Physiological State Recognition via HRV and Fractal Analysis Using AI and Unsupervised Clustering
Abstract
1. Introduction
- Absence of labeled data in real-life scenarios. In practical applications such as wearable devices and IoT-based systems, there are usually no predefined labels indicating the user’s physiological state (e.g., rest, stress, or exertion). This makes unsupervised learning a highly suitable choice for automatic classification.
- Individual variability and transitional states. Supervised models often lack the flexibility to account for inter-individual physiological differences and struggle to recognize new or intermediate states not represented in the training data. In contrast, unsupervised methods can identify natural groupings and dynamic transitions without the need for subjective categorization.
- Limitations of supervised models. While supervised techniques rely on clearly defined classes and high-quality annotations, these often require expert judgment or controlled laboratory environments. This restricts their applicability in autonomous, real-world health monitoring systems.
- Potential for anomaly detection. Clustering methods such as DBSCAN can detect outliers and transitional observations—capabilities that supervised learning typically lacks. This opens the door for early warning systems and adaptive feedback in personalized preventive healthcare.
- What latent structures can be uncovered in HRV and fractal metrics through dimensionality reduction techniques?
- Can unsupervised learning algorithms (e.g., K-Means, DBSCAN) differentiate physiological states without prior labeling?
- To what extent do AI-based visualizations support the detection of anomalies and natural groupings in HRV data?
2. Materials and Methods
2.1. Participants and Protocol
- V1—placed on the right chest, over the area of the right pectoral muscle;
- V—positioned centrally beneath the neck;
- V5—located below the left chest, near the left pectoral region;
- V3—placed horizontally aligned with V5, at the mid-chest level;
- N—attached to the right side of the abdomen, adjacent to the umbilicus.
2.2. Signal Preprocessing and Noise Removal
2.3. Determination and Extraction of HRV and Fractal Parameters
2.4. Feature Space Analysis Using PCA
2.4.1. Dimensionality Reduction via Principal Component Analysis
2.4.2. Principal Component Extraction
- Covariance matrix computation:
- Eigen decomposition:
- 3.
- Projection of data onto the first k eigenvectors:
2.4.3. Explained Variance and Component Interpretation
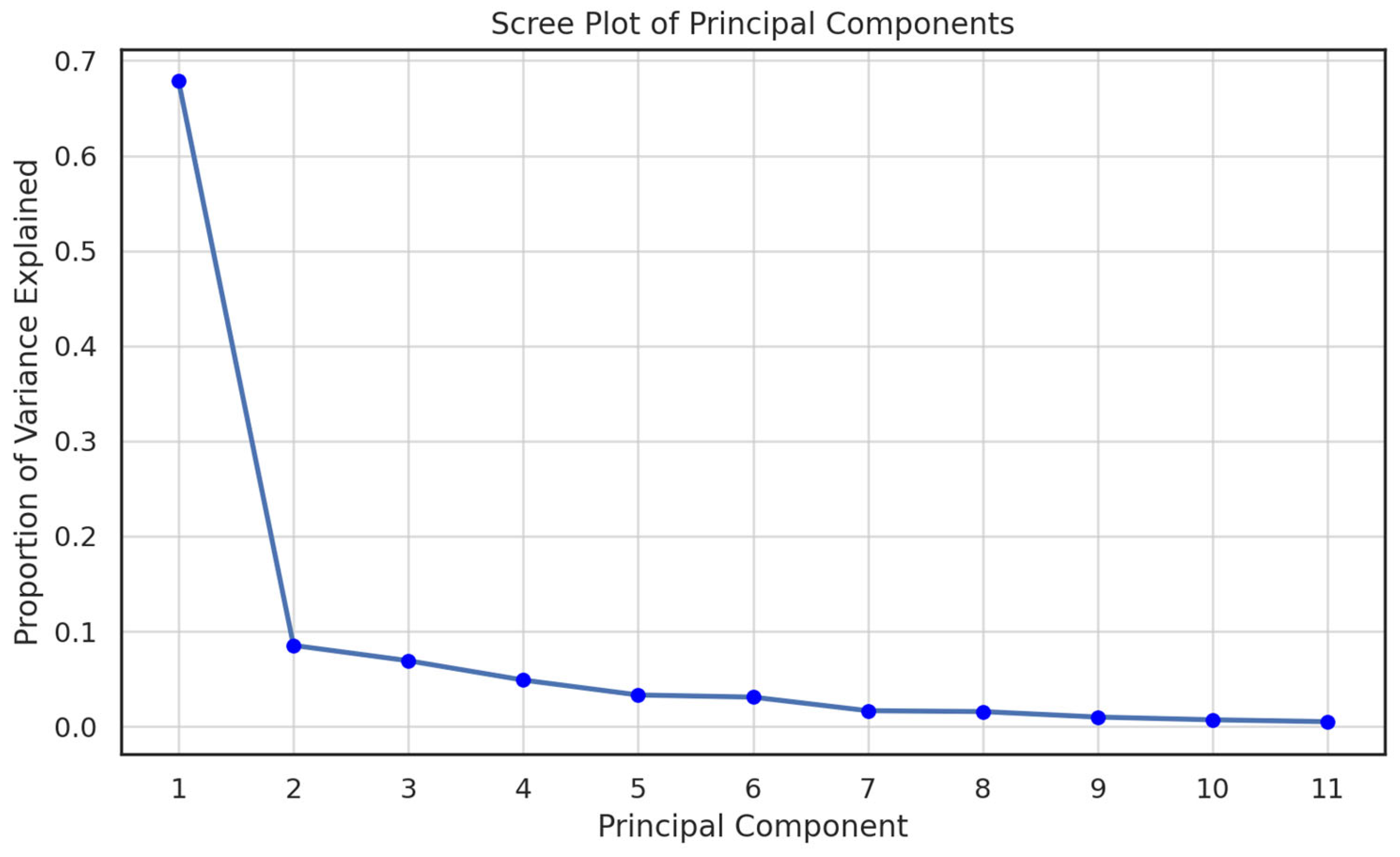
- Analysis of explained variance: Scree plot and cumulative curve
- Interpretation of PC1, PC2 and PC3
- Three-dimensional visualization
2.5. Clustering Algorithms
2.5.1. K-Means Clustering (k = 3)
2.5.2. Hierarchical Clustering (Ward Linkage)
2.5.3. DBSCAN
- A core point if it has at least minPts neighbors within ε;
- Reachable if it lies within ε of a core point;
- Noise otherwise.
3. Results
3.1. Statistical Analysis
3.2. Principal Component Analysis
- SampEn and SDNN (r = 0.88)—indicating a strong association between entropy and overall linear variability.
- RMSSD and SampEn (r = 0.86)—reflecting a high correlation between short-term variability and signal complexity.
- Hurst exponent and SampEn (r = 0.78)—supporting a shared fractal–entropic nature of the signal.
- Color-coded data points representing the three physiological states (rest, stress, load).
- Black vectors depicting the contribution of each HRV metric (SDNN, RMSSD, SampEn, Hurst exponent, FD, mean RR) to the first three principal components (PC1, PC2, PC3).
- Textual labels positioned in space according to the direction and magnitude of the vectors.
3.3. Cluster Analysis and Cumulative Explained Variance
3.4. Individual Clustering of Participants Using PCA-Transformed Components
3.5. DBSCAN Analysis for Physiological State Recognition
- The feasibility of unsupervised recognition of physiological states;
- The suitability of DBSCAN for biomedical data analysis, especially in the presence of individual variability;
- The importance of a personalized approach when interpreting HRV data.
3.6. Evaluated Characteristics
- Regarding latent structures in HRV and fractal features:
- 2.
- Regarding the ability of unsupervised algorithms to distinguish states:
- 3.
- Regarding the role of AI-based visualizations:
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shaffer, F.; Ginsberg, J.P. An overview of heart rate variability metrics and norms. Front. Public Health 2017, 5, 258. [Google Scholar] [CrossRef]
- Lapsa, D.; Janeliukstis, R.; Metshein, M.; Selavo, L. PPG and Bioimpedance-Based Wearable Applications in Heart Rate Monitoring—A Comprehensive Review. Appl. Sci. 2024, 14, 7451. [Google Scholar] [CrossRef]
- Natarajan, A.; Pantelopoulos, A.; Emir-Farinas, H.; Natarajan, P. Heart rate variability with photoplethysmography in 8 million individuals: A cross-sectional study. Lancet Digit. Health 2020, 2, e650–e657. [Google Scholar] [CrossRef] [PubMed]
- Aarthee, S.; Ramya, R.; Pramila, R.P.; Ezilarasan, M.R.; Suba, G.M. Advanced wearable health monitoring system with multi-sensor data and secure data management with blockchain technology. Int. J. Intell. Syst. Appl. Eng. 2024, 12, 808–813. [Google Scholar]
- Giles, D.; Draper, N.; Neil, W. Validity of the Polar V800 Heart Rate Monitor to Measure RR Intervals at Rest. Eur. J. Appl. Physiol. 2016, 116, 563–571. [Google Scholar] [CrossRef]
- Henriksen, A.; Svartdal, F.; Grimsgaard, S.; Hartvigsen, G.; Hopstock, L.A. Polar Vantage and Oura Physical Activity and Sleep Trackers: Validation and Comparison Study. JMIR Form. Res. 2022, 6, e27248. [Google Scholar] [CrossRef]
- Flatt, A.A.; Esco, M.R. Validity of the ithlete™ Smart Phone Application for Determining Ultra-Short-Term Heart Rate Variability. J. Hum. Kinet. 2013, 39, 85–92. [Google Scholar] [CrossRef]
- Plews, D.J.; Laursen, P.B.; Stanley, J.; Kilding, A.E.; Buchheit, M. Training Adaptation and Heart Rate Variability in Elite Endurance Athletes: Opening the Door to Effective Monitoring. Sports Med. 2013, 43, 773–781. [Google Scholar] [CrossRef]
- Zhang, Z. Photoplethysmography-Based Heart Rate Monitoring in Physical Activities via Joint Sparse Spectrum Reconstruction. IEEE Trans. Biomed. Eng. 2015, 62, 1902–1910. [Google Scholar] [CrossRef]
- Shin, H.S.; Lee, C.; Lee, M. Adaptive threshold method for the peak detection of photoplethysmographic waveform. Comput. Biol. Med. 2009, 39, 1145–1152. [Google Scholar] [CrossRef]
- Patro, K.K.; Jaya Manmadha Rao, M.; Jadav, A.; Rajesh Kumar, P. Noise Removal in Long-Term ECG Signals Using EMD-Based Threshold Method. In Data Engineering and Communication Technology; Reddy, K.A., Devi, B.R., George, B., Raju, K.S., Eds.; Lecture Notes on Data Engineering and Communications Technologies; Springer: Singapore, 2021; Volume 63, pp. 565–574. [Google Scholar] [CrossRef]
- Ouali, M.A.; Chafaa, K.; Ghanai, M.; Lorente, L.M.; Rojas, D.B. ECG Denoising Using Extended Kalman Filter. In Proceedings of the 2013 International Conference on Computer Applications Technology (ICCAT), Sousse, Tunisia, 20–22 January 2013; pp. 1–6. [Google Scholar] [CrossRef]
- Zhou, Y.; Hu, X.; Tang, Z.; Ahn, A.C. Denoising and Baseline Correction of ECG Signals Using Sparse Representation. In Proceedings of the 2015 IEEE Workshop on Signal Processing Systems (SiPS), Hangzhou, China, 14–16 October 2015; pp. 1–6. [Google Scholar] [CrossRef]
- Aktaruzzaman, M.; Sassi, R. Sample entropy parametric estimation for heart rate variability analysis. Comput. Cardiol. 2013, 40, 429–432. [Google Scholar] [CrossRef]
- Voss, A.; Schulz, S.; Schroeder, R.; Baumert, M.; Caminal, P. Methods derived from nonlinear dynamics for analysing heart rate variability. Philos. Trans. R. Soc. A 2009, 367, 277–296. [Google Scholar] [CrossRef]
- Kim, H.G.; Cheon, E.J.; Bai, D.S.; Lee, Y.H.; Koo, B.H. Stress and Heart Rate Variability: A Meta-Analysis and Review of the Literature. Psychiatry Investig. 2018, 15, 235–245. [Google Scholar] [CrossRef]
- Brockmann, L.; Hunt, K.J. Heart Rate Variability Changes with Respect to Time and Exercise Intensity during Heart-Rate-Controlled Steady-State Treadmill Running. Sci. Rep. 2023, 13, 8515. [Google Scholar] [CrossRef]
- May, R.; McBerty, V.; Zaky, A.; Gianotti, M. Vigorous Physical Activity Predicts Higher Heart Rate Variability among Younger Adults. J. Physiol. Anthropol. 2017, 36, 24. [Google Scholar] [CrossRef] [PubMed]
- Miyatsu, T.; Smith, B.M.; Koutnik, A.P.; Pirolli, P.; Broderick, T.J. Resting-State Heart Rate Variability after Stressful Events as a Measure of Stress Tolerance among Elite Performers. Front. Physiol. 2023, 13, 1070285. [Google Scholar] [CrossRef] [PubMed]
- Rennie, K.L.; Hemingway, H.; Kumari, M.; Brunner, E.; Malik, M.; Marmot, M. Effects of Moderate and Vigorous Physical Activity on Heart Rate Variability in a British Study of Civil Servants. Am. J. Epidemiol. 2003, 158, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Khatter, H.; Yadav, A.; Srivastava, A. Machine learning-based automated medical diagnosis for healthcare. In Proceedings of the 6th International Conference on Information Systems and Computer Networks (ISCON), Mathura, India, 3–4 March 2023; pp. 1–5. [Google Scholar]
- Oliveira, P.A.M.; Florez, H.; Astudillo, H. Clustering-Based Health Indicators for Health-Related Quality of Life. In Applied Informatics, Proceedings of the ICAI 2024, Viña del Mar, Chile, 24—26 October 2024; Communications in Computer and Information Science; Florez, H., Astudillo, H., Eds.; Springer: Cham, Switzerland, 2025; Volume 2237. [Google Scholar]
- Dai, Y.; Sun, S.; Che, L. Improved DBSCAN-based Data Anomaly Detection Approach for Battery Energy Storage Stations. J. Phys. Conf. Ser. 2022, 2351, 012025. [Google Scholar] [CrossRef]
- Oskooei, A.; Chau, S.M.; Weiss, J.; Sridhar, A.; Rodríguez Martínez, M.; Michel, B. DeStress: Deep Learning for Unsupervised Identification of Mental Stress in Firefighters from Heart-Rate Variability (HRV) Data. arXiv 2019, arXiv:1911.13213. [Google Scholar] [CrossRef]
- Iqbal, T.; Elahi, A.; Wijns, W.; Amin, B.; Shahzad, A. Improved Stress Classification Using Automatic Feature Selection from Heart Rate and Respiratory Rate Time Signals. Appl. Sci. 2023, 13, 2950. [Google Scholar] [CrossRef]
- Messaoud, I.B.; Thamsuwan, O. Heart Rate Variability-Based Stress Detection and Fall Risk Monitoring During Daily Activities: A Machine Learning Approach. Computers 2025, 14, 45. [Google Scholar] [CrossRef]
- Basri, A.M.; Turki, A.F. Evaluating Heart Rate Variability as a Biomarker for Autonomic Function in Parkinson’s Disease Rehabilitation: A Clustering-Based Analysis of Exercise-Induced Changes. Medicina 2025, 61, 527. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Vicente, A.; Hernando, D.; Marín-Puyalto, J.; Vicente-Rodríguez, G.; Garatachea, N.; Pueyo, E.; Bailón, R. Validity of the Polar H7 Heart Rate Sensor for Heart Rate Variability Analysis during Exercise in Different Age, Body Composition and Fitness Level Groups. Sensors 2021, 21, 902. [Google Scholar] [CrossRef] [PubMed]
- Georgieva-Tsaneva, G. Wavelet based interval varying algorithm for optimal non-stationary signal denoising. In ACM International Conference Proceeding Series; ACM: New York, NY, USA, 2019; pp. 200–206. ISBN 978-1-4503-7149-0. [Google Scholar] [CrossRef]
- Georgieva-Tsaneva, G.; Cheshmedzhiev, K.; Lebamovski, P. A Wavelet Based Hybrid Method for Time Interval Series Determining. In Proceedings of the International Conference on Computer Systems and Technologies 2024—CompSysTech’24; ACM International Conference Proceeding Series. Association for Computing Machinery: New York, NY, USA, 2024; pp. 137–142, ISBN 978-3-031-42134-1. [Google Scholar] [CrossRef]
- Georgieva-Tsaneva, G. QRS detection algorithm for long term Holter records. In CompSysTech ’13, Proceedings of the 14th International Conference on Computer Systems and Technologies, Ruse, Bulgaria, 27–28 June 2025; ACM: New York, NY, USA, 2013; pp. 112–119. ISBN 978-1-4503-2021-4. [Google Scholar] [CrossRef]
- Malik, M.; Camm, A.J.; Bigger, J.T.; Breithardt, G.; Cerutti, S.; Cohen, R.J.; Coumel, P.; Fallen, E.L.; Kennedy, H.L.; Kleiger, R.E.; et al. Heart rate variability. Standards of measurement, physiological interpretation, and clinical use. Eur. Heart J. 1996, 17, 354–381. [Google Scholar] [CrossRef]
- Richman, J.S.; Moorman, J.R. Physiological Time-Series Analysis Using Approximate Entropy and Sample Entropy. Am. J. Physiol. Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef]
- Korvin, G. Rescaled Range Analysis. Enc. Math. Geosci. 2022. Bassingthwaighte, J.B.; Raymond, G.M. Evaluating Rescaled Range Analysis for Time Series. Ann. Biomed. Eng. 1994, 22, 432–444. [Google Scholar] [CrossRef]
- Hinrikus, H.; Bachmann, M.; Karai, D.; Klonowski, W.; Lass, J.; Stepien, P.; Stepien, R.; Tuulik, V. Higuchi’s Fractal Dimension for Analysis of the Effect of External Periodic Stressor on Electrical Oscillations in the Brain. Med. Biol. Eng. Comput. 2011, 49, 585–591. [Google Scholar] [CrossRef]
- Nayak, S.K.; Pradhan, B.; Mohanty, B.; Sivaraman, J.; Ray, S.S.; Wawrzyniak, J.; Jarzębski, M.; Pal, K. A Review of Methods and Applications for a Heart Rate Variability Analysis. Algorithms 2023, 16, 433. [Google Scholar] [CrossRef]
- van Es, V.A.A.; Lopata, R.G.P.; Scilingo, E.P.; Nardelli, M. Contactless Cardiovascular Assessment by Imaging Photoplethysmography: A Comparison with Wearable Monitoring. Sensors 2023, 23, 1505. [Google Scholar] [CrossRef]
- Huang, Y.; Deng, Y. A Hybrid Model Utilizing Principal Component Analysis and Artificial Neural Networks for Driving Drowsiness Detection. Appl. Sci. 2022, 12, 6007. [Google Scholar] [CrossRef]
- Wang, J.S.; Lin, C.W.; Yang, Y.T.C. Using Heart Rate Variability Parameter-Based Feature Transformation Algorithm for Driving Stress Recognition. In Advanced Intelligent Computing, Proceedings of the 7th International Conference, ICIC 2011, Zhengzhou, China, 11–14 August 2011; Huang, D.S., Gan, Y., Bevilacqua, V., Figueroa, J.C., Eds.; Lecture Notes in Computer Science, 6838; Springer: Berlin/Heidelberg, Germany, 2011; pp. 569–576. [Google Scholar] [CrossRef]
- Retiti Diop Emane, C.; Song, S.; Lee, H.; Choi, D.; Lim, J.; Bok, K.; Yoo, J. Anomaly Detection Based on GCNs and DBSCAN in a Large-Scale Graph. Electronics 2024, 13, 2625. [Google Scholar] [CrossRef]
- Materko, W. Stratifying Autonomic Nervous System Regulation Patterns in Healthy Men: A Machine Learning Approach. Artif. Intell. Health 2025, 10, 025050006. [Google Scholar] [CrossRef]
- Lebamovski, P.; Gospodinova, E. Investigating Stress During a Virtual Reality Game Through Fractal and Multifractal Analysis of Heart Rate Variability. Appl. Syst. Innov. 2025, 8, 16. [Google Scholar] [CrossRef]
- Zamora-Justo, J.A.; Campos-Aguilar, M.; Beas-Jara, M.d.C.; Galván-Fernández, P.; Ponciano-Gómez, A.; Sigrist-Flores, S.C.; Jiménez-Flores, R.; Muñoz-Diosdado, A. Utility of Nonlinear Analysis of Heart Rate Variability in Early Detection of Metabolic Syndrome. Front. Physiol. 2025, 16, 1597314. [Google Scholar] [CrossRef] [PubMed]
- Borthakur, D.; Peltier, A.; Dubey, H.; Gyllinsky, J.; Mankodiya, K. SmartEAR: Smartwatch-Based Unsupervised Learning for Multi-Modal Signal Analysis in Opportunistic Sensing Framework. arXiv 2018, arXiv:1808.06473. [Google Scholar] [CrossRef]
- Schrumpf, F.; Bausch, G.; Sturm, M.; Fuchs, M. Similarity-Based Hierarchical Clustering of Physiological Parameters for the Identification of Health States—A Feasibility Study. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Republic of Korea, 11–15 July 2017; pp. 458–462. [Google Scholar] [CrossRef]
- Serantoni, C.; Zimatore, G.; Bianchetti, G.; Abeltino, A.; De Spirito, M.; Maulucci, G. Unsupervised Clustering of Heartbeat Dynamics Allows for Real-Time and Personalized Improvement in Cardiovascular Fitness. Sensors 2022, 22, 3974. [Google Scholar] [CrossRef]
- Georgieva-Tsaneva, G.; Cheshmedzhiev, K.; Tsanev, Y.-A.; Dechev, M.; Popovska, E. Healthcare Monitoring Using an Internet of Things-Based Cardio System. IoT 2025, 6, 10. [Google Scholar] [CrossRef]









| Parameter | Rest N = 22 [Mean ± sd] | Stress N = 22 [Mean ± sd] | Physical Activity (Load) N = 22 [Mean ± sd] |
|---|---|---|---|
| Mean RR (ms) | 842.16 ± 143.23 | 638.28 ± 126.17 | 589.42 ± 131.38 |
| SDNN (ms) | 161.43 ± 46.73 | 93.46 ± 45.68 | 118.27 ± 26.19 |
| RMSSD (ms) | 28.39 ± 7.24 | 9.86 ± 4.81 | 18.22 ± 5.93 |
| SampEn | 1.22 ± 0.12 | 1.72 ± 0.08 | 1.35 ± 0.18 |
| Hurst Exponent (H) | 0.72 ± 0.04 | 0.58 ± 0.05 | 0.63 ± 0.04 |
| Fractal Dimension (FD) | 1.18 ± 0.03 | 1.41 ± 0.04 | 1.36 ± 0.08 |
| LF (nu) | 37.21 ± 4.50 | 68.12 ± 6.42 | 54.00 ± 5.61 |
| HF (nu) | 63.42 ± 5.06 | 32.33 ± 4.22 | 46.00 ± 4.52 |
| LF/HF | 0.59 ± 0.11 | 2.12 ± 0.23 | 1.22 ± 0.14 |
| DFA α1 | 1.14 ± 0.06 | 0.88 ± 0.08 | 1.04 ± 0.07 |
| DFA α2 | 1.33 ± 0.05 | 0.86 ± 0.07 | 0.99 ± 0.06 |
| Parameter | Rest vs. Stress | Stress vs. Load | Rest vs. Load |
|---|---|---|---|
| Mean RR (ms) | <0.0001 | <0.05 | <0.0001 |
| SDNN (ms) | <0.001 | <0.05 | <0.001 |
| RMSSD (ms) | <0.0001 | <0.05 | <0.05 |
| SampEn | <0.0001 | <0.05 | <0.05 |
| Hurst Exponent (H) | <0.05 | <0.05 | <0.01 |
| Fractal Dimension (FD) | <0.0001 | <0.05 | <0.05 |
| LF (nu) | <0.001 | <0.01 | <0.001 |
| HF (nu) | <0.001 | <0.05 | <0.001 |
| LF/HF | <0.001 | <0.01 | <0.001 |
| DFA α1 | <0.0001 | <0.05 | <0.001 |
| DFA α2 | <0.0001 | <0.05 | <0.05 |
| Characteristics | Group 1 | Group 2 | Mean Difference | p-Value | 95% CI (Lower–Upper) | Significant Difference |
|---|---|---|---|---|---|---|
| Mean RR | Load | Rest | 261.79 | <0.0001 | 238.28–285.31 | Yes |
| Mean RR | Load | Stress | 24.36 | 0.79 | 48.84–95.87 | No |
| Mean RR | Rest | Stress | −189.44 | <0.0001 | −212.96–−165.92 | Yes |
| SDNN | Load | Rest | 20.79 | <0.0001 | 16.52–25.07 | Yes |
| SDNN | Load | Stress | −13.39 | <0.0001 | −17.67–−9.11 | Yes |
| SDNN | Rest | Stress | −34.18 | <0.0001 | −38.46–−29.91 | Yes |
| RMSSD | Load | Rest | −19.84 | <0.0001 | −24.23–−15.46 | Yes |
| RMSSD | Load | Stress | 15.09 | <0.0001 | 10.70–19.48 | Yes |
| RMSSD | Rest | Stress | 34.93 | <0.0001 | 30.54–39.32 | Yes |
| SampEn | Load | Rest | −0.08 | 0.26 | −0.19–0.039 | No |
| SampEn | Load | Stress | −0.33 | <0.0001 | −0.39–−0.27 | Yes |
| SampEn | Rest | Stress | −0.65 | <0.0001 | −0.71–−0.59 | Yes |
| H | Load | Rest | −0.09 | <0.0001 | −0.12–−0.06 | Yes |
| H | Load | Stress | 0.05 | <0.0001 | 0.02–0.08 | Yes |
| H | Rest | Stress | 0.14 | <0.0001 | 0.11–0.17 | Yes |
| FD | Load | Rest | −0.06 | <0.0001 | −0.08–−0.04 | Yes |
| FD | Load | Stress | 0.07 | <0.0001 | 0.05–0.09 | Yes |
| FD | Rest | Stress | 0.13 | <0.0001 | 0.11–0.15 | Yes |
| LFnu | Load | Rest | −15.79 | <0.0001 | −19.88–−11.71 | Yes |
| LFnu | Load | Stress | 16.57 | <0.0001 | 12.49–20.66 | Yes |
| LFnu | Rest | Stress | 32.37 | <0.0001 | 28.29–36.45 | Yes |
| HFnu | Load | Rest | 18.79 | <0.0001 | 15.41–22.18 | Yes |
| HFnu | Load | Stress | −12.58 | <0.0001 | −14.99–−9.18 | Yes |
| HFnu | Rest | Stress | −31.38 | <0.0001 | −34.77–−27.93 | Yes |
| LF/HF | Load | Rest | −0.67 | <0.0001 | −0.79–−0.55 | Yes |
| LF/HF | Load | Stress | 0.89 | <0.0001 | 0.78–1.01 | Yes |
| LF/HF | Rest | Stress | 1.56 | <0.0001 | 1.45–1.68 | Yes |
| DFA_α1 | Load | Rest | −0.21 | <0.0001 | −0.26–−0.14 | Yes |
| DFA_α1 | Load | Stress | 0.056 | <0.05 | 0.0005–0.12 | Yes |
| DFA_α1 | Rest | Stress | 0.26 | <0.05 | 0.21–0.32 | Yes |
| DFA_α2 | Load | Rest | −0.14 | <0.0001 | −0.19–−0.1 | Yes |
| DFA_α2 | Load | Stress | 0.16 | <0.0001 | 0.12–0.2 | Yes |
| DFA_α2 | Rest | Stress | 0.31 | <0.0001 | 0.26–0.35 | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Georgieva-Tsaneva, G.; Cheshmedzhiev, K.; Tsanev, Y.-A.; Dechev, M. Physiological State Recognition via HRV and Fractal Analysis Using AI and Unsupervised Clustering. Information 2025, 16, 718. https://doi.org/10.3390/info16090718
Georgieva-Tsaneva G, Cheshmedzhiev K, Tsanev Y-A, Dechev M. Physiological State Recognition via HRV and Fractal Analysis Using AI and Unsupervised Clustering. Information. 2025; 16(9):718. https://doi.org/10.3390/info16090718
Chicago/Turabian StyleGeorgieva-Tsaneva, Galya, Krasimir Cheshmedzhiev, Yoan-Aleksandar Tsanev, and Miroslav Dechev. 2025. "Physiological State Recognition via HRV and Fractal Analysis Using AI and Unsupervised Clustering" Information 16, no. 9: 718. https://doi.org/10.3390/info16090718
APA StyleGeorgieva-Tsaneva, G., Cheshmedzhiev, K., Tsanev, Y.-A., & Dechev, M. (2025). Physiological State Recognition via HRV and Fractal Analysis Using AI and Unsupervised Clustering. Information, 16(9), 718. https://doi.org/10.3390/info16090718








