Analysing the Performance and Interpretability of CNN-Based Architectures for Plant Nutrient Deficiency Identification
Abstract
1. Introduction
- Compared to previous studies, it presents an integrated approach that considers both the performance and explainability of CNN architectures when used for plant nutrient deficiency identification.
- As a first attempt, it focuses on comparing the explainability of two prominent XAI techniques, GRAD-CAM, and Shapley Additive exPlanations (SHAP) when used for plant nutrient deficiency identification.
2. Background and Related Work
2.1. Plant Nutrient Deficiency
2.2. Convolutional Neural Network
2.3. VGG16 Architecture
2.4. Inception-V3 Architecture
2.5. Explainable Artificial Intelligence
2.5.1. Shapley Additive exPlanations (SHAP)
- Local accuracy: at a minimum, the explanation model must reproduce the results of the original model [42]
- Consistency: Regardless of other features, the significance of a feature should not decrease if we change a model so that it depends more on that feature [42].
2.5.2. Gradient-Weighted Class Activation Mapping (Grad-CAM)
2.6. Related Work
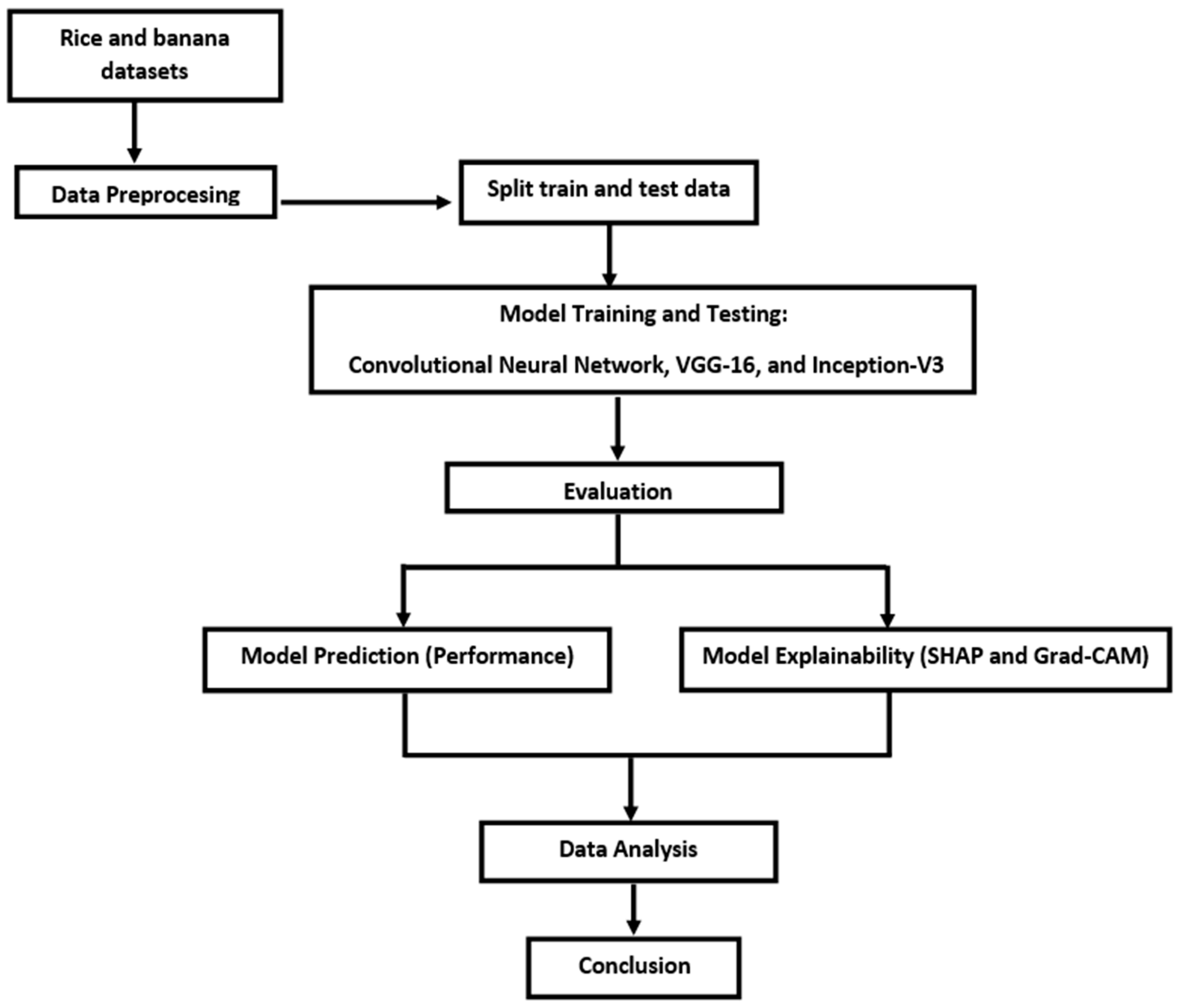
3. Methods
3.1. Data Acquisition
3.2. Image Pre-Processing
3.3. Data Training
Model Architectures
3.4. Evaluation Metrics
4. Results
4.1. Confusion Matrix
4.1.1. Rice Dataset
4.1.2. Banana Dataset
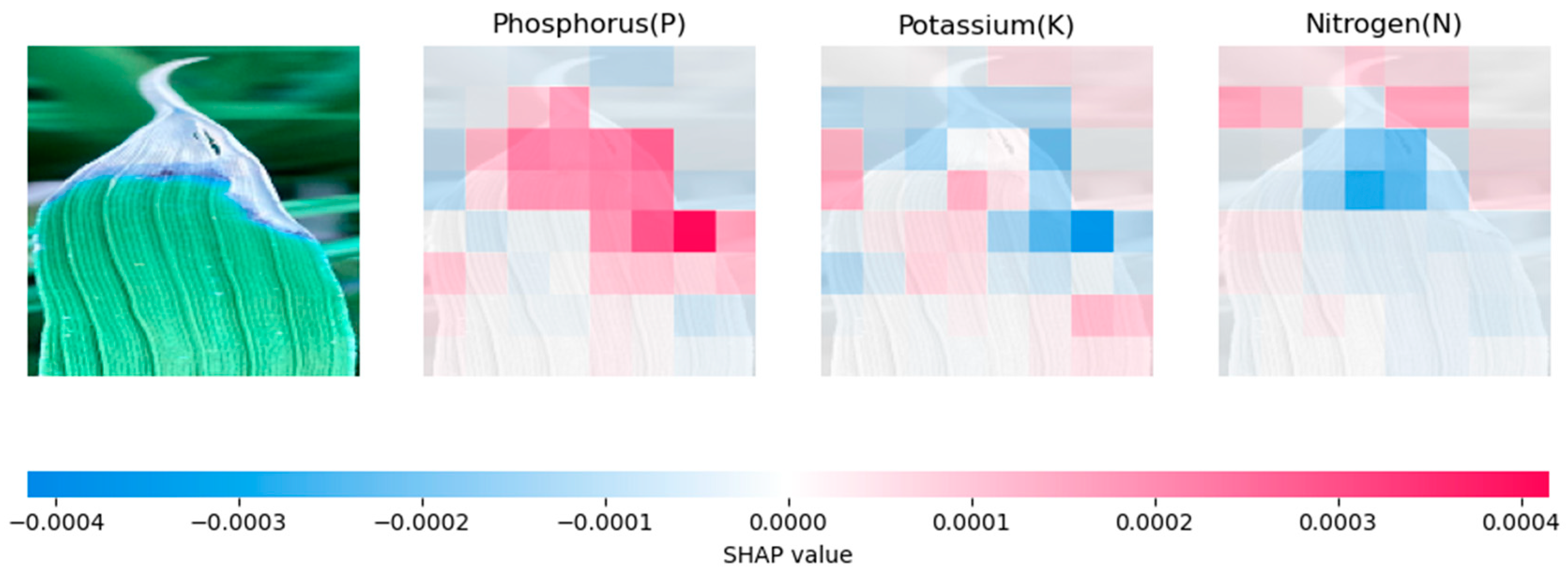
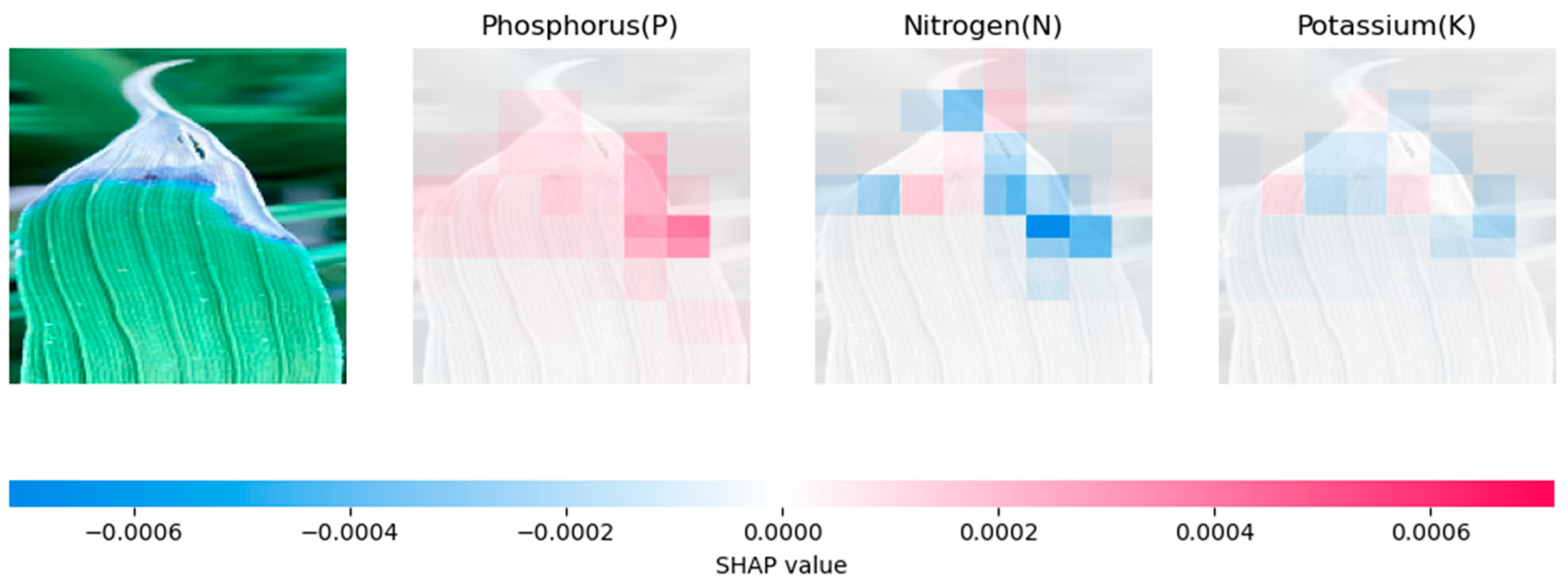
4.2. Model Explanation Using SHAP
4.2.1. Analysing Explainability of ML Models Using SHAP-Rice Dataset
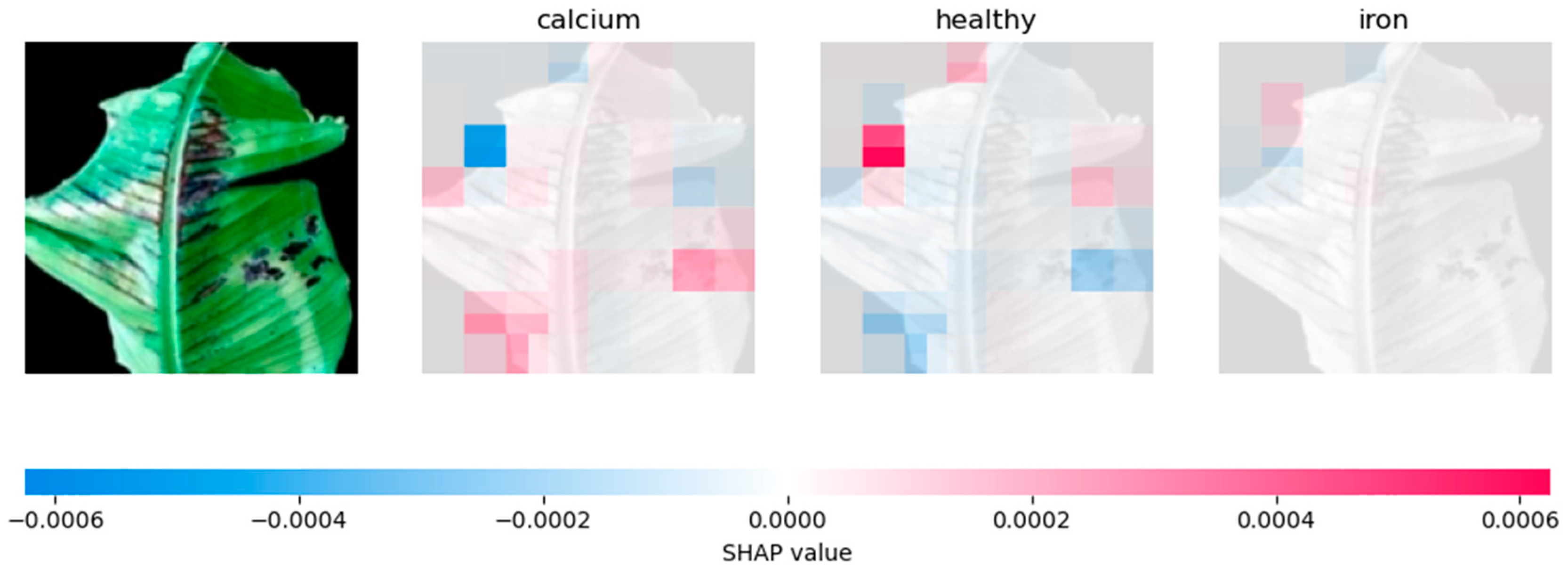
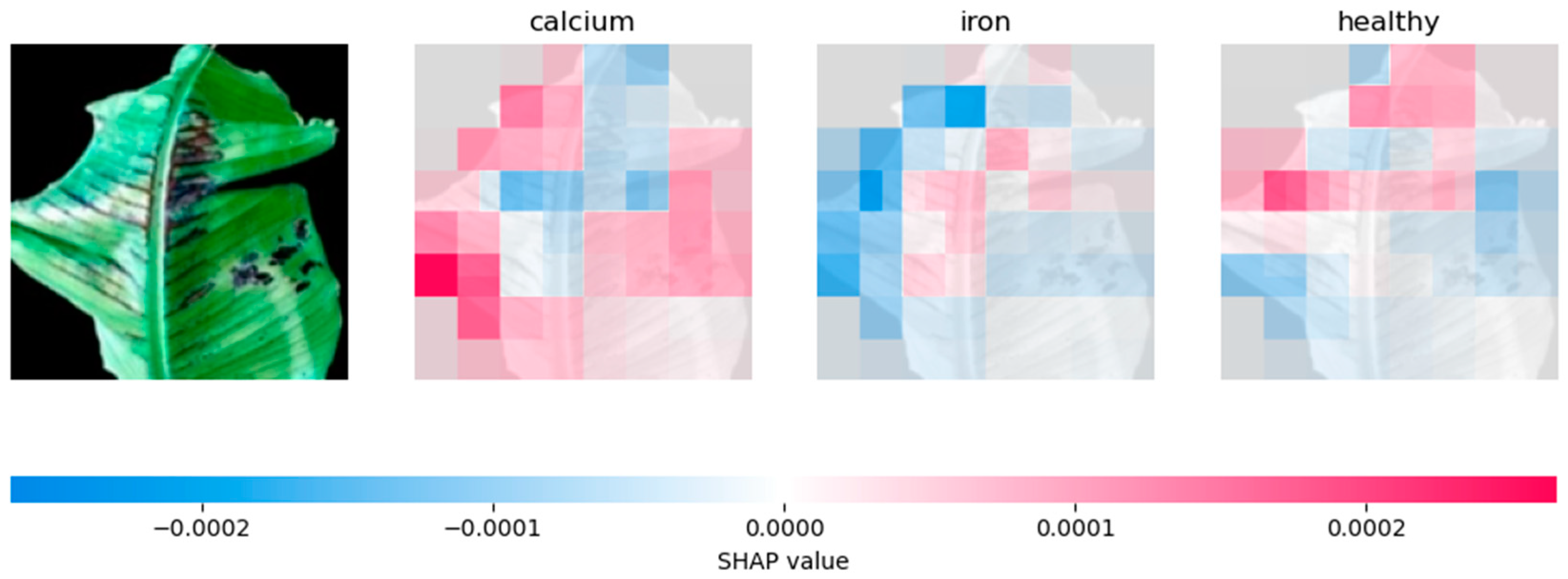
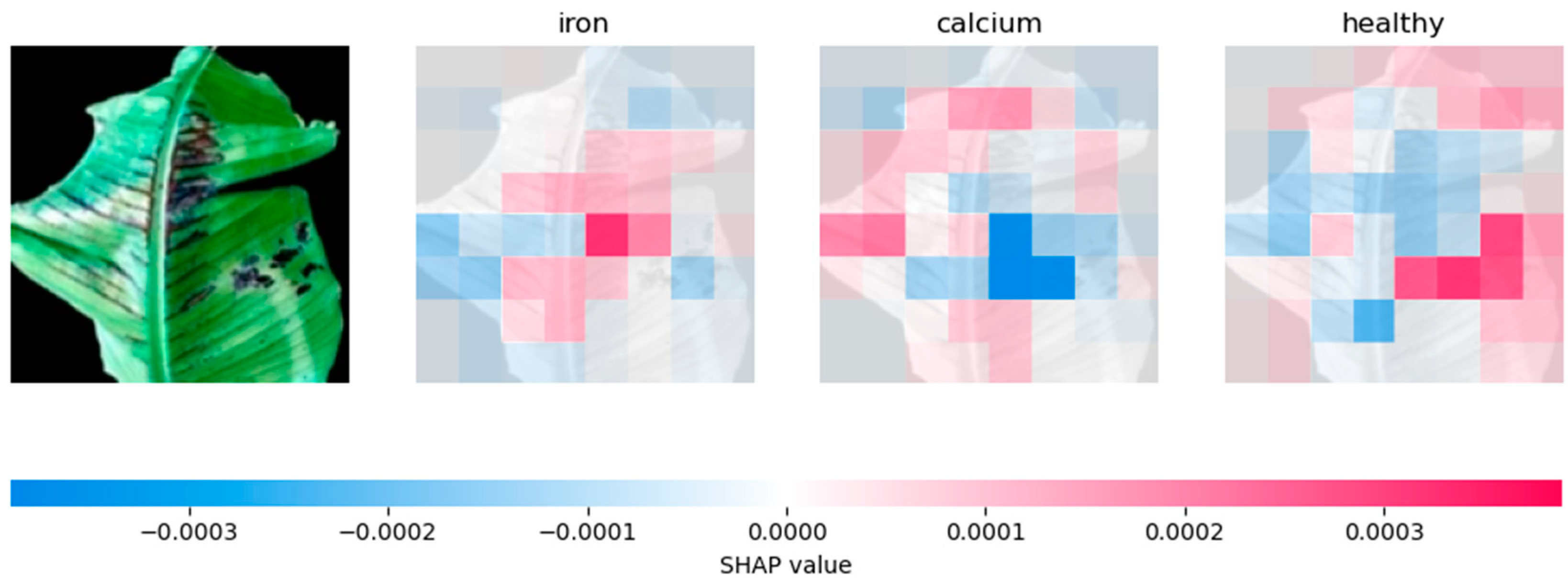
4.2.2. Analysing Explainability of ML Models Using SHAP-Banana Dataset
4.3. Model Explanation Using Grad-CAM
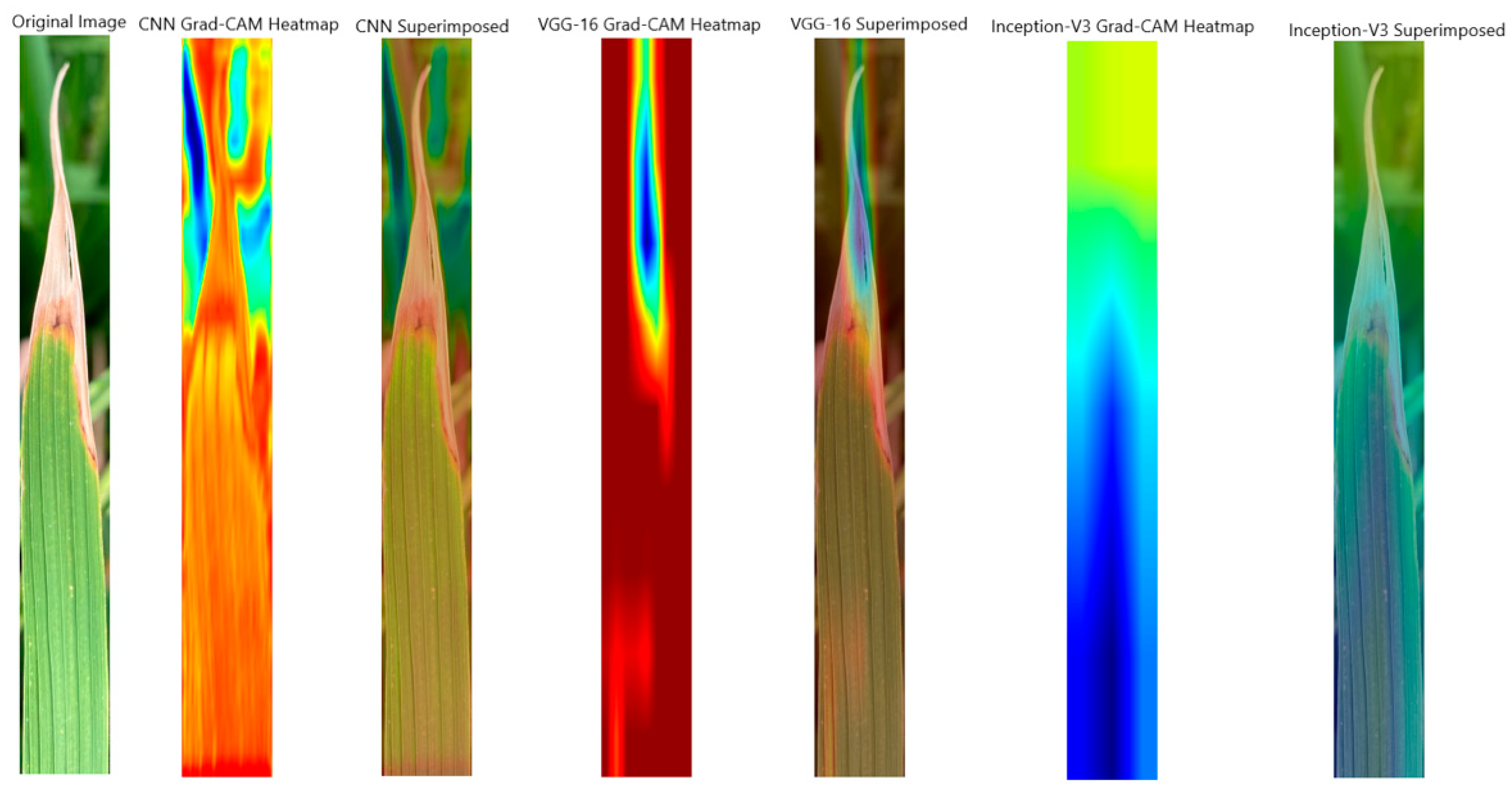
4.3.1. Analysing Explainability of ML Models Using Grad-CAM-Rice Dataset
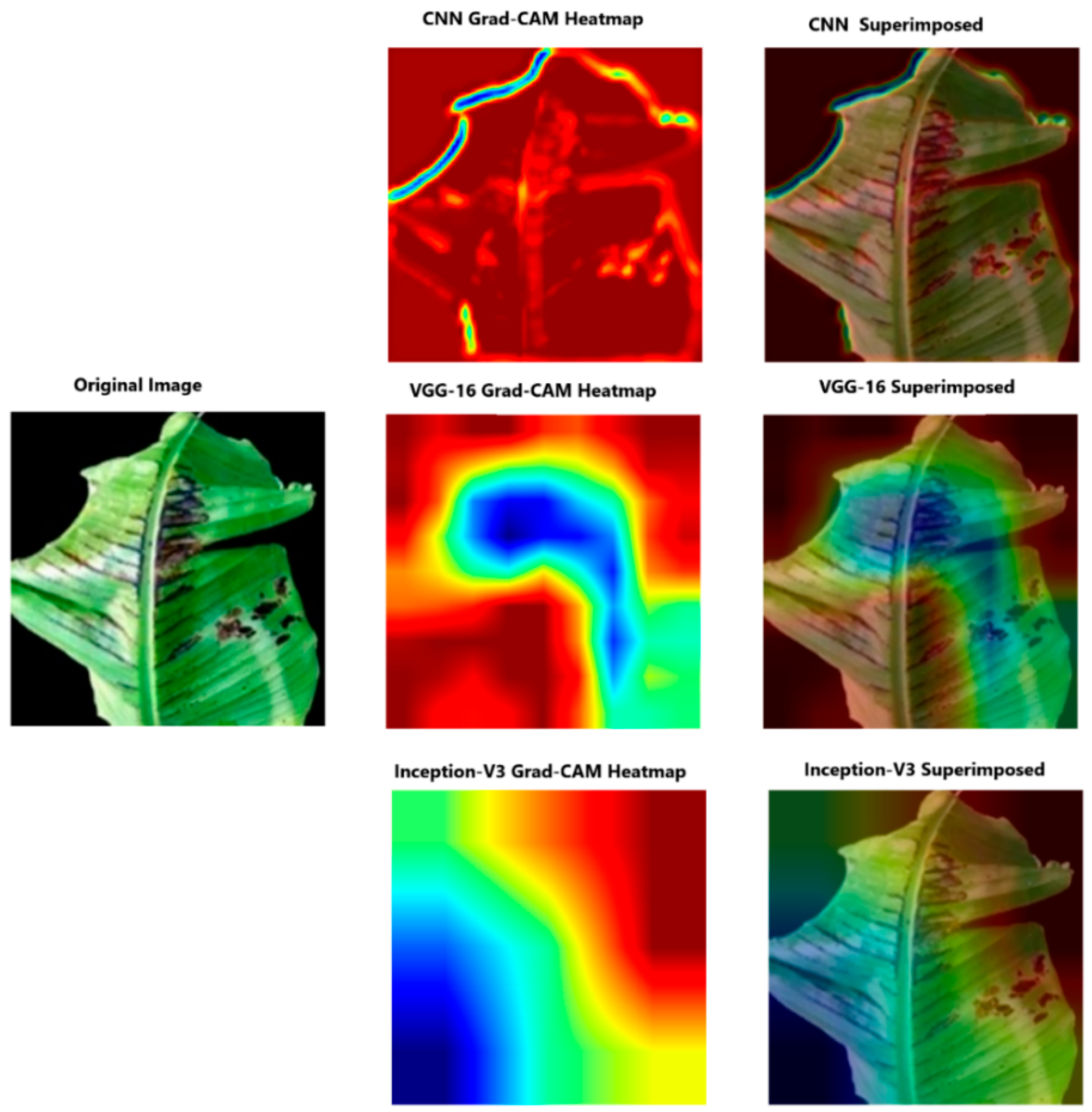
4.3.2. Analysing Explainability of ML Models Using Grad-CAM-Banana Dataset
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, X.; He, P.; Yang, F.; Ma, J.; Pampolino, M.F.; Johnston, A.M.; Zhou, W. Methodology of Fertilizer Recommendation Based on Yield Response and Agronomic Efficiency for Rice in China. Field Crops Res. 2017, 206, 33–42. [Google Scholar] [CrossRef]
- Ghosh, P.; Mondal, A.K.; Chatterjee, S.; Masud, M.; Meshref, H.; Bairagi, A.K. Recognition of Sunflower Diseases Using Hybrid Deep Learning and Its Explainability with AI. Mathematics 2023, 11, 2241. [Google Scholar] [CrossRef]
- Islam, S.; Reza, M.N.; Samsuzzaman, S.; Ahmed, S.; Cho, Y.J.; Noh, D.H.; Chung, S.-O.; Hong, S.J. Machine Vision and Artificial Intelligence for Plant Growth Stress Detection and Monitoring: A Review. Precis. Agric. Sci. Technol. 2024, 6, 33–57. [Google Scholar] [CrossRef]
- Liang, C.; Tian, J.; Liao, H. Proteomics Dissection of Plant Responses to Mineral Nutrient Deficiency. Proteomics 2013, 13, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Talukder MS, H.; Sarkar, A.K. Nutrients deficiency diagnosis of rice crop by weighted average ensemble learning. Smart Agric. Technol. 2023, 4, 100155. [Google Scholar] [CrossRef]
- Govindasamy, P.; Muthusamy, S.K.; Bagavathiannan, M.; Mowrer, J.; Jagannadham, P.T.; Maity, A.; Halli, H.M.; GK, S.; Vadivel, R.; TK, D.; et al. Nitrogen Use Efficiency—A Key to Enhance Crop Productivity under a Changing Climate. Front. Plant Sci. 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Siddique, A.B.; Shabala, S.; Zhou, M.; Zhao, C. Phosphorus Plays Key Roles in Regulating Plants’ Physiological Responses to Abiotic Stresses. Plants 2023, 12, 2861. [Google Scholar] [CrossRef] [PubMed]
- Kusanur, V.; Chakravarthi, V.S. Using Transfer Learning for Nutrient Deficiency Prediction and Classification in Tomato Plant. Int. J. Adv. Comput. Sci. Appl. 2021, 12, 784–790. [Google Scholar] [CrossRef]
- Yi, J.; Krusenbaum, L.; Unger, P.; Hüging, H.; Seidel, S.J.; Schaaf, G.; Gall, J. Deep Learning for Non-Invasive Diagnosis of Nutrient Deficiencies in Sugar Beet Using RGB Images. Sensors 2020, 20, 5893. [Google Scholar] [CrossRef]
- Kattenborn, T.; Leitloff, J.; Schiefer, F.; Hinz, S. Review on Convolutional Neural Networks (CNN) in Vegetation Remote Sensing. ISPRS J. Photogramm. Remote Sens. 2021, 173, 24–49. [Google Scholar] [CrossRef]
- Wang, C.; Li, C.; Han, Q.; Wu, F.; Zou, X. A Performance Analysis of a Litchi Picking Robot System for Actively Removing Obstructions, Using an Artificial Intelligence Algorithm. Agronomy 2023, 13, 2795. [Google Scholar] [CrossRef]
- Mazumder, M.K.A.; Mridha, M.F.; Alfarhood, S.; Safran, M.; Abdullah-Al-Jubair, M.; Che, D. A Robust and Light-Weight Transfer Learning-Based Architecture for Accurate Detection of Leaf Diseases across Multiple Plants Using Less Amount of Images. Front. Plant Sci. 2023, 14, 1321877. [Google Scholar] [CrossRef] [PubMed]
- Albattah, W.; Javed, A.; Nawaz, M.; Masood, M.; Albahli, S. Artificial Intelligence-Based Drone System for Multiclass Plant Disease Detection Using an Improved Efficient Convolutional Neural Network. Front. Plant Sci. 2022, 13, 808380. [Google Scholar] [CrossRef] [PubMed]
- Jafar, A.; Bibi, N.; Naqvi, R.A.; Sadeghi-Niaraki, A.; Jeong, D. Revolutionizing Agriculture with Artificial Intelligence: Plant Disease Detection Methods, Applications, and Their Limitations. Front. Plant Sci. 2024, 15, 1356260. [Google Scholar] [CrossRef] [PubMed]
- Taji, K.; Ghanimi, F. Enhancing Plant Disease Classification through Manual CNN Hyperparameter Tuning. Data Metadata 2023, 2, 112. [Google Scholar] [CrossRef]
- Dey, B.; Masum Ul Haque, M.; Khatun, R.; Ahmed, R. Comparative Performance of Four CNN-Based Deep Learning Variants in Detecting Hispa Pest, Two Fungal Diseases, and NPK Deficiency Symptoms of Rice (Oryza Sativa). Comput. Electron. Agric. 2022, 202, 107340. [Google Scholar] [CrossRef]
- Henna, S.; Alcaraz, J.M.L. From Interpretable Filters to Predictions of Convolutional Neural Networks with Explainable Artificial Intelligence. arXiv 2022, arXiv:2207.12958. [Google Scholar]
- Mccauley, A. Plant Nutrient Functions and Deficiency and Toxicity Symptoms. Nutr. Manag. Modul. 2009, 9, 1–16. [Google Scholar]
- Aleksandrov, V. Identification of Nutrient Deficiency in Plants by Artificial Intelligence. Acta Physiol. Plant 2022, 44, 29. [Google Scholar] [CrossRef]
- Sinha, D.; Tandon, P.K. An Overview of Nitrogen, Phosphorus and Potassium: Key Players of Nutrition Process in Plants. In Sustainable Solutions for Elemental Deficiency and Excess in Crop Plants; Springer: Singapore, 2020; pp. 85–117. ISBN 9789811586361. [Google Scholar]
- Beura, K.; Kohli, A.; Kumar, A.; Anupam, P.; Rajiv, D.; Shweta, R.; Nintu, S.; Mahendra, M.; Yanendra, S.; Singh, K.; et al. Souvenir, National Seminar on “Recent Developments in Nutrient Management Strategies for Sustainable Agriculture: The Indian Context; Bihar Agricultural University: Bhagalpur, India, 2022; ISBN 9789394490932. [Google Scholar]
- Andrianto, H.; Suhardi; Faizal, A.; Budi Kurniawan, N.; Praja Purwa Aji, D. Performance Evaluation of IoT-Based Service System for Monitoring Nutritional Deficiencies in Plants. Inf. Process. Agric. 2023, 10, 52–70. [Google Scholar] [CrossRef]
- Mattila, T.J.; Rajala, J. Do Different Agronomic Soil Tests Identify Similar Nutrient Deficiencies? Soil. Use Manag. 2022, 38, 635–648. [Google Scholar] [CrossRef]
- Mohd Adib, N.A.N.; Daliman, S. Conceptual Framework of Smart Fertilization Management for Oil Palm Tree Based on IOT and Deep Learning. In Proceedings of the IOP Conference Series: Earth and Environmental Science, Kelantan, Malaysia, 14–15 July 2021; IOP Publishing Ltd.: Bristol, UK, 2021; Volume 842. [Google Scholar]
- Nogueira De Sousa, R.; Moreira, L.A. Plant Nutrition Optimization: Integrated Soil Management and Fertilization Practices; TechOpen: Rijeka, Croatia, 2024. [Google Scholar]
- Hanif Hashimi, M.; Abad, Q.; Gul Shafiqi, S. A Review of Diagnostic Techniques of Visual Symptoms of Nutrients Deficiencies in Plant. Int. J. Agric. Res. 2023, 6, 1–9. [Google Scholar]
- Kamelia, L.; Rahman, T.K.B.A.; Saragih, H.; Haerani, R. The Comprehensive Review on Detection of Macro Nutrients Deficiency in Plants Based on the Image Processing Technique. In Proceedings of the 2020 6th International Conference on Wireless and Telematics, ICWT 2020, Yogyakarta, Indonesia, 3–4 September 2020; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2020. [Google Scholar]
- Ghosh, A.; Sufian, A.; Sultana, F.; Chakrabarti, A.; De, D. Fundamental Concepts of Convolutional Neural Network. In Recent Trends and Advances in Artificial Intelligence and Internet of Things; Springer: Cham, Switzerland, 2019; Volume 172, ISBN 9783030326449. [Google Scholar]
- Wang, H. On the Origin of Deep Learning. arXiv 2017, arXiv:1702.07800. [Google Scholar]
- Kamilaris, A.; Prenafeta-Boldú, F.X. A Review of the Use of Convolutional Neural Networks in Agriculture. J. Agric. Sci. 2018, 156, 312–322. [Google Scholar] [CrossRef]
- Canziani, A.; Paszke, A.; Culurciello, E. An Analysis of Deep Neural Network Models for Practical Applications. arXiv 2016, arXiv:1605.07678. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015; pp. 1–14. [Google Scholar]
- Khan, A.; Anabia, S.; Umme, Z.; Aqsa, S.Q. A Survey of the Recent Architectures of Deep Convolutional Neural Networks. Artif. Intell. Rev. 2020, 53, 5455–5516. [Google Scholar] [CrossRef]
- Leonardo, M.M.; Carvalho, T.J.; Rezende, E.; Zucchi, R.; Faria, F.A. Deep Feature-Based Classifiers for Fruit Fly Identification (Diptera: Tephritidae). In Proceedings of the 31st Conference on Graphics, Patterns and Images, SIBGRAPI 2018, Paraná, Brazil, 29 October–1 November 2018; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2019; pp. 41–47. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of Deep Learning: Concepts, CNN Architectures, Challenges, Applications, Future Directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Barredo, A.A.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; Garcia, S.; Gil-Lopez, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, Taxonomies, Opportunities and Challenges toward Responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef]
- Gerlings, J.; Shollo, A.; Constantiou, L. Reviewing the Need for Explainable Artificial Intelligence (XAI). arXiv 2021, arXiv:2012.01007. [Google Scholar] [CrossRef]
- Linardatos, P.; Papastefanopoulos, V.; Kotsiantis, S. Explainable Ai: A Review of Machine Learning Interpretability Methods. Entropy 2021, 23, 18. [Google Scholar] [CrossRef]
- Singh, H.; Roy, A.; Setia, R.K.; Pateriya, B. Estimation of Nitrogen Content in Wheat from Proximal Hyperspectral Data Using Machine Learning and Explainable Artificial Intelligence (XAI) Approach. Model. Earth Syst. Env. 2021, 8, 2505–2511. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.I. A Unified Approach to Interpreting Model Predictions. Adv. Neural Inf. Process Syst. 2017, 30, 4766–4775. [Google Scholar]
- Antwarg, L.; Miller, R.M.; Shapira, B.; Rokach, L. Explaining Anomalies Detected by Autoencoders Using SHAP. arXiv 2020, arXiv:1903.02407. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017. [Google Scholar]
- Panesar, A. Improving Visual Question Answering by Leveraging Depth and Adapting Explainability. In Proceedings of the 2022 31st IEEE International Conference on Robot and Human Interactive Communication (RO-MAN), Naples, Italy, 29 August–2 September 2022. [Google Scholar]
- Xu, Z.; Guo, X.; Zhu, A.; He, X.; Zhao, X.; Han, Y.; Subedi, R. Using Deep Convolutional Neural Networks for Image-Based Diagnosis of Nutrient Deficiencies in Rice. Comput. Intell. Neurosci. 2020, 2020, 7307252. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.T.; Choi, J.W.; Le, T.T.H.; Kim, J.W. A Comparative Study of Deep CNN in Forecasting and Classifying the Macronutrient Deficiencies on Development of Tomato Plant. Appl. Sci. 2019, 9, 1601. [Google Scholar] [CrossRef]
- Ibrahim, S.; Hasan, N.; Sabri, N.; Abu Samah, K.A.F.; Rahimi Rusland, M. Palm Leaf Nutrient Deficiency Detection Using Convolutional Neural Network (CNN). Int. J. Nonlinear Anal. Appl. 2022, 13, 1949–1956. [Google Scholar]
- Weeraphat, R. Nutrient Deficiency Symptoms in Rice. Available online: https://www.kaggle.com/datasets/guy007/nutrientdeficiencysymptomsinrice (accessed on 4 April 2023).
- Sunitha, P. Images of Nutrient Deficient Banana Plant Leaves, V1; Mendeley: London, UK, 2022. [Google Scholar] [CrossRef]
- Rakesh, S.; Indiramma, M. Explainable AI for Crop Disease Detection. In Proceedings of the 2022 4th International Conference on Advances in Computing, Communication Control and Networking (ICAC3N), Greater Noida, India, 16–17 December 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1601–1608. [Google Scholar]
- Mostafa, S.; Mondal, D.; Panjvani, K.; Kochian, L.; Stavness, I. Explainable Deep Learning in Plant Phenotyping. Front. Artif. Intell. 2023, 6, 1203546. [Google Scholar] [CrossRef]

| Title | Nutrient(s)/ Disease(s) | Plant Type | The Algorithm and Classifier Used | Findings (Accuracy) | XAI Technique |
|---|---|---|---|---|---|
| Ibrahim et al. [47] | Nitrogen, Potassium, Magnesium, Boron, Zinc and Manganese | Palm leaves | CNN | CNN: 94.2% | None |
| Talukder et al. [5] | Nitrogen, Phosphorus, and Potassium | Rice | InceptionV3, InceptionResNetV2, DenseNet121, DenseNet201, & DenseNet169 | DenseNet169: 96.6% | None |
| Xu et al. [45] | Nitrogen, manganese, calcium, magnesium, potassium, phosphorus, zinc, iron, and Sulfur | Rice | DCNN: Dense Net, ResNet, Inception-v3, and NasNet-large | DenseNet121: 97.44% | None |
| Singh et al. [40] | Nitrogen | Wheat | Six regression models (i.e., Random Forest) | Random Forest: R2 = 0.89 | SHAP |
| Our study | Nitrogen, Phosphorus, and Potassium | Rice, Banana | CNN, VGG-16, Inception-V3 | Inception-V3: Rice = 93% Banana = 92% | SHAP, Grad-CAM |
| Parameter | Settings |
|---|---|
| Cross-validation | k-fold (k = 10) |
| Epochs | 30 |
| Batch Size | 32 |
| Early Stopping | Patience = 10 |
| Initial learning rate | 0.001 |
| Optimiser | Adam |
| Loss | Categorical Cross-entropy |
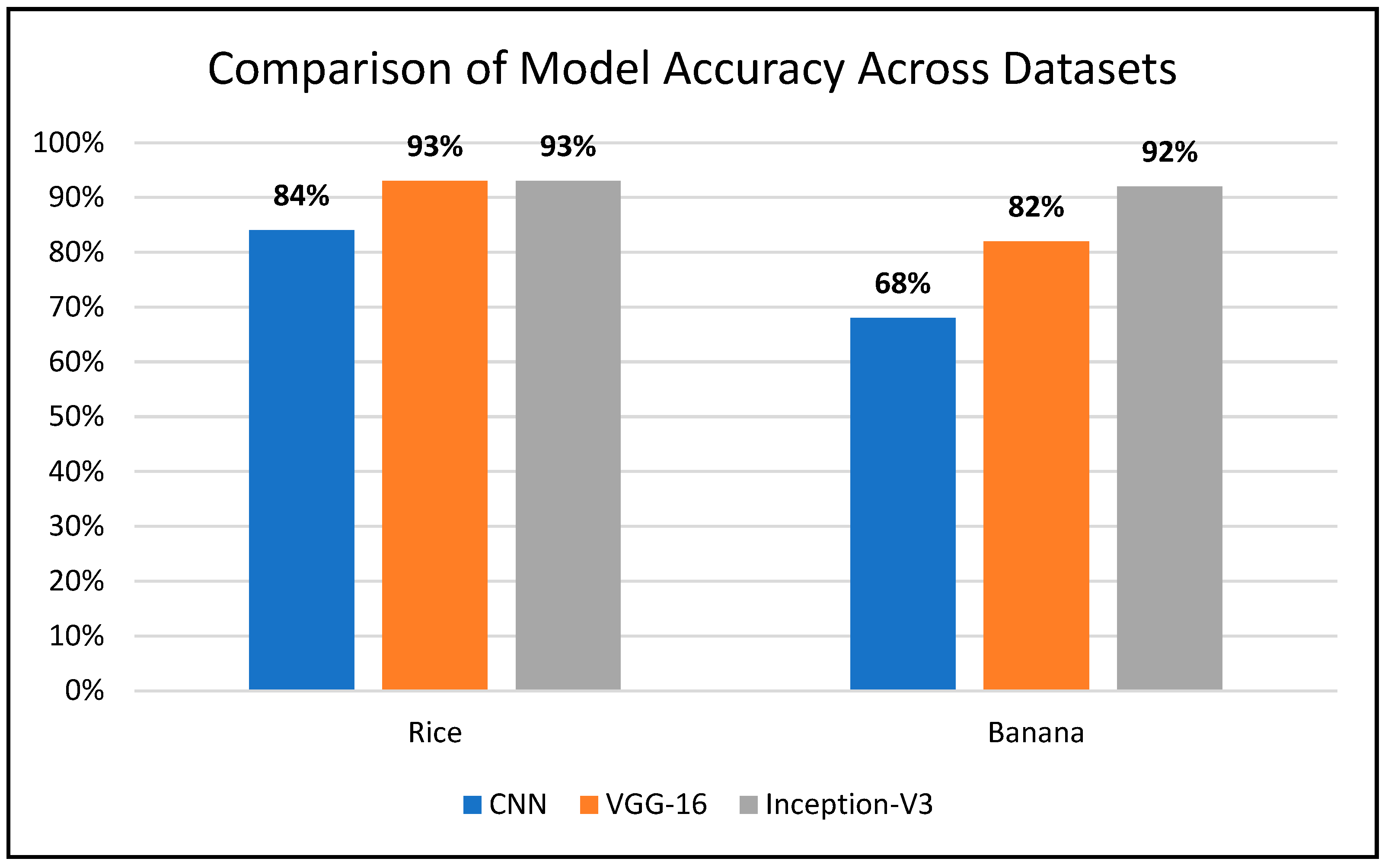
| Dataset | Classifiers | Accuracy | Precision | Recall | F1-Score | AUC |
|---|---|---|---|---|---|---|
| Rice | CNN | 84% | 84% | 84% | 84% | 93% |
| VGG-16 | 93% | 93% | 93% | 93% | 98% | |
| Inception-V3 | 93% | 93% | 93% | 93% | 98% | |
| Banana | CNN | 68% | 68% | 68% | 68% | 85% |
| VGG-16 | 82% | 81% | 82% | 81% | 92% | |
| Inception-V3 | 92% | 92% | 92% | 92% | 97% |
| True Labels | Nitrogen | Phosphorus | Potassium | |
|---|---|---|---|---|
| CNN | Nitrogen | 75 | 6 | 2 |
| Phosphorus | 7 | 59 | 4 | |
| Potassium | 6 | 13 | 60 | |
| VGG-16 | Nitrogen | 84 | 0 | 2 |
| Phosphorus | 2 | 57 | 5 | |
| Potassium | 4 | 4 | 74 | |
| Inception-V3 | Nitrogen | 94 | 3 | 1 |
| Phosphorus | 5 | 56 | 2 | |
| Potassium | 1 | 4 | 66 |
| True Labels | Calcium | Healthy | Iron | |
|---|---|---|---|---|
| CNN | Calcium | 109 | 50 | 28 |
| healthy | 48 | 132 | 14 | |
| iron | 20 | 8 | 129 | |
| VGG-16 | Calcium | 126 | 28 | 20 |
| healthy | 22 | 156 | 8 | |
| iron | 12 | 9 | 157 | |
| Inception-V3 | Calcium | 154 | 16 | 13 |
| healthy | 5 | 182 | 3 | |
| iron | 5 | 0 | 160 |
| Models | Rice Dataset | Banana Dataset |
|---|---|---|
| CNN | Relies heavily on prominent features, especially for phosphorus prediction | Influential regions on the sides of the leaf. |
| Inception-V3 | Relies on a broader set of features | Relies on broader context clues, resulting in a different pattern of SHAP values |
| VGG-16 | Similar to CNN, but with more evenly distributed feature contributions | Distributes SHAP values across the whole leaf. |
| Models | Rice Dataset | Banana Dataset |
|---|---|---|
| CNN | Grad-Cam heatmap focuses on the tip of the leaf. | Highlights banana leaf contours. |
| Inception-V3 | Highlights various broader areas. | Lacks precise overlap with the object of interest. |
| VGG-16 | Localises the defected region. | Focuses on the leaf itself. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mkhatshwa, J.; Kavu, T.; Daramola, O. Analysing the Performance and Interpretability of CNN-Based Architectures for Plant Nutrient Deficiency Identification. Computation 2024, 12, 113. https://doi.org/10.3390/computation12060113
Mkhatshwa J, Kavu T, Daramola O. Analysing the Performance and Interpretability of CNN-Based Architectures for Plant Nutrient Deficiency Identification. Computation. 2024; 12(6):113. https://doi.org/10.3390/computation12060113
Chicago/Turabian StyleMkhatshwa, Junior, Tatenda Kavu, and Olawande Daramola. 2024. "Analysing the Performance and Interpretability of CNN-Based Architectures for Plant Nutrient Deficiency Identification" Computation 12, no. 6: 113. https://doi.org/10.3390/computation12060113
APA StyleMkhatshwa, J., Kavu, T., & Daramola, O. (2024). Analysing the Performance and Interpretability of CNN-Based Architectures for Plant Nutrient Deficiency Identification. Computation, 12(6), 113. https://doi.org/10.3390/computation12060113







