Drought Stress-Related Gene Identification in Rice by Random Walk with Restart on Multiplex Biological Networks
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
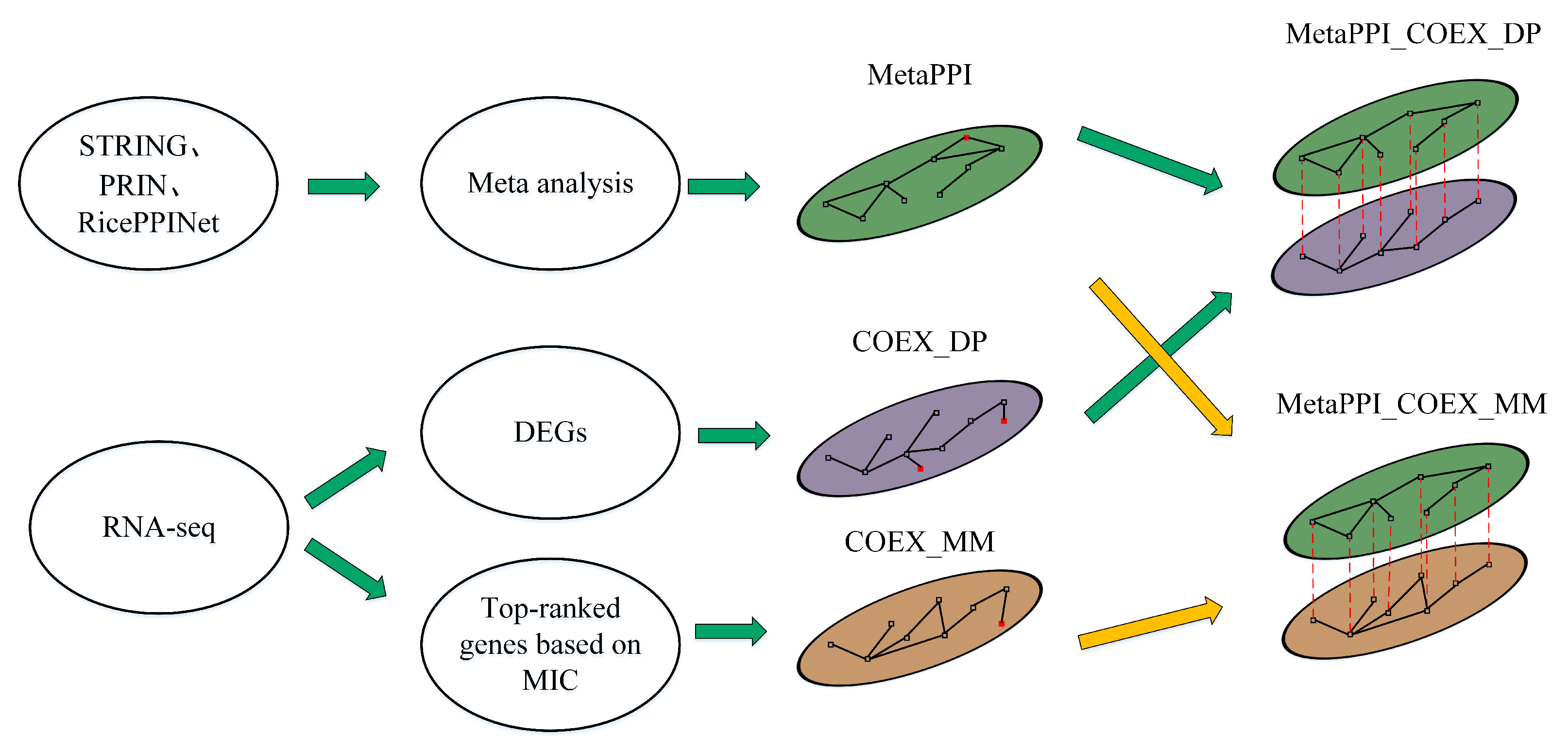
2.2. The Networks’ Construction
2.2.1. Protein–Protein Interaction Network
2.2.2. Gene Coexpression Network
2.2.3. Multiplex Network
2.3. Random Walk with Restart on Multiplex Networks
| Algorithm 1 Random Walk with Restart (RWR). |
|
2.4. Leave-One-Out Cross-Validation Strategy
| Algorithm 2 Leave-One-Out Cross-Validation Strategy. |
|
2.5. Association Analysis between Potential Genes and Known Drought Stress Genes
3. Results
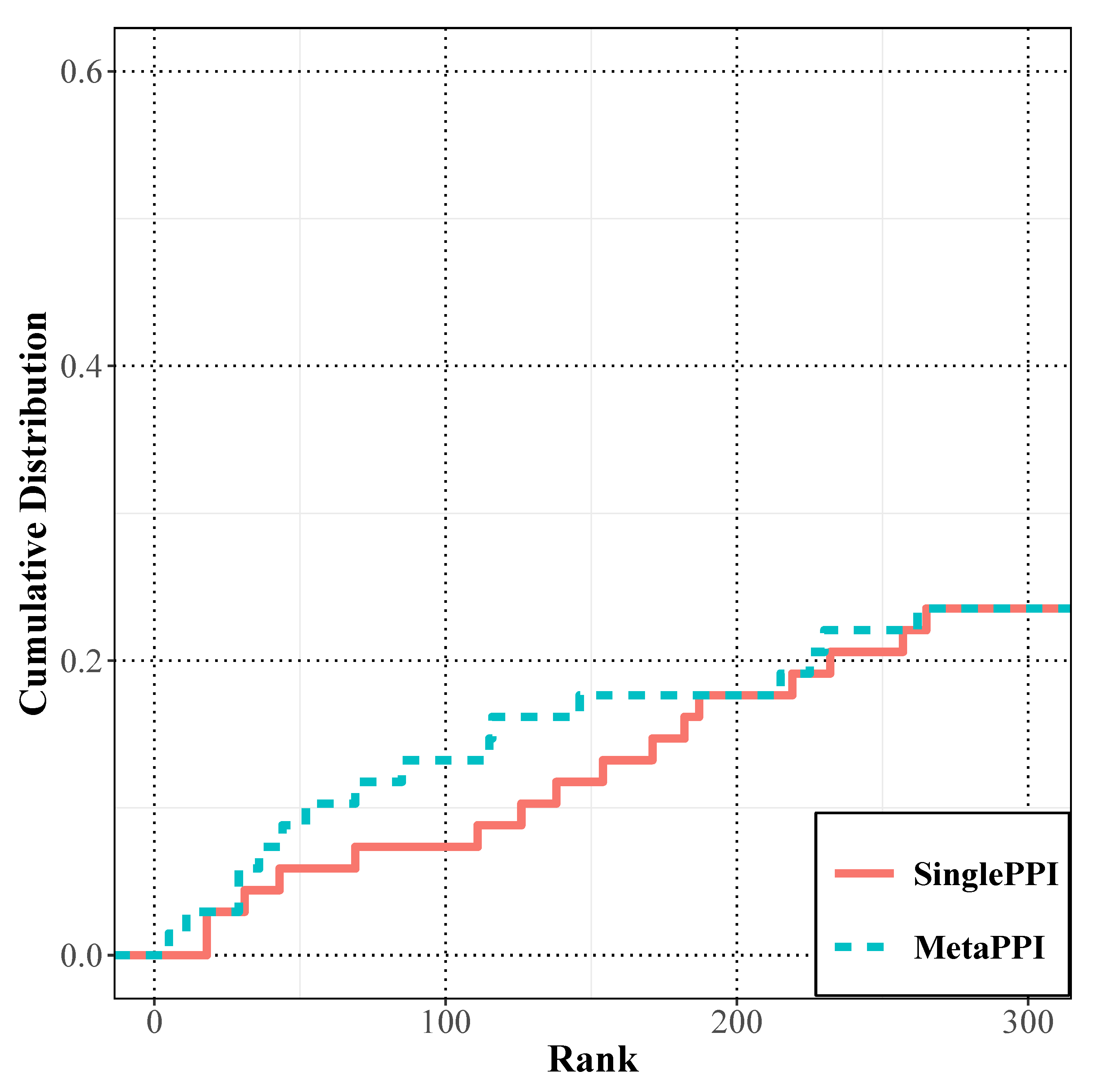
3.1. Prediction Performance Analysis of the RWR on the PPI Network
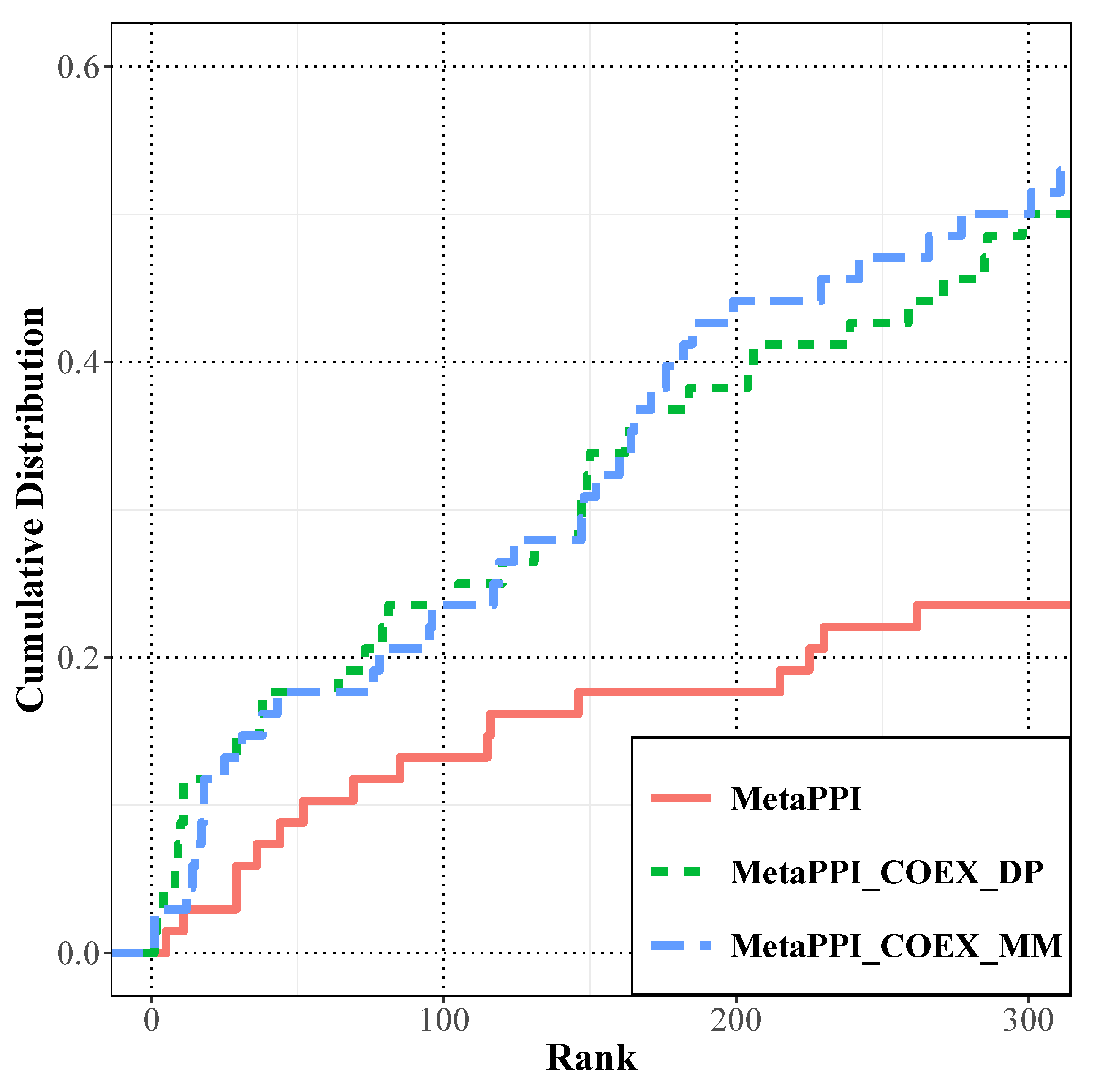
3.2. Prediction Performance Analysis of the RWR on the Multiplex Network
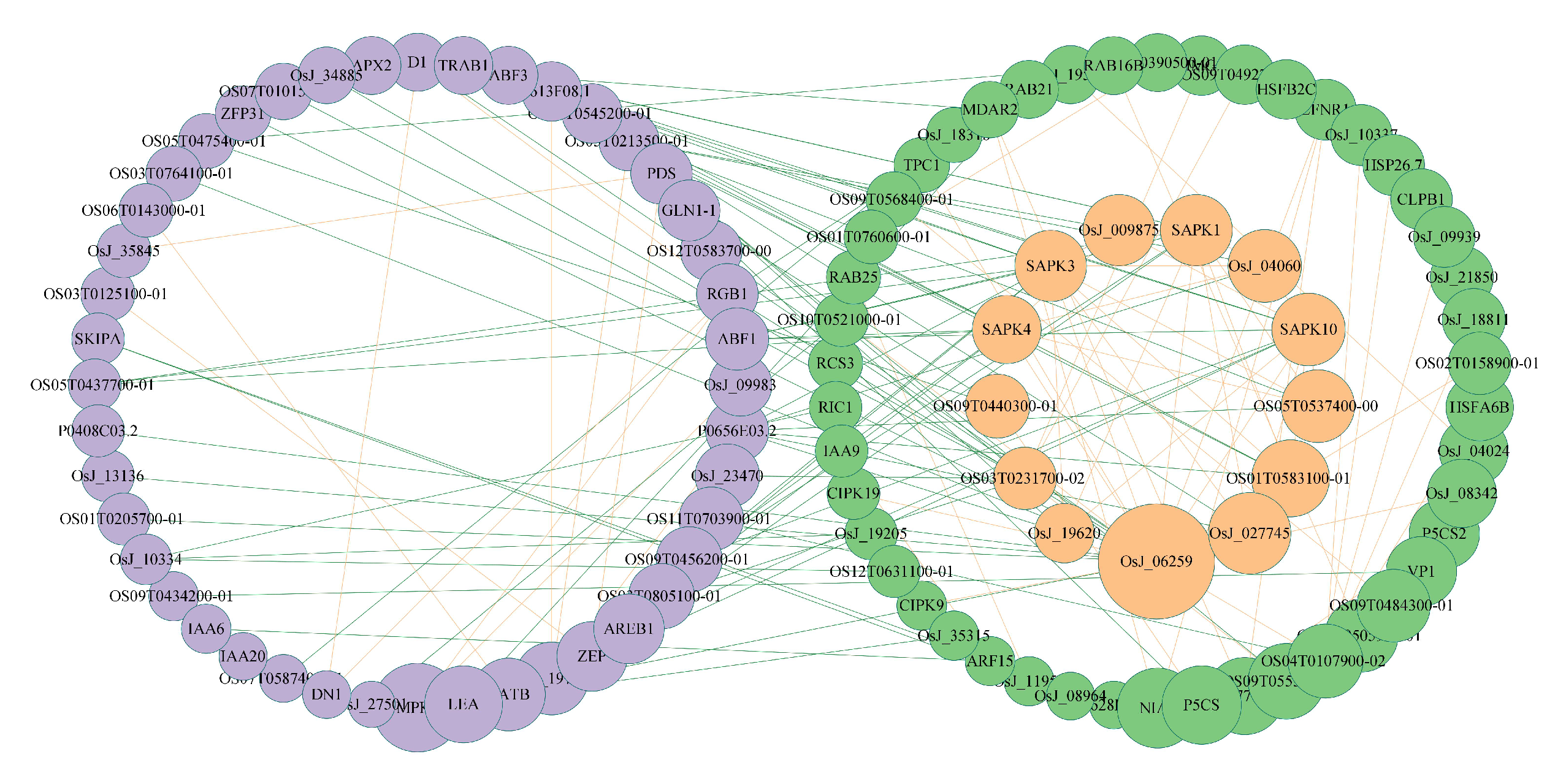
3.3. Obtaining Potential Genes Based on the RWR-M
3.4. Enrichment Analysis
3.5. Obtaining Candidate Genes Based on Association Analysis
3.6. Receiver Operating Characteristic Curve Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| GWAS | Genome-Wide Association Studies |
| PPI | Protein-Protein Interaction |
| RWR | Random Walk with Restart |
| RWR-M | Random Walk with Restart on multiplex network |
| WGCNA | Weighted gene co-expression network analysis |
| MIC | Maximal Information Coefficient |
| ROC | Receiver operating characteristics |
| SVM | Support vector machines |
| LOOCV | Leave-One-Out Cross-Validation |
| DEGs | Differentially expressed genes |
| SRA | Sequence Read Archive |
| CC | Cellular Component |
| BP | Biological Process |
| MF | Molecular Function |
References
- Zhang, Q. Successful Experience and Development Trend of China Hybrid Rice Seed Export and Enterprise Development Abroad. Chin. Rice. 2021, 27, 104–106. (In Chinese) [Google Scholar]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef]
- Bodily, P.M.; Fujimoto, M.S.; Page, J.T.; Clement, M.J.; Ebbert, M.T.; Ridge, P.G.; Alzheimer’s Disease Neuroimaging Initiative. A novel approach for multi-SNP GWAS and its application in Alzheimer’s disease. BMC Bioinform. 2016, 17, 268. [Google Scholar] [CrossRef] [PubMed]
- Peter, L.; Steve, H. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar]
- Yo-Han, Y.; Minjae, K.; Nalini, C.A.K.; Hong, W.J.; Ahn, H.R.; Lee, G.T.; Kang, S.; Suh, D.; Kim, J.O.; Kim, Y.J.; et al. Genome-Wide Transcriptome Analysis of Rice Seedlings after Seed Dressing with Paenibacillus yonginensis DCY84T and Silicon. Int. J. Mol. Sci. 2019, 20, 5883. [Google Scholar]
- Xiaobo, Z.; Chunjuan, L.; Shubo, W.; Tingting, Z.; Caixia, Y.; Shihua, S. Transcriptomic analysis and discovery of genes in the response of Arachis hypogaea to drought stress. Mol. Biol. Rep. 2021, 45, 119–131. [Google Scholar]
- Junxia, Y.; Tsutomu, T.; Toshihiro, O.; Hiroyuki, A.; Ikuko, T.; Eishin, O.; Hiroko, O.; Hatasu, K.; Toshiaki, H.; Wanyang, L.; et al. Combined linkage analysis and exome sequencing identifies novel genes for familial goiter. J. Hum. Genet. 2013, 58, 104–106. [Google Scholar]
- Alberto, V.; Laurent, T.; Claire, N.; Perrin, S.; Odelin, G.; Levy, N.; Cau, P.; Remy, E.; Baudot, A. Random walk with restart on multiplex and heterogeneous biological networks. Bioinformatics 2018, 35, 497–505. [Google Scholar]
- Li, Y.; Patra, J.C. Genome-wide inferring gene–phenotype relationship by walking on the heterogeneous network. Bioinformatics 2010, 26, 1219–1224. [Google Scholar] [CrossRef]
- Qu, J.; Wang, C.; Cai, S.; Zhao, W.D.; Cheng, X.L.; Ming, Z. Biased Random Walk With Restart on Multilayer Heterogeneous Networks for MiRNA–Disease Association Prediction. Front. Genet. 2021, 12, 1427. [Google Scholar] [CrossRef]
- Chauhan, V.K.; Dahiya, K. Problem formulations and solvers in linear SVM: A review. Artifical Intell. Rev. 2019, 52, 803–855. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018, 47, D607–D613. [Google Scholar] [CrossRef]
- Liu, S.; Liu, Y.; Zhao, J.; Cai, S.; Qian, H.M.; Zuo, K.; Zhao, L.; Zhang, L. A computational interactome for prioritizing genes associated with complex agronomic traits in rice (Oryza sativa). Plant J. 2017, 90, 177–188. [Google Scholar] [CrossRef]
- Gu, H.; Zhu, P.; Jiao, Y.; Meng, Y.; Chen, M. PRIN: A predicted rice interactome network. BMC Bioinform. 2011, 12, 161. [Google Scholar] [CrossRef]
- Hiroaki, S.; Shin, L.S.; Tsuyoshi, T.; Hisataka, N.; Jungsok, K.; Yoshihiro, K.; Hironobu, W.; Ching-chia, Y.; Masao, I.; Takashi, A.; et al. Rice Annotation Project Database (RAP-DB): An Integrative and Interactive Database for Rice Genomics. Plant Cell Physiol. 2012, 54, e6. [Google Scholar]
- Liu, Y.; Zhang, H.; Cao, D.; Li, L. Prediction of drought and salt stress-related genes in rice based on multi-platform gene expression data. Plant J. 2021, 47, 2423–2439. (In Chinese) [Google Scholar]
- Liu, S.; Wang, Z.; Zhu, R.; Wang, F.; Cheng, Y.; Liu, Y. Three Differential Expression Analysis Methods for RNA Sequencing: Limma, EdgeR, DESeq2. J. Vis. Exp. JoVE. 2021, 175, 177–188. [Google Scholar] [CrossRef]
- Xu, H.; Deng, Y. Dependent Evidence Combination Based on Shearman Coefficient and Pearson Coefficient. IEEE Access 2018, 6, 11634–11640. [Google Scholar] [CrossRef]
- Guo, Z.; Yu, B.; Hao, M.; Wang, W.; Zong, F. A novel hybrid method for flight departure delay prediction using Random Forest Regression and Maximal Information Coefficient. Aerosp. Sci. Technol. 2021, 116, 106822. [Google Scholar] [CrossRef]
- Cao, D.; Xu, N.; Chen, Y.; Zhang, H.Y.; Li, Y.; Yuan, Z.M. Construction of a Pearson- and MIC-Based Co-expression Network to Identify Potential Cancer Genes. Interdiscip. Sci. Comput. Life Sci. 2022, 14, 245–257. [Google Scholar] [CrossRef]
- Sun, G.; Li, J.; Dai, J.; Song, Z.; Lang, F. Feature selection for IoT based on maximal information coefficient. Future Gener. Comput. Syst. 2018, 89, 606–616. [Google Scholar] [CrossRef]
- Battiston, F.; Nicosia, V.; Latora, V. Structural measures for multiplex networks. Phys. Rev. E 2014, 89, 14. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Liu, J.; Nie, H.; Fu, Y.; Wan, L.; Kong, X. Random Walks: A Review of Algorithms and Applications. IEEE Trans. Emerg. Top. Comput. Intell. 2020, 4, 95–107. [Google Scholar] [CrossRef]
- Lei, X.; Bian, C. Integrating random walk with restart and k-Nearest Neighbor to identify novel circRNA-disease associatio. Sci. Rep. 2020, 10, 1943. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, L.; Wang, S.; Zhang, Y.H.; Kong, X.Y.; Huang, T.; Cai, Y.D. A computational method using the random walk with restart algorithm for identifying novel epigenetic factors. Mol. Genet Genom. 2018, 293, 293–301. [Google Scholar] [CrossRef]
- Liu, L.-L.; Zhang, S.-W. Advances in Predicting The Risk Pathogenic Genes With Random Walk. Prog. Biochem. Biophys. 2021, 48, 1184–1195. [Google Scholar]
- Belotti, F.; Peracchi, F. Fast leave-one-out methods for inference, model selection, and diagnostic checking. Stata J. 2020, 20, 785–804. [Google Scholar] [CrossRef]
- Carbon, S.; Ireland, A.; Mungall, C.J.; Shu, S.Q.; Marshall, B.; Lewis, S. AmiGO: Online access to ontology and annotation data. Bioinformatics 2008, 25, 288–289. [Google Scholar] [CrossRef]
- Otasek, D.; Morris, J.H.; Bouças, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef]
- Martínez-Camblor, P.; Pérez-Fernández, S.; Díaz-Coto, S. The area under the generalized receiver-operating characteristic curve. Int. J. Biostat. 2021, 18, 293–306. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Muller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Kamiyama, Y.; Hirotani, M.; Ishikawa, S.; Minegishi, F.; Katagiri, S.; Rogan, C.; Takahashi, F.; Nomoto, M.; Ishikawa, K.; Kodama, Y.; et al. Arabidopsis group C Raf-like protein kinases negatively regulate abscisic acid signaling and are direct substrates of SnRK2. Proc. Natl. Acad. Sci. USA 2021, 118, e2100073118. [Google Scholar] [CrossRef]
- Yoshiaki, K.; Sotaro, K.; Taishi, U. Growth Promotion or Osmotic Stress Response: How SNF1-Related Protein Kinase 2 (SnRK2) Kinases Are Activated and Manage Intracellular Signaling in Plants. Plants 2021, 10, 1443. [Google Scholar]
- Liu, Y.; Wang, B.; Li, J.; Sun, Z.; Chi, M.; Xing, Y.; Xu, B.; Yang, B.; Li, J.; Liu, J.; et al. A novel SAPK10-WRKY87-ABF1 biological pathway synergistically enhance abiotic stress tolerance in transgenic rice (Oryza sativa). Plant Physiol. Biochem. 2021, 168, 252–262. [Google Scholar]
- Singh, A.; Jha, S.K.; Bagri, J.; Pandey, G.K. ABA Inducible Rice Protein Phosphatase 2C Confers ABA Insensitivity and Abiotic Stress Tolerance in Arabidopsis. PLoS ONE 2015, 10, e0125168. [Google Scholar] [CrossRef]
- Min, M.K.; Kim, R.; Hong, W.J.; Jung, K.H.; Lee, J.Y.; Kim, B.G. OsPP2C09 Is a Bifunctional Regulator in Both ABA-Dependent and Independent Abiotic Stress Signaling Pathways. Int. J. Mol. Sci. 2021, 22, 393. [Google Scholar] [CrossRef]
- Miao, J.; Li, X.; Li, X.; Tan, W.; You, A.; Wu, S.; Tao, Y.; Chen, C.; Wang, J.; Zhang, D.; et al. OsPP2C09, a negative regulatory factor in abscisic acid signalling, plays an essential role in balancing plant growth and drought tolerance in rice. New Phytol. 2020, 227, 1417–1433. [Google Scholar] [CrossRef]
- Soma, F.; Takahashi, F.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Cellular Phosphorylation Signaling and Gene Expression in Drought Stress Responses: ABA-Dependent and ABA-Independent Regulatory Systems. Plants 2021, 10, 756. [Google Scholar] [CrossRef]
- Vavrdová, T.; Samaj, J.; Komis, G. Phosphorylation of Plant Microtubule-Associated Proteins During Cell Division. Front. Plant Sci. 2019, 10, 238. [Google Scholar] [CrossRef]
- Anna, K.; Izabela, W.; Ewa, K.; Maria, B.; Grazyna, D. SnRK2 Protein Kinases—Key Regulators of Plant Response to Abiotic Stresses. OMICS: J. Integr. Biol. 2011, 15, 859–872. [Google Scholar]
- Holappa, L.D.; Ronald, P.C.; Kramer, E.M. Evolutionary Analysis of Snf1-Related Protein Kinase2 (SnRK2) and Calcium Sensor (SCS) Gene Lineages, and Dimerization of Rice Homologs, Suggest Deep Biochemical Conservation across Angiosperms. Front. Plant Sci. 2017, 8, 395. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, J.-B.; Wei, N.; Liu, Z.-H.; Li, Y.; Zheng, Y.; Li, X.-B. A type-2C protein phosphatase (GhDRP1) participates in cotton (Gossypium hirsutum) response to drought stress. Plant Mol. Biol. 2021, 107, 499–517. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Lee, J.Y.; Lee, Y.; Kim, T.; Lee, S. Comprehensive survey of the VxG phi L motif of PP2Cs from Oryza sativa reveals the critical role of the fourth position in regulation of ABA responsiveness. Plant Mol. Biol. 2019, 101, 455–469. [Google Scholar] [CrossRef] [PubMed]
| GO Term | Number of Genes | Ontology | Description | p-Value |
|---|---|---|---|---|
| GO:0009266 | 18 | BP | Response to temperature stimulus | |
| GO:0009628 | 22 | BP | Response to abiotic stimulus | |
| GO:0009408 | 11 | BP | Response to heat | |
| GO:0009631 | 6 | BP | Cold acclimation | |
| GO:0009414 | 11 | BP | Response to water deprivation | |
| GO:0009719 | 22 | BP | Response to endogenous stimulus | |
| GO:0019222 | 44 | BP | Regulation of metabolic process | |
| GO:0031072 | 5 | MF | Heat shock protein binding | |
| GO:0106307 | 8 | MF | Protein threonine phosphatase activity | |
| GO:0042578 | 10 | MF | Phosphoric ester hydrolase activity | |
| GO:0106306 | 8 | MF | Protein serine phosphatase activity | |
| GO:0017111 | 11 | MF | Nucleoside–triphosphatase activity | |
| GO:0005737 | 60 | CC | Cytoplasm | |
| GO:0043231 | 80 | CC | Intracellular membrane-bounded organelle | |
| GO:0043229 | 83 | CC | Intracellular organelle |
| Candidate Genes | Name in STRING | Annotation | The Chromosome Location |
|---|---|---|---|
| Os10g0564500 | SAPK3 | Serine/threonine-protein kinase SAPK3; may play a role in the signal transduction of the hyperosmotic response | 10 |
| Os02g0281000 | OsJ_06259 | Os02g0281000 protein | 2 |
| Os03g0268600 | PP2C30; OsJ_009875 | Probable protein phosphatase 2C 30; belongs to the PP2C family | 3 |
| Os05g0537400 | PP2C50 | Probable protein phosphatase 2c 50; protein phosphatase involved in abscisic acid (ABA) signaling. Together with PYL3 and SAPK10, may form an ABA signaling module involved in the stress response | 5 |
| Os01g0846300 | PP2C09; OsJ_04060 | Probable protein phosphatase 2c 9; belongs to the PP2C family | 1 |
| Os01g0869900 | SAPK4 | Serine/threonine-protein kinase sapk4; may play a role in the signal transduction of the hyperosmotic response | 1 |
| Os03g0390200 | SAPK1 | Serine/threonine-protein kinase SAPK1; may play a role in the signal transduction of the hyperosmotic response | 3 |
| Os03g0610900 | SAPK10 | Serine/threonine-protein kinase SAPK10; may play a role in the signal transduction of the hyperosmotic response | 3 |
| Os03g0231700 | OS03T0231700-02 | Os03g0231700 protein; squalene monooxygenase, putative, expressed | 3 |
| Os05g0572700 | OsJ_19620 | Probable protein phosphatase 2C 51; belongs to the PP2C family | 5 |
| Os09g0325700 | OsJ_027745; PP108 | Probable protein phosphatase 2C 68; belongs to the PP2C family | 9 |
| Os01g0583100 | OS01T0583100-01; PP2C06 | Probable protein phosphatase 2C 6; belongs to the PP2C family | 1 |
| Os09g0440300 | OS09T0440300-01 | Aldehyde dehydrogenase-like protein; belongs to the aldehyde dehydrogenase family | 9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, L.; Zhang, H.; Cao, D.; Xu, Y.; Li, L.; Ning, Z.; Zhu, L. Drought Stress-Related Gene Identification in Rice by Random Walk with Restart on Multiplex Biological Networks. Agriculture 2023, 13, 53. https://doi.org/10.3390/agriculture13010053
Zhu L, Zhang H, Cao D, Xu Y, Li L, Ning Z, Zhu L. Drought Stress-Related Gene Identification in Rice by Random Walk with Restart on Multiplex Biological Networks. Agriculture. 2023; 13(1):53. https://doi.org/10.3390/agriculture13010053
Chicago/Turabian StyleZhu, Liu, Hongyan Zhang, Dan Cao, Yalan Xu, Lanzhi Li, Zilan Ning, and Lei Zhu. 2023. "Drought Stress-Related Gene Identification in Rice by Random Walk with Restart on Multiplex Biological Networks" Agriculture 13, no. 1: 53. https://doi.org/10.3390/agriculture13010053
APA StyleZhu, L., Zhang, H., Cao, D., Xu, Y., Li, L., Ning, Z., & Zhu, L. (2023). Drought Stress-Related Gene Identification in Rice by Random Walk with Restart on Multiplex Biological Networks. Agriculture, 13(1), 53. https://doi.org/10.3390/agriculture13010053






