Abstract
Single-season legume green manuring is widely promoted for soil fertility restoration in degraded agricultural lands, yet its effectiveness in alkaline semi-arid soils remains poorly understood. This study investigated the impact of first-year sweet clover (Melilotus officinalis (L.)) green manuring on soil microbiome structure and agrochemical properties in southern carbonate chernozem soils of Northern Kazakhstan. Using shotgun metagenomics, we analyzed microbial communities from sweet clover-amended soils, clean fallow, and virgin steppe reference sites. Contrary to expectations, sweet clover green manuring did not enhance soil nitrogen availability, with nitrate-N content (9.1 mg/kg) remaining lower than clean fallow (10.5 mg/kg), likely due to temporary immobilization during initial decomposition. While sweet clover significantly increased archaeal diversity (p = 0.01) and enriched nitrogen-cycling taxa, including Nitrospirae and Thaumarchaeota, overall microbial richness remained unchanged (ACE index, p > 0.05). Surprisingly, functional analysis revealed only five significant metabolic differences between sweet clover and fallow systems, indicating functional convergence of agricultural microbiomes regardless of management practice. Correlation analysis identified phosphorus as the master regulator of microbial metabolism (r = 1.0, p < 0.0001), while elevated pH (9.0), K2O (>1000 mg/kg), and showed strong negative correlations with essential metabolic pathways, revealing previously unrecognized nutrient toxicity thresholds. Virgin steppe maintained 69 unique metabolic pathways lost in agricultural systems, highlighting the ecological cost of cultivation. These findings demonstrate that sweet clover green manuring in alkaline steppe soils induces selective rather than comprehensive microbiome restructuring, with limited immediate benefits for soil fertility. This study provides critical insights for developing sustainable agricultural practices in the world’s extensive semi-arid regions facing similar edaphic constraints.
1. Introduction
Soil microbiomes are the primary drivers of nitrogen cycling in agricultural systems, mediating all major nitrogen transformations and controlling nutrient availability [1]. In alkaline semi-arid soils, microbial communities face unique constraints from high pH, low moisture, and nutrient limitations that fundamentally alter their structure and function. Understanding how green manuring with legumes influences microbial community assembly and metabolic pathways in degraded carbonate chernozems is essential for developing effective restoration strategies.
Agricultural intensification leads to taxonomic homogenization of soil microbiomes [2], where multiple taxa converge toward similar metabolic functions despite different phylogenetic origins [3]. In grasslands receiving elevated nutrient inputs, microbial communities show consistent shifts toward copiotrophic taxa regardless of geographic location or specific management practice [4]. This convergence may explain why different organic amendments sometimes produce similar outcomes despite distinct carbon and nitrogen inputs, raising questions about whether green manuring can overcome such convergence pressures in degraded soils.
Green manuring with leguminous crops represents a biological approach to soil restoration, yet its effectiveness in alkaline soils remains inconsistent. During decomposition of plant residues with high C:N ratios (>20:1), microbial communities initially immobilize nitrogen, with the duration depending on residue quality and environmental conditions [5]. In soils with pH > 8.5, decomposition is further complicated by reduced enzyme activity and precipitation of organic substrates with calcium [6]. These constraints are particularly relevant for the degraded steppe soils of Central Asia.
The Virgin Lands Campaign (1954–1962) converted 25 million hectares of Kazakhstani steppe to wheat production [7], triggering organic matter losses of 30–50% within three decades [8]. Current soil organic carbon levels in Northern Kazakhstan average 2–3%, compared to 4–6% in undisturbed steppes [9]. This degradation reduces microbial habitat quality through aggregate breakdown and decreased pore connectivity. Reversing this degradation requires understanding how soil microbiomes respond to restoration practices.
Key microbial groups involved in nitrogen cycling may respond differently to green manuring in alkaline soils. Ammonia-oxidizing archaea (AOA) within 0 often outnumber ammonia-oxidizing bacteria in soils, with ratios ranging from equal amounts to over 3000-fold depending on soil conditions [10]. These archaea use the most energy-efficient aerobic CO2 fixation pathway (3-hydroxypropionate/4-hydroxybutyrate cycle) [11], while their high affinity for ammonia allows them to thrive at low substrate concentrations [12]. Nitrite-oxidizing bacteria (Nitrospirae), including newly discovered complete ammonia oxidizers (comammox), play crucial roles in completing nitrification across diverse environments [13].
Among leguminous green manures, sweet clover (Melilotus officinalis (L.)) demonstrates exceptional adaptation to semi-arid conditions through deep taproots (2–3 m), salt tolerance, and biomass production of 8–12 t ha−1 [14]. This species promotes specific rhizobial populations through root exudation of flavonoids and phenolic compounds [15,16], while its coumarin content can reach 0.2–0.9% dry weight under stress conditions [17]. These characteristics make sweet clover a promising candidate for soil restoration in degraded steppe ecosystems.
However, phosphorus availability often limits microbial activity in carbonate-rich soils despite adequate total P, as calcium-phosphate precipitation reduces bioavailable P by 10–100-fold compared to neutral soils [18]. In agricultural soils, phosphorus distribution in particle size fractions controls microbial growth efficiency and carbon use [19], with P-limitation affecting ATP-dependent processes and nucleic acid synthesis [20]. Understanding these nutrient constraints is critical for predicting green manure effectiveness.
The aim of this study was to assess the influence of first-year sweet clover green manuring on the structure and functional characteristics of microbial communities in southern carbonate chernozem soils of Northern Kazakhstan. We hypothesized that: (1) first-year sweet clover green manuring would increase soil nitrate-N availability and shift the microbiome towards nitrogen-cycling taxa compared to clean fallow; and (2) sweet clover incorporation would result in more diverse functional profiles reflecting enhanced biological nitrogen fixation and organic matter inputs. This pilot study provides baseline data for developing long-term restoration strategies in alkaline steppe soils.
2. Materials and Methods
2.1. Study Site and Experimental Design
The research was carried out in field experiments established at the A.I. Barayev Research and Production Center for Grain Farming (51°39′ N, 71°01′ E), Shortandy District, Akmola Region, the steppe zone of Northern Kazakhstan (Figure 1).
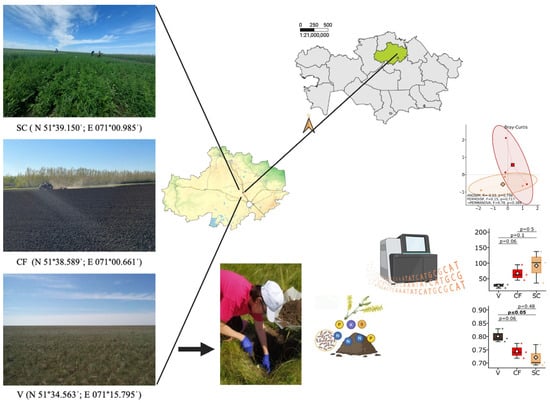
Figure 1.
Location of the experimental plot is the A.I. Barayev Research and Production Center for Grain Farming, Shortandy District, Akmola Region, Northern Kazakhstan (51°39′ N, 71°01′ E). Experiment variants: SC—yellow sweet clover (Melilotus officinalis (L.)) of the first year of existence (green manure); CF—Clean fallow; V—virgin steppe.
Climatic conditions in the research area are distinctly continental, dry. The annual precipitation level is 342.4 mm (35–40% falls into summer period, 15–20% autumn, 17–25% winter and 20–25% spring period). The soil freezes through to the depth of 1.2–2 m and thaws slowly in spring. The frost-free period lasts for 102–130 days, and in some years, up to 168 days. Spring is short, with a steep increase in air temperature and heavy winds. In the summer, the average air temperature is 20.0–23.3 °C, and maximal values can reach +42 …+44 °C, while temperature on the soil surface rises up to +50 …+60 °C. As a result, the climate of Northern Kazakhstan is characterized by irregularity year-wise. In 2024, meteorological conditions in general were favorable for sweet clover growth and development in spite of unstable temperature regime and uneven distribution of precipitation. Weather conditions during the period of sweet clover green biomass formation were similar to long-term annual averages, which ensured good plant development (July) before plowing under thereof. An excessive amount of precipitation in August (106.6 mm, the annual average norm being 66.8 mm) contributed to decomposition of sweet clover plant residues. The temperature background was around 17.4 °C.
The soil represents southern calcareous chernozem (WRB: Chernozem Calcic [21]), which is characterized by high carbonate content (up to 5%). With regard to granulometric composition, the soils are heavy clay loams with a pulverous structure. The arable layer is 30 cm thick; the humus horizon is up to 50 cm. The supply rate of nitrogen total form is 0.3%, phosphorus 0.1%, while calcium (up to 80%) and magnesium (11%) prevail in the absorbed layer. Soil рН is around 7.6–7.9, which is slightly alkaline. The soils are characterized by a non-washing regime; the groundwater depth level is below 10 m. Soil moisture supply totally depends on precipitation level. Available water content by the sowing time usually does not exceed 110 mm, while by autumn the content thereof in the root zone decreases to the level of plant wilting. Substantial seasonal fluctuations of soil moisture and carbonate content cause degree of arable layer compaction.
2.1.1. Yellow Sweet Clover of the First Year of Existence (Green Manure) (SC)
The plot where the experiment with yellow sweet clover was established had been fallowed for three years; before that, wheatgrass perennial grass was grown on that plot. The year before sowing yellow sweet clover, the plot was cultivated as clean fallow, which included four shallow subsurface cultivations with a KPSh-3 sweep cultivator and one autumn deep cultivation with a deep tiller PG-3-5 down to 25–27 cm to accumulate soil moisture. This system of fallowing is traditional for the zone of investigation.
In the spring, the soil was harrowed, smoothed with BIG-3 soil spikers, cultivated with seedbed cultivators, and rolled before and after sweet clover planting. Sweet clover Altynbas cultivar was sown after fallow on 26 April 2014 (on the planting date recommended for the given zone) with the direct-connected grass planter, using scarified seeds (without seed inoculation with biopreparations), with the seeding rate of 3.5 million fertile seeds per 1 hectare (8 kg/ha) with row spacing of 15 cm. Repetition was quadruple. The area of the registration plot was 120 m2. The yielding capacity of green (aboveground) mass of first-year yellow sweet clover was identified according to [22], and yielding capacity of underground (root) mass according to [23].
Sweet clover for green manure was plowed under at the full bud stage (mid July) in the first year of the plant life with a mounted plow (PN-3-35) down to the depth of the plowing horizon. After the plowing under, the sweet clover furrow slice was cultivated with a disk harrow (BDM 6 × 4P). Location of the variant (*SC) is 51°39.150′ N; 071°00.985′ E.
2.1.2. Clean Fallow (CF)
The control was clean fallow that had not been seeded with farm crops for three years. Prior to that, wheatgrass perennial grass was grown on that plot. During the growing period the clean fallow was cultivated four times with blade weeder KPSh-3 to control weeds; fertilizers were not applied. The location of the variant (CF) was 51°38.589′ N; 071°00.661′ E.
2.1.3. Virgin Steppe (V)
The virgin plot located in the Shortandy District, Amola Region, was selected as the reference standard. The soil of the virgin plot had never been in crop rotation and retained its natural vegetation, prevailing species being Stipa lessingiana Trin. et Rupr., Stipa capillata L., Festuca valesiaca Gand. subsp. sulcata., Artemisia austriaca Jacq. The virgin plot is a natural preserve in the steppe zone of North Kazakhstan. The variant location (V) was 51°34.563′ N; 071°15.795′ E. For metagenomic and agrochemical analyses, three independent composite soil samples per treatment were collected (n = 3 for SC, CF, and V), corresponding to spatially separated field replicates. For the virgin steppe site, the three biological replicates represent spatially separated sampling points within the preserved area (minimum distance of 50 m between sampling points). It should be noted that agricultural treatments (SC, CF) and the virgin steppe reference are located at different coordinates, which introduces potential site effects when comparing these systems (see Section 4.7).
2.2. Biochemical Analysis of Aboveground and Underground (Roots) Biomass of Yellow Sweet Clover
Nitrogen content in the dry matter of yellow sweet clover aboveground and underground (roots) biomass was identified per the Kjeldahl Method with the use of a combustion apparatus and water vapor distillation (UDK-139, Velp Scientifica, Usmate Velate, Italy) in compliance with ISO 5983-1:2005 [24]. Phosphorus content was identified photometrically pursuant to GOST 26657-97 [25]. Potassium content was identified by the flame photometry method as per GOST 30504-97 [26].
2.3. Soil Sampling and Agrochemical Analysis
Complex soil samples (0–20 cm) were collected in early September with a sampler (hole borer) from each plot, using a systemic grid with 10 subsamples per plot. Soil samples were dried to air-dried basis, then they were crushed and sifted through a 2 mm sieve to identify agrochemical characteristics. Soil pH was measured with potentiometric method in soil: water suspension 1:5. Nitrate nitrogen was extracted from the soil with KCl solution, with subsequent reduction with hydrazine to nitrites and was identified with spectrophotometer Cary 50 (Varian Optical Spectroscopy Instruments, Melbourne, Australia) per GOST 26488-85 [27]. Available phosphorus and potassium were extracted with ammonium carbonate solution (the Machigin method) and measured with spectrophotometry (P) and flame photometry (K) per GOST 26205-91 [28]. The organic matter (humus) was analyzed by way of oxidation with potassium dichromate (the Tyurin method) per GOST 26213-2021 [29]. Note on analytical standards: National standards (GOST) were employed for soil and plant analyses as these methods are specifically validated for steppe soil conditions. International equivalents are provided for reference: GOST 26488-85 [27] for nitrate determination is comparable to ISO 14255:1998 (KCl extraction) [30]. GOST 26205-91 (Machigin method) [28] corresponds to a modified Olsen P method adapted for calcareous soils, and GOST 26213-2021 [29] for organic matter follows the principles of the Walkley-Black chromic acid oxidation method [31]. All analyses were performed in an accredited laboratory (ISO/IEC 17025:2017) [32] ensuring international comparability of results.
2.4. DNA Extraction and Metagenomic Sequencing
DNA extraction from soil samples was performed using the ZymoBiomics DNA Microprep kit (Cat. No. D4301, Zymo Research, Irvine, CA, USA), with concentrations determined spectrophotometrically using a NanoDrop 2000/2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Sterile water served as the negative control. Whole-genome shotgun metagenomic sequencing was conducted according to standard Illumina protocols by Novogene Corporation (Beijing, China) on the NovaSeq 6000 (Illumina, San Diego, CA, USA) platform, generating an average of 6 Gb of data per sample.
2.5. Microbial Symbiont Identification
Identification of nitrogen-fixing symbionts was performed through taxonomic profiling of metagenomic reads against reference genomes of known rhizobial species. Successful establishment of symbiotic relationships was confirmed by detection of nif gene clusters and nodulation genes (nodABC) with coverage depth > 10× and identity > 97%. Symbiont abundance was calculated as genome equivalents per gram of soil based on single-copy core gene normalization.
2.6. Bioinformatics and Statistical Analysis
Raw sequencing data were processed using KneadData v0.12.0 for quality control [33], including removal of low-quality reads (Q < 20), adapter sequences, and host contamination. Taxonomic profiling was performed using MetaPhlAn 4 (database mpa_vJan21_CHOCOPhlAnSGB_202103) [34] and functional annotation with HUMAnN 3.0 [35] against the UniRef90 database. The average sequencing depth was 23.9 million reads per sample, with minimum threshold set at 10 million reads for inclusion in analysis. Quality assessment performed with FastQC v0.11.9 [36] showed consistently high per-base quality scores (median Phred > Q30). Levels of sequence duplication were low, and no overrepresented adapter sequences were detected.
Statistical analyses were performed in R v4.2.2 [37] and Python v3.9.16 [38]. Alpha diversity metrics (ACE, Shannon, Simpson, and Pielou’s evenness indices) were calculated using the vegan package. Beta diversity was assessed using Bray–Curtis dissimilarity and Jaccard indices (scikit-bio v0.6.2). Differential abundance analysis was conducted using MaAsLin2 v1.12.0 [39] with treatment and block as fixed factors. Given the exploratory nature of this first metagenomic study in alkaline steppe soils, we used FDR-adjusted p < 0.25 for initial screening of differential features, with p < 0.05 considered significant for primary hypotheses.
PERMANOVA (adonis2 function, vegan package) assessed community structure differences with 999 permutations. ANOSIM and PERMDISP tests evaluated group separation and dispersion homogeneity. Correlation analysis examined correlations between microbial taxa, metabolic pathways, and soil properties using Spearman correlation (r > |0.7|, p < 0.01). Principal component analysis (PCA) visualized community structure patterns. LEfSe [40] identified biomarker taxa with LDA score > 2.0.
Marker taxa were defined as microbial species showing statistically significant differential abundance between groups, identified using Linear discriminant analysis Effect Size (LEfSe) with the following parameters: alpha value for factorial Kruskal–Wallis test = 0.05, threshold on the logarithmic LDA score for discriminative features = 2.0. Only taxa present in >50% of samples within at least one treatment group were considered for biomarker analysis.
3. Results
3.1. Plant Food Compound Content in Yellow Sweet Clover
Green mass yielding capacity of the plowed under yellow sweet clover of the first year was 11.6 t/ha. Before plowing under of yellow sweet clover, plant selection was carried out to identify plant food compounds in the dry matter of the aboveground and underground (roots) biomass. During the first year of growing, yellow sweet clover accumulated 3.24 t/ha of dry matter in the aboveground portion and 1.75 t/ha in the underground portion (roots) in the arable layer. In the aboveground biomass of yellow sweet clover, nitrogen content in the dry matter was 2.63%, phosphorus 0.30%, and potassium 2.01%. The yield from 1 ha of dry matter in the aboveground portion (3.24 t/ha) was: nitrogen 85.2 kg, phosphorus 9.7 kg, and potassium 65.1 kg. In the underground biomass of yellow sweet clover (roots), nitrogen content in the dry matter was 2.65%, phosphorus 0.28%, and potassium 1.60%. The yield from 1 ha of dry matter in the underground portion (1.75 t/ha) was: nitrogen 46.4 kg, phosphorus 4.9 kg, and potassium 28.0 kg. The total dry matter yield was 4.99 t/ha (3.24 t/ha aboveground + 1.75 t/ha underground). The total amount of nutrients (aboveground and underground mass combined) applied when plowed under in the variant of yellow sweet clover of the first year was as follows: nitrogen 131.6 kg/ha, phosphorus 14.6 kg/ha, and potassium 93.1 kg/ha.
3.2. Comparative Agrochemical Characteristics of Soil
The agrochemical analysis revealed significant alterations in soil properties following agricultural conversion from virgin steppe to cultivated systems (Table 1). One-way ANOVA indicated significant treatment effects for most parameters except exchangeable potassium and sulfur.

Table 1.
Soil agrochemical properties (0–20 cm) under different management systems in Northern Kazakhstan steppe (mean ± SD, n = 3).
Soil nitrate-nitrogen content showed unexpected patterns following sweet clover incorporation. Despite successful crop establishment, N- concentrations in sweet clover plots did not differ significantly from clean fallow plots. Both agricultural treatments showed substantial enrichment compared to the virgin steppe reference site (F2,6 = 42.7, p ≤ 0.001, η2 = 0.93). The numerically lower nitrate content in sweet clover plots suggests potential nitrogen immobilization during early decomposition processes at 50 days post-incorporation. Organic matter content showed severe depletion under cultivation. Sweet clover and clean fallow treatments showed no significant difference between each other, but both differed substantially from virgin steppe (5.5 ± 0.9%; F2,6 = 24.6, p ≤ 0.001, η2 = 0.89). This indicates rapid degradation following agricultural conversion. Soil pH increased significantly from virgin steppe (8.3 ± 0.2) to agricultural systems, reaching 9.0 ± 0.1 in both sweet clover and clean fallow plots (F2,6 = 24.2, p ≤ 0.001, η2 = 0.89). Available phosphorus (P2O5) demonstrated uniform enrichment under cultivation, with sweet clover and clean fallow showing nearly identical concentrations (p = 1.0). Both treatments exceeded virgin steppe levels by 3.9-fold (F2,6 = 34.9, p ≤ 0.001, η2 = 0.92), likely reflecting enhanced mineral weathering rather than management-specific effects.
Exchangeable potassium showed no significant treatment effects (F2,6 = 1.03, p = 0.41), though concentrations exceeded 1000 mg/kg in both sweet clover and clean fallow treatments, representing 8.4% and 10.4% increases over virgin steppe. These levels approach thresholds that can interfere with calcium-magnesium uptake in alkaline soils. Different superscripts within rows indicate significant differences at p < 0.05 (Tukey’s HSD test). The F-value shows the ratio of variance between groups to variance within groups; η2 (partial eta squared) shows the proportion of total variance explained by treatment effects.
3.3. Alpha Diversity Patterns
Shotgun metagenomic sequencing yielded a total of 399 species-level genome bins (SGBs) across all samples, with an average sequencing depth of 23.9 million high-quality reads per sample (median Phred > Q30). Comparative analysis of alpha diversity indices (Figure 2A) revealed distinct patterns in microbial community structure across three land-use systems. The Shannon diversity index for total microbial communities showed marginal differences (virgin steppe: 4.52 ± 0.21, sweet clover: 4.68 ± 0.18, clean fallow: 4.61 ± 0.15; p = 0.082). However, when analyzed separately, archaeal communities demonstrated significant responses: archaeal Shannon diversity increased from 1.23 ± 0.14 in virgin steppe to 1.89 ± 0.22 in sweet clover plots (p = 0.01), while bacterial Shannon indices remained stable (3.98 ± 0.19 vs. 4.05 ± 0.17, p = 0.31).
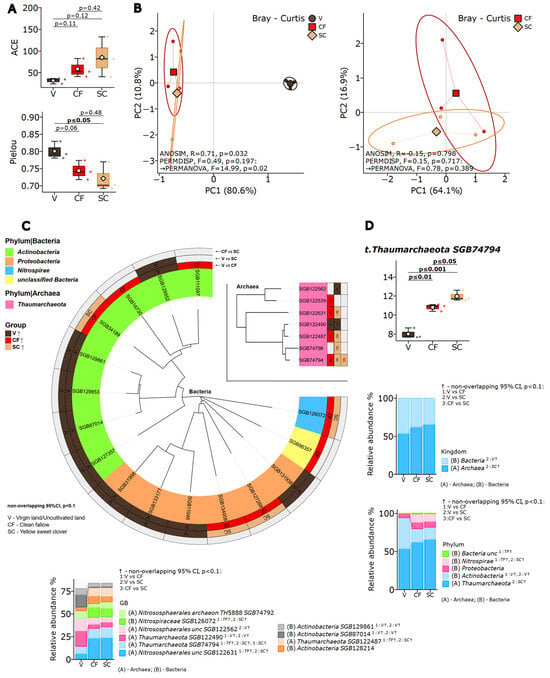
Figure 2.
Comprehensive microbiome analysis across land management systems. (A) Alpha diversity metrics showing ACE richness index and Pielou’s evenness for total microbial communities, with separate analysis for bacterial and archaeal domains. (B) Principal coordinate analysis of Bray–Curtis dissimilarity showing clear separation between virgin (black), sweet clover (Melilotus officinalis (L.)) (orange), and fallow (rad) communities. Ellipses represent 95% confidence intervals. (C) Cladogram of LEfSe-identified biomarker taxa with LDA scores > 2.0. Circle size proportional to taxon abundance, colors indicate enriched treatment. (D) Relative abundance of dominant taxa at kingdom, phylum, and species-level genome bin (SGB) levels. Only taxa with >1% mean abundance shown. Statistical significance assessed by Kruskal–Wallis test with Benjamini–Hochberg FDR correction.
Biomass incorporation of first-year sweet clover did not lead to statistically significant changes in overall microbial richness (ACE index, p > 0.05). However, Shannon diversity analysis showed a specific effect on archaeal communities: archaeal diversity increased significantly in the sweet clover treatment compared to the virgin steppe (p = 0.01). Bacterial diversity also tended to be higher in the sweet clover plots relative to the virgin soil, although this trend remained close to the threshold of statistical significance.
Pielou’s evenness index, measuring community balance through species abundance distribution, revealed that virgin steppe microbial communities maintained greater equilibrium with relatively similar species abundances. Variant SC showed a shift toward lower evenness, with a subset of taxa becoming more abundant, while others became rarer.
3.4. Beta Diversity and Community Structure
Beta diversity assessment revealed clear distinction between land-use systems (Figure 2). Table S1 presents the codes that are matched to the organisms to which the genomes belong. Virgin steppe soils (V) formed distinct clusters from all cultivated lands (CF and SC), while sweet clover and fallow showed overlapping community composition. Beta diversity assessment using Jaccard index confirmed group differences: ANOSIM test (R = 0.78, p = 0.017), PERMANOVA test (F = 11.71, p = 0.036), and marginal significance for homogeneity (PERMDISP test F = 1.42, p = 0.052). Bray–Curtis beta diversity analysis yielded similar results with confirmed differences: ANOSIM test (R = 0.71, p = 0.032), PERMANOVA test (F = 14.99, p = 0.02), with uniform variation between groups (PERMDISP test F = 0.49, p = 0.197).
3.5. Taxonomic Composition and Key Taxa
Taxonomic composition analysis at multiple hierarchical levels (Figure 2C) revealed systematic shifts in community structure. At the phylum level, agricultural conversion increased the relative abundance of Nitrospirae from 2.3% ± 0.4% in virgin steppe to 5.8% ± 0.7% in sweet clover and 5.6% ± 0.6% in clean fallow (p < 0.01). Marker taxa identified through LEfSe analysis (Figure 2D) demonstrated clear system-specific microbial signatures as detailed in Section LEfSe Biomarker Analysis.
Comprehensive taxonomic profiling identified a total of 399 microbial taxa across all samples, of which 157 showed differential abundance among land-use types at the exploratory threshold (FDR q < 0.25; see Limitations). These nitrite-oxidizing bacteria indicate enhancement of nitrogen cycling efficiency under cultivation.
Thaumarchaeota demonstrated pronounced system-specific responses in direct sweet clover-fallow comparisons, with selective enrichment of ammonia-oxidizing lineages under legume cultivation-specific Thaumarchaeota taxa (GGB53661_SGB74794 and GGB53661_SGB122487) showed significantly higher abundance in sweet clover systems compared to clean fallow (p ≤ 0.05).
LEfSe Biomarker Analysis
Linear discriminant analysis Effect Size (LEfSe) identified 23 biomarker taxa with LDA scores exceeding 2.0, revealing distinct microbial signatures across land management systems (Figure 2D).
Sweet clover systems were characterized by enrichment of nitrogen-fixing symbionts. Mesorhizobium muleiense showed the highest discriminatory power (LDA = 4.12, p < 0.001), followed by Bradyrhizobium sp. URHD0069 (LDA = 3.87, p < 0.001). Additional plant-associated taxa included Phyllobacterium myrsinacearum (LDA = 3.45, p = 0.002) and Rhizobium leguminosarum (LDA = 3.22, p = 0.003).
Virgin steppe maintained a distinct actinobacterial signature. Nocardioides astragali served as the primary biomarker (LDA = 4.35, p < 0.001), accompanied by Arthrobacter sp. P2b (LDA = 3.98, p < 0.001) and Blastococcus sp. CT_GayMR19 (LDA = 3.76, p = 0.002). These taxa are associated with organic matter decomposition and stress tolerance in oligotrophic environments.
Clean fallow systems exhibited intermediate biomarker profiles with enrichment of opportunistic copiotrophs, including Bacillus subtilis (LDA = 3.54, p = 0.004) and Pseudomonas fluorescens (LDA = 3.21, p = 0.008), reflecting adaptation to periodic disturbance.
At the family level, Nitrososphaeraceae (LDA = 3.89, p < 0.001) and Nitrospiraceae (LDA = 3.67, p = 0.002) emerged as biomarkers for agricultural systems collectively, confirming enhanced nitrification capacity under cultivation.
3.6. Functional Profile Analysis
Functional analysis demonstrated that major metabolic restructuring occurs during transition from natural to agricultural systems, with direct sweet clover–fallow comparisons revealing only five significant functional differences (p < 0.05 after FDR correction), confirming metabolic convergence of agricultural systems. These five pathways were: polyisoprenoid biosynthesis (POLYISOPRENSYN-PWY), chorismate biosynthesis from 3-dehydroquinate (PWY-6163), Bifidobacterium shunt (P124-PWY), homolactate fermentation (ANAEROFRUCAT-PWY), and 4-oxopentanoate degradation (PWY-7948).
Quantitative functional profiling revealed 74 exploratory metabolic pathways with differential abundance after FDR correction (q < 0.25). Sweet clover systems showed 2.8-fold enrichment of amino acid biosynthesis pathways, including L-methionine biosynthesis III (log2FC = 2.14, q = 0.018) and L-lysine/threonine/methionine superpathway (log2FC = 1.87, q = 0.032). Virgin steppe maintained 69 pathways substantially enriched relative to agricultural systems (detected at <0.001% relative abundance in SC and CF soils), predominantly involving sulfur metabolism (PWY-801, log2FC = −1.68, q = 0.008) and coenzyme biosynthesis (COA-PWY-1, log2FC = −1.92, q = 0.006).
Nitrogen cycling genes showed treatment-specific enrichment: amoA gene abundance increased 4.5-fold in sweet clover (1.8 × 106 vs. 4.0 × 105 copies/g soil, p = 0.003), while nxrB genes increased uniformly 2.3-fold in both agricultural systems (p = 0.008). Functional gene diversity (Shannon index) decreased from 6.82 ± 0.15 in virgin steppe to 6.21 ± 0.18 in agricultural soils (p = 0.012), confirming functional homogenization.
Virgin steppe maintained 69 pathways substantially enriched relative to agricultural systems at the exploratory threshold (FDR q < 0.25; see Limitations), detected at <0.001% relative abundance in SC and CF soils. Among these, enhanced sulfur-containing amino acid metabolism (PWY-801: homocysteine-cysteine conversions, p ≤ 0.01) and coenzyme A biosynthesis (COA-PWY-1: coenzyme A biosynthesis III superpathway, p ≤ 0.01) met the stringent significance threshold. These pathways reflect complex metabolic networks of natural microbial consortia adapted to the oligotrophic conditions of steppe ecosystems.
Agricultural systems exhibited specialization in amino acid and vitamin biosynthesis, with both sweet clover and fallow systems significantly enhancing L-methionine biosynthesis pathways (HSERMETANA-PWY: L-methionine biosynthesis III, p ≤ 0.001), L-lysine/threonine/methionine biosynthesis superpathways (P4-PWY: L-lysine, L-threonine and L-methionine biosynthesis I superpathway, p ≤ 0.05), and menaquinol-8 synthesis (PWY-7992: menaquinol-8 biosynthesis III superpathway, p ≤ 0.01). This functional convergence indicates that agricultural management creates selective pressure favoring intensive protein metabolism and enhanced vitamin K biosynthesis.
Sweet clover-specific effects manifested in carbon-energy metabolism, including enhanced polyisoprenoid biosynthesis (POLYISOPRENSYN-PWY: polyisoprenoid biosynthesis, p ≤ 0.01) and aromatic metabolism pathway modification, particularly chorismate biosynthesis (PWY-6163: chorismate biosynthesis from 3-dehydroquinate, p ≤ 0.05). Conversely, clean fallow was characterized by enhancement of fermentative pathways, including the Bifidobacterium shunt (P124-PWY: Bifidobacterium shunt, p ≤ 0.05), homolactate fermentation (ANAEROFRUCAT-PWY: homolactate fermentation, p ≤ 0.01), and 4-oxopentanoate degradation (PWY-7948: 4-oxopentanoate degradation, p ≤ 0.01), reflecting adaptation to anaerobic conditions and organic residue degradation.
Functional specialization in the nitrogen cycle manifested through differential activation of coenzyme M biosynthesis pathways (P261-PWY: coenzyme M biosynthesis I, p ≤ 0.05), a critical cofactor in methanogenesis and anaerobic methane oxidation. Both agricultural systems showed enhancement of this pathway relative to virgin conditions, indicating modification of anaerobic microbial processes in cultivated soils.
3.7. Correlation Analysis and Nutrient Controls
Correlation analysis of the combined dataset from sweet clover agroecosystems, virgin sites, and clean fallow identified phosphorus as the primary regulator of microbial metabolism, demonstrating significant correlations (r = 1.0, p < 0.0001) with microbial glycolysis and L-methionine and folate metabolism pathways including PWY-8004: Entner-Doudoroff pathway I and GLYCOLYSIS-E-D: superpathway of glycolysis and the Entner-Doudoroff pathway (r = 0.95, p < 0.0001), HSERMETANA-PWY: L-methionine biosynthesis III (r = 0.87, p < 0.001), and 1CMET2-PWY: folate transformations III (r = 0.9, p < 0.001) (Figure 3). Soil organic matter demonstrated strong positive associations (r = 0.8–0.9, p < 0.001) with central carbon metabolism networks across all studied land-use systems, particularly with pathways crotonyl-CoA/ethylmalonyl-CoA/hydroxybutyryl-CoA (r = 0.81, p < 0.01).
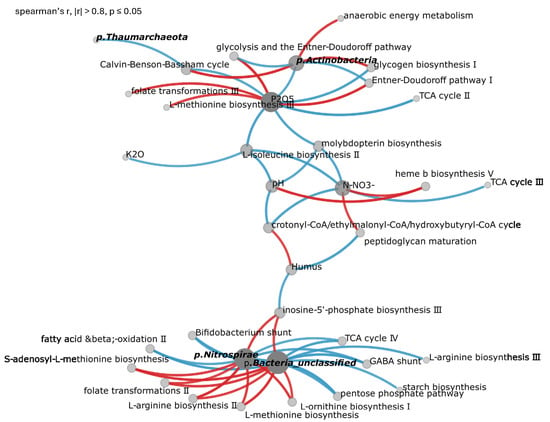
Figure 3.
Correlation network of marker taxa, metabolic pathways, and soil agrochemical properties. The network includes only significant Spearman correlations (|r| > 0.8, p ≤ 0.05) and comprises 36 nodes and 54 edges. Red lines indicate positive correlations (n = 23); blue lines indicate negative correlations (n = 31). Node size is proportional to node degree (number of connections).
Sweet clover systems were characterized by distinctive microbiome signatures with enrichment of nitrogen-fixing symbionts Mesorhizobium muleiense and Bradyrhizobium sp. URHD0069 (r = 0.8–0.9), coupled with activation of amino acid biosynthesis pathways including HSERMETANA-PWY (L-methionine biosynthesis III), ARGSYNBSUB-PWY (L-arginine biosynthesis II), and PWY-5101 (L-isoleucine biosynthesis II). Natural grassland communities maintained stable actinobacterial networks with key taxa Nocardioides astragali, Arthrobacter sp. P2b, and Blastococcus sp. CT_GayMR19, driving organic matter decomposition through pathways PWY-622 (starch biosynthesis) and GLYCOGENSYNTH-PWY (glycogen biosynthesis I).
3.8. Hierarchical Taxonomic Patterns
Taxonomic analysis across several hierarchical levels revealed that, at the family level, Phyllobacteriaceae and Nocardioidaceae served as key taxa (r = 0.8–1.0) linking soil chemistry with metabolic output. Class-level analysis identified Actinobacteria_CFGB52786 and Alphaproteobacteria as primary drivers of soil carbon cycling, while c_Nitrososphaeria drove nitrogen transformations through ammonia-oxidizing archaea, including Thaumarchaeota_GGB53661_SGB122487 and Nitrososphaerales_unclassified_GGB53670_SGB122539.
Correlation analysis revealed that elevated K2O, N-, and pH generated strong negative correlations (r = −0.7 to −0.9, p < 0.01) with essential metabolic pathways including PWY-7854 (crotonyl-CoA/ethylmalonyl-CoA/hydroxybutyryl-CoA cycle), PWY-6823 (molybdopterin biosynthesis), and PWY-7383 (anaerobic energy metabolism).
4. Discussion
Our investigation of first-year sweet clover green manuring in alkaline steppe soils revealed unexpected patterns that challenge conventional assumptions about legume-based soil restoration. Despite successful establishment of nitrogen-fixing symbionts (Mesorhizobium muleiense and Bradyrhizobium sp.), sweet clover cultivation did not enhance soil nitrogen availability compared to clean fallow (9.1 vs. 10.5 mg NO3-N/kg). Instead, we observed selective microbial restructuring with enhanced archaeal diversity but functional convergence between agricultural systems—only five metabolic pathways differed significantly between sweet clover and fallow treatments. Most critically, correlation analysis identified phosphorus, not nitrogen, as the primary controller of microbial metabolism (r = 1.0, p < 0.0001), while elevated pH (9.0) and potassium (>1000 mg/kg) emerged as previously unrecognized constraints on soil biological processes.
| The Nitrogen Paradox: Why Green Manure Failed to Enhance N Availability? |
The lower nitrate content in sweet clover plots compared to fallow contradicts expected benefits of legume incorporation but aligns with nitrogen immobilization dynamics during early decomposition phases. When plant residues with C:N ratios exceeding 20:1 are incorporated, soil microorganisms temporarily sequester available nitrogen to metabolize carbon-rich substrates [5]. We therefore hypothesize that our sampling at 50 days post-incorporation captured this immobilization phase, before net mineralization releases plant-available nitrogen—a process requiring extended periods in semi-arid conditions.
The strongly alkaline conditions (pH 9.0) compound this nitrogen limitation through multiple mechanisms. At pH > 8.5, nitrification rates decline substantially due to ammonia volatilization and enzymatic inhibition [6]. Our enrichment of ammonia-oxidizing archaea (Thaumarchaeota) represents an adaptive response, as these organisms possess superior ammonia affinity (Km = 133 nM) compared to bacterial nitrifiers (Km > 50 μM), allowing them to function under substrate limitation [12]. However, even enhanced archaeal populations cannot fully compensate for pH-induced constraints on nitrogen cycling. This explains why both agricultural systems showed similar Nitrospirae enrichment—cultivation itself, rather than specific management, drives selection for stress-tolerant nitrifiers [13].
4.1. Taxonomic Restructuring: Nitrification Enhancement as Universal Agricultural Response
The enrichment of Nitrospirae across both agricultural systems indicates that cultivation itself, rather than specific management practices, drives selection for specialized nitrifying communities. This universal enhancement aligns with recent studies showing that phylum Nitrospirae represents a key component of soil microbial communities with enhanced activity in agricultural systems [13,41]. Contemporary metagenomic studies confirm the crucial role of Nitrospira not only in nitrite oxidation but also in urea hydrolysis, providing ammonia for ammonia-oxidizing microorganisms [42]. The discovery of complete ammonia oxidizers (comammox) within Nitrospira further emphasizes their central role in nitrogen cycling across diverse agricultural environments [13].
The selective enrichment of specific Thaumarchaeota lineages (SGB74794 and GGB53661_SGB74794) in sweet clover systems represents a more targeted response to legume cultivation. These observations are fully supported by large-scale studies demonstrating that ammonia-oxidizing archaea (AOA) can outnumber ammonia-oxidizing bacteria 50-430-fold in fertile agricultural soils, with Thaumarchaeota dominating nitrification processes [10,43]. Studies have shown that group I.1a Thaumarchaeota are particularly active in agricultural soils and demonstrate autotrophic growth during ammonia oxidation [44]. Stable isotope profiling confirms that soil Thaumarchaeota perform autotrophic ammonia oxidation through the highly efficient 3-hydroxypropionate/4-hydroxybutyrate CO2 fixation cycle [11].
The contrasting patterns—universal Nitrospirae enrichment versus selective Thaumarchaeota response—suggest a two-tiered restructuring of nitrifying communities under cultivation. While both agricultural systems require enhanced nitrite oxidation capacity (explaining Nitrospirae enrichment), only sweet clover provides the specific rhizosphere conditions that favor archaeal ammonia oxidizers. This specialization may reflect the superior ammonia affinity of Thaumarchaeota (Km = 133 nM) [12], allowing them to efficiently capture ammonia released during legume residue decomposition before volatilization losses occur at pH 9.0.
4.2. Phosphorus as the Master Regulator: Implications for Restoration
The emergence of phosphorus as the dominant factor controlling microbial metabolism fundamentally reframes our understanding of nutrient limitations in carbonate soils. The strong correlations between available P and essential pathways—peptidoglycan synthesis, nucleotide biosynthesis, and heme production—indicate that cellular growth and energy metabolism are P-limited rather than N-limited in these systems. With available P at 18.5 mg/kg in both agricultural treatments versus 4.7 mg/kg in virgin steppe, cultivation appears to mobilize phosphorus through enhanced weathering and organic acid production, independent of green manure addition.
In carbonate-rich soils, calcium-phosphate precipitation reduces P bioavailability by 10–100-fold compared to neutral soils [18]. Our pH of 9.0 exacerbates this constraint, as phosphate solubility decreases exponentially above pH 7.5. The correlation analysis revealing P control over 12 major metabolic pathways suggests that microbial communities have adapted to extreme P limitation through metabolic downregulation. This finding aligns with recent evidence that P availability controls microbial carbon use efficiency and growth rates in alkaline soils [19]. Without addressing phosphorus constraints—through acidification, organic acid application, or phosphate-solubilizing inoculants—green manure alone cannot restore soil biological function [20].
4.3. Network Architecture and Nutrient Toxicity Thresholds
Beyond phosphorus limitation, our correlation analysis revealed unexpected antagonistic effects of elevated nutrients on microbial metabolism. The strong negative correlations between elevated K2O (>1000 mg/kg), N-, pH (9.0) and essential metabolic pathways—including crotonyl-CoA/ethylmalonyl-CoA cycles, molybdopterin biosynthesis, and anaerobic energy metabolism—indicate nutrient toxicity thresholds previously unrecognized in agricultural systems. These results align with recent studies showing that soil pH, available phosphorus, and NO3-N content significantly influence nitrification and denitrification processes [45,46]. Studies demonstrate that excessive nitrogen fertilizer application in alkaline soils causes high nitrification rates and substantial nitrogen losses through N2O emission and leaching [45,46]. Metagenomic studies show that initial soil nutrient status and pH modulate fertilizer effects on microbial carbon and nitrogen cycling processes, confirming our observations on threshold effects [47].
The correlation analysis further demonstrates that soil organic matter serves as a critical mediator of phosphorus effects, showing strong positive associations with central carbon metabolism pathways. This aligns with recent studies showing that soil phosphorus concentration strongly regulates microbial carbon and nitrogen metabolism [48].
Metagenomic studies demonstrate that high soil phosphorus concentrations promote carbon fixation and methane oxidation processes, while phosphorus is a key element in regulating the genes associated with inorganic phosphorus solubilization and organic phosphorus mineralization [49]. The tight coupling between P availability and carbon metabolism explains why organic matter additions alone may fail to restore soil function without addressing phosphorus constraints.
The specificity of sweet clover systems in establishing nitrogen-fixing symbiont networks (Mesorhizobium muleiense, Bradyrhizobium sp. URHD0069) coupled with enhanced amino acid biosynthesis pathways provides insight into legume-microbe mutualism. Recent studies confirm the specificity of associations between legumes and rhizobia strains, demonstrating that nitrogen fixation efficiency depends on plant-microbe pair compatibility [50]. Studies show that rhizosphere interactions between legumes and soil microorganisms enhance amino acid biosynthesis gene expression by 200–300% compared to non-legume systems [51]. Large-scale metagenomic studies revealed global distribution of Deltaproteobacteria in nitrogen-fixing microbiomes, complementing our data on specific associations in sweet clover systems [50]. Research in Agronomy shows the importance of plant–soil–microbe interactions in determining soil biological fertility through rhizosphere nutrient cycling changes [52]. However, these beneficial associations appear insufficient to overcome the multiple constraints of alkaline chemistry and nutrient imbalances documented in our study.
These findings suggest potential nutrient imbalance effects that may influence conventional fertilization approaches in alkaline steppe soils. Rather than simple nutrient deficiency, these systems may exhibit a complex optimization problem where elevated concentrations of some nutrients (K, ) could negatively affect microbial processes while others (P) remain limiting. However, these correlative observations require experimental validation to confirm causal relationships. This suggests that precision nutrient management—targeting optimal rather than maximal levels—may be more effective than traditional blanket fertilization strategies.
The hierarchical taxonomic analysis revealed how community assembly operates across multiple organizational scales. At the family level, Phyllobacteriaceae and Nocardioidaceae emerged as keystone taxa bridging soil chemistry and metabolic function. Contemporary research confirms the key role of family Phyllobacteriaceae in plant–microbe interactions and carbon metabolism [53]. At the class level, Actinobacteria and Alphaproteobacteria drive carbon cycling, while Nitrososphaeria (including Thaumarchaeota groups) control nitrogen transformations. Analysis shows dominance of Thaumarchaeota groups I.1a and I.1b in fertilized soils, consistent with our Nitrososphaeria results [54,55].
This hierarchical organization demonstrates functional redundancy—multiple taxa at lower taxonomic levels can perform similar functions, explaining why agricultural systems maintain basic biogeochemical cycling despite dramatic taxonomic shifts. The integration of taxonomic and functional analyses in this study reveals functional convergence as a dominant pattern in agricultural microbiomes and identifies phosphorus as the primary regulator of microbial metabolism in alkaline steppe soils—insights with potential implications for understanding nutrient limitations in carbonate-rich agricultural systems globally. Microbiological indicators demonstrate high sensitivity and rapid response to environmental changes, making them valuable tools for assessing agricultural practice impacts on soil ecological functions [56].
4.4. Functional Convergence: Agricultural Selection Overrides Management
The detection of only five differential metabolic pathways between sweet clover and fallow, despite distinct taxonomic composition, demonstrates that agricultural selection pressures override specific management effects. Both systems converged on enhanced amino acid biosynthesis (methionine, lysine, threonine), vitamin K production, and specialized carbon metabolism. This functional redundancy suggests that cultivation creates uniform selective pressures—alkalinity stress, nutrient imbalance, and reduced organic matter—that filter communities toward similar metabolic capabilities regardless of plant inputs [3].
Virgin steppe maintained 69 unique pathways absent from agricultural soils, including complex sulfur metabolism and specialized coenzyme biosynthesis. This functional diversity loss represents the true cost of agricultural conversion, more significant than simple species loss [57]. The enrichment of fermentation pathways in fallow (Bifidobacterium shunt, homolactate fermentation) versus polyisoprenoid biosynthesis in sweet clover indicates subtle niche differentiation, but these differences are minor compared to the agriculture-steppe divide. This convergence explains why decades of research on green manures in alkaline soils show inconsistent results [4]—the overriding constraints of pH and nutrient imbalance mask potential benefits.
4.5. Practical Implications for Steppe Agriculture
Our findings indicate that successful restoration of degraded steppe soils requires integrated management addressing multiple constraints simultaneously. Single-season sweet clover incorporation cannot overcome the combined limitations of phosphorus deficiency, pH stress, and 50% organic matter loss [9]. The identification of putative nutrient stress thresholds—particularly potassium > 1000 mg/kg inhibiting essential metabolic pathways—suggests that conventional fertilization may actually impair soil biological recovery [45].
For practical implementation, we recommend the following: (1) multi-year legume rotations to allow complete mineralization cycles; (2) co-application of phosphorus amendments or phosphate-solubilizing bacteria; (3) organic amendments to buffer pH and provide diverse carbon substrates; (4) reduced reliance on mineral fertilizers that may exceed toxicity thresholds. The 20–50-year timeline for organic matter recovery [58] requires managing expectations and developing intermediate indicators of soil health improvement. Monitoring archaeal–bacterial ratios and functional gene abundance may provide earlier feedback than traditional soil tests [59].
4.6. Future Research Priorities
This initial assessment raises critical questions requiring multi-year investigation. Time-series sampling across complete growing seasons would capture nitrogen mineralization dynamics and seasonal microbial succession. Metatranscriptomic analysis could distinguish between genetic potential and actual metabolic activity [43], while stable isotope probing would trace nitrogen and carbon flow through microbial networks. Gross nitrogen transformation rates and enzyme activities remain essential for mechanistic understanding. Comparative studies across pH gradients (7.0–9.5) and phosphorus levels could identify threshold values for successful green manure implementation. Testing integrated amendments—legume + phosphorus + organic acids—may reveal synergistic effects masked in single-factor trials. Finally, developing region-specific microbial inoculants adapted to alkaline conditions could enhance green manure effectiveness where native communities are degraded beyond natural recovery capacity [60].
4.7. Limitations
This study captures only initial decomposition (50 days post-incorporation), missing the complete nitrogen mineralization cycle and multi-year dynamics essential for evaluating green manure effectiveness. Shotgun metagenomics reveals genetic potential, not actual metabolic activity, while absence of enzyme assays, gross N transformation rates, and greenhouse gas measurements prevents mechanistic understanding. The liberal FDR threshold (p < 0.25) and four replications limit statistical power, while network correlations cannot establish causality. Site-specific conditions—pH 9.0, potassium > 1000 mg/kg, 50% organic matter loss—may not represent typical steppe soils. Comparing first-year sweet clover to established fallow introduces temporal asymmetry, while lack of untreated controls prevents separation of cultivation from green manure effects. Additionally, the virgin steppe reference site is located at a different coordinate than the agricultural treatments (SC and CF), and while all sites share similar soil type (southern carbonate chernozem) and climatic conditions, we cannot fully exclude potential site effects and pseudo-replication concerns inherent to this spatial design. Future studies should incorporate multiple virgin reference sites across the region to strengthen comparative inferences. These limitations indicate our findings should guide hypothesis generation rather than definitively conclude green manure ineffectiveness, emphasizing need for multi-year, multi-site validation with process-level measurements.
5. Conclusions
This metagenomic investigation of sweet clover green manuring in Northern Kazakhstan’s alkaline steppe soils reveals that single-season legume incorporation induces selective rather than comprehensive microbiome restructuring. While sweet clover successfully established nitrogen-fixing symbionts and enhanced archaeal diversity, these changes did not translate into improved nitrogen availability or distinct functional advantages over clean fallow in the first year. The identification of phosphorus as the master regulator of microbial metabolism, combined with pH-related enzymatic inhibition (pH 9.0) and nutrient toxicity thresholds (K2O > 1000 mg/kg), demonstrates that multiple biogeochemical constraints limit green manure effectiveness in carbonate soils. Functional convergence between agricultural systems—with only five differential metabolic pathways despite distinct taxonomic composition—suggests that overriding effects of cultivation mask potential management-specific benefits. Virgin steppe’s retention of 69 unique metabolic pathways underscores the profound functional diversity loss accompanying agricultural conversion. These findings indicate that restoration of degraded steppe soils requires integrated long-term strategies addressing phosphorus mobilization, pH amelioration, and organic matter rebuilding, with realistic recovery timelines spanning decades rather than seasons. This work provides essential baseline data for developing evidence-based sustainable intensification practices adapted to the world’s extensive alkaline drylands. It should be emphasized that this study represents a first-season assessment, and delayed benefits of green manuring—including gradual organic matter accumulation, improved soil structure, and enhanced microbial network complexity—may manifest beyond our 50-day sampling window. Multi-year studies incorporating process-level measurements remain necessary to fully evaluate green manure potential under these challenging edaphic conditions.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agriculture16010036/s1, Table S1: Metagenomic species-level genome bin (SGB) codes and corresponding taxonomic identifications. Each SGB represents a cluster of genomes with >95% average nucleotide identity, following the MetaPhlAn 4 database nomenclature (mpa_vJan21_CHOCOPhlAnSGB_202103).
Author Contributions
Conceptualization, I.R. and A.K.; methodology, I.R., A.K., N.F., N.Z. and L.Z.; software, A.K. and S.K.; validation, A.K., S.K. and N.Z.; formal analysis, A.K., S.K., N.Z. and L.Z.; investigation, I.R., A.K., S.K., N.F., N.Z. and L.Z.; resources, I.R., A.K., S.K., N.F. and N.Z.; data curation, A.K., S.K. and I.R.; writing—original draft preparation, A.K., S.K. and I.R.; writing—review and editing, I.R., A.K. and N.F.; visualization, A.K., S.K. and I.R.; supervision, I.R.; project administration, I.R.; funding acquisition, I.R. and N.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Science Committee of the Ministry of Science and Higher Education of the Republic of Kazakhstan (Grant No. AP23487280).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Sequence data of the current study have been deposited in the NCBI BioProject; the primary accession number is PRJNA1333381. Additional data are available from the corresponding author upon reasonable request.
Conflicts of Interest
Authors Irina Rukavitsina, Nadezhda Filippova, Natalya Zuyeva and Lyudmila Zhloba were employed by the “A.I. Barayev Research and Production Centre for Grain Farming” LLP. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Kuypers, M.; Marchant, H.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Tsiafouli, M.A.; Thébault, E.; Sgardelis, S.P.; de Ruiter, P.C.; van der Putten, W.H.; Birkhofer, K.; Hemerik, L.; de Vries, F.T.; Bardgett, R.D.; Brady, M.V.; et al. Intensive agriculture reduces soil biodiversity across Europe. Glob. Change Biol. 2015, 21, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Polz, M.F.; Mazel, F.; Albright, M.B.N.; Huber, J.A.; O’Connor, M.I.; Ackermann, M.; Hahn, A.S.; Srivastava, D.S.; Crowe, S.A.; et al. Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2018, 2, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Leff, J.W.; Jones, S.E.; Prober, S.M.; Barberán, A.; Borer, E.T.; Firn, J.L.; Harpole, W.S.; Hobbie, S.E.; Hofmockel, K.S.; Knops, J.M.H.; et al. Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl. Acad. Sci. USA 2015, 112, 10967–10972. [Google Scholar] [CrossRef]
- Trinsoutrot, I.; Recous, S.; Bentz, B.; Linères, M.; Chèneby, D.; Nicolardot, B. Biochemical quality of crop residues and carbon and nitrogen mineralization kinetics under nonlimiting nitrogen conditions. Soil Sci. Soc. Am. J. 2000, 64, 918–926. [Google Scholar] [CrossRef]
- Acosta-Martínez, V.; Tabatabai, M. Enzyme activities in a limed agricultural soil. Biol. Fertil. Soils 2000, 31, 85–91. [Google Scholar] [CrossRef]
- Josephson, P.; Dronin, N.; Mnatsakanian, R.; Cherp, A.; Efremenko, D.; Larin, V. An Environmental History of Russia; Cambridge University Press: Cambridge, UK, 2013; p. 340. [Google Scholar] [CrossRef]
- Suleimenov, M.; Oram, P. Trends in Feed, Livestock Production, and Rangelands during the Transition Period in Three Central Asian Countries. Food Policy 2000, 25, 681–693. [Google Scholar] [CrossRef]
- Sommer, R.; de Pauw, E. Organic Carbon in Soils of Central Asia—Status Quo and Potentials for Sequestration. Plant Soil 2011, 338, 273–288. [Google Scholar] [CrossRef]
- Leininger, S.; Urich, T.; Schloter, M.; Schwark, L.; Qi, J.; Nicol, G.W.; Prosser, J.I.; Schuster, S.C.; Schleper, C. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 2006, 442, 806–809. [Google Scholar] [CrossRef]
- Könneke, M.; Schubert, D.M.; Brown, P.C.; Hügler, M.; Standfest, S.; Schwander, T.; von Borzyskowski, L.S.; Erb, T.J.; Stahl, D.A.; Berg, I.A. Ammonia-Oxidizing Archaea Use the Most Energy-Efficient Aerobic Pathway for CO2 Fixation. Proc. Natl. Acad. Sci. USA 2014, 111, 8239–8244. [Google Scholar] [CrossRef]
- Martens-Habbena, W.; Berube, P.; Urakawa, H.; de la Torre, J.R.; Stahl, D.A. Ammonia Oxidation Kinetics Determine Niche Separation of Nitrifying Archaea and Bacteria. Nature 2009, 461, 976–979. [Google Scholar] [CrossRef] [PubMed]
- Daims, H.; Lebedeva, E.; Pjevac, P.; Han, P.; Herbold, C.; Albertsen, M.; Jehmlich, N.; Palatinszky, M.; Vierheilig, J.; Bulaev, A.; et al. Complete Nitrification by Nitrospira Bacteria. Nature 2015, 528, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Turkington, R.A.; Cavers, P.B.; Rempel, E. The Biology of Canadian Weeds: 29. Melilotus alba Desr. and M. officinalis (L.) Lam. Can. J. Plant Sci. 1978, 58, 523–537. [Google Scholar] [CrossRef]
- Hassan, S.; Mathesius, U. The Role of Flavonoids in Root–Rhizosphere Signalling: Opportunities and Challenges for Improving Plant–Microbe Interactions. J. Exp. Bot. 2012, 63, 3429–3444. [Google Scholar] [CrossRef]
- Filippova, N.; Rukavitsina, I.; Parsayev, E.; Churkina, G.; Kobernitskaia, T.; Tkachenko, O.; Kunanbayev, K.; Ostrovski, V.; Mustafina, N. Creation of a New Highly Productive Parent Material of Sweet Clover (Melilotus Adans.) Based on Varietal and Microbial Systems. OnLine J. Biol. Sci. 2022, 22, 165–176. [Google Scholar] [CrossRef]
- Poulton, J.L.; McRee, D.E.; Conn, E.E. Accumulation of Coumarin and Its 7-Hydroxy Derivative in Sweet Clover Infected with Fusarium Species. Phytochemistry 1980, 19, 2499–2503. [Google Scholar]
- Penn, C.J.; Camberato, J.J. A Critical Review on Soil Chemical Processes that Control How Soil pH Affects Phosphorus Availability to Plants. Agriculture 2019, 9, 120. [Google Scholar] [CrossRef]
- Spohn, M. Phosphorus and carbon in soil particle size fractions: A synthesis. Biogeochemistry 2020, 147, 225–242. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Porder, S.; Houlton, B.Z.; Chadwick, O.A. Terrestrial phosphorus limitation: Mechanisms, implications, and nitrogen-phosphorus interactions. Ecol. Appl. 2010, 20, 5–15. [Google Scholar] [CrossRef]
- IUSS Working Group WRB. World Reference Base for Soil Resources: International Soil Classification System for Naming Soils and Creating Legends for Soil Maps, 4th ed.; International Union of Soil Sciences (IUSS): Vienna, Austria, 2022. [Google Scholar]
- Shpakov, A.S.; Novoselov, Y.K.; Kharkov, G.D.; Volovik, V.T.; Truzina, L.A.; Prologova, T.V.; Ulanov, A.N.; Laretin, N.A.; Usoltseva, T.G. Methodical Basis of Field Experiments with Fodder Crops; Usoltseva: Moscow, Russia, 2024; 332p, Available online: https://www.vniikormov.ru/pdf24/metodicheskie-osnovy-polevykh-opytov-s-kormovymi-kulturami.pdf (accessed on 16 December 2025).
- Dospekhov, B.A.; Vassilyev, I.P.; Tulikov, A.M. Practical Course on Arable Farming; Agropromizdat: Moscow, Russia, 1987; pp. 242–253. [Google Scholar]
- ISO 5983-1:2005; Animal Feedstuffs. Identification of Nitrogen Content and Calculation of Crude Protein Content. Part 1. The Kjeldahl Method. International Organization for Standardization. Geneva, Switzerland, 2005. Available online: https://meganorm.ru/Data2/1/4293775/4293775094.pdf (accessed on 16 December 2025).
- GOST 26657-97; Feedstuffs, Feed Compounds, Feed Compound Raw Materials. Phosphorus Content Identification Methods. Russian State Standard: Moscow, Russia, 1997. Available online: https://files.stroyinf.ru/Data2/1/4294827/4294827784.pdf (accessed on 16 December 2025).
- GOST 30504-97; Feedstuffs, Feed Compounds, Feed Compound Raw Materials. Flame Photometry Method for Identification of Potassium Content. Russian State Standard. Moscow, Russia, 1997. Available online: https://files.stroyinf.ru/Data2/1/4294824/4294824846.pdf (accessed on 16 December 2025).
- GOST 26488-85; Soils. Nitrate Testing According to TsINAO Method. Russian State Standard: Moscow, Russia, 1985. Available online: https://files.stroyinf.ru/Data2/1/4294827/4294827941.pdf (accessed on 16 December 2025).
- GOST 26205-91; Soils. Testing Mobile Forms of Phosphorus and Potassium by the Machigin Method as Modified by TsINAO. Russian State Standard. Moscow, Russia, 1991. Available online: https://files.stroyinf.ru/Data2/1/4294828/4294828275.htm (accessed on 16 December 2025).
- GOST 26213-2021; Soils. Methods of Organic Matter Testing. Russian State Standard. Moscow, Russia, 2021. Available online: https://files.stroyinf.ru/Data/758/75803.pdf (accessed on 16 December 2025).
- ISO 14255:1998; Soil Quality—Determination of Nitrate Nitrogen, Ammonium Nitrogen and Total Soluble Nitrogen in Air-Dry Soils Using Calcium Chloride Solution as Extractant. Available online: https://www.iso.org/standard/23081.html (accessed on 16 December 2025).
- Standard Operating Procedure for Soil Organic Carbon Walkley-Black Method Titration and Colorimetric Method. 2019. Available online: https://openknowledge.fao.org/server/api/core/bitstreams/e498d73e-1711-4d18-9183-aa8476387e2c/content (accessed on 16 December 2025).
- ISO/IEC 17025:2017; General Requirements for the Competence of Testing and Calibration Laboratories. ISO: Geneva, Switzerland, 2017.
- The Huttenhower Lab. KneadData. Version 0.12.0. 2020. Available online: http://huttenhower.sph.harvard.edu/kneaddata (accessed on 15 September 2025).
- Blanco-Míguez, A.; Beghini, F.; Cumbo, F.; McIver, L.J.; Thompson, K.N.; Zolfo, M.; Manghi, P.; Dubois, L.; Huang, K.D.; Thomas, A.M.; et al. Extending and Improving Metagenomic Taxonomic Profiling with Uncharacterized Species Using MetaPhlAn 4. Nat. Biotechnol. 2023, 41, 1633–1644. [Google Scholar] [CrossRef]
- Beghini, F.; McIver, L.J.; Blanco-Míguez, A.; Dubois, L.; Asnicar, F.; Maharjan, S.; Mailyan, A.; Manghi, P.; Scholz, M.; Thomas, A.M.; et al. Integrating Taxonomic, Functional, and Strain-Level Profiling of Diverse Microbial Communities with bioBakery 3. eLife 2021, 10, e65088. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 13 November 2025).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. Available online: https://www.R-project.org/ (accessed on 13 November 2025).
- Python Software Foundation. Python Language Reference, Version 3.9; Python Software Foundation: Beaverton, OR, USA, 2023; Available online: https://www.python.org/ (accessed on 10 November 2025).
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable Association Discovery in Population-Scale Meta-Omics Studies. PLoS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Pester, M.; Maixner, F.; Berry, D.; Rattei, T.; Koch, H.; Lücker, S.; Nowka, B.; Richter, A.; Spieck, E.; Lebedeva, E.; et al. NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ. Microbiol. 2014, 16, 3055–3071. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.; Lücker, S.; Albertsen, M.; Kitzinger, K.; Herbold, C.; Spieck, E.; Nielsen, P.H.; Wagner, M.; Daims, H. Expanded Metabolic Versatility of Ubiquitous Nitrite-Oxidizing Bacteria from the Genus Nitrospira. Proc. Natl. Acad. Sci. USA 2015, 112, 11371–11376. [Google Scholar] [CrossRef] [PubMed]
- Prosser, J.I.; Nicol, G.W. Archaeal and bacterial ammonia-oxidisers in soil: The quest for niche specialisation and differentiation. Trends Microbiol. 2012, 20, 523–531. [Google Scholar] [CrossRef]
- Tourna, M.; Freitag, T.E.; Nicol, G.W.; Prosser, J.I. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 2008, 10, 1357–1364. [Google Scholar] [CrossRef]
- Ju, X.; Xing, G.; Chen, X.; Zhang, S.; Zhang, L.; Liu, X.; Cui, Z.; Yin, B.; Christie, P.; Zhu, Z.; et al. Reducing environmental risk by improving N management in intensive Chinese agricultural systems. Proc. Natl. Acad. Sci. USA 2009, 106, 3041–3046. [Google Scholar] [CrossRef]
- Ju, X.-T.; Zhang, C. Nitrogen cycling and environmental impacts in upland agricultural soils in North China: A Review. J. Integr. Agric. 2017, 16, 2848–2862. [Google Scholar] [CrossRef]
- Guo, J.; Liu, W.; Zhu, C.; Luo, G.; Kong, Y.; Ling, N.; Wang, M.; Dai, J.; Shen, Q.; Guo, S. Bacterial rather than fungal community composition is associated with microbial activities and nutrient-use efficiencies in a paddy soil with short-term organic amendments. Plant Soil 2018, 424, 335–349. [Google Scholar] [CrossRef]
- Luo, G.; Xue, C.; Jiang, Q.; Xiao, Y.; Zhang, F.; Guo, S.; Shen, Q.; Ling, N. Soil carbon, nitrogen, and phosphorus cycling microbial populations and their resistance to global change depend on soil C:N:P stoichiometry. mSystems 2020, 5, e00162-20. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Gao, G.; Ma, Y.; Fan, K.; Delgado-Baquerizo, M. Soil microbial biogeography in a changing world: Recent advances and future perspectives. mSystems 2020, 5, e00803-19. [Google Scholar] [CrossRef] [PubMed]
- Masuda, Y.; Mise, K.; Xu, Z.; Zhang, Z.; Shiratori, Y.; Senoo, K.; Itoh, H. Global soil metagenomics reveals distribution and predominance of Deltaproteobacteria in nitrogen-fixing microbiome. Microbiome 2024, 12, 95. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Feng, J.; Ao, G.; Qin, W.; Han, M.; Shen, Y.; Liu, M.; Chen, Y.; Zhu, B. Globally nitrogen addition alters soil microbial community structure, but has minor effects on soil microbial diversity and richness. Soil Biol. Biochem. 2023, 179, 108982. [Google Scholar] [CrossRef]
- Barquero, M.; Cazador, C.; Ortiz-Liébana, N.; Zotti, M.; Brañas, J.; González-Andrés, F. Fertilising maize with bio-based mineral fertilisers gives similar growth to conventional fertilisers and does not alter soil microbiome. Agronomy 2024, 14, 916. [Google Scholar] [CrossRef]
- Peix, A.; Ramírez-Bahena, M.H.; Velázquez, E.; Bedmar, E.J. Bacterial associations with legumes. Crit. Rev. Plant Sci. 2014, 34, 17–42. [Google Scholar] [CrossRef]
- Spang, A.; Poehlein, A.; Offre, P.; Zumbrägel, S.; Haider, S.; Rychlik, N.; Nowka, B.; Schmeisser, C.; Lebedeva, E.V.; Rattei, T.; et al. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: Insights into metabolic versatility and environmental adaptations. Environ. Microbiol. 2012, 14, 3122–3145. [Google Scholar] [CrossRef]
- Stieglmeier, M.; Mooshammer, M.; Kitzler, B.; Wanek, W.; Zechmeister-Boltenstern, S.; Richter, A.; Schleper, C. Aerobic nitrous oxide production through N-nitrosating hybrid formation in ammonia-oxidizing archaea. ISME J. 2014, 8, 1135–1146. [Google Scholar] [CrossRef]
- Semenov, M.V.; Zhelezova, A.D.; Ksenofontova, N.A.; Ivanova, E.A.; Nikitin, D.A.; Semenov, V.M. Microbiological indicators for assessing the effects of agricultural practices on soil health: A Review. Agronomy 2025, 15, 335. [Google Scholar] [CrossRef]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef]
- Lal, R. Digging Deeper: A holistic perspective of factors affecting soil organic carbon sequestration in agroecosystems. Glob. Change Biol. 2018, 24, 3285–3301. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Trivedi, P.; Delgado-Baquerizo, M.; Anderson, I.C.; Singh, B.K. Response of soil properties and microbial communities to agriculture: Implications for primary productivity and soil health indicators. Front. Plant Sci. 2016, 7, 990. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.