Abstract
In the face of global climate change, crop pests and diseases have emerged on a large scale, with diverse species lasting for long periods and exerting wide-ranging impacts. Identifying crop pests and diseases efficiently and accurately is crucial in enhancing crop yields. Nonetheless, the complexity and variety of scenarios render this a challenging task. In this paper, we propose a fine-grained crop disease classification network integrating the efficient triple attention (ETA) module and the AttentionMix data enhancement strategy. The ETA module is capable of capturing channel attention and spatial attention information more effectively, which contributes to enhancing the representational capacity of deep CNNs. Additionally, AttentionMix can effectively address the label misassignment issue in CutMix, a commonly used method for obtaining high-quality data samples. The ETA module and AttentionMix can work together on deep CNNs for greater performance gains. We conducted experiments on our self-constructed crop disease dataset and on the widely used IP102 plant pest and disease classification dataset. The results showed that the network, which combined the ETA module and AttentionMix, could reach an accuracy as high as 98.2% on our crop disease dataset. When it came to the IP102 dataset, this network achieved an accuracy of 78.7% and a recall of 70.2%. In comparison with advanced attention models such as ECANet and Triplet Attention, our proposed model exhibited an average performance improvement of 5.3% and 4.4%, respectively. All of this implies that the proposed method is both practical and applicable for classifying diseases in the majority of crop types. Based on classification results from the proposed network, an install-free WeChat mini program that enables real-time automated crop disease recognition by taking photos with a smartphone camera was developed. This study can provide an accurate and timely diagnosis of crop pests and diseases, thereby providing a solution reference for smart agriculture.
1. Introduction
Crop diseases have a significant negative impact on the yield and quality of agricultural production. Large-scale diseases can destroy large numbers of crops, resulting in severe crop yield reduction [1]. Therefore, timely detection of and intervention in crop diseases is essential to improve food yields. Traditional investigation into crop pests and diseases relies on experienced manpower, which is time consuming and laborious and faces difficulties in ensuring accuracy. With the continuous expansion of the application of artificial intelligence and big data technology in the field of agriculture, more and more research efforts are beginning to focus on the automatic identification of crop pests and diseases based on machine learning.
Early methods for crop disease image recognition relied on a priori knowledge to extract hand-designed features and use classifiers such as SVM (support vector machine) for disease classification. Guo et al. [2] used a Bayesian approach to identify downy mildew, anthracnose, powdery mildew, and gray mold infections using texture and color features. The average accuracy for the four diseases was 88.48%. Zhang et al. [3] used the k-means clustering method to separate infection sites in cucumber leaves. However, these methods are only designed for specific scenarios and are less effective in the actual natural environment. With the continuous in-depth research into computer vision, deep convolutional neural networks (CNNs) have achieved great success in many computer tasks [4,5,6,7,8]. So far, some remarkable achievements have been made in crop disease classification research based on deep CNNs. Lu et al. [9] used a deep multi-instance learning approach to design an automated diagnosis system for wheat diseases, and the accuracy of the model exceeded 95%. CGDR [10] captures information on diverse features of tomato leaf disease in different dimensions and sensory fields using a multi-branching structure of comprehensive grouped differentiation residuals. Deng et al. [11] segmented the diseased regions of tomato leaves by their proposed MC-UNet. Zhou et al. [12] proposed a residual distillation transformer architecture and obtained 92% classification accuracy on four categories of rice leaf diseases: white leaf blight, brown spot, rice blight, and brown fly. Hasan et al. [13] have introduced a novel CNN architecture. It is relatively small in scale yet shows promising performance, enabling the prediction of rice leaf diseases with moderate accuracy and lower time complexity.
These works are of great value for understanding crop disease identification but have the following three limitations: (1) The majority of existing work use public datasets for training, and these data have uniform backgrounds and light intensities, and most of the disease sites are concentrated in the leaf area. As a result, these methods have a weak generalization ability in real-world growing environments. (2) There are still many difficulties in the identification of some fine-grained crop diseases due to inter-class similarities and the complex background of real environments in the field. (3) Most of the currently available research is at the laboratory stage, without the proposed crop disease identification models having been applied in a real environment.
To address these issues, the purpose of this research was to design a framework that supports the identification of crop diseases in a real growing environment. In the proposed fine-grained crop disease classification model, an ETA module and an AttentionMix were designed and introduced as the key parts of the deep CNNs for better detection performance. In terms of field application, we also developed a crop disease identification WeChat mini program based on the WeChat public platform. By embedding the proposed crop disease recognition model in the mini program, we can achieve crop disease recognition by just using smartphones to take images.
To sum up, the main contributions of this paper include the following:
- (1)
- We propose an effective triple attention module to efficiently extract channel attention and spatial attention information from crop disease images.
- (2)
- An AttentionMix data augmentation strategy is proposed to avoid the loss of object information due to random cuts in CutMix.
- (3)
- We build a large-scale crop disease dataset containing images of five crops—wheat, rice, rape, corn, and apple—with images taken in real field conditions.
- (4)
- We develop a crop disease identification WeChat mini program to achieve disease identification using images taken with smartphones.
- (5)
- Extensive experiments on the crop disease dataset and the common pest and disease dataset are employed to demonstrate the advanced nature of our proposed method.
2. Related Works
2.1. Attention Mechanism
In recent years, the attention mechanism, which can help the model to better extract the key features in the images, has improved in recognition accuracy and is increasingly used in image feature extraction. A large number of deep CNN and attention mechanism algorithms have been successfully applied in computer vision, bringing new opportunities for the classification of crop pests and diseases. Gao et al. [14] proposed a dual -ranch, efficient channel attention (DECA) module by improving SENet [15], which uses the dual-branch 1D convolution operation to filter effective feature information. The disease recognition accuracies of the AI Challenger 2018 dataset [16], PlantVillage [17] dataset, and self-collected cucumber disease dataset were 86.35%, 99.74%, and 98.54%, respectively. Chen et al. [18] proposed a hybrid attention module called spatially efficient channel attention by realizing the serial connection of spatial attention and efficient channel attention. When combined with neural networks, this module achieved a classification accuracy of 87.28% in classifying some data from the crop disease dataset in the 2018 AI_Challenge competition. Huang et al. [19] introduced the Inception module based on the residual network (ResNet18) and utilized its multi-scale convolutional kernel structure to extract the disease features of different scales, improving the richness of features. Wang et al. [20] resolved the interference problems caused by the two attention mechanisms in CBAM [21] by implementing a parallel connection of channel attention and spatial attention. In contrast to the original YOLOv5 model, which achieved an accuracy rate of 82%, the accuracy of the improved model was boosted by 5%.
SENet [15] introduced an SE module that learns weights for feature channels, thereby emphasizing important channels and suppressing less important ones through an attention mechanism. CBAM [21] generates attentional feature maps in both the channel and spatial dimensions, serially. However, the dimensionality reduction in its channel attention has a negative impact on model predictions. ECANet [22] improves SENet by using 1D convolution for local cross-channel interaction without dimensionality reduction. Triplet Attention [23] highlights the independence of CBAM’s channel and spatial attention, applying rotation operations in three branches to capture cross-channel interactions between the channel dimension C and the spatial dimensions W/H. More details of the above models are shown in Figure 1. There, σ represents the activation function, and, in this paper, the activation function used is ReLU. ⨀ denotes the broadcast element-wise multiplication, and ⊕ represents the broadcast element-wise addition. The usages of these symbols remain consistent in all the illustrations within this paper. Nevertheless, it fails to fully capture the attention information of input features. Our proposed ETA module computes the channel attention in each of the three branches of Triplet Attention using 1D convolution before performing subsequent operations, thus improving the extraction of both channel and spatial attention and the feature representation.
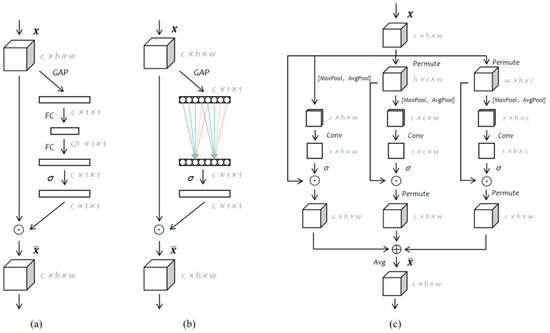
Figure 1.
Comparisons with different attention modules: (a) Squeeze Excitation (SE) module; (b) ECA module; (c) Triplet Attention module.
2.2. Data Augmentation
The acquisition of training data is labor intensive, and certain data are scarce. Data augmentation, which includes methods such as rotation, clipping, and so on, boosts the data quantity and improves the model performance but introduces noise. Multi-sample synthesis produces higher-quality samples. CutMix [24], a widely recognized method, cuts a part of an image, fills the region with random data from the training set, and assigns labels proportionally. It enhances recognition by recognizing objects in a local view and adding sample information, but the randomness may cause the loss of object information and label errors (as shown in Figure 2). We propose AttentionMix, which convolves the network’s output feature map to obtain an attention map. Based on this attention map, the salient regions of the image are identified, and these salient regions are synthesized with other images to avoid label errors resulting from the loss of target information.

Figure 2.
Visual comparisons of CutMix and AttentionMix. The random cropping operation of CutMix may result in the loss of object information, which in turn leads to incorrect label assignment. The proposed AttentionMix can effectively address this issue by extracting salient regions of the image based on neural networks. The red box represents the cropping areas that maybe given by both methods.
3. Methods
In this section, we first describe our crop disease dataset and then introduce the efficient triple attention (ETA) module and AttentionMix data augmentation strategy proposed in this article. Finally, the development process of the crop disease recognition mini program is provided.
The ETA module and AttentionMix can work together on a deep CNN for greater performance improvement. Figure 3 illustrates our proposed architecture for fine-grained crop disease classification. The backbone network with the ETA module can extract the features of crops in the input image more efficiently. In addition, based on the feature maps output from the backbone network, AttentionMix can more accurately acquire salient regions in the image, effectively avoiding label assignment errors caused by the loss of object information when mixing with other images to generate new sample data. The mix of images is re-fed into the backbone to participate in network training along with the raw image. This doubles the dataset expansion.
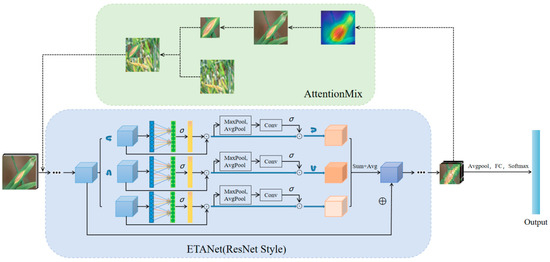
Figure 3.
Architecture of our disease classification method.
3.1. Crop Disease Datasets
Crop disease identification is a task that requires fine-grained visual classification. Traditional disease identification methods rely on rich experience, and it is difficult to achieve accurate identification and timely prevention. Deep convolutional neural networks can automatically detect the disease from images but require a large number of training samples. There are several publicly available datasets for disease detection; however, the scale of these datasets is relatively small. PlantVillage [17] contains 14 plant species with a total of 26 kinds of different diseases. PlantVillage only includes diseases on leaf parts, and each image has single leaf on a simple background, which means it is unable to meet practical application in the field. Rice Leaf Disease [25] contains images of four types of diseases: bacterial blight, blast, brown spot, and tungro varieties. Therefore, we built a large-scale crop disease dataset. Specifically, in an open-air natural-light environment, we used a digital camera to collect JPEG format images of five crop diseases of wheat, rice, rape, corn, and apple. When taking pictures, we adjusted the camera to place the diseased part of the crop in the center of the image as much as possible. We collected a total of 64 diseases of these five crop categories, with disease sites including leaves, fruits, roots, branches, and leaf sheaths. The disease types were classified according to expert guidance. The dataset currently has a total of 27,027 images, which are stored in the PASCAL VOC [26] dataset format. Figure 4 shows some image samples.

Figure 4.
Examples of our crop disease dataset.
There were 9 categories of wheat diseases, 18 categories of rice diseases, 8 categories of rape diseases, 12 categories of corn diseases, and 17 categories of apple diseases, for a total of 27,027 images. Figure 5 shows specific information about the five-crop disease dataset, including the name of each disease and the corresponding number of images. In the network training phase, we used 80% of the images as the training set and 20% of the images as the test set.
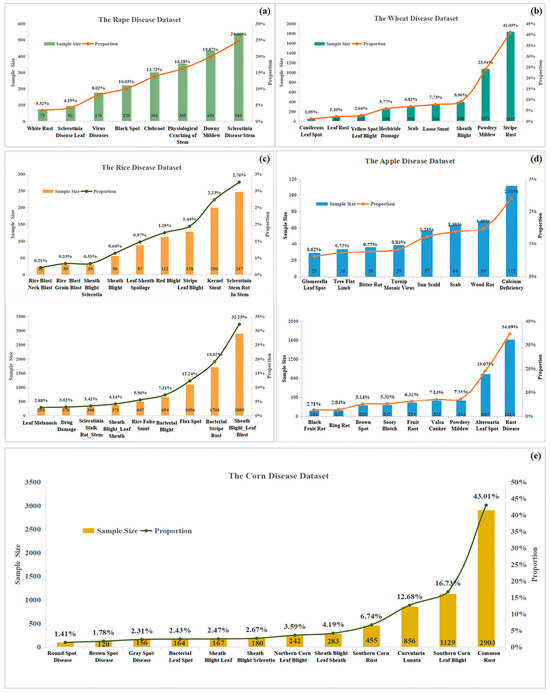
Figure 5.
Overview of crop disease datasets. (a) Rape disease dataset, (b) wheat disease dataset, (c) rice disease dataset, (d) apple disease dataset, and (e) corn disease dataset.
3.2. Efficient Triplet Attention Module
We discussed the working of ECANet and Triplet Attention in Section 1. To address the shortcomings in their work, we proposed a more efficient feature extraction module that considers both channel attention and spatial attention. The diagram of this module can be viewed in Figure 6. Given an input tensor , it passes into the three branches of the module. From top to bottom, the first branch is responsible for establishing the interaction between the H-dimension and the C-dimension. The input tensor is rotated 90° counterclockwise along the H axis to obtain the rotation tensor . For the rotation tensor , the global average pool (GAP) [27] is first used for each feature channel, and then a one-dimensional convolution is used to capture cross-channel interaction information without dimensionality reduction, and finally the sigmoid function is used to generate the channel weights . To avoid adjusting the value of through cross-validation, we determine adaptively by using as follows:
where indicates the nearest odd number of , is the channel dimension (i.e., number of filters), and we set and to 2 and 1 throughout all the experiments. The rotation tensor is multiplied with the channel weights by the broadcast element to obtain the channel attention feature . is then passed through the MaxPool and AvgPool operations, respectively, and is subsequently reduced to , which is of the shape (). is passed through a standard convolutional layer of kernel size k × k and a BN [28] layer to obtain an intermediate output with a shape of (). The sigmoid function is used to generate attention weights for this intermediate output, which is then applied to the tensor to obtain the result . The final output is rotated 90° clockwise along the H axis to keep it consistent with the input shape.
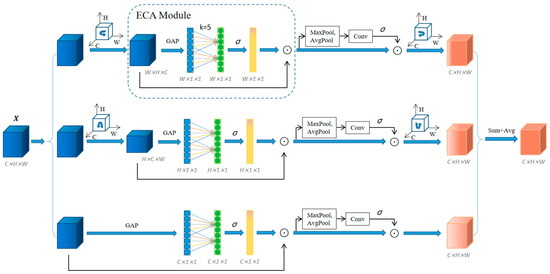
Figure 6.
Illustration of the efficient triplet attention (ETA) module.
The second branch is responsible for establishing the interaction between the C-dimension and the W-dimension. It rotates the input tensor by 90° counterclockwise along the W axis, and the other operations are the same as the first branch. The third branch is responsible for establishing the interaction between the H and W dimensions. This branch does not require rotation of the input tensor and performs the extraction of channels and spatial attention directly. Finally, the tensor output of the three branches is summed and averaged to obtain the final output of the module. Like other attention modules, the ETA module can be easily applied to deep CNNs, and the resulting network is denoted by ETANet.
3.3. AttentionMix
CutMix demonstrated that data augmentation of mixed images can significantly improve the generalization of models in image recognition tasks, but random cut-based image mixing may suffer from label misassignment due to loss of object information. We proposed the AttentionMix method to improve the above deficiencies. Specifically, in the training phase, we define and to be the training image and its label. is the image to be mixed, and is its label. The attention map is obtained by passing the feature map to a convolutional layer with a convolutional kernel size of . A channel is randomly selected from the attention map. A threshold is chosen, and then a bounding box is found in the original image that completely encloses the part larger than the threshold. The attention cropping image of the training image and its label can be obtained based on the attention cropping . is obtained based on the size of the training image, and the scale rate resizes to . and are denoted as follows:
where is sampled from the uniform distribution , and , denote the upper and lower bounds of the sampling range.
is pasted into a random region of to obtain a mixed image , and the label of can be denoted as follows:
where is the scale ratio of to , and is denoted as follows:
The obtained mixed images effectively avoid the problem of target information loss due to random cutting and make image fusion more robust. Feeding the mixed image into the network to participate in training with the input image can effectively improve the recognition performance of the network. Figure 7 illustrates our proposed AttentionMix data augmentation method.
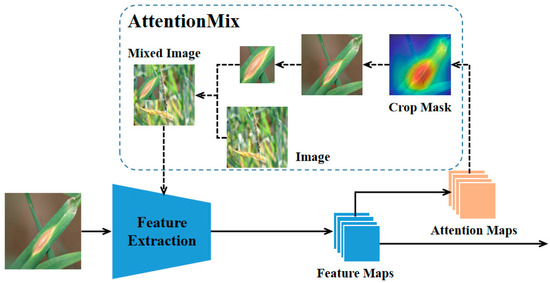
Figure 7.
Illustration of the AttentionMix image data augmentation method.
3.4. Crop Disease Identification WeChat Mini Program
The WeChat mini program has the advantages of not requiring installation, being ready to go after use, being opened at any time when needed, not taking up mobile phone memory, and having a functional implementation that is completely comparable to APP. Additionally, the total number of WeChat users worldwide has exceeded 1.26 billion people, which is very beneficial to the application and promotion of the WeChat mini program. It is extremely suitable for responding to the requirements of the current task from both the user and development perspectives.
This paper develops a crop disease identification mini program based on the WeChat public platform to achieve the disease identification of crops by taking photos with smartphones. Figure 8 shows the architecture of the disease identification of the mini program. The crop disease identification system includes two parts: the front end and the back end. The front end is developed by WeChat developer tools and is mainly responsible for page interaction and data display. The back end is responsible for business logic implementation and returning the correct data. It is developed through the Django framework, which is used to receive requests from the front end, process data, and return responses. The front end acquires a disease image by calling the mobile phone camera or selecting it from the photo album through the WeChat mini program and sends it to the back end through the POST request. The back end receives the image and feeds it into the trained crop disease recognition model to identify the category of disease in the image. The recognition results are stored in MySQL database and returned to the front end for display.
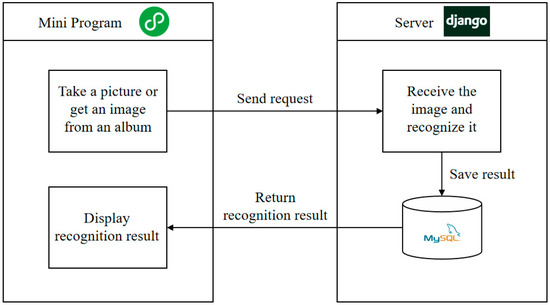
Figure 8.
Architecture of the crop disease identification mini program.
The crop disease identification mini program consists of three main parts (Figure 9): the identification module, the disease library module, and the home module. The recognition module, as the core function of the mini program, is responsible for the acquisition of disease images of five crops, namely wheat, rice, rape, corn, and apple; the recognition of disease images; and the display of the analysis results. Users can view the detailed information of a certain disease by entering the disease name or selecting the disease category. Disease profiles, characteristics, and preventive measures can be viewed to further understand the disease. Users can login to the app by authorizing their WeChat account information (username and WeChat avatar).
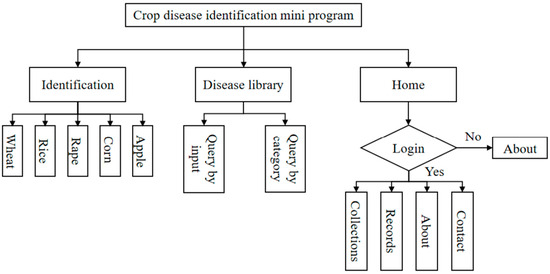
Figure 9.
Crop disease identification mini program framework.
The mini program was developed using a combination of HTML5, CSS3, and JavaScript technologies. The key functionality of the program was implemented through the integration of the trained crop disease recognition model with the WeChat API. The operation process of the mini program is as follows: when the user launches the program, they are presented with a simple and intuitive interface that allows them to either take a photo using the smartphone camera or select an image from the photo album. Once the image is selected, it is uploaded to the server, where it is processed by the recognition model. The recognition result is then returned to the mini program and displayed to the user within a short response time, typically less than 2 s. However, during the testing phase, we also identified some limitations. For example, the program may experience slower performance on older smartphone models with limited processing power. To address this issue, we are planning to optimize the code and explore techniques such as model compression to reduce the computational load and improve the overall performance of the mini program. The mini program, serving as a powerful tool in smart agriculture, enables farmers to monitor crop diseases anytime and anywhere, quickly obtaining identification results, and contributes to improving the quality and efficiency of agricultural production.
4. Experiments and Results
In this section, we first present the details of the experimental implementation and then give experimental results and analyses for the crop disease dataset we created as well as for the IP102 [29] pest dataset. Finally, to further validate our results, we provide visualization results of sample images to demonstrate the ability of our approach to capture more accurate feature representations.
4.1. Experimental Settings
To ensure the fairness of the experiments, we evaluated all the models involved using ResNet-50 as the backbone network and set the same hyperparameters. Specifically, the input image size was first scaled to 512 × 512 and then randomly cropped to 448 × 448. The models were optimized using stochastic gradient descent (SGD) with a weight decay of 1 × 10−5, a momentum of 0.9, and a small batch size of 16. The initial learning rate was set to 1 × 10−3 and decayed by 0.9 every two periods. All the experiments were performed using one NVIDIA 2080Ti GPU to accelerate the training network.
4.2. Comparison Using the Crop Disease Datasets
We evaluated our proposed method on the crop disease dataset. ResNet-50 was used as the baseline, and ImageNet-pretrained provided by PyTorch [30] was used for each model participating in the evaluation. Table 1 presents the prediction results of the proposed ETANet and other methods on our crop disease dataset. When the basic model ResNet was not equipped with additional attention mechanisms, its performance on each crop dataset was relatively weak. After introducing attention mechanisms, the model performance significantly improved. ECANet introduced attention in the channel dimension and showed excellent performance on each crop dataset. For instance, the accuracy rate on the wheat dataset reached 94.7%, which was 4.3% higher than that of ResNet. This indicates that, by re-weighting the channel features, the model can focus on more discriminative feature channels and enhance its ability to capture different crop disease features, reducing interference from irrelevant information. Triplet Attention focuses on the spatial dimension and also brings significant gains. It achieved an accuracy rate of 98.2% on the rice dataset, which is an obvious advantage compared with ResNet-50’s 92.8%. ETANet combined channel and spatial attention mechanisms and demonstrated the most outstanding performance. On the corn dataset, it achieved an accuracy rate as high as 96.1%. Looking at all crop datasets comprehensively, it completely outperformed ECANet and Triplet Attention, which only adopted single attention dimensions. This fully indicates that ETANet optimizes feature extraction and utilization in different dimensions, enabling the model to have a deeper understanding of the complex feature patterns of crop diseases and further enhancing the accuracy and robustness of recognition.

Table 1.
Classification accuracy (%) of different methods on crop disease datasets.
By comparing the performance of ResNet-50 with and without data augmentation strategies, as shown in Table 2, it becomes clear that both CutMix and AttentionMix substantially enhance its capabilities. Take the wheat dataset as an example. The accuracy of ResNet-50 [31] by itself (as seen in Table 1) was 90.4%. However, when combined with CutMix, the accuracy of CutMix + ResNet-50 soared to 99.3%, and further, AttentionMix + ResNet-50 elevated it to 99.4%. Such remarkable progress suggests that these data-augmentation techniques broaden the diversity of the training data, allowing the model to capture more comprehensive and variant features related to crop diseases. CutMix, with its unique image mixing method, enriches the data distribution. Meanwhile, AttentionMix, by virtue of its advanced feature manipulation, refines the data more effectively, thereby reducing overfitting and enhancing generalization abilities. For ETANet, its combination with data augmentation strategies also yields significant benefits. In the corn dataset, the accuracy of standalone ETANet (derived from Table 1) was 96.1%. However, when paired with CutMix, that is, CutMix + ETANet, the accuracy climbed to 96.8% and, with AttentionMix, AttentionMix + ETANet achieved an accuracy of 96.9%. ETANet coordinates well with data augmentation as it can better focus on the augmented features, further strengthening the discriminative power of the model. It is capable of precisely extracting and leveraging the crucial information from the augmented data, thus attaining a higher accuracy rate in disease identification.

Table 2.
Comparison of classification accuracy (%) of CutMix and AttentionMix on crop disease datasets.
4.3. Comparison on the IP102 Dataset
To verify the robustness and generalization of the method proposed in this paper, we also evaluated it on the common pest and disease dataset IP102 [29]. IP102 is a crop pest dataset for target classification and detection tasks. The 102 refers to 102 pest categories. It contains more than 75,000 images of eight crops, including rice, corn, wheat, sugar beet, alfalfa, grapes, citrus, and mango, which show a natural long-tailed distribution. The first five crops are field crops, and the last three are economic crops.
Similarly, we provide the classification results of ECANet, Triplet Attention, and our method on the IP102 crop pest dataset using ResNet-50 as a baseline. The performance of CutMix and AttentionMix on this dataset is also compared. The detailed classification accuracy is given in Table 3.

Table 3.
Classification results of different methods on the IP102 dataset.
Among the base models, ResNet-50 had an accuracy of 68.4% and a recall of 52.7% on the IP102 dataset, indicating limitations in feature extraction and a tendency to misclassify or miss samples. ECANet and Triplet Attention performed slightly better, while ETANet, with its dual attention mechanism, achieved an accuracy of 69.7% and a recall of 63.1%, showing stronger capabilities in capturing features. The CutMix strategy significantly improved the performance of all models. For example, CutMix + ResNet-50 boosted the accuracy to 72.6% and the recall to 63.5%. It expanded the diversity of data, reduced overfitting and helped the model adapt to complex pest images. The combination with attention models yielded even better results. The AttentionMix strategy brought about a remarkable leap. AttentionMix + ETANet led with an accuracy of 78.7% and a recall of 70.2%. Its carefully designed data augmentation and model synergy work excellently, accurately focusing on key features.
Given that the IP102 dataset has numerous pest categories, complex backgrounds and unbalanced samples, base models struggle to handle all aspects. CutMix alleviates some problems, while AttentionMix, in combination with ETANet and others, addresses them specifically. It pays attention to rare pests and strengthens feature extraction, achieving high accuracy and recall rates, thus providing an effective solution for pest identification.
4.4. Visualization
To further validate our proposed method, we provide the Grad-CAM [32] results for some sample images. Grad-CAM calculates the weights of each channel of the feature map using the back-propagation gradient of the network to obtain the heat map. Based on the heat map, we can visualize the regions of interest in the network. As shown in Figure 10, ETANet can capture more accurate and relevant target boundaries from the image samples. In turn, AttentionMix can acquire more accurate targets when cutting and mixing images to avoid label misallocation. Compared with other attention algorithms, our method can help to improve the performance of deep neural networks more effectively.
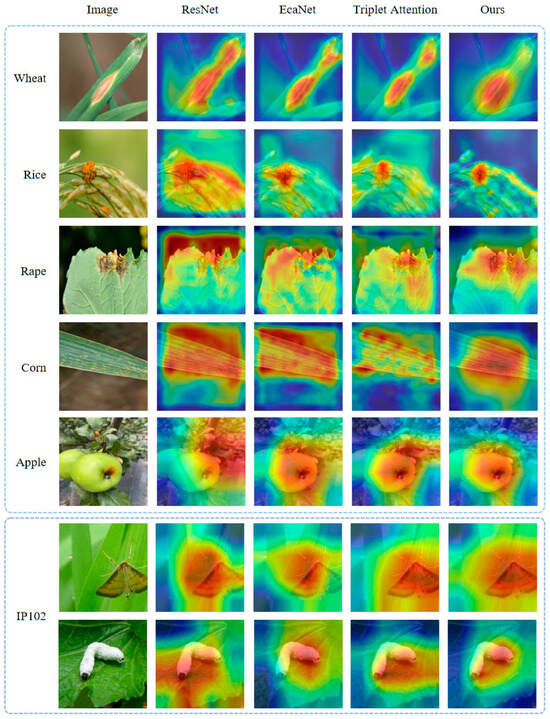
Figure 10.
Visualization of Grad-CAM results.
5. Conclusions
Crop disease recognition in natural environments faces difficulties such as high light contrast, multiple locations of disease appearance, and slight differences between disease classes, thereby making crop disease recognition challenging. In this paper, by analyzing the shortcomings in the existing work, we mainly focused on the following aspects.
In natural environments, crop disease recognition is filled with numerous challenges. The high contrast in lighting, the diverse locations at which diseases can appear and the subtle differences among disease categories all add to the difficulty of precisely identifying crop diseases. In this study, a fine-grained crop disease classification network that combines the efficient triple attention (ETA) module and the AttentionMix data augmentation strategy was proposed. This effectively deals with many of the problems in existing crop disease recognition work and offers strong support for pest and disease control in smart agriculture.
The ETA module, which has a unique three-branch structure, efficiently gathers channel and spatial attention information. It achieves this by circumventing the limitations of traditional methods in feature extraction. On the crop disease dataset, ETANet had an average 4.2% increase in accuracy compared with ResNet. This clearly shows its ability to effectively capture complex disease feature patterns and improve the model’s representational skills. AttentionMix successfully fixed the problem of incorrect label assignment caused by random cropping in CutMix. Experiments showed that AttentionMix had a 0.4% increase in accuracy compared to CutMix. This significantly increased the amount of high-quality training data and made the model better at generalizing. The combined effect of the ETA module and AttentionMix allowed the model to achieve great results on both the crop disease dataset and the difficult IP102 dataset. On the IP102 dataset, the AttentionMix + ETANet combination reached an accuracy of 78.7% and a recall of 70.2%. This clearly shows an advantage over other advanced methods and strongly proves the high precision and robustness of this combination in identifying pests and diseases in complex situations.
The crop disease identification WeChat mini-program made from the research findings allows for real-time automatic identification just by taking photos with a smartphone. By bringing together web and mini-program development technologies with the deep learning model, it gives farmers a convenient, non-destructive, and fast disease diagnosis tool. This greatly improves the timeliness and accuracy of disease prevention and control in agricultural production.
Because some diseases do not occur often, the crop disease dataset has a long-tailed distribution. This means that there are relatively few collected images. In the future, we will keep looking into datasets with long-tailed distributions to make the model better at classifying the tail classes in which data are limited. At the same time, we will combine other types of data, like hyperspectral image data, for disease recognition and use the recognition model in equipment like agricultural inspection robots. This will help make crop disease prevention and control more automated and precise; this is also an area of research that we are interested in.
In summary, the results of this study are not only beneficial in model performance but also have high practical use. They provide a useful solution for disease management in smart agriculture. Future research will help the technology in this area move forward and develop even further.
Author Contributions
Y.Z.: Conceptualization, Methodology, Code writing, Writing—original draft. N.Z.: Visualization, Supervision, Writing—review and editing. X.C.: Resources. J.Z.: Data curation, Data processing. W.D.: Data curation, Data processing. T.S.: Resources, Supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Central Public-Interest Scientific Institution Basal Research Fund (No. Y2022QC17), the Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-CAE-202302, CAAS-ASTIP-2024-AII), and the Agricultural Independent Innovation of Jiangsu Province (CX(24)1021).
Data Availability Statement
The datasets presented in this article are not readily available because the data are part of an ongoing study. Requests to access the datasets should be made to the corresponding author, and a description of the use of the data should be provided.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Li, L.; Zhang, S.; Wang, B. Plant disease detection and classification by deep learning—A review. IEEE Access 2021, 9, 56683–56698. [Google Scholar] [CrossRef]
- Guo, P.; Liu, T.; Li, N. Design of automatic recognition of cucumber disease image. Inf. Technol. J. 2014, 13, 2129. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, X.; You, Z.; Zhang, L. Leaf image based cucumber disease recognition using sparse representation classification. Comput. Electron. Agric. 2017, 134, 135–141. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, PMLR 2019, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Tan, M.; Le, Q. Efficientnetv2: Smaller models and faster training. In Proceedings of the International Conference on Machine Learning, PMLR 2021, online, 18–24 July 2021; pp. 10096–10106. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Redmon, J. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Lu, E.; Cole, F.; Dekel, T.; Zisserman, A.; Freeman, W.T.; Rubinstein, M. Omnimatte: Associating objects and their effects in video. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual, 19–25 June 2021; pp. 4507–4515. [Google Scholar]
- Li, M.; Zhou, G.; Chen, A.; Li, L.; Hu, Y. Identification of tomato leaf diseases based on LMBRNet. Eng. Appl. Artif. Intell. 2023, 123, 106195. [Google Scholar] [CrossRef]
- Deng, Y.; Xi, H.; Zhou, G.; Chen, A.; Wang, Y.; Li, L.; Hu, Y. An effective image-based tomato leaf disease segmentation method using MC-UNet. Plant Phenomics 2023, 5, 0049. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhong, Y.; Zhou, S.; Song, J.; Xiang, W. Rice leaf disease identification by residual-distilled transformer. Eng. Appl. Artif. Intell. 2023, 121, 106020. [Google Scholar] [CrossRef]
- Hasan, M.; Rahman, T.; Uddin, A.F.M.S.; Galib, S.M.; Akhond, M.R.; Uddin, J.; Hossain, A. Enhancing rice crop management: Disease classification using convolutional neural networks and mobile application integration. Agriculture 2023, 13, 1549. [Google Scholar] [CrossRef]
- Gao, R.; Wang, R.; Feng, L.; Li, Q.; Wu, H. Dual-branch, efficient, channel attention-based crop disease identification. Comput. Electron. Agric. 2021, 190, 106410. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Wu, J.; Zheng, H.; Zhao, B.; Li, Y.; Yan, B.; Liang, R.; Wang, W.; Zhou, S.; Lin, G.; Fu, Y.; et al. Large-scale datasets for going deeper in image understanding. In Proceedings of the 2019 IEEE International Conference on Multimedia and Expo (ICME), IEEE 2019, Shanghai, China, 8–12 July 2019; pp. 1480–1485. [Google Scholar]
- Hughes, D.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Chen, Z.; Cao, M.; Ji, P.; Ma, F. Research on Crop Disease Classification Algorithm Based on Mixed Attention Mechanism. J. Phys. Conf. Ser. 2021, 1961, 012048. [Google Scholar] [CrossRef]
- Huang, L.; Luo, Y.; Yang, X.; Yang, G.; Wang, D. Crop Disease Recognition Based on Attention Mechanism and Multi-scale Residual Network. Trans. Chin. Soc. Agric. Mach. 2021, 52, 264–271. [Google Scholar]
- Wang, X.; Dong, Q.; Yang, G. YOLOv5 Improved by Optimized CBAM for Crop Pest Identification. Comput. Syst. Appl. 2023, 32, 261–268. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Wang, Q.; Wu, B.; Zhu, P.; Zuo, W.; Hu, Q. ECA-Net: Efficient channel attention for deep convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020; pp. 11534–11542. [Google Scholar]
- Misra, D.; Nalamada, T.; Arasanipalai, A.U.; Hou, Q. Rotate to attend: Convolutional triplet attention module. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Virtual, 5–9 January 2021; pp. 3139–3148. [Google Scholar]
- Yun, S.; Han, D.; Oh, S.J.; Chun, S.; Choe, J.; Yoo, Y. Cutmix: Regularization strategy to train strong classifiers with localizable features. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of South Korea, 27 October–2 November 2019; pp. 6023–6032. [Google Scholar]
- Sethy, P.K.; Barpanda, N.K.; Rath, A.K.; Behera, S.K. Deep feature based rice leaf disease identification using support vector machine. Comput. Electron. Agric. 2020, 175, 105527. [Google Scholar] [CrossRef]
- Everingham, M.; Van Gool, L.; Williams, I.C.K.; Winn, J.; Zisserman, A. The pascal visual object classes (voc) challenge. Int. J. Comput. Vis. 2010, 88, 303–338. [Google Scholar] [CrossRef]
- Lin, M.; Chen, Q.; Yan, S. Network in network. arXiv 2013, arXiv:1312.4400. [Google Scholar]
- Ioffe, S. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv 2015, arXiv:1502.03167. [Google Scholar]
- Wu, X.; Zhan, C.; Lai, Y.K.; Cheng, M.; Yang, J. Ip102: A large-scale benchmark dataset for insect pest recognition. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 8787–8796. [Google Scholar]
- Paszke, A.; Gross, S.; Chintala, S.; Chnan, G.; Yang, E.; DeVito, Z.; Lin, Z.; Desmaison, A.; Antiga, L.; Lerer, A. Automatic Differentiation in Pytorch. 2017. Available online: https://openreview.net/forum?id=BJJsrmfCZ (accessed on 5 January 2025).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).