obtained from these simulations plays an important role in unraveling the intricate relationship between structure and function in the target, shedding light on the essence of protein–ligand interactions. Also, MD simulations contribute to enhancing the enrichment factor of virtual screening through the provision of a more precise ranking of hit compounds, coupled with enthalpy-based estimates of binding energy calculations [132]. The utilization of the MM/PBSA method subsequent to MD simulations facilitates the quantitative assessment of binding affinity between a protein and a ligand. Herein, the contributing energy terms which include van der Waals (vdW), electrostatic, polar solvation, SASA, and the enthalpy-based estimates of binding free energies of the various protein–ligand complexes were computed using the g_mmpbsa tool [93]. For each complex, the energy contributions per residue were also assessed to identify hotspot residues within the binding sites, highlighting their significance in facilitating ligand binding.
3.9.1. Enthalpy-Based Estimates of Binding Energy of the Protein–Ligand Complexes
An enthalpy-based binding free energy of −82.862 kJ/mol was observed for the EBNA1–VK-1727 complex (Table 5). Compounds ZINC000085568136, ZINC000095913339, ZINC000095909247, ZINC000095913339, ZINC000013380012, and ZINC000070454124 demonstrated stronger binding affinity to EBNA1 than the known inhibitor, with energies of −149.73, −146.08, −129.4, −124.99, −116.9, and −115.73 kJ/mol, respectively (Table 5). ZINC000014588133 demonstrated comparable energy to VK-1727 (−82.04 kJ/mol), while ZINC000085631200 had a relatively weaker binding affinity to EBNA1 (−22.3 kJ/mol) [Table 5]. ZINC000085543530 and ZINC000085628525 demonstrated unfavorable affinity to EBNA1 with positive energy values of 46.4 and 24.17 kJ/mol, respectively, and were thus eliminated as potential EBNA1 binders (Table 5).

Table 5.
Energy contributions from MM/PBSA calculations for the protein–ligand complexes. All energy values are in kJ/mol.
For the HPV16 E6 protein, the two known inhibitors, baicalein and gossypetin, had binding energies of −85.543 and −81.853 kJ/mol, respectively. The strong binding affinities observed herein for baicalein and gossypetin support their experimental activity against the HPV E6 protein, as reported previously [59]. Compounds ZINC000070454124, ZINC000013380012, ZINC000085597263, ZINC000085568136, and ZINC000095909247 demonstrated higher binding affinity to the HPV16 E6 protein than the two known inhibitors, with energy values of −154.8, −137.34, −115.49, −110.66, and −91.62 kJ/mol, respectively (Table 5). ZINC000014588133 also demonstrated a binding energy of −65.69 kJ/mol. However, compounds ZINC000085628525, ZINC000085631200, ZINC000085543530, and ZINC000095913339 demonstrated positive (repulsive) energies of 142.2, 157.75, 176.95, and 487.96 kJ/mol, respectively (Table 5). Due to the high energy values of compounds ZINC000085628525, ZINC000085631200, ZINC000085543530, and ZINC000095913339, they cannot be considered HPV16 E6 binders.
For the HPV18 E6 protein, baicalein and gossypetin demonstrated good binding energies of −104.82 and −108.87 kJ/mol, respectively (Table 5), which are consistent with experimental studies that prove their anti-E6 activity [59]. Compounds ZINC000085597263 (−164.17 kJ/mol), ZINC000085568136 (−157.83 kJ/mol), ZINC000070454124 (−145.09 kJ/mol), ZINC000013380012 (−143.562 kJ/mol), ZINC000095909247 (−112.69 kJ/mol), and ZINC000014588133 (−108.64 kJ/mol) demonstrated comparable or better binding energies to the HPV18 E6 protein (Table 5). As observed for the HPV16 E6 protein, compounds ZINC000085543530, ZINC000085628525, ZINC000085631200, and ZINC000095913339 also had positive binding energies of 7.62, 51.18, 60.06, and 260.9 kJ/mol, respectively (Table 5). This suggests that these four compounds are not effective binders of the E6 protein of both HPV subtypes (HPV16 and HPV18).
Based on the MM/PBSA analyses of the EBNA1, HPV16 E6, and HPV18 E6 protein in complex with the ligands, six compounds, namely, ZINC000013380012, ZINC000070454124, ZINC000014588133, ZINC000085568136, ZINC000095909247, and ZINC000085597263, demonstrated very strong affinity to the three proteins and are worthy of further experimental testing to ascertain their potential anti-cervical cancer activity (Table 5).
3.9.2. Per-Residue Energy Contributions
The per-residue energy contributions of the complexes via MM/PBSA calculations were also assessed (Figures S10–S12). The per-residue energy decomposition helps us understand the energetic aspects of molecular interactions within a complex. This approach breaks down the total binding free energy of a molecular system into contributions from individual residues. Per-residue energy decompositions provide insights into the particular amino acids or residues that play a substantial role in the binding affinity between proteins and ligands and promote the stability of the complex. Previous studies have suggested that residues that contribute energies above 5 or below −5 kJ/mol can be considered critical for ligand binding [133]. In this study, for the known inhibitor, VK-1727, no EBNA1 residue contributed above 5 or below −5 kJ/mol to its binding. However, noteworthy contributions were observed for residues Lys477, Ile481, and Leu582, contributing favorably to VK-1727 binding with energies of −3.96, −4.95, and −3.81 kJ/mol, respectively, while Asp581 contributed a positive energy of 3.82 kJ/mol (Figure S10j). Based on the energy contributions in the EBNA1–VK-1727 complex, residues with contributions above 3 or below −3 kJ/mol are identified as critical for ligand binding.
For the complexes of EBNA1 with ZINC000013380012, ZINC000070454124, ZINC000014588133, and ZINC000095909247, all the positive (unfavorable) energies contributed by EBNA1 residues were found to be less than 3 kJ/mol. For the EBNA1–ZINC000013380012 complex, residues Lys477, Asn480, Ile481, Lys586, Pro587, Pro589, and Thr590 demonstrated favorable energy values of −8.71, −4.22, −6.37, −5.04, −3.24, −4.34, and −3.87 kJ/mol, respectively (Figure S10a). In the EBNA1–ZINC000070454124 complex, Pro476, Lys477, Asn480, Ile481, Lys586, and Pro589 exhibited favorable energies of −6.87, −9.52, −3.34, −3.13, −3.10, and −3.02 kJ/mol, respectively (Figure S10b). The EBNA1–ZINC000014588133 complex revealed four residues, Lys477, Asn480, Lys586, and Pro587 contributing favorably to ligand binding, with energies of −4.95, −5.27, −7.72, and −12.20 kJ/mol, respectively (Figure S10c). For the EBNA1–ZINC000095909247 complex, favorable energies were observed for residues Lys477, Ile481, Asn519, Leu582, Pro589, and Thr590, with values of −6.05, −6.81, −3.34, −3.09, −4.43, and −3.60 kJ/mol, respectively (Figure S10e). For the EBNA1–ZINC000085568136 complex, residues Ser467, His468, Met469, Ile481, Thr515, Lys586, Pro587, Pro589, and Thr590 contributed negative (favorable) energies of −3.24, −4.01, −7.06, −4.56, −3.98, −5.01, −15.45, −4.78, and −4.00 kJ/mol, respectively, while Lys477 had an unfavorable energy of 6.23 kJ/mol (Figure 9). For the EBNA1–ZINC000085597263 complex, residues Met469, Gln471, Lys477, Asn480, Arg486, and Pro587 contributed favorable energies of −7.42, −4.89, −6.72, −6.44, −3.12, and −5.03 kJ/mol (Figure S10f). Residues Glu483 and Lys586 were also observed to contribute repulsive (positive or unfavorable) energies of 4.22 and 5.64 kJ/mol, respectively (Figure S10f).
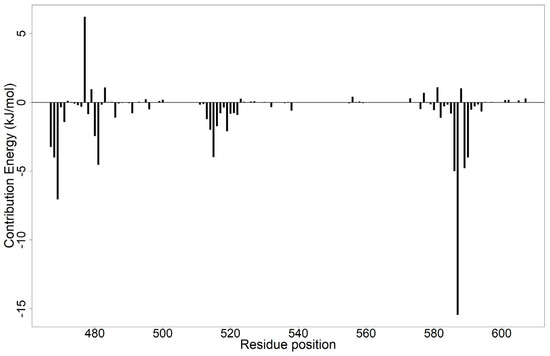
Figure 9.
Per-residue energy decomposition plot of EBNA1–ZINC000085568136 complex. Residues Ser467, His468, Met469, Ile481, Thr515, Lys586, Pro587, Pro589, and Thr590 contributed favorably to ligand binding, while Lys477 contributed unfavorably.
For the EBNA1–ZINC000085631200 (Figure S10d), EBNA1–ZINC000095913339 (Figure S10g), EBNA1–ZINC000085628525 (Figure S10h), and EBNA1–ZINC000085543530 (Figure S10i) complexes, similar results were observed for the per-residue energy contributions. A total of 15 residues, including Ser467, Lys477, Arg486, Arg491, Arg496, Lys514, Arg521, Arg522, Arg532, Arg538, Arg555, Lys576, Lys580, Lys586, and Arg594, were observed to contribute unfavorably to ligand binding. Conversely, residues Gln471, Glu479, Asn480, Ile481, Glu483, Gly484, Glu495, Asp499, Glu500, Ser513, Ser516, Leu520, Glu556, Ile558, Glu573, Asp577, Asp581, Leu582, Val583, Pro587, Pro589, Thr590, Ile593, Asp601, Asp602, Asp605, and Pro607 contributed favorably to the binding of these four compounds. These four compounds interacted simultaneously with residues in both binding pockets of the EBNA1 DNA-binding domain [77]. All the selected compounds were observed to interact with Lys477 and Lys586. For the EBNA1–ligand complexes, residues Lys477, Asn480, Ile481, Gly484, Leu582, Thr585, Lys586, Pro587, and Thr590 interacted with >4 out of the 11 compounds and are suggested to be hotspot residues. Previous molecular docking studies also identified EBNA1 residues Lys477, Ile481, Leu582, and Lys586 as hotspot residues crucial for ligand binding [80].
For the HPV16 E6 protein complexed with baicalein and ZINC000095909247, all the contributing energies were below 3 kJ/mol. For the HPV16 E6–baicalein complex, residues Val31, Tyr32, Val53, Val62, and Tyr70 contributed −3.98, −3.76, −3.40, −3.17, and −4.81 kJ/mol, respectively (Figure S11k). Residues Gln6, Val53, and Tyr70 contributed favorable energies of −6.29, −3.05, and −4.20 kJ/mol, respectively for the HPV16 E6–ZINC000095909247 complex (Figure S11f). For the HPV16 E6–gossypetin complex, residues Lys11, Leu50, Arg102, Gln107, Arg129, Arg131, and Trp132 contributed favorably to binding with energies of −5.06, −3.69, −4.16, −5.33, −4.63, −8.05, and −3.44 kJ/mol, respectively, while Asp49 and Glu75 contributed unfavorable energies of 8.75 and 7.57 kJ/mol, respectively (Figure S11l). For the HPV16 E6–ZINC000013380012 complex, residues Lys11, Leu50, Cys51, Val53, Val62, Leu67, Tyr70, Gln107, and Arg131 contributed favorable energies of −3.59, −10.40, −3.85, −3.66, −5.08, −4.52, −3.32, −3.32, and −15.28 kJ/mol, respectively, while Asp49 and Glu75 contributed unfavorably, with energies of 4.33 and 17.12 kJ/mol, respectively (Figure S11a).
For the HPV16 E6–ZINC000070454124 complex, Lys11, Leu50, Cys51, Val53, Val62, Leu100, Arg102, Gln107, Arg131, and Trp132 contributed favorable energies of −4.23, −7.36, −4.65, −3.20, −3.05, −4.48, −10.10, −4.03, −24.09, and −4.06 kJ/mol, respectively, while Asp49 and Glu75 contributed unfavorable energies of 9.73 and 14.41 kJ/mol, respectively (Figure S11b). For the HPV16 E6–ZINC000014588133 complex, Arg10, Lys11, Arg77, Arg129, and Arg131 contributed favorable energies of −6.76, −3.81, −3.99, −6.48, and −3.32 kJ/mol, respectively, while Glu75 had unfavorable energy of 5.30 kJ/mol (Figure S11c). With regards to the HPV16 E6–ZINC000085568136 complex, residues Arg10, Val31, Tyr32, Leu50, Cys51, Val53, Arg55, Ser74, Arg77, and Arg131 contributed favorable energies of −4.25, −4.67, −4.93, −3.05, −3.33, −10.06, −5.46, −3.05, −6.05, and −7.26 kJ/mol, respectively, while Glu7 and Glu75 contributed unfavorable energies of 3.13 and 12.09 kJ/mol, respectively (Figure S11d). For the HPV16 E6–ZINC000085597263 complex, residues Tyr32, Cys33, Lys34, Arg55, Gln107, and Arg131 contributed favorable energies of −3.06, −4.43, −8.20, −4.65, −3.90, and −12.78 kJ/mol, respectively, while Glu75 contributed unfavorable energy of 9.51 kJ/mol (Figure S11g). For the HPV16 E6 systems the four ligands that had high (positive) enthalpy-based binding energies with the protein, 25 residues were observed to contribute unfavorable energies to ligand binding, while 23 residues promoted ligand binding (Figure S11e,h–j). Residues Lys11, Leu50, Cys51, Val53, Gln107, Arg131, and Glu75 are hotspot residues critical for ligand binding to the E6 protein of HPV16. This corroborates the findings from previous studies that suggested Cys51, Leu50, Arg102, Arg131, Leu67, Val62, and Gln107 as key residues in E6 inhibition, while suggesting Leu50 and Cys51 as crucial residues for high affinity ligand binding [106].
For HPV18 E6 protein complexed with baicalein and gossypetin, all residues contributed energies <3 kJ/mol. For the HPV18 E6–baicalein complex, residues Val1033, Leu1052, Phe1053, Val1055, Ile1069, and Tyr1072 contributed favorably to baicalein’s binding with energies of −3.91, −3.41, −3.79, −4.86, −10.85, and −6.52 kJ/mol, respectively (Figure S12k). For the HPV18 E6–gossypetin complex, residues Lys1013, Lys1050, Leu1052, Phe1053, Ile1069, Tyr1072, Arg1104, and Gln1109 contributed favorable energies of −3.71, −3.72, −9.61, −5.98, −7.43, −4.42, −3.68, and −4.71 kJ/mol, respectively (Figure S12l). For the HPV18 E6–ZINC000013380012 complex, residues Lys1050, Leu1052, Phe1053, Val1055, Ile1069, Tyr1072, Arg1076, and Arg1104 contributed favorable energies of −4.03, −7.11, −4.83, −3.92, −9.88, −4.88, −3.97, and −4.56 kJ/mol, respectively, while Asp1051 contributed unfavorable energy of 3.70 kJ/mol (Figure S12a). For the HPV18 E6–ZINC000070454124 complex, residues Lys1013, Leu1052, Phe1053, Val1055, Ile1069, Leu1102, Arg1104, Gln1109, Lys1110, and His1133 contributed favorable energies of −6.84, −8.23, −12.95, −6.77, −5.92, −3.96, −9.19, −4.60, −3.18, and −4.32 kJ/mol, respectively, while Asp1051 contributed unfavorable energy of 4.86 kJ/mol (Figure S12b). For the HPV18 E6–ZINC000014588133 complex, residues Lys1050, Leu1052, Phe1053, Ile1069, Tyr1072, and Arg1076 contributed favorable energies of −3.30, −6.19, −4.25, −11.10, −5.00, and −6.82 kJ/mol, respectively, while Arg1135 contributed unfavorable energy of 3.50 kJ/mol (Figure S12c).
For the HPV18 E6–ZINC000085568136 complex, residues Tyr1012, Lys1013, Lys1050, Leu1052, Phe1053, Val1055, Ile1069, Tyr1072, and Arg1104 contributed favorable energies of −4.56, −6.54, −3.35, −10.65, −16.68, −4.74, −7.05, −4.49, and −9.11 kJ/mol, respectively, while Asp1051 contributed unfavorable energy of 7.21 kJ/mol (Figure S12d). For the HPV18 E6–ZINC000095909247 complex, residues Lys1050, Leu1052, Ile1069, Tyr1072, Arg1074, Arg1076, Arg1104, and Gln1109 contributed favorable energies of −4.19, −4.81, −6.37, −5.63, −3.15, −7.75, −6.17, and −4.21 kJ/mol, respectively, while Asp1051 and Asp1070 contributed favorable energies of 3.46 and 3.15 kJ/mol, respectively (Figure S12f). For the HPV18 E6–ZINC000085597263 complex, residues Lys1013, Val1033, Tyr1034, Lys1050, Leu1052, Val1055, Cys1068, Ile1069, Tyr1072, Arg1104, and His1133 contributed favorable energies of −4.69, −5.33, −3.60, −3.34, −5.77, −5.46, −3.69, −7.62, −8.73, −7.08, and −5.85 kJ/mol, respectively, while Asp1051 and Arg1135 contributed unfavorable energies of 6.08 and 5.60 kJ/mol, respectively (Figure S12g).
For the four HPV18 E6 protein–ligand complexes that demonstrated positive enthalpy-based binding energies [ZINC000085631200 (Figure S12e), ZINC000095913339 (Figure S12h), ZINC000085628525 (Figure S12i), and ZINC000085543530 (Figure S12j)], 23 residues were observed to contribute unfavorably to ligand binding, while 26 residues contributed favorable energies. From the per-residue energy decomposition analysis of the HPV18 E6–ligand complexes, residues Lys1013 (Lys13), Lys1050 (Lys50), Asp1051, Leu1052 (Leu52), Phe1053 (Phe53), Val1055 (Val55), Ile1069 (Ile69), Tyr1072 (Tyr72), and Arg1104 (Arg104) contributed to binding of most of the compounds and can be considered as hotspot residues crucial for ligand binding. The canonical HPV18 E6 sequence numberings are in brackets, while those of the PDB structure (6SJV) used are written without brackets.
3.10. Structural Similarity Search of the Potential Candidates
The literature was searched to identify the existing use of the six top compounds in the treatment of viruses, cancers, and related complications or diseases. Structural similarity searches were also performed via DrugBank to identify analogs of the top six compounds [ZINC000013380012 (broussonol B), ZINC000070454124, ZINC000014588133, ZINC000085568136, ZINC000095909247, and ZINC000085597263] (Figure 10) [94,95]. Structural similarity searches help to elucidate potential biological activities by revealing connections between chemical structures and bioactive properties [134,135,136]. This approach facilitates the identification of analogs, enabling the recognition of shared pharmacophores and mechanisms of action. The DrugBank-based similarity searches for the top compounds, using a similarity score threshold of 0.7, revealed analogous drugs, with the exception of ZINC000070454124 and ZINC000085597263.
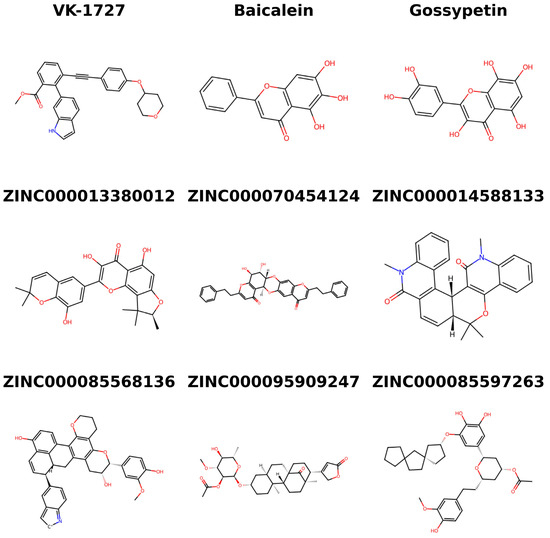
Figure 10.
Chemical structures of the three known inhibitors (VK-1727, baicalein, and gossypetin) and the six potential anti-cervical cancer compounds identified in this study.
Compound ZINC000013380012 (broussonol B) is found in Broussonetia papyrifera, commonly known as paper mulberry and B. kazinoki (kozo) [137]. The cytotoxic activity of the methanolic extracts of leaf, bark, and fruit of B. papyrifera was investigated in relation to MCF-7, HeLa, and HepG2 cell lines [138]. The leaf extract exhibited significant cytotoxic activity against MCF-7 and HeLa cell lines, with IC50 values of 105 μg/mL and 110 μg/mL, confirmed by MTT assay (IC50 values: 87.5 μg/mL and 106.2 μg/mL) [138]. The bark extract demonstrated notable activity against HeLa cells (IC50: 75.3 μg/mL and 88.3 μg/mL), while both leaf and bark extracts displayed moderate activity against HepG2 cells. Conversely, the fruit extract showed insignificant cytotoxicity across all tested cell lines [138]. Another study also reported that polyphenols extracted from B. papyrifera stem bark induced apoptosis in HepG2 cells through mitochondria-mediated mechanisms, achieved by the deactivation of ERK and AKT signaling pathways [139]. Broussonol B has also been previously shown via MTT assay to possess low cytotoxicity against A549 and HCT-8 human tumor cell lines [137].
Furthermore, a structural similarity search revealed that ZINC000013380012 is similar to troxerutin (0.717), icariin (0.707), and monoxerutin (0.703). Troxerutin has been previously reported to exhibit a growth inhibitory effect on HeLa cells, with an effective IC50 of 320 mg/mL after 24 h of treatment [140]. This inhibitory effect was associated with increased levels of apoptosis-related proteins (Bax, cleaved caspase-3, and TNF-α) and a decrease in anti-apoptotic Bcl-2 protein expression [140]. The Bcl-2 inhibitor ABT-737 has been shown to be beneficial in treating post-transplant EBV-related B lymphoproliferative disorders [110]. Troxerutin also induced oxidative stress by elevating malondialdehyde concentration while reducing the activity levels of glutathione peroxidase and superoxide dismutase [140]. The anticancer properties of icariin have been documented in the literature [141,142,143,144,145,146]. The anticancer properties of icariin against human HeLa cervical cancer cell line and normal HCvEpC cells were assessed previously [146]. Icariin demonstrated dose-dependent inhibition of HeLa cell growth, with an IC50 of 20 µM, while exhibiting minimal toxic effects on normal HCvEpC cells [146]. Like troxerutin [140], the anticancer effects of icariin were attributed to the induction of apoptosis, marked by caspase 3 and 9 cleavage, the upregulation of pro-apoptotic Bax, and the downregulation of anti-apoptotic Bcl-2 [146]. Additionally, icariin triggered the generation of reactive oxygen species (ROS), reduced mitochondrial membrane potential, and inhibited the mTOR/PI3K/AKT signaling cascade in HeLa cells, indicating its potent anticancer activity [146].
Another study investigated the antitumor effects of icariin in U14 tumor-bearing mice and SiHa cells [147]. Icariin exhibited a dose-dependent suppression of tumor tissue growth in vivo and significantly reduced the viability of SiHa cells in vitro. In SiHa cells, icariin was found to inhibit migration, invasion capacity, and the expression of inflammatory markers such as TGF-β1, TNF-α, IL-6, IL-17A, and IL-10 [147]. Additionally, icariin promoted apoptosis in cervical cancer cells by downregulating key apoptotic markers, including Ki67, survivin, Bcl-2, c-Myc, and upregulating P16, P53, and Bax levels, both in vivo and in vitro [147]. The study further revealed that icariin exerted its effects through the impairment of the TLR4/MyD88/NF-κB and Wnt/β-catenin pathways [147]. In another study, the role of icariin in modulating malignant behaviors by targeting the miR-875-5p/MDM4 axis in cervical cancer was investigated [148]. The study revealed downregulation of miR-875-5p in cervical cancer cells, promoting malignant phenotypes and autophagy and inhibiting apoptosis [148]. Conversely, the overexpression of miR-875-5p demonstrated opposing effects. MiR-875-5p was identified as a sponge for MDM4, and elevated MDM4 expression attenuated the impact of miR-875-5p mimic on autophagy and epithelial–mesenchymal transition. However, icariin was found to counteract the inhibitory effects of miR-875-5p inhibitor on autophagy and xenograft tumor growth [148]. These suggest that ZINC000013380012 may serve as a promising lead molecule for the development of cervical cancer therapy, offering a potential alternative to current treatment modalities.
Compound ZINC000014588133 (melicobisquinolinone B) is found in the leaves and branches of Melicope denhamii [149] and M. pteleifolia [150]. Melicobisquinolinone B has previously been shown to possess a weak anti-proliferative activity against DLD-1 human colon cancer cells [151], which suggests the need for further investigations into its potential as an anticancer agent or its optimization for enhanced efficacy. ZINC000014588133 is structurally similar to vintafolide, vinorelbine, and anhydrovinblastine, with scores of 0.704, 0.701, and 0.7, respectively. Vintafolide, antigen drug conjugate [152], has been shown to be effective in ovarian cancer by targeting the folate receptor [153,154,155,156]. Vinorelbine is used for non-small cell lung cancer treatment [157,158,159,160] and has also proven to be active against cervical cancers with moderate toxicity [161,162]. Anhydrovinblastine serves as the precursor for vinblastine [163,164], an anticancer drug with a broad spectrum of activity [165,166]. Moreover, ester anhydrovinblastine derivatives have been reported to demonstrate potent cytotoxicity against human non-small cell lung cancer (A549) and cervical epithelial adenocarcinoma (HeLa) cell lines [167].
Compound ZINC000085568136 is structurally similar to thiostrepton, with a score of 0.7. Thiostrepton is a known FOXM1 inhibitor [30,168,169,170,171]. In nude mice models, knockdown of HPV E6 and FOXM1 or treatment with thiostrepton significantly reduced metastatic lung tumor nodules in HPV-positive cancer cells [30], making ZINC000085568136 an interesting candidate to test against FOXM1, E6, and cervical cancer cell lines. Compound ZINC000095909247 with IUPAC name [(2R,3S,4R,5S,6S)-2-[[(1R,4R,6S,9S,10R,13R,14R)-9,13-dimethyl-17-oxo-14-(5-oxo-2H-furan-3-yl)-6-tetracyclo [11.3.1.01,10.04,9]heptadecanyl]oxy]-5-hydroxy-4-methoxy-6-methyloxan-3-yl] acetate, identified in the literature as “2-O-acetyl-cerleaside A”, is found in the seeds of Cerbera manghas and C. odollam [172,173]. 2-O-acetyl-cerleaside A has been previously shown to be cytotoxic against oral human epidermoid carcinoma, human breast cancer, and human small cell lung cancer (NCI-H187) cell lines with ED50 values of 7.56, 4.62, and 7.42 μg/mL, respectively [173]. In light of the observed moderate to strong anticancer properties of ZINC000095909247, it is prudent that its efficacy against cervical cancer be evaluated and validated.
Compound ZINC000095909247 is structurally similar to peruvoside (0.842), lanatoside C (0.837), cymarin (0.828), acetyldigitoxin (0.808), acetyldigoxin (0.808), ouabain (0.798), deslanoside (0.797), digoxin (0.783), digitoxin (0.783), metildigoxin (0.783), gitoformate (0.767), oleandrin (0.759), and P-57AS3 (0.722). These compounds, with the exception of P-57AS3, are cardiac glycosides, which are used as therapeutic agents for heart ailments to enhance the force of myocardial contractions while simultaneously reducing the frequency of these contractions [174]. However, P57AS3 exhibits similarities to the steroidal core found in cardiac glycosides [175]. Cardiac glycosides have been reported to inhibit the expression of EBV early antigen, with EC50 values less than 60 nM [176]. Cardiac glycosides have also shown potent anticancer activities, and there is a growing interest in repurposing them for cancer treatments [177,178,179,180,181]. Lanatoside C has specifically exhibited anti-proliferative and cytotoxic effects on cervical cancer cells [118]. Lanatoside C induces apoptosis through JAK2-STAT6 signaling inhibition [118]. Digitoxin also displayed potent cytotoxicity against HeLa cervical cancer cells with an IC50 value of 28 nM after 48 h of exposure [182]. Following a 48 h exposure to digoxin, the viability of both human lung (A549) and cervical cancer (HeLa) cell lines were adversely affected in a concentration-dependent manner, resulting in IC50 values of 31 nM for A549 cells and 151 nM for HeLa cells [183]. The anti-EBV and broad-spectrum anticancer properties, as well as specific anti-cervical cancer activities observed in these cardiac glycosides, make ZINC000095909247 an interesting candidate to probe further.
4. Conclusions
High-risk HPV types 16 and 18 are known to cause cervical cancer in women. EBV is also believed to contribute to cervical cancer progression by elevating HPV E6 level, which in turn, suppresses p53 production. Cervical cancers are the fourth most prevalent cancers in women globally and a leading cause of death. Finding drug candidates that can inhibit cervical cancer growth and progression will be a major breakthrough. This study therefore sought to use a robust computational drug discovery pipeline to shortlist natural compounds that could target E6 proteins of HPV16 and HPV18 simultaneously. The shortlisted compounds that exhibited favorable docking scores against both HPV E6 proteins were subjected to another round of scrutiny by reassessing their binding affinity against the EBNA1 protein. This step aimed to identify potential inhibitors capable of targeting both HPV E6 and EBNA1, with the ultimate goal of providing therapeutic benefits for cervical cancer patients experiencing HPV/EBV co-infection. The good candidates were further subjected to ADMET profiling and toxicity testing. Biological activity predictions supported the antiviral and anticancer activities of the most promising candidates, setting the stage for MD simulations and MM/PBSA calculations. The biological activity predictions revealed that the compounds possess anticancer properties and may function as p53 expression enhancers, as well as apoptosis agonists. These properties are crucial for cervical cancer drugs, as p53 downregulation is mediated by HPV E6 in cervical cancer patients. The MD simulations and MM/PBSA computations revealed that six compounds, comprising ZINC000013380012 (broussonol B), ZINC000070454124, ZINC000014588133, ZINC000085568136, ZINC000095909247, and ZINC000085597263, demonstrated better stability and high affinity to the three oncoproteins (EBNA1, HPV16 E6, and HPV18 E6) studied herein. Broussonol B has previously demonstrated cytotoxicity against A549 and HCT-8 human tumor cell lines [137], warranting cervical cancer testing. The shortlisted compounds are interesting candidates to probe further for their anti-HPV, anti-EBV, and anti-cervical cancer properties.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/computation12060112/s1: Figure S1: Protein–ligand interaction map of EBNA1 in complex with (a) ZINC000013380012; (b) ZINC000070454124; (c) ZINC000014588133; (d) ZINC000085631200; (e) ZINC000095909247; (f) ZINC000085597263; (g) ZINC000095913339; (h) ZINC000085628525; and (i) ZINC000085543530. The green dotted lines indicate hydrogen bonds, while the red arcs with spikes represent hydrophobic interactions; Figure S2: Protein–ligand interaction map of HPV16 E6 in complex with (a) ZINC000013380012; (b) ZINC000070454124; (c) ZINC000014588133; (d) ZINC000085568136; (e) ZINC000085631200; (f) ZINC000095909247; (g) ZINC000095913339; (h) ZINC000085628525; (i) ZINC000085543530; and (j) Gossypetin. The green dotted lines indicate hydrogen bonds, while the red arcs with spikes represent hydrophobic interactions; Figure S3: Protein–ligand interaction map of HPV18 E6 in complex with (a) ZINC000013380012; (b) ZINC000070454124; (c) ZINC000014588133; (d) ZINC000085568136; (e) ZINC000085631200; (f) ZINC000095909247; (g) ZINC000095913339; (h) ZINC000085628525; (i) ZINC000085543530; and (j) Gossypetin. The green dotted lines indicate hydrogen bonds, while the red arcs with spikes represent hydrophobic interactions; Figure S4: RMSD plots for the unbound HPV16 E6 protein and HPV16 E6–ligand complexes. The RMSD plots are shown for apo-HPV16 E6, baicalein, and gossypetn, as well as (a) ZINC000013380012, ZINC000070454124, ZINC000014588133, ZINC000085568136, and ZINC000085631200 and (b) ZINC000095909247, ZINC000085597263, ZINC000095913339, ZINC000085628525, and ZINC000085543530; Figure S5: RMSD plots for the unbound HPV18 E6 protein and HPV18 E6–ligand complexes. The RMSD plots are shown for apo-HPV18 E6, baicalein, and gossypetn, as well as (a) ZINC000013380012, ZINC000070454124, ZINC000014588133, ZINC000085568136, and ZINC000085631200 and (b) ZINC000095909247, ZINC000085597263, ZINC000095913339, ZINC000085628525, and ZINC000085543530; Figure S6: Rg plots for the unbound HPV16 E6 protein and HPV16 E6–ligand complexes. The Rg plots are shown for apo-HPV16 E6, baicalein, and gossypetn, as well as (a) ZINC000013380012, ZINC000070454124, ZINC000014588133, ZINC000085568136, and ZINC000085631200 and (b) ZINC000095909247, ZINC000085597263, ZINC000095913339, ZINC000085628525, and ZINC000085543530; Figure S7: Rg plots for the unbound HPV18 E6 protein and HPV18 E6–ligand complexes. The Rg plots are shown for apo-HPV18 E6, baicalein, and gossypetn, as well as (a) ZINC000013380012, ZINC000070454124, ZINC000014588133, ZINC000085568136, and ZINC000085631200 and (b) ZINC000095909247, ZINC000085597263, ZINC000095913339, ZINC000085628525, and ZINC000085543530; Figure S8: RMSF plot for the unbound HPV16 E6 and HPV16 E6–ligand complexes after 100 ns MD simulation. The unbound HPV16 E6 protein, HPV16 E6 complexed with baicalein, gossypetin, ZINC000013380012, ZINC000070454124, ZINC000014588133, ZINC000085568136, ZINC000085631200, ZINC000095909247, ZINC000085597263, ZINC000095913339, ZINC000085628525, and ZINC000085543530 are colored black, red, green, blue, yellow, brown, grey, violet, cyan, magenta, orange, indigo, and maroon, respectively; Figure S9: RMSF plot for the unbound HPV18 E6 and HPV18 E6–ligand complexes after 100 ns MD simulation. The unbound HPV18 E6 protein, HPV18 E6 complexed with baicalein, gossypetin, ZINC000013380012, ZINC000070454124, ZINC000014588133, ZINC000085568136, ZINC000085631200, ZINC000095909247, ZINC000085597263, ZINC000095913339, ZINC000085628525, and ZINC000085543530 are colored black, red, green, blue, yellow, brown, grey, violet, cyan, magenta, orange, indigo, and maroon, respectively; Figure S10: Per-residue energy decomposition plots of the various complexes showing the energy contributions of each amino acid residue for EBNA1 in complex with (a) ZINC000013380012; (b) ZINC000070454124; (c) ZINC000014588133; (d) ZINC000085631200; (e) ZINC000095909247; (f) ZINC000085597263; (g) ZINC000095913339; (h) ZINC000085628525; (i) ZINC000085543530; and (j) VK-1727; Figure S11: Per-residue energy decomposition plots of the various complexes showing the energy contributions of each amino acid residue for HPV16 E6 in complex with (a) ZINC000013380012; (b) ZINC000070454124; (c) ZINC000014588133; (d) ZINC000085568136; (e) ZINC000085631200; (f) ZINC000095909247; (g) ZINC000085597263; (h) ZINC000095913339; (i) ZINC000085628525; (j) ZINC000085543530; (k) baicalein; and (l) gossypetin; Figure S12: Per-residue energy decomposition plots of the various complexes showing the energy contributions of each amino acid residue for HPV18 E6 in complex with (a) ZINC000013380012; (b) ZINC000070454124; (c) ZINC000014588133; (d) ZINC000085568136; (e) ZINC000085631200; (f) ZINC000095909247; (g) ZINC000085597263; (h) ZINC000095913339; (i) ZINC000085628525; (j) ZINC000085543530; (k) baicalein; and (l) gossypetin; Table S1: DockThor docking scores of the known inhibitors and shortlisted compounds against the three oncoproteins, EBNA1, HPV16 E6, andHPV18 E6; Table S2: Clusters generated from the first 80 ns and the last 20 ns segments of each MD simulation using a cut-off of 0.2 nm; Table S3: Comparative analysis of RMSD values between clusters extracted from the initial 80 ns and final 20 ns segments of MD simulations. “First*” and “Last*” denote clusters derived from the “first 80 ns” and “last 20 ns” intervals, respectively, with “*” representing the specific cluster number.
Author Contributions
W.A.M.III and E.B. conceptualized the project. E.B. and W.A.M.III predominantly undertook all the computational analysis with inputs from C.N.A., M.V., P.O.S. and S.K.K. W.A.M.III and E.B. co-wrote the first draft. The final manuscript was written through contributions of all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Acknowledgments
The authors are grateful to the editor and the reviewers for their contributions.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Schiller, J.T.; Lowy, D.R. An Introduction to Virus Infections and Human Cancer. In Recent Results in Cancer Research. Fortschritte der Krebsforschung. Progres dans les Recherches sur le Cancer; Springer: Berlin/Heidelberg, Germany, 2021; Volume 217, pp. 1–11. [Google Scholar]
- Shih, W.-L.; Fang, C.-T.; Chen, P.-J. Anti-Viral Treatment and Cancer Control. In Recent Results in Cancer Research; Springer: Berlin/Heidelberg, Germany, 2014; pp. 269–290. ISBN 0080-0015. [Google Scholar]
- Shi, Y.; Peng, S.-L.; Yang, L.-F.; Chen, X.; Tao, Y.-G.; Cao, Y. Co-Infection of Epstein-Barr Virus and Human Papillomavirus in Human Tumorigenesis. Chin. J. Cancer 2016, 35, 16. [Google Scholar] [CrossRef] [PubMed]
- Trama, J.P.; Trikannad, C.; Yang, J.J.; Adelson, M.E.; Mordechai, E. High-Risk HPV Genotype Distribution According to Cervical Cytology and Age. Open Forum Infect. Dis. 2022, 9, ofac595. [Google Scholar] [CrossRef] [PubMed]
- Ampofo, A.G.; Boyes, A.W.; Asibey, S.O.; Oldmeadow, C.; Mackenzie, L.J. Prevalence and Correlates of Modifiable Risk Factors for Cervical Cancer and HPV Infection among Senior High School Students in Ghana: A Latent Class Analysis. BMC Public Health 2023, 23, 340. [Google Scholar] [CrossRef] [PubMed]
- Okunade, K.S. Human Papillomavirus and Cervical Cancer. J. Obstet. Gynaecol. 2020, 40, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Ploner, A.; Elfström, K.M.; Wang, J.; Roth, A.; Fang, F.; Sundström, K.; Dillner, J.; Sparén, P. HPV Vaccination and the Risk of Invasive Cervical Cancer. N. Engl. J. Med. 2020, 383, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Illah, O.; Olaitan, A. Updates on HPV Vaccination. Diagnostics 2023, 13, 243. [Google Scholar] [CrossRef] [PubMed]
- Wirtz, C.; Mohamed, Y.; Engel, D.; Sidibe, A.; Holloway, M.; Bloem, P.; Kumar, S.; Brotherton, J.; Reis, V.; Morgan, C. Integrating HPV Vaccination Programs with Enhanced Cervical Cancer Screening and Treatment, a Systematic Review. Vaccine 2022, 40, A116–A123. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.K.; Abe, S.K.; Thilagaratnam, S.; Haruyama, R.; Pathak, R.; Jayasekara, H.; Togawa, K.; Bhandari, A.K.C.; Shankar, A.; Nessa, A.; et al. Towards Elimination of Cervical Cancer—Human Papillomavirus (HPV) Vaccination and Cervical Cancer Screening in Asian National Cancer Centers Alliance (ANCCA) Member Countries. Lancet Reg. Health–West. Pac. 2023, 39, 100860. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Kombe Kombe, A.J.; Li, B.; Zahid, A.; Mengist, H.M.; Bounda, G.-A.; Zhou, Y.; Jin, T. Epidemiology and Burden of Human Papillomavirus and Related Diseases, Molecular Pathogenesis, and Vaccine Evaluation. Front. Public Health 2021, 8, 552028. [Google Scholar] [CrossRef] [PubMed]
- Echelman, D.; Feldman, S. Management of Cervical Precancers: A Global Perspective. Hematol. Oncol. Clin. N. Am. 2012, 26, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Pang, C.L.; Thierry, F. Human Papillomavirus Proteins as Prospective Therapeutic Targets. Microb. Pathog. 2013, 58, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Jee, B.; Yadav, R.; Pankaj, S.; Shahi, S.K. Immunology of HPV-Mediated Cervical Cancer: Current Understanding. Int. Rev. Immunol. 2021, 40, 359–378. [Google Scholar] [CrossRef] [PubMed]
- Graham, S.V.; Faizo, A.A.A. Control of Human Papillomavirus Gene Expression by Alternative Splicing. Virus Res. 2017, 231, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Hakizimana, J.K.; Timchenko, V.N.; Shakmaeva, M.A.; Kaplina, T.A.; Subbotina, M.D.; Bannova, S.L.; Fedorova, A.V.; Sukhovetskaya, V.F.; Pavlova, E.B.; Pavlova, N.V. EBV Mononucleosis in Children in Modern Conditions. Child. Infect. 2020, 19, 23–28. [Google Scholar] [CrossRef]
- Hjalgrim, H.; Askling, J.; Rostgaard, K.; Hamilton-Dutoit, S.; Frisch, M.; Zhang, J.-S.; Madsen, M.; Rosdahl, N.; Konradsen, H.B.; Storm, H.H.; et al. Characteristics of Hodgkin’s Lymphoma after Infectious Mononucleosis. N. Engl. J. Med. 2003, 349, 1324–1332. [Google Scholar] [CrossRef] [PubMed]
- Balfour, H.H.; Dunmire, S.K.; Hogquist, K.A. Infectious Mononucleosis. Clin. Transl. Immunol. 2015, 4, e33. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Tang, K.; Qian, J.; Deng, H.; Zeng, M.; Zheng, S.; Ding, K.; Du, Y.; Sun, R. Immunotherapy for EBV-Associated Nasopharyngeal Carcinoma. Crit. Rev. Oncog. 2018, 23, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Duan, X.; Chen, X.; Zhan, M.; Peng, H.; Meng, Y.; Li, X.; Li, X.-Y.; Pang, G.; Dou, X. Immunotherapeutic Approaches in EBV-Associated Nasopharyngeal Carcinoma. Front. Immunol. 2023, 13, 1079515. [Google Scholar] [CrossRef]
- Orem, J.; Sandin, S.; Mbidde, E.; Mangen, F.W.; Middeldorp, J.; Weiderpass, E. Epstein-Barr Virus Viral Load and Serology in Childhood Non-Hodgkin’s Lymphoma and Chronic Inflammatory Conditions in Uganda: Implications for Disease Risk and Characteristics. J. Med. Virol. 2014, 86, 1796–1803. [Google Scholar] [CrossRef]
- Kanda, T.; Yajima, M.; Ikuta, K. Epstein-Barr Virus Strain Variation and Cancer. Cancer Sci. 2019, 110, 1132–1139. [Google Scholar] [CrossRef] [PubMed]
- Pagano, J.; Whitehurst, C.; Andrei, G. Antiviral Drugs for EBV. Cancers 2018, 10, 197. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.Y.; Krumm, A.; Schubach, W.H. Promoter-Specific Targeting of Human SWI-SNF Complex by Epstein-Barr Virus Nuclear Protein 2. J. Virol. 2000, 74, 8893–8903. [Google Scholar] [CrossRef] [PubMed]
- Klinke, O.; Feederle, R.; Delecluse, H.-J. Genetics of Epstein–Barr Virus MicroRNAs. Semin. Cancer Biol. 2014, 26, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Andrei, G.; Trompet, E.; Snoeck, R. Novel Therapeutics for Epstein–Barr Virus. Molecules 2019, 24, 997. [Google Scholar] [CrossRef] [PubMed]
- de Lima, M.A.P.; Neto, P.J.N.; Lima, L.P.M.; Gonçalves Júnior, J.; Teixeira Junior, A.G.; Teodoro, I.P.P.; Facundo, H.T.; da Silva, C.G.L.; Lima, M.V.A. Association between Epstein-Barr Virus (EBV) and Cervical Carcinoma: A Meta-Analysis. Gynecol. Oncol. 2018, 148, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, Q.; Gupta, I.; Vranic, S.; Al Moustafa, A.-E. Human Papillomaviruses and Epstein–Barr Virus Interactions in Colorectal Cancer: A Brief Review. Pathogens 2020, 9, 300. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-M.; Cheng, Y.-W.; Wang, Y.-C.; Wu, T.-C.; Chen, C.-Y.; Lee, H. Up-Regulation of FOXM1 by E6 Oncoprotein through the MZF1/NKX2-1 Axis Is Required for Human Papillomavirus–Associated Tumorigenesis. Neoplasia 2014, 16, 961–971. [Google Scholar] [CrossRef]
- Diab, A.; Gem, H.; Swanger, J.; Kim, H.Y.; Smith, K.; Zou, G.; Raju, S.; Kao, M.; Fitzgibbon, M.; Loeb, K.R.; et al. FOXM1 Drives HPV+ HNSCC Sensitivity to WEE1 Inhibition. Proc. Natl. Acad. Sci. USA 2020, 117, 28287–28296. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Hong, X.; Wei, Z.; Xie, M.; Li, W.; Liu, G.; Guo, H.; Yang, J.; Wei, W.; Zhang, S. Ubiquitination of the HPV Oncoprotein E6 Is Critical for E6/E6AP-Mediated P53 Degradation. Front. Microbiol. 2019, 10, 2483. [Google Scholar] [CrossRef]
- Martinez-Zapien, D.; Ruiz, F.X.; Poirson, J.; Mitschler, A.; Ramirez, J.; Forster, A.; Cousido-Siah, A.; Masson, M.; Vande Pol, S.; Podjarny, A.; et al. Structure of the E6/E6AP/P53 Complex Required for HPV-Mediated Degradation of P53. Nature 2016, 529, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Clemente-Soto, A.; Salas-Vidal, E.; Milan-Pacheco, C.; Sánchez-Carranza, J.; Peralta-Zaragoza, O.; González-Maya, L. Quercetin Induces G2 Phase Arrest and Apoptosis with the Activation of P53 in an E6 Expression-independent Manner in HPV-positive Human Cervical Cancer-derived Cells. Mol. Med. Rep. 2019, 19, 2097–2106. [Google Scholar] [CrossRef] [PubMed]
- Soumia, M.; Hajji, H.; El Mzibri, M.; Younes, F.Z.; Mohammed, B.; Mohamed, B.; Benaissa, M. In-Silico Molecular Modeling Studies to Identify Novel Potential Inhibitors of HPV E6 Protein. Vaccines 2022, 10, 1452. [Google Scholar] [CrossRef] [PubMed]
- Hoseini Tabatabaie, F.; Hosseini, S.Y.; Hashemi, S.M.A.; Safaie, A.; Sarvari, J. A Preliminary Sequence Analysis of the Epstein-Barr Virus Nuclear Antigen 1 (EBNA1) Carboxy-Terminal Region in Cervical and Ovarian Cancers. Iran. J. Pathol. 2023, 18, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.S.; Hung, S.C.; Kieff, E. Epstein-Barr Virus Nuclear Antigen 1 Activates Transcription from Episomal but Not Integrated DNA and Does Not Alter Lymphocyte Growth. Proc. Natl. Acad. Sci. USA 2001, 98, 15233–15238. [Google Scholar] [CrossRef] [PubMed]
- Xin, S.; Du, S.; Liu, L.; Xie, Y.; Zuo, L.; Yang, J.; Hu, J.; Yue, W.; Zhang, J.; Cao, P.; et al. Epstein-Barr Virus Nuclear Antigen 1 Recruits Cyclophilin A to Facilitate the Replication of Viral DNA Genome. Front. Microbiol. 2019, 10, 149. [Google Scholar] [CrossRef] [PubMed]
- Alipour, A.H.; Hashemi, S.M.A.; Moattari, A.; Farhadi, A.; Sarvari, J. Epstein-Barr Virus Nuclear Antigen 1 Increases the Expression of Viral Oncogenes and Cellular Genes in the HeLa Cell Line. Int. J. Mol. Cell. Med. 2022, 11, 346–356. [Google Scholar] [CrossRef]
- Tan, Z.; Zhang, S. Past, Present, and Future of Targeting Ras for Cancer Therapies. Mini-Rev. Med. Chem. 2016, 16, 345–357. [Google Scholar] [CrossRef]
- Zhang, S.; Tan, Z. Editorial (Thematic Issue: The Elephant in the Room: Targeting Ras for Therapeutic Development). Mini-Rev. Med. Chem. 2016, 16, 343–344. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Chaudhai, R.; Zhang, S. Polypharmacology in Drug Development: A Minireview of Current Technologies. ChemMedChem 2016, 11, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Kefale, B.; Engidaw, M.T.; Tesfa, D.; Molla, M.; Tegegne, G.T. Medication-Related Problems among Patients with Cervical Cancers at Oncology Centers of University of Gondar Comprehensive Specialized Hospital: A Hospital-Based Retrospective Study. J. Oncol. Pharm. Pract. 2024, 30, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Ryszkiewicz, P.; Malinowska, B.; Schlicker, E. Polypharmacology: Promises and New Drugs in 2022. Pharmacol. Rep. 2023, 75, 755–770. [Google Scholar] [CrossRef] [PubMed]
- Jamir, E.; Sarma, H.; Priyadarsinee, L.; Kiewhuo, K.; Nagamani, S.; Sastry, G.N. Polypharmacology Guided Drug Repositioning Approach for SARS-CoV2. PLoS ONE 2023, 18, e0289890. [Google Scholar] [CrossRef] [PubMed]
- Tomaselli, D.; Lucidi, A.; Rotili, D.; Mai, A. Epigenetic Polypharmacology: A New Frontier for Epi-drug Discovery. Med. Res. Rev. 2020, 40, 190–244. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Lou, H.-X. Strategies to Diversify Natural Products for Drug Discovery. Med. Res. Rev. 2018, 38, 1255–1294. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Li, J.; Chen, A.; Li, Y.; Xia, M.; Guo, P.; Yao, S.; Chen, S. Fucosterol Exhibits Selective Antitumor Anticancer Activity against HeLa Human Cervical Cell Line by Inducing Mitochondrial Mediated Apoptosis, Cell Cycle Migration Inhibition and Downregulation of m-TOR/PI3K/Akt Signalling Pathway. Oncol. Lett. 2018, 15, 3458–3463. [Google Scholar] [CrossRef]
- Choudhari, A.S.; Suryavanshi, S.A.; Kaul-Ghanekar, R. The Aqueous Extract of Ficus Religiosa Induces Cell Cycle Arrest in Human Cervical Cancer Cell Lines SiHa (HPV-16 Positive) and Apoptosis in HeLa (HPV-18 Positive). PLoS ONE 2013, 8, e70127. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, C.; Arul Murugan, N.; Priyakumar, U.D. Structure-Based Drug Repurposing: Traditional and Advanced AI/ML-Aided Methods. Drug Discov. Today 2022, 27, 1847–1861. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.R.; Joshi, H.V.; Shah, U.A.; Patel, J.K. A Review on Computational Software Tools for Drug Design and Discovery. Indo Glob. J. Pharm. Sci. 2022, 12, 53–81. [Google Scholar] [CrossRef]
- Chen, C.Y.-C.C. TCM Database@Taiwan: The World’s Largest Traditional Chinese Medicine Database for Drug Screening in Silico. PLoS ONE 2011, 6, e15939. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M.; et al. RCSB Protein Data Bank: Powerful New Tools for Exploring 3D Structures of Biological Macromolecules for Basic and Applied Research and Education in Fundamental Biology, Biomedicine, Biotechnology, Bioengineering and Energy Sciences. Nucleic Acids Res. 2021, 49, D437–D451. [Google Scholar] [CrossRef] [PubMed]
- Rose, P.W.; Prlić, A.; Altunkaya, A.; Bi, C.; Bradley, A.R.; Christie, C.H.; Di Costanzo, L.; Duarte, J.M.; Dutta, S.; Feng, Z.; et al. The RCSB Protein Data Bank: Integrative View of Protein, Gene and 3D Structural Information. Nucleic Acids Res. 2017, 45, D271–D281. [Google Scholar] [CrossRef] [PubMed]
- Yip, K.M.; Fischer, N.; Paknia, E.; Chari, A.; Stark, H. Atomic-Resolution Protein Structure Determination by Cryo-EM. Nature 2020, 587, 157–161. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, J.; Suzek, T.O.; Zhang, J.; Wang, J.; Bryant, S.H. PubChem: A Public Information System for Analyzing Bioactivities of Small Molecules. Nucleic Acids Res. 2009, 37, W623–W633. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 Update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 Update: Improved Access to Chemical Data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef]
- Malecka, K.A.; Fera, D.; Schultz, D.C.; Hodawadekar, S.; Reichman, M.; Donover, P.S.; Murphy, M.E.; Marmorstein, R. Identification and Characterization of Small Molecule Human Papillomavirus E6 Inhibitors. ACS Chem. Biol. 2014, 9, 1603–1612. [Google Scholar] [CrossRef]
- Messick, T.E.; Smith, G.R.; Soldan, S.S.; McDonnell, M.E.; Deakyne, J.S.; Malecka, K.A.; Tolvinski, L.; van den Heuvel, A.P.J.; Gu, B.-W.; Cassel, J.A.; et al. Structure-Based Design of Small-Molecule Inhibitors of EBNA1 DNA Binding Blocks Epstein-Barr Virus Latent Infection and Tumor Growth. Sci. Transl. Med. 2019, 11, aau5612. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Sterling, T.; Irwin, J.J. ZINC 15—Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Shoichet, B.K. ZINC—A Free Database of Commercially Available Compounds for Virtual Screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Kwofie, S.; Broni, E.; Yunus, F.; Nsoh, J.; Adoboe, D.; Miller, W.; Wilson, M. Molecular Docking Simulation Studies Identifies Potential Natural Product Derived-Antiwolbachial Compounds as Filaricides against Onchocerciasis. Biomedicines 2021, 9, 1682. [Google Scholar] [CrossRef] [PubMed]
- Sander, T.; Freyss, J.; Von Korff, M.; Rufener, C. DataWarrior: An Open-Source Program for Chemistry Aware Data Visualization and Analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Rappe, A.K.; Casewit, C.J.; Colwell, K.S.; Goddard, W.A.; Skiff, W.M. UFF, a Full Periodic Table Force Field for Molecular Mechanics and Molecular Dynamics Simulations. J. Am. Chem. Soc. 1992, 114, 10024–10035. [Google Scholar] [CrossRef]
- Artemova, S.; Jaillet, L.; Redon, S. Automatic Molecular Structure Perception for the Universal Force Field. J. Comput. Chem. 2016, 37, 1191–1205. [Google Scholar] [CrossRef] [PubMed]
- Kondapuram, S.K.; Sarvagalla, S.; Coumar, M.S. Docking-Based Virtual Screening Using PyRx Tool: Autophagy Target Vps34 as a Case Study. In Molecular Docking for Computer-Aided Drug Design Fundamentals, Techniques, Resources and Applications; Academic Press: Cambridge, MA, USA, 2021; pp. 463–477. [Google Scholar] [CrossRef]
- Dallakyan, S.; Arthur, J. Olson Small Molecule Library Screening by Docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Fu, A.; Zhang, L. Progress in Molecular Docking. Quant. Biol. 2019, 7, 83–89. [Google Scholar] [CrossRef]
- Pinzi, L.; Rastelli, G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, H.; Yu, K.; Jin, Z. Molecular Docking-Based Computational Platform for High-Throughput Virtual Screening. CCF Trans. High Perform. Comput. 2022, 4, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, EfficientOptimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Barret, R. Lipinski’s Rule of Five. In Therapeutical Chemistry; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Plinski, E.F.; Plinska, S. Veber’s Rules in Terahertz Light. Res. Sq. 2020, preprint. [Google Scholar] [CrossRef]
- Lee, E.K.; Kim, S.Y.; Noh, K.-W.; Joo, E.H.; Zhao, B.; Kieff, E.; Kang, M.-S. Small Molecule Inhibition of Epstein–Barr Virus Nuclear Antigen-1 DNA Binding Activity Interferes with Replication and Persistence of the Viral Genome. Antivir. Res. 2014, 104, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Thompson, S.; Schultz, D.C.; Zhu, W.; Jiang, H.; Luo, C.; Lieberman, P.M. Discovery of Selective Inhibitors against Ebna1 via High Throughput in Silico Virtual Screening. PLoS ONE 2010, 5, e10126. [Google Scholar] [CrossRef] [PubMed]
- Messick, T.E.; Tolvinski, L.; Zartler, E.R.; Moberg, A.; Frostell, Å.; Smith, G.R.; Reitz, A.B.; Lieberman, P.M. Biophysical Screens Identify Fragments That Bind to the Viral DNA-Binding Proteins EBNA1 and LANA. Molecules 2020, 25, 1760. [Google Scholar] [CrossRef] [PubMed]
- Jakhmola, S.; Jonniya, N.A.; Sk, M.F.; Rani, A.; Kar, P.; Jha, H.C. Identification of Potential Inhibitors against Epstein–Barr Virus Nuclear Antigen 1 (EBNA1): An Insight from Docking and Molecular Dynamic Simulations. ACS Chem. Neurosci. 2021, 12, 3060–3072. [Google Scholar] [CrossRef] [PubMed]
- Guedes, I.A.; Mü Ller Pereira Da Silva, M.; Galheigo, M.; Krempser, E.; Silva De Magalhã Es, C.; Correa Barbosa, H.J.; Dardenne, L.E. DockThor-VS: A Free Platform for Receptor-Ligand Virtual Screening. J. Mol. Biol. 2024, in press. [Google Scholar] [CrossRef]
- Guedes, I.A.; Costa, L.S.C.; dos Santos, K.B.; Karl, A.L.M.; Rocha, G.K.; Teixeira, I.M.; Galheigo, M.M.; Medeiros, V.; Krempser, E.; Custódio, F.L.; et al. Drug Design and Repurposing with DockThor-VS Web Server Focusing on SARS-CoV-2 Therapeutic Targets and Their Non-Synonym Variants. Sci. Rep. 2021, 11, 5543. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Filimonov, D.A.; Lagunin, A.A.; Gloriozova, T.A.; Rudik, A.V.; Druzhilovskii, D.S.; Pogodin, P.V.; Poroikov, V.V. Prediction of the Biological Activity Spectra of Organic Compounds Using the Pass Online Web Resource. Chem. Heterocycl. Compd. 2014, 50, 444–457. [Google Scholar] [CrossRef]
- Parasuraman, S. Prediction of Activity Spectra for Substances. J. Pharmacol. Pharmacother. 2011, 2, 52–53. [Google Scholar] [CrossRef] [PubMed]
- Lagunin, A.; Stepanchikova, A.; Filimonov, D.; Poroikov, V. PASS: Prediction of Activity Spectra for Biologically Active Substances. Bioinformatics 2000, 16, 747–748. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Dodda, L.S.; Cabeza de Vaca, I.; Tirado-Rives, J.; Jorgensen, W.L. LigParGen Web Server: An Automatic OPLS-AA Parameter Generator for Organic Ligands. Nucleic Acids Res. 2017, 45, W331–W336. [Google Scholar] [CrossRef] [PubMed]
- Dodda, L.S.; Vilseck, J.Z.; Tirado-Rives, J.; Jorgensen, W.L. 1.14*CM1A-LBCC: Localized Bond-Charge Corrected CM1A Charges for Condensed-Phase Simulations. J. Phys. Chem. B 2017, 121, 3864–3870. [Google Scholar] [CrossRef]
- DeLano, W.L. PyMOL: An Open-Source Molecular Graphics Tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Kumar, R.; Lynn, A. G_mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Cheng, D.; Shrivastava, S.; Tzur, D.; Gautam, B.; Hassanali, M. DrugBank: A Knowledgebase for Drugs, Drug Actions and Drug Targets. Nucleic Acids Res. 2008, 36, D901–D906. [Google Scholar] [CrossRef] [PubMed]
- Caballero Alfonso, A.Y.; Chayawan, C.; Gadaleta, D.; Roncaglioni, A.; Benfenati, E. A KNIME Workflow to Assist the Analogue Identification for Read-Across, Applied to Aromatase Activity. Molecules 2023, 28, 1832. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.-C.; Senese, S.; Li, C.-M.; Hu, Q.; Huang, Y.; Damoiseaux, R.; Torres, J.Z. Large-Scale Chemical Similarity Networks for Target Profiling of Compounds Identified in Cell-Based Chemical Screens. PLoS Comput. Biol. 2015, 11, e1004153. [Google Scholar] [CrossRef] [PubMed]
- Nabati, F.; Moradi, M.; Mohabatkar, H. In Silico Analyzing the Molecular Interactions of Plant-Derived Inhibitors against E6AP, P53, and c-Myc Binding Sites of HPV Type 16 E6 Oncoprotein. Mol. Biol. Res. Commun. 2020, 9, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Zanier, K.; Stutz, C.; Kintscher, S.; Reinz, E.; Sehr, P.; Bulkescher, J.; Hoppe-Seyler, K.; Travé, G.; Hoppe-Seyler, F. The E6AP Binding Pocket of the HPV16 E6 Oncoprotein Provides a Docking Site for a Small Inhibitory Peptide Unrelated to E6AP, Indicating Druggability of E6. PLoS ONE 2014, 9, e112514. [Google Scholar] [CrossRef] [PubMed]
- Dhamodharan, P.; Ponnusamy, N.; Odumpatta, R.; Lulu, S.; Arumugam, M. Computational Investigation of Marine Bioactive Compounds against E6 Oncoprotein of Human Papilloma Virus-HPV16. J. Appl. Pharm. Sci. 2018, 8, 023–032. [Google Scholar] [CrossRef][Green Version]
- Kumar, S.; Jena, L.; Sahoo, M.; Kakde, M.; Daf, S.; Varma, A.K. In Silico Docking to Explicate Interface between Plant-Originated Inhibitors and E6 Oncogenic Protein of Highly Threatening Human Papillomavirus 18. Genom. Inform. 2015, 13, 60. [Google Scholar] [CrossRef] [PubMed]
- Fazeli, M.; Sarvazad, H.; Rahnejat, N.; Rostampour, R.; Rad, M.G.; Eskandari-Roozbahani, N. Investigation of Potential Antiviral Natural Products with an Effect on HPV18 E6 Protein by Molecular Docking Method. Funct. Foods Health Dis. 2021, 11, 586. [Google Scholar] [CrossRef]
- Atabaki, V.; Pourahmad, J.; Hosseinabadi, T. Phytochemical Compounds from Jurinea Macrocephala Subsp.Elbursensis and Their Cytotoxicity Evaluation. S. Afr. J. Bot. 2021, 137, 399–405. [Google Scholar] [CrossRef]
- Chang, M.W.; Lindstrom, W.; Olson, A.J.; Belew, R.K. Analysis of HIV Wild-Type and Mutant Structures via in Silico Docking against Diverse Ligand Libraries. J. Chem. Inf. Model. 2007, 47, 1258–1262. [Google Scholar] [CrossRef]
- Indari, O.; Kumar Singh, A.; Tiwari, D.; Chandra Jha, H.; Nath Jha, A. Deciphering Antiviral Efficacy of Malaria Box Compounds against Malaria Exacerbating Viral Pathogens- Epstein Barr Virus and SARS-CoV-2, an in Silico Study. Med. Drug Discov. 2022, 16, 100146. [Google Scholar] [CrossRef] [PubMed]
- Kolluru, S.; Momoh, R.; Lin, L.; Mallareddy, J.R.; Krstenansky, J.L. Identification of Potential Binding Pocket on Viral Oncoprotein HPV16 E6: A Promising Anti-Cancer Target for Small Molecule Drug Discovery. BMC Mol. Cell Biol. 2019, 20, 30. [Google Scholar] [CrossRef] [PubMed]
- Jamkhande, P.G.; Pathan, S.K.; Wadher, S.J. In Silico PASS Analysis and Determination of Antimycobacterial, Antifungal, and Antioxidant Efficacies of Maslinic Acid in an Extract Rich in Pentacyclic Triterpenoids. Int. J. Mycobacteriol. 2016, 5, 417–425. [Google Scholar] [CrossRef]
- Stiasny, A.; Freier, C.P.; Kuhn, C.; Schulze, S.; Mayr, D.; Alexiou, C.; Janko, C.; Wiest, I.; Dannecker, C.; Jeschke, U.; et al. The Involvement of E6, P53, P16, MDM2 and Gal-3 Inthe Clinical Outcome of Patients with Cervical Cancer. Oncol. Lett. 2017, 14, 4467–4476. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thandra, K.C.; Barsouk, A.; Saginala, K.; Padala, S.A.; Barsouk, A.; Rawla, P. Epidemiology of Non-Hodgkin’s Lymphoma. Med. Sci. 2021, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Robert, A.; Pujals, A.; Favre, L.; Debernardi, J.; Wiels, J. The BCL-2 Family Protein Inhibitor ABT-737 as an Additional Tool for the Treatment of EBV-Associated Post-Transplant Lymphoproliferative Disorders. Mol. Oncol. 2020, 14, 2520–2532. [Google Scholar] [CrossRef] [PubMed]
- Chabay, P. Advances in the Pathogenesis of EBV-Associated Diffuse Large B Cell Lymphoma. Cancers 2021, 13, 2717. [Google Scholar] [CrossRef] [PubMed]
- Hongyu, L.; Hongling, S.; Qian, X.; Dongrui, D.; Shixuan, W.; Yunping, L.; Ding, M. Expression of Pin1 and Ki67 in Cervical Cancer and Their Significance. J. Huazhong Univ. Sci. Technol. Med. Sci. 2006, 26, 120–122. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, S.; Zhu, T.; Zhou, J.; Xu, Q.; Lu, Y.; Ma, D. Pin1 Contributes to Cervical Tumorigenesis by Regulating Cyclin D1 Expression. Oncol. Rep. 2006, 16, 491–496. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, T.; Liu, Z.; Shi, F.; Wang, J. Pin1 Modulates Chemo-Resistance by up-Regulating FoxM1 and the Involvements of Wnt/β-Catenin Signaling Pathway in Cervical Cancer. Mol. Cell. Biochem. 2016, 413, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Chen, J.; Zhang, T.; Liu, N. LPCAT1 Functions as an Oncogene in Cervical Cancer through Mediating JAK2/STAT3 Signaling. Exp. Cell Res. 2022, 421, 113360. [Google Scholar] [CrossRef] [PubMed]
- Morgan, E.L.; Macdonald, A. JAK2 Inhibition Impairs Proliferation and Sensitises Cervical Cancer Cells to Cisplatin-Induced Cell Death. Cancers 2019, 11, 1934. [Google Scholar] [CrossRef] [PubMed]
- Arellano, A.R.; Lopez-Pulido, E.I.; Martínez-Neri, P.A.; Chávez, C.E.; Lucano, R.G.; Fafutis-Morris, M.; Aguilar-Lemarroy, A.; Muñoz-Valle, J.F.; Pereira-Suárez, A.L. STAT3 Activation Is Required for the Antiapoptotic Effects of Prolactin in Cervical Cancer Cells. Cancer Cell Int. 2015, 15, 83. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Chen, L.; Shao, J.; Jiang, C.; Zhao, Y.; Li, Y.; Ke, H.; Zhang, R.; Zhu, J.; Yu, M. Lanatoside C Inhibits Human Cervical Cancer Cell Proliferation and Induces Cell Apoptosis by a Reduction of the JAK2/STAT6/SOCS2 Signaling Pathway. Oncol. Lett. 2021, 22, 740. [Google Scholar] [CrossRef] [PubMed]
- Senba, M.; Buziba, N.; Mori, N.; Fujita, S.; Morimoto, K.; Wada, A.; Toriyama, K. Human Papillomavirus Infection Induces NF-ΚB Activation in Cervical Cancer: A Comparison with Penile Cancer. Oncol. Lett. 2011, 2, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Akbarpour Arsanjani, A.; Abuei, H.; Behzad-Behbahani, A.; Bagheri, Z.; Arabsolghar, R.; Farhadi, A. Activating Transcription Factor 3 Inhibits NF-κB P65 Signaling Pathway and Mediates Apoptosis and Cell Cycle Arrest in Cervical Cancer Cells. Infect. Agent Cancer 2022, 17, 62. [Google Scholar] [CrossRef] [PubMed]
- Nees, M.; Geoghegan, J.M.; Hyman, T.; Frank, S.; Miller, L.; Woodworth, C.D. Papillomavirus Type 16 Oncogenes Downregulate Expression of Interferon-Responsive Genes and Upregulate Proliferation-Associated and NF-ΚB-Responsive Genes in Cervical Keratinocytes. J. Virol. 2001, 75, 4283–4296. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, Y.; Sui, L.; Wang, J.; Li, F. Phenyllactic Acid Promotes Cell Migration and Invasion in Cervical Cancer via IKK/NF-ΚB-Mediated MMP-9 Activation. Cancer Cell Int. 2019, 19, 241. [Google Scholar] [CrossRef] [PubMed]
- James, M.A.; Lee, J.H.; Klingelhutz, A.J. Human Papillomavirus Type 16 E6 Activates NF-ΚB, Induces CIAP-2 Expression, and Protects against Apoptosis in a PDZ Binding Motif-Dependent Manner. J. Virol. 2006, 80, 5301–5307. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Huang, L.; Gong, J.; Shi, C.; Wang, Z.; Ye, B.; Xuan, A.; He, X.; Long, D.; Zhu, X.; et al. NF-ΚB Pathway Link with ER Stress-Induced Autophagy and Apoptosis in Cervical Tumor Cells. Cell Death Discov. 2017, 3, 17059. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xu, L.; Li, E.; Dong, G. Application of Molecular Dynamics Simulation in Biomedicine. Chem. Biol. Drug Des. 2022, 99, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Allu, A.S.; Tiriveedhi, V. Cancer Salt Nostalgia. Cells 2021, 10, 1285. [Google Scholar] [CrossRef] [PubMed]
- Kuzmanic, A.; Zagrovic, B. Determination of Ensemble-Average Pairwise Root Mean-Square Deviation from Experimental B-Factors. Biophys. J. 2010, 98, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Kufareva, I.; Abagyan, R. Methods of Protein Structure Comparison. In Methods in Molecular Biology (Clifton, N.J.); NIH Public Access: Bethesda, MD, USA, 2011; Volume 857, pp. 231–257. [Google Scholar]
- Koliński, M.; Dec, R.; Dzwolak, W. Multiscale Modeling of Amyloid Fibrils Formed by Aggregating Peptides Derived from the Amyloidogenic Fragment of the A-Chain of Insulin. Int. J. Mol. Sci. 2021, 22, 12325. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, M.Y.; Bogatyreva, N.S.; Galzitskaya, O.V. Radius of Gyration as an Indicator of Protein Structure Compactness. Mol. Biol. 2008, 42, 623–628. [Google Scholar] [CrossRef]
- Shityakov, S.; Skorb, E.V.; Nosonovsky, M. Topological Bio-Scaling Analysis as a Universal Measure of Protein Folding. R. Soc. Open Sci. 2022, 9, 220160. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shi, D.; Zhou, S.; Liu, H.; Liu, H.; Yao, X. Molecular Dynamics Simulations and Novel Drug Discovery. Expert Opin. Drug Discov. 2018, 13, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Kwofie, S.; Dankwa, B.; Enninful, K.; Adobor, C.; Broni, E.; Ntiamoah, A.; Wilson, M. Molecular Docking and Dynamics Simulation Studies Predict Munc18b as a Target of Mycolactone: A Plausible Mechanism for Granule Exocytosis Impairment in Buruli Ulcer Pathogenesis. Toxins 2019, 11, 181. [Google Scholar] [CrossRef] [PubMed]
- Parvaiz, N.; Ahmad, F.; Yu, W.; MacKerell, A.D.; Azam, S.S. Discovery of Beta-Lactamase CMY-10 Inhibitors for Combination Therapy against Multi-Drug Resistant Enterobacteriaceae. PLoS ONE 2021, 16, e0244967. [Google Scholar] [CrossRef] [PubMed]
- Nasser, M.; Yusof, U.K.; Salim, N. Deep Learning Based Methods for Molecular Similarity Searching: A Systematic Review. Processes 2023, 11, 1340. [Google Scholar] [CrossRef]
- Safizadeh, H.; Simpkins, S.W.; Nelson, J.; Li, S.C.; Piotrowski, J.S.; Yoshimura, M.; Yashiroda, Y.; Hirano, H.; Osada, H.; Yoshida, M.; et al. Improving Measures of Chemical Structural Similarity Using Machine Learning on Chemical–Genetic Interactions. J. Chem. Inf. Model. 2021, 61, 4156–4172. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.-C.C.; Wang, S.S.; Wu, Y.; Chen, R.-Y.Y.; Yu, D.-Q.Q. Five New Diprenylated Flavonols from the Leaves of Broussonetia Kazinoki. J. Nat. Prod. 2001, 64, 1206–1209. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.N.; Ramakrishnaiah, H.; Krishna, V.; Radhika, M. Cytotoxic Activity of Broussonetia Papyrifera(l.) Vent on MCF-7, HeLa and HepG2 Cell Lines. Int. J. Pharm. Pharm. Sci. 2014, 6, 339–342. [Google Scholar]
- Dou, C.-Z.; Liu, Y.-F.; Zhang, L.-L.; Chen, S.-H.; Hu, C.-Y.; Liu, Y.; Zhao, Y.-T. Polyphenols from Broussonetia Papyrifera Induce Apoptosis of HepG2 Cells via Inactivation of ERK and AKT Signaling Pathways. Evid.-Based Complement. Altern. Med. 2021, 2021, 8841706. [Google Scholar] [CrossRef] [PubMed]
- Hassanshahi, J.; Mirzahosseini-pourranjbar, A.; Hajializadeh, Z.; Kaeidi, A. Anticancer and Cytotoxic Effects of Troxerutin on HeLa Cell Line: An in-Vitro Model of Cervical Cancer. Mol. Biol. Rep. 2020, 47, 6135–6142. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.Y.; Ding, D.N.; Wang, Y.R.; Liu, S.X.; Peng, C.; Shen, F.; Zhu, X.Y.; Li, C.; Tang, L.P.; Han, F.J. Icariin as a Potential Anticancer Agent: A Review of Its Biological Effects on Various Cancers. Front. Pharmacol. 2023, 14, 1216363. [Google Scholar] [CrossRef]
- Song, L.; Chen, X.; Mi, L.; Liu, C.; Zhu, S.; Yang, T.; Luo, X.; Zhang, Q.; Lu, H.; Liang, X. Icariin-induced Inhibition of SIRT6/NF-κB Triggers Redox Mediated Apoptosis and Enhances Anti-tumor Immunity in Triple-negative Breast Cancer. Cancer Sci. 2020, 111, 4242–4256. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.-L.; Chan, K.-G.; Pusparajah, P.; Saokaew, S.; Duangjai, A.; Lee, L.-H.; Goh, B.-H. Anti-Cancer Properties of the Naturally Occurring Aphrodisiacs: Icariin and Its Derivatives. Front. Pharmacol. 2016, 7, 191. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Liu, H.; Long, M.; Song, L.; Meng, Z.; Lin, S.; Zhang, Y.; Qin, J. Icariin Attenuates the Tumor Growth by Targeting MiR-1-3p/TNKS2/Wnt/β-Catenin Signaling Axis in Ovarian Cancer. Front. Oncol. 2022, 12, 940926. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jiang, K.; Zhao, F. Icariin Regulates the Proliferation and Apoptosis of Human Ovarian Cancer Cells through MicroRNA-21 by Targeting PTEN, RECK and Bcl-2. Oncol. Rep. 2015, 33, 2829–2836. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Xie, T.; Liu, W. Icariin Inhibits the Growth of Human Cervical Cancer Cells by Inducing Apoptosis and Autophagy by Targeting MTOR/PI3K/AKT Signalling Pathway. JBUON 2019, 24, 990–996. [Google Scholar]
- Li, C.; Yang, S.; Ma, H.; Ruan, M.; Fang, L.; Cheng, J. Influence of Icariin on Inflammation, Apoptosis, Invasion, and Tumor Immunity in Cervical Cancer by Reducing the TLR4/MyD88/NF-ΚB and Wnt/β-Catenin Pathways. Cancer Cell Int. 2021, 21, 206. [Google Scholar] [CrossRef] [PubMed]
- Duan, P.; Cheng, J.; Mao, R.; Wang, R.; Jin, Y.; Li, C. Icariin-Mediated MiR-875-5p Inhibits Autophagy and Epithelial-Mesenchymal Transition by Regulation of MDM4 in Cervical Cancer. J. Biomed. Nanotechnol. 2022, 18, 2708–2720. [Google Scholar] [CrossRef]
- Nakashima, K.; Oyama, M.; Ito, T.; Witono, J.R.; Darnaedi, D.; Tanaka, T.; Murata, J.; Iinuma, M. Melicodenines A and B, Novel Diels–Alder Type Adducts Isolated from Melicope Denhamii. Tetrahedron Lett. 2011, 52, 4694–4696. [Google Scholar] [CrossRef]
- Kamperdick, C.; Van, N.H.; Van Sung, T.; Adam, G. Bisquinolinone Alkaloids from Melicope Ptelefolia. Phytochemistry 1999, 50, 177–181. [Google Scholar] [CrossRef]
- Nakashima, K.; Oyama, M.; Ito, T.; Akao, Y.; Witono, J.R.; Darnaedi, D.; Tanaka, T.; Murata, J.; Iinuma, M. Novel Quinolinone Alkaloids Bearing a Lignoid Moiety and Related Constituents in the Leaves of Melicope Denhamii. Tetrahedron 2012, 68, 2421–2428. [Google Scholar] [CrossRef]
- Chelariu-Raicu, A.; Mahner, S.; Moore, K.N.; Lorusso, D.; Coleman, R.L. Integrating Antibody Drug Conjugates in the Management of Gynecologic Cancers. Int. J. Gynecol. Cancer 2023, 33, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Graybill, W.S.; Coleman, R.L. Vintafolide: A Novel Targeted Agent for Epithelial Ovarian Cancer. Futur. Oncol. 2014, 10, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Vergote, I.; Leamon, C.P. Vintafolide: A Novel Targeted Therapy for the Treatment of Folate Receptor Expressing Tumors. Ther. Adv. Med. Oncol. 2015, 7, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Ledermann, J.A.; Canevari, S.; Thigpen, T. Targeting the Folate Receptor: Diagnostic and Therapeutic Approaches to Personalize Cancer Treatments. Ann. Oncol. 2015, 26, 2034–2043. [Google Scholar] [CrossRef]
- Assaraf, Y.G.; Leamon, C.P.; Reddy, J.A. The Folate Receptor as a Rational Therapeutic Target for Personalized Cancer Treatment. Drug Resist. Updat. 2014, 17, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Piccirillo, M.C.; Daniele, G.; Di Maio, M.; Bryce, J.; De Feo, G.; Del Giudice, A.; Perrone, F.; Morabito, A. Vinorelbine for Non-Small Cell Lung Cancer. Expert Opin. Drug Saf. 2010, 9, 493–510. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Kenmotsu, H.; Yamanaka, T.; Nakamura, S.; Tsuboi, M. Randomized Phase III Study of Cisplatin With Pemetrexed and Cisplatin With Vinorelbine for Completely Resected Nonsquamous Non–Small-Cell Lung Cancer: The JIPANG Study Protocol. Clin. Lung Cancer 2018, 19, e1–e3. [Google Scholar] [CrossRef] [PubMed]
- Nobili, S.; Lavacchi, D.; Perrone, G.; Vicini, G.; Tassi, R.; Landini, I.; Grosso, A.; Roviello, G.; Mazzanti, R.; Santomaggio, C.; et al. Vinorelbine in Non-Small Cell Lung Cancer: Real-World Data From a Single-Institution Experience. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2020, 28, 237–248. [Google Scholar]
- Douillard, J.-Y.; Tribodet, H.; Aubert, D.; Shepherd, F.A.; Rosell, R.; Ding, K.; Veillard, A.-S.; Seymour, L.; Le Chevalier, T.; Spiro, S.; et al. Adjuvant Cisplatin and Vinorelbine for Completely Resected Non-Small Cell Lung Cancer: Subgroup Analysis of the Lung Adjuvant Cisplatin Evaluation. J. Thorac. Oncol. 2010, 5, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Coronel, J.A.; Cetina, L.D.C.; Cantú, D.; Cerezo, O.; Hernández, C.S.; Rivera, L.; Chacón, A.P.; Duenas-Gonzalez, A. A Randomized Comparison of Cisplatin and Oral Vinorelbine as Radiosensitizers in Aged or Comorbid Locally Advanced Cervical Cancer Patients. Int. J. Gynecol. Cancer 2013, 23, 884–889. [Google Scholar] [CrossRef] [PubMed]
- Lacava, J.A.; Leone, B.A.; Machiavelli, M.; Romero, A.O.; Perez, J.E.; Elem, Y.L.; Ferreyra, R.; Focaccia, G.; Suttora, G.; Salvadori, M.A.; et al. Vinorelbine as Neoadjuvant Chemotherapy in Advanced Cervical Carcinoma. J. Clin. Oncol. 1997, 15, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Endo, T.; Goodbody, A.; Vukovic, J.; Misawa, M. Biotransformation of Anhydrovinblastine to Vinblastine by a Cell-Free Extract of Catharanthus Roseus Cell Suspension Cultures. Phytochemistry 1987, 26, 3233–3234. [Google Scholar] [CrossRef]
- McLauchlan, W.R.; Hasan, M.; Baxter, R.L.; Ian Scott, A. Conversion of Anhydrovinblastine to Vinblastine by Cell-Free Homogenates of Catharanthus Roseus Cell Suspension Cultures. Tetrahedron 1983, 39, 3777–3780. [Google Scholar] [CrossRef]
- Dhyani, P.; Quispe, C.; Sharma, E.; Bahukhandi, A.; Sati, P.; Attri, D.C.; Szopa, A.; Sharifi-Rad, J.; Docea, A.O.; Mardare, I.; et al. Anticancer Potential of Alkaloids: A Key Emphasis to Colchicine, Vinblastine, Vincristine, Vindesine, Vinorelbine and Vincamine. Cancer Cell Int. 2022, 22, 206. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Velasco, A.; Flores-Tafoya, P.D.J.; Fragoso-Serrano, M.; Leitão, S.G.; Pereda-Miranda, R. Resin Glycosides from Operculina Hamiltonii and Their Synergism with Vinblastine in Cancer Cells. J. Nat. Prod. 2022, 85, 2385–2394. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-K.; Shao, Y.; Ding, H.; Hu, L.-H. Synthesis and Structure−Activity Relationship Studies of Cytotoxic Ester and Ether Anhydrovinblastine Derivatives. J. Nat. Prod. 2008, 71, 1669–1676. [Google Scholar] [CrossRef] [PubMed]
- Kongsema, M.; Wongkhieo, S.; Khongkow, M.; Lam, E.W.F.; Boonnoy, P.; Vongsangnak, W.; Wong-Ekkabut, J. Molecular Mechanism of Forkhead Box M1 Inhibition by Thiostrepton in Breast Cancer Cells. Oncol. Rep. 2019, 42, 953–962. [Google Scholar] [CrossRef] [PubMed]
- Hegde, N.S.; Sanders, D.A.; Rodriguez, R.; Balasubramanian, S. The Transcription Factor FOXM1 Is a Cellular Target of the Natural Product Thiostrepton. Nat. Chem. 2011, 3, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, P.; Chen, L.; Chen, H. Down-Regulation of FoxM1 by Thiostrepton or Small Interfering RNA Inhibits Proliferation, Transformation Ability and Angiogenesis, and Induces Apoptosis of Nasopharyngeal Carcinoma Cells. Int. J. Clin. Exp. Pathol. 2014, 7, 5450–5460. [Google Scholar] [PubMed]
- Jiang, L.; Wu, X.; Wang, P.; Wen, T.; Yu, C.; Wei, L.; Chen, H. Targeting FoxM1 by Thiostrepton Inhibits Growth and Induces Apoptosis of Laryngeal Squamous Cell Carcinoma. J. Cancer Res. Clin. Oncol. 2015, 141, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Cheenpracha, S.; Karalai, C.; Rat-a-pa, Y.; Ponglimanont, C.; Chantrapromma, K. New Cytotoxic Cardenolide Glycoside from the Seeds of Cerbera manghas. Chem. Pharm. Bull. 2004, 52, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Laphookhieo, S.; Cheenpracha, S.; Karalai, C.; Chantrapromma, S.; Ratapa, Y.; Ponglimanont, C.; Chantrapromma, K. Cytotoxic Cardenolide Glycoside from the Seeds of Cerbera odollam. Phytochemistry 2004, 65, 507–510. [Google Scholar] [CrossRef]
- Karaś, K.; Sałkowska, A.; Dastych, J.; Bachorz, R.A.; Ratajewski, M. Cardiac Glycosides with Target at Direct and Indirect Interactions with Nuclear Receptors. Biomed. Pharmacother. 2020, 127, 110106. [Google Scholar] [CrossRef] [PubMed]
- Stowell, J.D. Novel Ingredients for Weight Loss: New Developments. In Novel Food Ingredients for Weight Control; Elsevier: Amsterdam, The Netherlands, 2007; pp. 218–233. ISBN 9781845690304. [Google Scholar]
- Cai, J.; Zhang, B.-D.; Li, Y.-Q.; Zhu, W.-F.; Akihisa, T.; Kikuchi, T.; Xu, J.; Liu, W.-Y.; Feng, F.; Zhang, J. Cardiac Glycosides from the Roots of Streblus Asper Lour. with Activity against Epstein-Barr Virus Lytic Replication. Bioorg. Chem. 2022, 127, 106004. [Google Scholar] [CrossRef] [PubMed]
- Calderón-Montaño, J.M.; Burgos-Morón, E.; Orta, M.L.; Maldonado-Navas, D.; García-Domínguez, I.; López-Lázaro, M. Evaluating the Cancer Therapeutic Potential of Cardiac Glycosides. Biomed Res. Int. 2014, 2014, 794930. [Google Scholar] [CrossRef] [PubMed]
- Ayogu, J.I.; Odoh, A.S. Prospects and Therapeutic Applications of Cardiac Glycosides in Cancer Remediation. ACS Comb. Sci. 2020, 22, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Kumavath, R.; Paul, S.; Pavithran, H.; Paul, M.K.; Ghosh, P.; Barh, D.; Azevedo, V. Emergence of Cardiac Glycosides as Potential Drugs: Current and Future Scope for Cancer Therapeutics. Biomolecules 2021, 11, 1275. [Google Scholar] [CrossRef] [PubMed]
- Schneider, N.; Cerella, C.; Simões, C.M.O.; Diederich, M. Anticancer and Immunogenic Properties of Cardiac Glycosides. Molecules 2017, 22, 1932. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Wang, F.; Tian, D.; Huang, L.; Streator, E.; Zhu, J.; Kurihara, H.; He, R.; Yao, X.; Zhang, Y.; et al. Cardiac Glycosides Inhibit Cancer through Na/K-ATPase-Dependent Cell Death Induction. Biochem. Pharmacol. 2020, 182, 114226. [Google Scholar] [CrossRef] [PubMed]
- Gan, H.; Qi, M.; Chan, C.; Leung, P.; Ye, G.; Lei, Y.; Liu, A.; Xue, F.; Liu, D.; Ye, W.; et al. Digitoxin Inhibits HeLa Cell Growth through the Induction of G2/M Cell Cycle Arrest and Apoptosis In Vitro and In Vivo. Int. J. Oncol. 2020, 57, 562–573. [Google Scholar] [CrossRef] [PubMed]
- Pereira, D.G.; Rendeiro, M.M.; Cortes, V.F.; Barbosa, L.A.; Quintas, L.E.M. Antagonistic Anticancer Effect of Paclitaxel and Digoxin Combination. J. Cell. Biochem. 2019, 120, 13107–13114. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).