Abstract
Haemoglobinopathies are common monogenic disorders with diverse clinical manifestations, partly attributed to the influence of modifier genes. Recent years have seen enormous growth in the amount of genetic data, instigating the need for ranking methods to identify candidate genes with strong modifying effects. Here, we present the first evidence-based gene ranking metric (IthaScore) for haemoglobinopathy-specific phenotypes by utilising curated data in the IthaGenes database. IthaScore successfully reflects current knowledge for well-established disease modifiers, while it can be dynamically updated with emerging evidence. Protein–protein interaction (PPI) network analysis and functional enrichment analysis were employed to identify new potential disease modifiers and to evaluate the biological profiles of selected phenotypes. The most relevant gene ontology (GO) and pathway gene annotations for (a) haemoglobin (Hb) F levels/Hb F response to hydroxyurea included urea cycle, arginine metabolism and vascular endothelial growth factor receptor (VEGFR) signalling, (b) response to iron chelators included xenobiotic metabolism and glucuronidation, and (c) stroke included cytokine signalling and inflammatory reactions. Our findings demonstrate the capacity of IthaGenes, together with dynamic gene ranking, to expand knowledge on the genetic and molecular basis of phenotypic variation in haemoglobinopathies and to identify additional candidate genes to potentially inform and improve diagnosis, prognosis and therapeutic management.
1. Introduction
Haemoglobinopathies are inherited disorders of haemoglobin (Hb) accounting for over 330,000 annual affected births worldwide. With 5.2% of the global population estimated to carry a potentially pathogenic gene, haemoglobinopathies are the most common monogenic disorders and a serious global public health problem [1]. They are endemic and prevalent in former malaria regions in the Mediterranean, sub-Saharan Africa, the Middle East and South-East Asia, but demographic events, such as global population mobility and migration, have contributed to their spread in all parts of the world [2,3]. As rare disorders in regions with traditionally low incidence and a growing public health burden in resource-limited countries, haemoglobinopathies pose major challenges for health professionals to efficiently diagnose, treat and care for patients [4].
The Hb protein complex comprises two α-like globin chains, encoded by genes in the α-globin locus (Chromosome: 16, Accession: NG_000006), and two β-like globin chains, encoded by genes in the β-globin locus (Chromosome: 11, Accession: NG_000007). The molecular pathology of haemoglobinopathies is traced to genetic defects in the two globin gene clusters, with more than 2000 different mutant alleles reported to date on the IthaGenes database of the ITHANET community portal [5,6]. These mutations can be grouped into those that impair globin chain synthesis, causing thalassaemia syndromes, and those that alter the structure of the Hb protein, causing structural haemoglobinopathies [7]. The pathophysiology and clinical manifestations of haemoglobinopathies are extremely varied with a range of acute and chronic complications that severely impair the quality of life and survival of patients, including iron overload, cardiac siderosis, liver fibrosis, viral hepatitis and endocrine dysfunction for transfusion-dependent thalassaemia, and painful crisis, stroke, acute chest syndrome, pulmonary hypertension, leg ulcers and priapism for sickle cell disease (SCD) [7,8]. Notably, the clinical management and treatment of haemoglobinopathies is challenging as patients with identical genetic defects often present different symptoms, which can even vary in severity over time.
A better understanding of genotype-phenotype correlations and the mechanisms underlying the clinical heterogeneity of haemoglobinopathies not only can improve the management of treatment but can also provide a better chance for the development of personalised medicine. Such knowledge can also enable the identification of affected individuals with a risk for increased disease severity towards early intervention with targeted and preventive care. To this end, β-thalassaemia and SCD, as the commonest of the β-haemoglobinopathies, have been investigated extensively to uncover the genetic determinants in interpatient phenotypic variability. The two best-characterised modifiers are co-inheritance of α-thalassaemia [9,10] and persistence of foetal haemoglobin (Hb F) production [11]. While elevated Hb F levels have no clinical benefit to adults not affected by a haemoglobinopathy, they have been demonstrated to ameliorate disease severity [12,13]. A large number of genome-wide analyses across diverse ethnic populations identified three quantitative trait loci (QTL) modulating Hb F levels: a promoter variant of the Gγ-globin gene (XmnI-HBG2), the HBS1L-MYB intergenic region (HMIP) and BCL11A, which together explain up to 50% of the genetic variation in Hb F [14,15]. Over the past few years, large-scale genome-wide association studies (GWAS) of improved power uncovered additional loci with modest effects on Hb F levels [16,17,18].
Nevertheless, these well-documented modifiers cannot explain the clinical diversity observed among haemoglobinopathy patients. Facilitated by the advent of technology, recent studies have identified variants associated with laboratory and clinical markers of disease severity, such as albuminuria and elevated glomerular filtration rate (GFR) for early renal disease [19], serum lactate dehydrogenase (LDH) for haemolysis [20], abnormal transcranial Doppler velocities for stroke [21] and elevated tricuspid regurgitant jet velocities for cardiopulmonary complications [22,23] (for a comprehensive review see [24]). Measurement of such markers would help risk-stratify patients to direct care, assist with early screening and diagnosis of symptoms, adjust dosing regimens for safe and effective drug therapy, and optimise personalised treatment prior to irreversible tissue damage and organ failure [25,26]. The widespread use of genomic tools provided vast (and still expanding) accumulation of data from association studies with a plethora of publications reporting on significant associations for numerous phenotypes in β-thalassaemia and SCD.
In the past, data on genetic modifiers of haemoglobinopathies had been scattered across hundreds of published papers, with previous efforts to collect and analyse such data restricted to comprehensive review articles [27,28,29,30] and databases without future updating and annotation [31]. Due to the large volume of literature and the amount of time required to screen and collect relevant data, important information was bound to remain inaccessible to the broad scientific community. Over the past few years, the ITHANET community portal has been curating and annotating disease-modifying genes and variants [5,6], using rigorous literature monitoring. Gene-to-phenotype associations are manually reviewed from the literature by individual assessment and annotated in the IthaGenes database of the portal. With 312 modifier genes and over 600 disease-modifying variants collected from over 450 eligible publications currently annotated in IthaGenes, ITHANET is the first knowledgebase to provide a comprehensive, continuously updated collection of information on genetic modifiers of haemoglobinopathies.
Although such gene-phenotype associations have been freely available on IthaGenes and elsewhere for a few years, the utilisation and analysis of the data has been challenging, owing to the lack of a robust measure to rank available evidence for each gene-phenotype relationship. While experimental validation is an effective approach to deduce strong genetic modifiers from a large number of candidates, it can be laborious and expensive. Alternatively, computational or mathematical methods for gene ranking enable quick assessment of large gene lists to identify top candidates. In fact, several methods have been implemented in the past to evaluate and rank the role of genes in the pathogenicity of different diseases [32,33,34,35,36]. However, similar evidence-based approaches to rank disease-modifying genes have been challenging due to the less prominent role of such genes in disease severity compared to the well-established disease-causing genes and the fact that each modifier gene may influence clinical manifestations for only a small fraction of patients. Moreover, functional analysis of such data and its biological and clinical interpretation have been difficult, and strongly depend on bioinformatics expertise [37].
The present work demonstrates how data organised in IthaGenes can be used by experimental and computational scientists alike to unravel complex gene-phenotype relationships and to explore their relevance in the development of new models of care and therapy for haemoglobinopathies. Specifically, an evidence-based gene ranking algorithm is developed and implemented to study the functional profile of genes that have been linked to modulation of the clinical manifestation and progression of haemoglobinopathies. In addition, functional enrichment analysis, with a focus on protein–protein interaction (PPI) networks as well as pathway and gene ontology (GO) analysis, is utilised to provide insights into the molecular pathology of these diseases and to identify novel target genes for further investigation. Importantly, the analysis revealed functional relationships between curated target genes for selected phenotypes, forming well-connected networks with roles in multiple mechanisms implicated in haemoglobinopathy-specific phenotypes.
2. Methods
2.1. Data Collection and Preprocessing
The data on disease modifiers were retrieved from the IthaGenes database, which provides a continuously updated, publicly available collection of disease-modifying genes and variations. The content of the IthaGenes database is collected from published peer-reviewed literature using PubMed, through automatic weekly searches for haemoglobinopathy-specific keywords, previously described in Kountouris et al. (2014) [5]. In brief, the titles and abstracts of retrieved publications are screened manually by the IthaGenes Curation Team and, if relevant, the full text is thoroughly examined to extract information on the relationships between genes, variants and phenotypes. The references from each publication are manually filtered to expand information on previously reported gene–phenotype relationships and to identify new disease-modifying variants and genes. Consequently, the final list of articles utilised in IthaGenes describes studies aiming to unravel genotype-phenotype relationships relevant to haemoglobinopathies and include GWAS, linkage, candidate gene, case-level and functional studies. Statistically significant associations (p value <0.05) or experimental evidence are then extracted from the articles and used for gene and/or variant annotation in IthaGenes. Each genetic modifier is linked to at least one phenotypic term mapped with standardised annotations curated by the human phenotype ontology (HPO) [38,39]. Those with poor phenotype definitions or terms not contained in HPO are annotated by terms that best describe the clinical characteristics of the study population or laboratory risk factors investigated. Moreover, genetic modifiers are linked to data from existing public databases (e.g., National Centre for Biotechnology Information (NCBI) Gene, Online Mendelian Inheritance in Man (OMIM), Universal Protein Resource (UniProt), Single Nucleotide Polymorphism Database (dbSNP)) and receive a multitude of additional manual annotations, such as gene function, the role in disease and the effect on phenotypes.
As part of this study, the dataset retrieved from IthaGenes was further processed to identify studies that report on the same piece of evidence, e.g., reviews reporting associations described in original studies that had already been included in the dataset. Such duplicated evidence was removed from the dataset, whereas the quality of each study was also further annotated and evaluated by collecting information about the type and design of study, reported p values and confidence intervals and use of multiple testing, if needed. The final dataset used for the current study comprises 493 unique gene-phenotype relationships, derived from a total of 312 genes and 59 phenotypes, with data on β-thalassaemia and SCD analysed together as pooled data for β-haemoglobinopathies.
2.2. Development of an Evidence-Based Approach for Gene Ranking
The volume of available evidence for each gene–phenotype relationship in the dataset is represented quantitatively with three different scores, namely Association Score, Variant Score and Experimental Score, using a point system to reflect the strength of each piece of evidence. Similar approaches have been developed in the past to quantify existing evidence for gene-disease relationships [36,40,41], but, to our knowledge, this is the first effort to develop an evidence-based framework for modifier genes in a Mendelian disorder. The point system used for each individual score is shown in Table 1 and briefly described below.

Table 1.
The point system used to score available evidence and to calculate the three individual scores (Association Score, Variant Score and Experimental Score) involved in the calculation of IthaScore. The point system was based on a similar approach described in References [40,46].
The Association Score (AS) represents the sum of points derived from statistically significant associations for each gene-phenotype relationship. For every study in the dataset, the most significant variant of each gene for a given phenotype was selected to represent the strength of the gene-phenotype relationship. Three different evidence levels were considered to score each study for a given phenotype as follows: (a) case-level studies and association studies reporting statistically significant associations with a p value of <0.05, scored with 0.5 point, (b) association studies with at least one variant with a p value of <0.001, scored with 1 point, and (c) association studies with at least one variant with a p value of <10−5, scored with 1.5 points. To avoid possible bias from multiple case-level studies (under the lowest evidence level above), a maximum of four case-level studies (i.e., a total of 2 points, with 0.5 point awarded for each case study) were considered for each gene–phenotype relationship. In addition, all association studies were evaluated qualitatively to detect studies with weak methodology (e.g., lacking multiple comparison procedures and confidence intervals). Such cases remained in the dataset to avoid reduction of the evidence pool, but their AS was reduced by a penalty of 25%. Subsequently, the sum of all points from different independent studies was calculated for each gene–phenotype relationship.
The Variant Score (VS) represents the number of variants identified in each gene–phenotype pair and are curated in IthaGenes, the largest database of modifiers relevant to haemoglobinopathies. In each gene-phenotype relationship, a single point was awarded for every variant in the database.
The Experimental Score (ES) represents the sum of all points derived from experimental evidence available for each gene–phenotype relationship. Given that the implication of modifier genes in the pathology of haemoglobinopathies needs to be validated by experiments that support a role for that gene with respect to the phenotype under study, a point system similar to the work of Strande et al. [40] was employed to divide experimental evidence into three main categories: gene function (biochemical function, protein interaction and expression), functional alteration, and model systems (model organisms and phenotypic rescue). Experimental studies on gene function, functional alteration and model systems received 1, 1.5, and 2 points, respectively. The sum of all points derived from experimental evidence was subsequently calculated for each gene–phenotype relationship.
A maximum allowed sum of points was set for each individual score in order to count for multiple replication studies establishing a gene–phenotype, but, at the same time, to avoid overrepresentation (i.e., very high scores) of well-established disease modifier genes in our analysis, such as BCL11A and KLF1. The maximum allowed scores for AS, VS and ES were set to 8, 20 and 6, respectively. All individual scores for each gene-phenotype pair were subsequently normalised to be canonical (from 0 to 1) by dividing the total score by the maximum allowed sum of points.
The overall score, called IthaScore, is calculated with the formula below, using a weighted sum of all individual scores and reflects the available evidence for each gene-phenotype relationship. The weights have been selected to represent both the strength of each evidence type, but also the volume of available evidence in the dataset. Therefore, a stronger weight is used for association studies that represent the overwhelming fraction of evidence in the dataset, with around 85% of scores derived from association studies and 15% from functional studies.
2.3. Functional Enrichment Analysis
Functional enrichment analysis was performed for each phenotype in the dataset using their corresponding gene lists. Specifically, the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database v.11.0 [42] was used for the construction of PPI networks, followed by GO term enrichment and pathway analysis (Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome). Only functional enrichment terms with a p value of 10–5 or lower (after false discovery rate (FDR) correction), as provided by STRING, were considered for further investigation. The Human Genome Organisation (HUGO) Gene Nomenclature Committee (HGNC) approved gene symbols were used as input data, thereby excluding intergenic regions from functional enrichment and network analysis. Connections (edges) between proteins (nodes) were predicted at a high confidence cut off of ≥0.7 using all types of evidence available in STRING, while the top five additional interactors with the initial gene set were also included in the analysis. High-resolution bitmaps of the PPI networks were displayed and exported from STRING. In addition, GOnet [43] was used to investigate and visualise relationships between specific gene lists and statistically significant GO terms. The Comparative Toxicogenomics Database (CTD) MyVenn tool [44] was utilised to identify common genes between different phenotype-specific gene list in the dataset, specifically “Hb F levels”, “F-cell numbers” and “Hb F response to hydroxyurea”. The Cytoscape software, version 3.7.2 [45], was used to visualise gene-phenotype relationships in a network format.
3. Results and Discussion
3.1. Exploratory Analysis of Modifier Gene Lists
The dataset was analysed to identify genes and genomic locations involved in several phenotypes relevant to haemoglobinopathies, visualised in Figure 1. Although, as expected, the majority of genes (219 out of 312 genes) were assigned to a single phenotypic term, numerous genes were linked to multiple phenotypes. Notably, eleven genes are assigned to five or more phenotypic terms, of which HMIP, BCL11A and NOS3 ranked top with 10, 11 and 13 phenotypic terms, respectively. Such multiple assignments are expected due to common pathophysiological mechanisms in many disease complications (e.g., haemolysis and vaso-occlusion), thus involving similar sets of genes. The full list of genes ordered by the number of phenotypic annotations is shown in Supplementary Table S1.

Figure 1.
Gene distribution per phenotype annotations. Pie diagram shows the distribution of genes (n (%), for a total of 312 genes) based on the number of assigned phenotypic terms (1, 2, 3, 4, and ≥5).
Figure 2 shows a summary of the 59 phenotypic terms in the dataset ordered by the number of genes and variant annotations. The number of genes and variants differed among phenotypes, with “Hb F levels” (82 genes, 299 variants) being the most prevalent term, followed by “Hb F response to hydroxyurea” (42 genes, 69 variants). Other frequently assigned terms involved clinical descriptions relevant to haemolysis and vaso-occlusion, including: ”stroke”, “osteonecrosis/avascular necrosis”, “pulmonary arterial hypertension”, “pain”, “acute chest syndrome” and ”leg ulcers”. In addition, 36 phenotypic terms were assigned to five or fewer genes, of which thirteen were annotated with a single gene bearing a few variants.
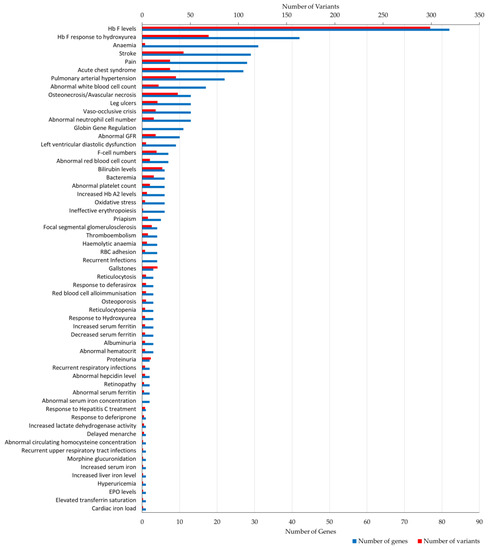
Figure 2.
Number of gene and variant annotations per phenotypic term. Bar plot illustrates the number of genes (bottom x-axis, shown in blue) and variants (top x-axis, shown in red) assigned to each phenotypic term (total of 59) stored in IthaGenes. GFR, glomerular filtration rate; RBC, red blood cell; EPO, erythropoietin.
3.2. Evidence-Based Gene Ranking
The gene ranking analysis integrated a scoring metric, called IthaScore, where each gene-phenotype interaction was assigned a combined score of various evidence measures (see Methods for details on the calculation of IthaScore). Overall, 483 gene-phenotype interactions were identified and scored, with IthaScore ranging from 0.023 to 0.875. The entire gene list as well as their scores and ranking are shown in Supplementary Table S2. Higher gene scores indicate a greater likelihood that genes would have an effect on the phenotypes investigated. Figure 3 shows the distribution of IthaScore for all gene-phenotype relationships, while Table 2 shows the top scoring genes for each of the 59 phenotypic terms in the dataset. Importantly, most of the gene-phenotype pairs have a low IthaScore, thus highlighting that available evidence for those relationships is currently weak and that additional studies are needed before such associations are considered reliable.
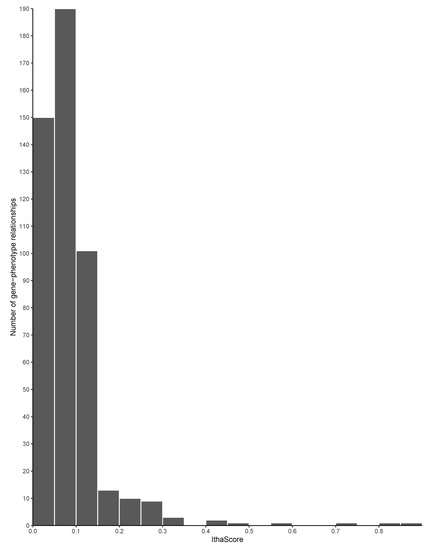
Figure 3.
Distribution of IthaScore. Histogram shows the distribution of IthaScore in the range 0–1 for 483 gene-phenotype interactions.

Table 2.
The gene with the highest IthaScore for each phenotype. “Phenotype ID” column indicates the identifier assigned to each phenotype throughout this work.
In contrast, our approach is validated by its ability to produce a high IthaScore for well-established disease modifiers, particularly those involved in Hb F production and modulation, such as BLC11A, HMIP and KLF1. This is demonstrated in Table 3, which lists the ten gene-phenotype pairs with the highest IthaScore, with nine of them involved in Hb F modulation. In addition, our method successfully highlights, with a high IthaScore, the well-established role of UGT1A1 in bilirubin metabolism, especially since genetic variations in UGT1A1 constitute major risk factors for unconjugated hyperbilirubinemia [47].

Table 3.
Top 10 gene-phenotype interactions with the highest IthaScore. “Phenotype ID” column represents the identifier assigned to each phenotype throughout this work.
A gene-phenotype network was constructed and shown in Figure 4 depicting gene-phenotype relationships with an IthaScore of at least 0.1 to show significant relationships and, also, allow clearer interpretation and visualisation. The edge weights represent the strength of the relationship based on the calculated IthaScore, while each phenotype is labelled with a unique identifier, as defined in Table 2, for better visualisation of the network.
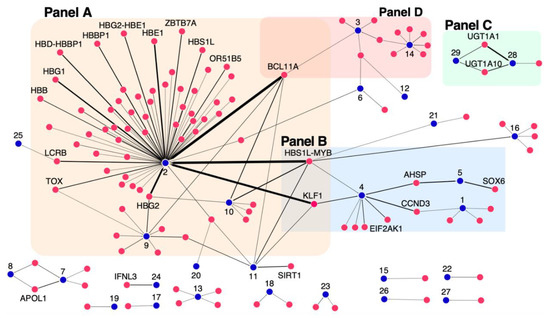
Figure 4.
Network diagram of gene–phenotype interactions. The network depicts relationships between genes and phenotypes of haemoglobinopathies. Genes (red nodes) are connected to phenotypes (blue nodes) by edges. The thickness of the edges represents the corresponding IthaScore, where a stronger edge indicates a greater weight for the gene-phenotype relationship. Only gene–phenotype relationships with gene scores ≥0.1 are displayed on the network for better visualisation, while gene names are only shown for gene scores ≥0.2. The “Phenotype ID” shown in Table 2 is used to label phenotypic nodes, as follows: (1) Abnormal red blood cell count, (2) Hb F levels, (3) Stroke, (4) Anaemia, (5) Ineffective erythropoiesis, (6) Osteonecrosis/Avascular necrosis, (7) Focal segmental glomerulosclerosis, (8) Proteinuria, (9) Hb F response to hydroxyurea, (10) F-cell numbers, (11) Globin gene regulation, (12) Bacteremia, (13) Abnormal white blood cell count, (14) Acute chest syndrome, (15) Osteoporosis, (16) Abnormal platelet count, (17) Left ventricular diastolic dysfunction, (18) Pain, (19) Abnormal serum iron concentration, (20) Hyperuricemia, (21) Abnormal hematocrit, (22) Increased serum ferritin, (23) Vaso-occlusive crisis, (24) Response to Hepatitis C treatment, (25) Increased Hb A2 levels, (26) Erythropoietin (EPO) levels, (27) Haemolytic anaemia, (28) Bilirubin levels and (29) Gallstones.
Naturally, phenotype “Hb F levels” (node 2 in Figure 4, Panel A) is a clear hub in the network showing both the highest number of connected genes (with IthaScore ≥0.1) and the strongest connections, i.e., the highest IthaScore in the network. High scoring loci in the “Hb F levels” phenotype, such as BCL11A and HMIP, have also weaker connections with other phenotypes in the network, although this can be an indirect effect in the disease severity due to the well-established role of high Hb F as a disease-modifying factor.
As the level of Hb F is a major predictor of survival in haemoglobinopathies, genetic markers that modulate Hb F production have been investigated extensively. Similar to numerous studies reported to date [16,48,49], the top-ranked genes for interaction with “Hb F levels” include BCL11A (0.875), HMIP (0.825), KLF1 (0.711) and HBG2 (0.6). Another sensitive biological indicator of Hb F is the abundance of Hb F-containing erythrocytes (F cells) [11]. In this analysis and in line to published work [50], BCL11A, HMIP, KLF1 and HBG2 are ranked as the leading modifiers of “F cell numbers” (node 10). Moreover, hydroxyurea (HU), as a potent pharmacological inducer of Hb F, is used in the treatment of SCD, although with highly variable degrees of clinical response [51]. The search for genetic modifiers of “Hb F response to HU” (node 9) identified associations to BCL11A and HBG2 as the most robust [52]. Although both genes drew top scores following ranking, less prominent Hb F-promoting loci, including SAR1A, MAP3K5, NOS1 and ARG2, emerged as promising predictors of drug response based on the calculated IthaScore. While many of the Hb F-promoting loci are also associated with Hb F response to HU, the absence of strong Hb F modulators, such as KLF1 and HMIP, from loci associated with Hb F response to HU suggests that some mechanisms of HU-induced Hb F may differ from mechanisms of endogenous Hb F regulation (candidate mechanisms of HU-induced Hb F are summarised in Pule et al. [53]).
Other smaller subnetworks shown in Figure 4 highlight the role of different genes in other disease phenotypes, specifically including (a) anaemia (node 4), ineffective erythropoiesis (node 5) and abnormal red blood cell (RBC) count (node 1), (b) bilirubin metabolism (node 28) and gallstone formation (node 29), and (c) phenotypes/complications related to vaso-occlusion and/or haemolysis like acute chest syndrome (ACS) (node 14) and stroke (node 3). The above phenotype groups are highlighted in Panels B, C and D of Figure 4 respectively, and discussed below.
Ineffective erythropoiesis is a hallmark of β-thalassaemia characterised by excess free alpha haemoglobin (α-Hb) pool in erythroid precursors, which leads to their premature destruction within the bone marrow, resulting in abnormal counts of RBCs in circulation and, thus, to anaemia [54]. Supported predominantly by functional evidence, SOX6 and AHSP were identified as the leading modifiers of “ineffective erythropoiesis” (node 5), while AHSP also achieved high IthaScore for interaction with “anaemia” (node 4). In fact, AHSP is a candidate molecular chaperone for free α-Hb and a critical modulator of β-thalassaemia [55]. Additionally, CCND3 had a high IthaScore for interaction with “anaemia” and “abnormal RBC counts” (node 1), which is in line with its role in controlling cell cycle progression and differentiation during haematopoiesis and thereby RBC size and count [56].
One of the best-known genetic modifiers of bilirubin metabolism and cholelithiasis in haemoglobinopathies is the UGT1A locus [27]. As expected, and illustrated in Panel C of Figure 4, members of the UGT1A family, namely UGT1A10, UGT1A6 and UGT1A1, were among the top-ranked genes for interactions with “bilirubin levels” (node 28) and “gallstone” formation (node 29).
Haemolysis and vaso-occlusive phenomena are fundamental features of SCD affecting a variety of tissues and organs [57]. Here, we present candidate genes that could potentially influence two of the most important complications of SCD: ACS (node 14) and stroke (node 3) (Panel D of Figure 4). ACS is a vaso-occlusive crisis of the pulmonary vasculature and one of the leading causes of hospitalisation among SCD patients [58] and has been associated with effects of endothelial nitric oxide (eNOS) metabolism, inflammation, cell adhesion, hypoxia and endothelial damage [59]. As expected, high-scoring genes for “acute chest syndrome” (node 14) included EDN1 and NOS3, as well as genes involved in the TGF-β signalling pathway, namely TGFBR3, SMAD1 and SMAD7. Although stroke is one of the most disabling complications, the factors that lead to stroke remain elusive [60]. The top-scoring genes for “stroke” (node 3) included ENPP1, TGFBR3, ADCY9, BCL11A and BMP6.
3.3. Functional Enrichment Analysis for Selected Phenotypes
Towards understanding the biological meaning behind large lists of genes for specific phenotypes and in search for their mechanisms of action, functional enrichment analysis focused on identification of enriched GO terms, specifically biological process (BP) and molecular function (MF), as well as associated pathways (from KEGG and Reactome). Only enriched GO terms and biological pathways with an FDR <10−5 were considered. Those associated with a low gene count in the database were more specific, thus giving a greater biological meaning. Given that a complete functional enrichment analysis for each of the 59 phenotypes is beyond the scope of this work, we demonstrate the results of the analysis for three selected phenotypes related to different pathophysiological mechanisms and of different gene set sizes: (a) Hb F levels in relation with Hb F response to HU, (b) response to iron chelators and (c) stroke.
3.3.1. Hb F Levels and Hb F Response to Hydroxyurea
The discovery of genetic markers for the upregulation of Hb F in patients with β-thalassaemia and SCD has been a major ongoing research effort for decades, resulting in a large volume of data in the literature. Drawing information from studies showing a positive correlation between Hb F levels and the number of F cells [61], the gene sets of these two phenotypes were pooled for simplicity (from here on referred to as phenotype “Hb F levels/F-cells”). Additionally, the major benefit of hydroxyurea (HU) on disease severity is directly related to its effect on Hb F production [62]. The large number of reported genes made it challenging to establish informative GO term and pathway rankings with relevance to the “Hb F levels/F-cells” phenotype, instigating the need for further gene set enrichment analysis. As to remove noisy information from the analysis and to identify candidate genes that regulate fetal γ-globin genes and also modulate HU-induced Hb F levels, genes that were common between phenotypes “Hb F levels/F-cells” and “Hb F response to HU” were identified (11 genes) and used as input data for analysis. These included ARG2, ASS1, BCL11A, FLT1, HBE1, HBG2, MAP3K5, NOS1, SAR1A, TOX and VEGFA. Five additional interactors were allowed in the network to identify the most significant interactions to the initial protein list and achieve a meaningful size for network analysis (16 nodes total), shown in Figure 5A. Interestingly, these interactors contained five additional proteins without prior connotation to the above Hb F-related phenotypes, except for the VEGF receptor KDR (kinase insert domain receptor). These new candidate proteins included ASL (argininosuccinate lyase), OTC (ornithine carbamoyltransferase), PGF (placental growth factor) and VEGFB (vascular endothelial growth factor B). In addition, three of the proteins (MAP3K5, SAR1A and TOX) were not engaged in any interactions with the high confidence interaction score 0.7 in STRING.
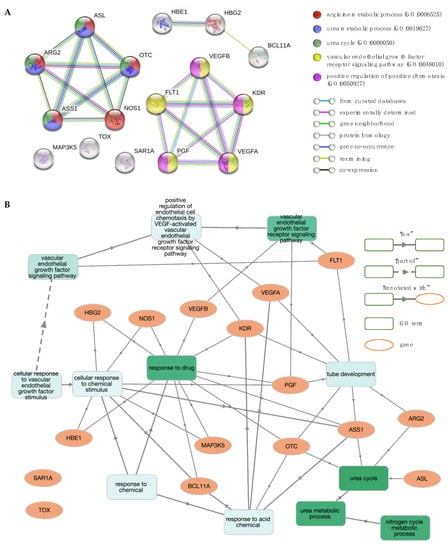
Figure 5.
Network and enrichment analysis for Hb F levels and Hb F response to hydroxyurea (HU). (A) The protein–protein interaction (PPI) network contains 16 nodes (proteins; circles) connected by edges (protein–protein interactions). The most significant gene ontology (GO) biological process (BP) terms are shown. (B) GO BP enrichment analysis using GOnet (q value ≤0.01; p value ≤7.88 × 10–6). Green colour intensifies as the significance level of enrichment decreases.
The PPI network and the subsequent functional enrichment analysis of the final protein list resulted in two distinct clusters (Figure 5A). One cluster included five proteins, namely ARG2, ASL, ASS1, NOS1 and OTC, that are annotated with GO terms and pathways involved in nitrogen metabolism, including GO terms “urea cycle” (GO:0000050), “urea metabolic process” (GO:0019627) and “arginine metabolic process” (GO:0006525) and pathways “urea cycle” (HSA-70635) and “arginine biosynthesis” (hsa00220). The second cluster contained five proteins, namely FLT1, KDR, PGF, VEGFA and VEGFB, that are linked to functional terms related to the VEGF-VEGFR system, including “positive regulation of angiogenesis” (GO:0045766), “vascular endothelial growth factor receptor signalling pathway” (GO:0048010), and pathways involved in vascular endothelial growth factor (VEGF) ligand-receptor interactions (VEGF binds to VEGFR leading to receptor dimerisation “HSA-195399” and MAPK signalling pathway “hsa04010”). Overall, significant GO terms and pathways were consistent between them, with Figure 5B illustrating interactions between genes and GO term enrichment analysis.
Notably, three of the query proteins (ARG2, ASS1 and NOS1) and two of the new interactors (ASL and OTC) are involved in the urea cycle and the L-arginine biosynthesis sub-pathway (Figure 6). Specifically, argininosuccinase (ASL) catalyses the production of arginine from arginosuccinate, while ornithine carbamoyltransferase (OTC) catalyses the production of citrulline, an intermediate substrate in the pathway of arginine synthesis. Drawing information from studies investigating the factors that are implicated in a variable Hb F response to HU treatment, there is strong evidence to suggest that the arginine-dependent nitric oxide (NO) pathway is involved in the induction of Hb F [63,64,65]. NO is a signalling agent produced from the metabolism of L-arginine by the enzyme nitric oxide synthase (NOS) [66]. The underlying mechanism involves NO-mediated activation of soluble guanylate cyclase (sGC) and subsequent signalling via the sGC/cyclic guanosine monophosphate (cGMP)-dependent protein kinase (PKG) pathway [67]. Considering that this effect can also be mediated by other NO donor substrates, it is important to explore ASL and OTC as potential mechanisms by which drug-mediated NO production could be therapeutic or prognostic of drug efficacy.
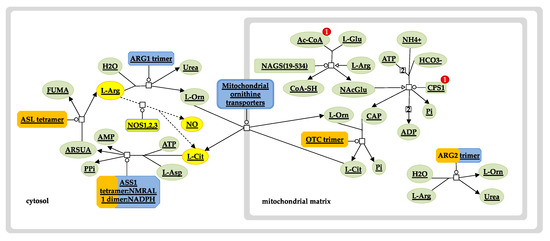
Figure 6.
Urea cycle and nitric oxide pathway. Diagram depicts enzymes and intermediates of the urea cycle (solid lines) and the nitric oxide (NO) pathway (dashed lines, yellow nodes). Overrepresentation of the Hb F-related gene set used in the analysis is shown in orange. The size of the orange strip increases with the level of gene representation in the query set. The urea cycle pathway was exported from the Reactome pathway database and edited to include and highlight the role of NO shown in yellow. Ac-CoA, acetyl coenzyme A; AMP, adenosine monophosphate; ARG1, arginase 1; ARG2, arginase 2; ARSUA, argininosuccinate; ASL, arginosuccinate lyase; ASS1, arginosuccinate synthase; ATP, adenosine triphosphate; CAP, carbamoyl phosphate; CoA-SH, coenzyme A; CPS1, carbamoyl phosphate synthase 1; FUMA, fumarate; L-Arg, L-arginine; L-Asp, L-aspartate; L-Cit, L-citrulline; L-Glu, L-glutamine; L-Orn, L-ornithine; NAcGlu, N-acetylglutamic acid; NAGS, N-acetylglutamate synthase; NOS1,2,3, nitric oxide synthase 1 (neuronal, nNOS), 2 (inducible, iNOS), 3 (endothelial, eNOS); OTC, ornithine transcarbamylase; Pi, inorganic phosphate; PPi, inorganic pyrophosphate.
Also associated with Hb F levels and Hb F response to HU were proteins involved in vasculogenesis and angiogenesis, namely VEGFA (vascular endothelial growth factor A), FLT1 (vascular endothelial growth factor receptor 1, VEGFR1) and the new interactors VEGFB (vascular endothelial growth factor B) and PGF (placenta growth factor). The mechanism by which these genes influence Hb F production is still unclear, yet a growing amount of evidence implicates an effect on the process of erythropoiesis [68,69,70,71]. Notably, additional studies will be necessary to identify the functional role of VEGF signalling and other potent factors on erythropoiesis, as well as their effect on globin gene transcription programs.
3.3.2. Response to Iron Chelators
Deferiprone and deferasirox are standard drugs for iron chelation therapy in transfusion-dependent anaemias. Decreasing excess accumulation of iron through the use of chelation reduces damage to critical organs [72]. However, patients show different rates of adherence and drug-related toxicities, indicating that genetic factors may influence the way drugs are metabolised [73,74]. To identify potential molecular pathways related to response to chelation therapy with deferiprone and deferasirox, genes associated with phenotypes “response to deferiprone” and “response to deferasirox” were pooled (ABCC2, CYP1A1, CYP1A2 and UGT1A6) and used as input for the STRING database. Figure 7A shows the PPI network, including five additional interactors, namely UGT1A3, UGT1A4, UGT1A8, UGT1A9 and AHR, and Figure 7B illustrates the interactions between genes and GO term enrichment analysis.
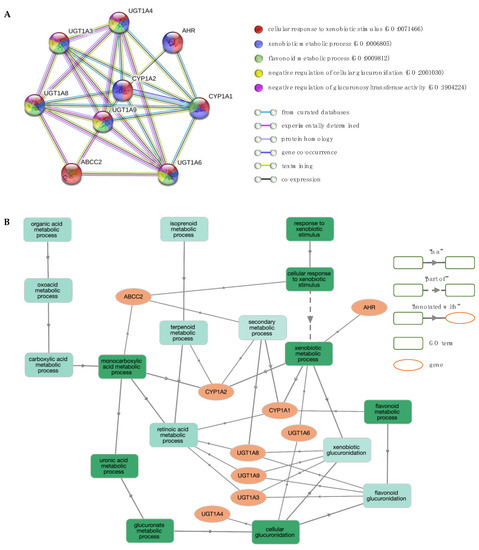
Figure 7.
Network and enrichment analysis for response to iron chelators. (A) The PPI network contains nine nodes (proteins; circles) connected by edges (protein–protein interactions). The most significant GO BP terms are shown. (B) GO BP enrichment analysis using GOnet (q value ≤0.01; p value ≤5.3 × 10–9). Green colour intensifies as the significance level of enrichment decreases.
The enriched GO BP terms indicate gene functions mainly associated with (a) xenobiotic metabolism, including “cellular response to xenobiotic stimulus” (GO:0071466), “xenobiotic metabolic process” (GO:0006805) and “flavonoid metabolic process” (GO:0009812), (b) glucuronidation, such as “negative regulation of cellular glucuronidation” (GO:2001030), “negative regulation of glucuronosyltransferase activity” (GO:1904224) and “xenobiotic glucuronidation” (GO:0052697), and (c) fatty acid metabolism, including “monocarboxylic acid metabolic process” (GO:0032787), “negative regulation of fatty acid metabolic process” (GO:0045922), and “omega-hydroxylase P450 pathway” (GO:0097267). Pathways involved in “retinol metabolism” (hsa00830), “metabolism of xenobiotics by cytochrome P450” (hsa00980) and “glucuronidation” (HSA-156588) were also deemed enriched. Notably, only the UGT1 locus is associated with the glucuronidation pathway.
These findings are in line with published work demonstrating that deferiprone and deferasirox are mainly metabolised by glucuronidation [73,75], a major pathway of xenobiotic biotransformation (phase II metabolism, conjugation) catalysed by uridine 5’-diphospho-glucuronyltransferases (UGT). Members of the UGT1 family are the most important in terms of drug metabolism and are found primarily in the liver [76]. Cytochrome P450 (CYP) is another family of xenobiotic-metabolising enzymes (phase I metabolism, functionalisation) [77], of which only two members (CYP1A1 and CYP1A2) appear in the PPI network. Both CYP1 proteins interact with the aryl hydrocarbon receptor (AHR), a xenobiotic receptor that regulates the activation of CYP1A1, CYP1A2 and several other genes, including UGT1A4, UGT1A6 and UGT1A9 [76,78,79].
Glucuronidation of deferasirox is mainly mediated by UGT1A1 and to a lesser extent by UGT1A3, with minor contributions from UGT1A7 and UGT1A9, and trace activities by several other UGTs (UGT1A4, UGT1A6, UGT1A8, UGT1A10, UGT2B4, UGT2B7, UGT2B15 and UGT2B17) [80]. Oxidative metabolism by CYP enzymes (CYP1A1, CYP1A2 and CYP2D6) has a minor contribution to the elimination process [81]. Deferasirox and its glucuronide metabolites are eliminated mainly by hepatobiliary transport via multidrug-resistance protein 2 (MRP2) [82]. MRP2, also known as ABCC2, is an anion transporter expressed at important pharmacological barriers, such as the canalicular membrane of hepatocytes, with an important role in the elimination of xenobiotic substrates [83]. Moreover, glucuronidation of deferiprone is catalysed almost exclusively by the UGT1A6 in hepatic tissues with subsequent excretion in the urine. Several other UGTs (UGT1A7, UGT1A8, UGT1A9, UGT1A10, UGT2B7, and UGT2B15) exhibit trace activities and are not expected to impact the formation of glucuronide metabolites [75,84,85].
The results of the functional enrichment analysis indicate that the proteins involved in the metabolism and transport of deferiprone and deferasirox may also influence response to therapy. Specifically, new candidate modifiers include UGT1A4, UGT1A8 and UGT1A9, which exhibited low metabolic clearance of these drugs with in vitro animal tissue models. As drug metabolism and interactions are species-specific [86] and given that drug-metabolising enzymes have different rates of maturation at different developmental stages [87,88], further studies are needed to unravel their role in the biotransformation of iron chelators as to better serve patients. Overall, our analysis revealed new genes as candidate pharmacogenetic biomarkers of deferiprone and deferasirox efficacy that seek further investigation.
3.3.3. Stroke
Stroke is one of the most devastating complications of SCD affecting up to 11% of patients with sickle cell anaemia (Hb SS) and sickle β0-thalassaemia under 18 years of age without intervention [89,90,91]. Sibship studies demonstrated that stroke has an inherited component and is, therefore, genetically modifiable [92]. However, stroke is a complex process with variability in lesion size, location and etiology, and, thus, unlikely to be modified by a single gene [60]. Genetic susceptibility appears to be guided by many genes with small effect sizes [93]. The dataset consisted of 28 modifiers with diverse functions, including inflammation (TNF, TGFBR3, IL4R, BMP6, CCL2, LTC4S and IL6), adhesion (VCAM1, TEK, SELP, CSF2, LDLR and ECE1), coagulation (ANXA2 and F5), signal transduction (ADYC9, ADRB2 and AGT), cell survival (MET), oxidative stress (HMOX1 and PON1) and transcriptional regulation (ERG, HDAC9 and BCL11A) [21]. The high genetic heterogeneity reflects the complexity of stroke pathogenicity.
The enrichment analysis of the GO terms and biological pathways was carried out on 33 proteins (Figure 8A), of which IL4 (interleukin 4), IKBKG (inhibitor of nuclear factor kappa B kinase regulatory subunit gamma), RIPK1 (receptor interacting serine/threonine kinase 1), TRADD (TNFRSF1A associated via death domain) and TRAF2 (TNF receptor associated factor 2) were new interactors at the confidence interaction score ≥0.7 (Figure 8B). Interestingly, IKBKG, RIPK1, TRADD and TRAF2 formed a discreet and distinct cluster that linked to the rest of the network via interaction with TNF (tumour necrosis factor). TNF is a pro-inflammatory cytokine produced by brain cells with presence in all stages of brain injury by stroke. It plays a central role during cerebral ischemia and exerts both damaging and protective functions via interaction with the TNF receptor superfamily member 1A (TNFRSF1A, also known as TNFR1). The DD domain of the TNFR1 binds TRADD, which in turn recruits TRAF2, RIPK1 and Fas-associated via death domain (FADD). Binding of TRAF2 with cellular inhibitor of apoptosis proteins (cIAPs) facilitates NF-κB activation and induction of NF-κB-regulated anti-apoptotic factors. The protein IKBKG forms part of the IκB kinase (IKK) complex involved in the activation of NF-κB. On the other hand, activation of RIPK1 and FADD-interacting initiator caspase [FADD-like interleukin-1β-converting enzyme (FLICE)/caspase-8] lead to necrotic or apoptotic cell death (for review see [94,95]). Other potent pro-inflammatory cytokines with a significant impact on stroke pathology include interleukin 1 (IL-1), IL-4, IL-6, IL-8, IL-10 and IL-17 [96,97].
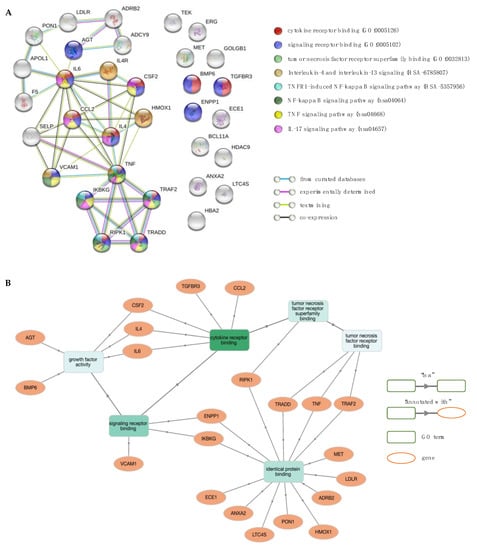
Figure 8.
Network and enrichment analysis for stroke. (A) The PPI network contains 33 nodes (proteins; circles) connected by edges (protein–protein interactions; horizontal lines). Coloured nodes highlight proteins associated with significant molecular function (MF) terms and biological pathways. (B) GO MF enrichment analysis using GOnet (q value ≤0.01; p value ≤1.94 × 10–5). Green colour intensifies as the significance level of enrichment decreases.
Functional enrichment analysis revealed 146 significant GO BP terms (p value <10–5), of which the top five were annotations to general terms in GO hierarchy (e.g., “positive regulation of multicellular organismal process”—GO:0051240, “response to organic substance”—GO:0010033 and “response to oxygen-containing compound”—GO:1901700). Enrichment analysis of GO MF terms yielded seven significant terms, of which the more relevant (in order of increasing FDR) were “cytokine receptor binding” (GO:0005126), “signalling receptor binding” (GO:0005102), “cytokine activity” (GO:0005125) and “tumour necrosis factor receptor superfamily binding” (GO:0032813). The most prominent pathways involved “TNF signalling pathway” (hsa04668), “IL-17 signalling pathway” (hsa04657), “NF-kappa B signalling pathway” (hsa04064), “IL-4 and IL-13 signalling” (HSA-6785807) and “TNFR1-induced NFkappaB signalling pathway” (HSA-5357956). Overall, analysis showed that the most relevant GO terms and biological pathways are associated with cytokine signalling and cascade inflammatory reactions.
Moreover, results indicate that some of the candidate stroke modifiers in haemoglobinopathies are shared by stroke victims in the general population (e.g., CCL2, F5, IL-6, SELP, TGFBR3, TNF, and VCAM1). Large studies have been conducted to identify genes affecting stroke risk in the general population, resulting in the development of several biomarker panels that aim to risk stratify patients according to stroke type and to provide prognostic information for targeted interventions (biomarker panels are summarized in [98]). As many biomarkers are not disease-specific, diagnostic sensitivity and specificity is compromised [99,100,101]. The present work reveals the most prevalent biomarkers for stroke known to date in haemoglobinopathies. The stroke phenotype in IthaGenes comprises different stroke sub-types, such as large and small vessel types. Based on published reports of a variable genetic component across different types of stroke [60] and towards the development of a comprehensive account on stroke genetics, future work will focus on investigating genetic modifiers for each type of stroke separately. Knowledge on stroke biomarkers specific to haemoglobinopathies could serve as a guiding tool to assess future risk and to elucidate potential stroke pathways towards more effective personalised therapy.
4. Conclusions
Haemoglobinopathies are a heterogeneous group of Hb disorders characterised by diverse phenotypic manifestations. Despite considerable progress in accumulating knowledge on the genetic architecture of these phenotypes, the role of modulating genes on phenotypic expression is largely unclear. Deciphering gene–phenotype interactions is a crucial step in understanding disease pathology. The present work aims to highlight potential genes and molecular pathways that could explain the pathogenesis and complexity of haemoglobinopathy-specific phenotypes.
Using data from the IthaGenes database, a gene scoring algorithm (IthaScore) was developed to assist in evidence-based ranking of genetic modifiers for disease phenotypes. Gene scores were based on manual curation using a point system to collate and grade heterogeneous information and replication studies for each gene–phenotype relationship, with quantitative and qualitative evaluation of available evidence. IthaScore will be dynamically recalculated with emerging evidence for existing or new phenotype relationships and provides a measure of the volume and quality of evidence for such relationships. It does not provide any information about the size of the disease-modifying effect of any gene on the corresponding phenotype but can be a useful tool for gene ranking specific to haemoglobinopathies and their relevant phenotypes. This algorithm was validated in part by its ability to rank well-established genetic modifiers with high scores, such as the major QTLs (BCL11A, HMIP and HBG2) of Hb F production, one of the greatest markers and best-studied modulators of disease severity.
To our knowledge, this is the first study integrating literature curation, gene ranking and functional enrichment analysis to evaluate candidate genetic modifiers for haemoglobinopathies. While we demonstrate the capacity of this approach to identify novel information, we urge caution when utilising the presented results due to potential limitations. Several of gene-phenotype findings are reported once without further replication, or they exhibit inconsistencies across studies due to differences in data collection and processing approaches. This is reflected by the low IthaScore calculated for the majority of gene-phenotype relationships. Moreover, most of published whole-genome scans do not always identify the true disease-modifying QTLs across large genomic regions, while findings may also come from studies with a small sample size and/or limited phenotyping, which are prone to noise [102,103]. Although IthaGenes harbours the largest collection of literature and continuously updated data on genetic modifiers for currently 59 phenotypes, additional genes may influence phenotypes that are not haemoglobinopathy-specific but are relatively common in the general population, such as stroke and osteoporosis.
In conclusion, the functional enrichment analysis for three phenotypes specific to β-thalassaemia and/or SCD provides preliminary proof that IthaGenes, as a comprehensive and scalable knowledgebase of genetic modifiers in haemoglobinopathies, together with dynamic gene scoring, can be used to unravel the molecular underpinnings of phenotypic diversity and identify new genes with plausible influence on haemoglobinopathy-specific phenotypes. Our findings add to current scientific knowledge and set the basis for future investigations. Research towards the discovery of phenotype-specific biomarkers will inform affected individuals about their health risk and allow them to thoughtfully consider their treatment options particularly with regards to stem cell transplantation and gene therapy, which offer the promise of complete cure albeit at a risk. Overall, the characterisation of candidate modifiers presents a novel and exciting opportunity to identify stratification biomarkers that help define treatment subgroups of patients in the frame of personalised medicine, as well as new diagnostic and therapeutic gene targets.
Supplementary Materials
The following are available online at https://www.mdpi.com/2077-0383/8/11/1927/s1, Table S1: Phenotypic term annotations for candidate modifier genes, Table S2: Gene ranking.
Author Contributions
C.S., M.K. and P.K. conceived and designed the study; C.S. and A.M. collected and curated data; P.P. guided phenotype annotations and clinical interpretation; P.K. developed the gene ranking algorithm; S.T. and P.K. performed bioinformatics analyses; C.S., S.T., A.M. and P.K. prepared figures; C.S. wrote the manuscript; all authors have read and approved the final version of the manuscript.
Funding
This work was funded by the Cyprus Research Promotion Foundation (EXCELLENCE/1216/256) and the Cyprus Institute of Neurology and Genetics.
Acknowledgments
We extend special thanks to the following scientists that contributed or curated data: Carsten W. Lederer, Pavlos Fanis and Ioanna Kousiappa. We also thank the Cyprus Institute of Neurology and Genetics for computer equipment and for hosting ITHANET.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Modell, B.; Darlison, M. Global epidemiology of haemoglobin disorders and derived service indicators. Bull. World Health Organ. 2008, 86, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Weatherall, D.J.; Clegg, J.B. Inherited haemoglobin disorders: An increasing global health problem. Bull. World Health Organ. 2001, 79, 704–712. [Google Scholar] [PubMed]
- Piel, F.B.; Tatem, A.J.; Huang, Z.; Gupta, S.; Williams, T.N.; Weatherall, D.J. Global migration and the changing distribution of sickle haemoglobin: A quantitative study of temporal trends between 1960 and 2000. Lancet Glob. Health 2014, 2, e80–e89. [Google Scholar] [CrossRef]
- Henderson, S.; Timbs, A.; McCarthy, J.; Gallienne, A.; Van Mourik, M.; Masters, G.; May, A.; Khalil, M.S.M.; Schuh, A.; Old, J. Incidence of haemoglobinopathies in various populations—The impact of immigration. Clin. Biochem. 2009, 42, 1745–1756. [Google Scholar] [CrossRef] [PubMed]
- Kountouris, P.; Lederer, C.W.; Fanis, P.; Feleki, X.; Old, J.; Kleanthous, M. IthaGenes: An Interactive Database for Haemoglobin Variations and Epidemiology. PLoS ONE 2014, 9, e103020. [Google Scholar] [CrossRef] [PubMed]
- Kountouris, P.; Stephanou, C.; Bento, C.; Fanis, P.; Elion, J.; Ramesar, R.S.; Zilfalil, B.A.; Robinson, H.M.; Traeger-Synodinos, J.; Human Variome Project Global Globin 2020 Challenge; et al. ITHANET: Information and database community portal for haemoglobinopathies. bioRxiv 2017, 209361. [Google Scholar] [CrossRef]
- Galanello, R.; Origa, R. Beta-thalassemia. Orphanet J. Rare Dis. 2010, 5, 11. [Google Scholar] [CrossRef]
- Rees, D.C.; Williams, T.N.; Gladwin, M.T. Sickle-cell disease. Lancet 2010, 376, 2018–2031. [Google Scholar] [CrossRef]
- Sripichai, O.; Munkongdee, T.; Kumkhaek, C.; Svasti, S.; Winichagoon, P.; Fucharoen, S. Coinheritance of the different copy numbers of alpha-globin gene modifies severity of beta-thalassemia/Hb E disease. Ann. Hematol. 2008, 87, 375–379. [Google Scholar] [CrossRef]
- Higgs, D.R.; Aldridge, B.E.; Lamb, J.; Clegg, J.B.; Weatherall, D.J.; Hayes, R.J.; Grandison, Y.; Lowrie, Y.; Mason, K.P.; Serjeant, B.E.; et al. The Interaction of Alpha-Thalassemia and Homozygous Sickle-Cell Disease. N. Engl. J. Med. 1982, 306, 1441–1446. [Google Scholar] [CrossRef]
- Thein, S.L.; Menzel, S. Discovering the genetics underlying foetal haemoglobin production in adults. Br. J. Haematol. 2009, 145, 455–467. [Google Scholar] [CrossRef]
- Powars, D.R.; Weiss, J.N.; Chan, L.S.; Schroeder, W.A. Is there a threshold level of fetal hemoglobin that ameliorates morbidity in sickle cell anemia? Blood 1984, 63, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Musallam, K.M.; Sankaran, V.G.; Cappellini, M.D.; Duca, L.; Nathan, D.G.; Taher, A.T. Fetal hemoglobin levels and morbidity in untransfused patients with β-thalassemia intermedia. Blood 2012, 119, 364–367. [Google Scholar] [CrossRef] [PubMed]
- Thein, S.L.; Menzel, S.; Lathrop, M.; Garner, C. Control of fetal hemoglobin: New insights emerging from genomics and clinical implications. Hum. Mol. Genet. 2009, 18, R216–R223. [Google Scholar] [CrossRef] [PubMed]
- Galarneau, G.; Palmer, C.D.; Sankaran, V.G.; Orkin, S.H.; Hirschhorn, J.N.; Lettre, G. Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat. Genet. 2010, 42, 1049–1051. [Google Scholar] [CrossRef]
- Uda, M.; Galanello, R.; Sanna, S.; Lettre, G.; Sankaran, V.G.; Chen, W.; Usala, G.; Busonero, F.; Maschio, A.; Albai, G.; et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of β-thalassemia. Proc. Natl. Acad. Sci. USA 2008, 105, 1620–1625. [Google Scholar] [CrossRef]
- Mtatiro, S.N.; Singh, T.; Rooks, H.; Mgaya, J.; Mariki, H.; Soka, D.; Mmbando, B.; Msaki, E.; Kolder, I.; Thein, S.L.; et al. Genome Wide Association Study of Fetal Hemoglobin in Sickle Cell Anemia in Tanzania. PLoS ONE 2014, 9, e111464. [Google Scholar] [CrossRef]
- Liu, L.; Pertsemlidis, A.; Ding, L.-H.; Story, M.D.; Steinberg, M.H.; Sebastiani, P.; Hoppe, C.; Ballas, S.K.; Pace, B.S. Original Research: A case-control genome-wide association study identifies genetic modifiers of fetal hemoglobin in sickle cell disease. Exp. Biol. Med. 2016, 241, 706–718. [Google Scholar] [CrossRef]
- Schaefer, B.A.; Flanagan, J.M.; Alvarez, O.A.; Nelson, S.C.; Aygun, B.; Nottage, K.A.; George, A.; Roberts, C.W.; Piccone, C.M.; Howard, T.A.; et al. Genetic Modifiers of White Blood Cell Count, Albuminuria and Glomerular Filtration Rate in Children with Sickle Cell Anemia. PLoS ONE 2016, 11, e0164364. [Google Scholar] [CrossRef]
- Aguiar, L.; Matos, A.; Gil, Â.; Afonso, C.; Almeida, S.; Braga, L.; Lavinha, J.; Kjollerstrom, P.; Faustino, P.; Bicho, M.; et al. Sickle cell anemia—Nitric oxide related genetic modifiers of hematological and biochemical parameters. Clin. Hemorheol. Microcirc. 2016, 64, 957–963. [Google Scholar] [CrossRef]
- Flanagan, J.M.; Frohlich, D.M.; Howard, T.A.; Schultz, W.H.; Driscoll, C.; Nagasubramanian, R.; Mortier, N.A.; Kimble, A.C.; Aygun, B.; Adams, R.J.; et al. Genetic predictors for stroke in children with sickle cell anemia. Blood 2011, 117, 6681–6684. [Google Scholar] [CrossRef]
- Desai, A.A.; Zhou, T.; Ahmad, H.; Zhang, W.; Mu, W.; Trevino, S.; Wade, M.S.; Raghavachari, N.; Kato, G.J.; Peters-Lawrence, M.H.; et al. A Novel Molecular Signature for Elevated Tricuspid Regurgitation Velocity in Sickle Cell Disease. Am. J. Respir. Crit. Care Med. 2012, 186, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Jacob, S.A.; Novelli, E.M.; Isenberg, J.S.; Garrett, M.E.; Chu, Y.; Soldano, K.; Ataga, K.I.; Telen, M.J.; Ashley-Koch, A.; Gladwin, M.T.; et al. Thrombospondin-1 Gene Polymorphism is Associated with Estimated Pulmonary Artery Pressure in Patients with Sickle Cell Anemia. Am. J. Hematol. 2017, 92, E31–E34. [Google Scholar] [CrossRef] [PubMed]
- Rees, D.C.; Gibson, J.S. Biomarkers in sickle cell disease. Br. J. Haematol. 2012, 156, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Seyhan, A. Biomarkers in drug discovery and development. Eur. Pharm. Rev. 2010, 5, 19–25. [Google Scholar]
- Kalpatthi, R.; Novelli, E.M. Measuring success: Utility of biomarkers in sickle cell disease clinical trials and care. Hematology 2018, 2018, 482–492. [Google Scholar] [CrossRef]
- Thein, S.L. Genetic modifiers of the beta-haemoglobinopathies. Br. J. Haematol. 2008, 141, 357–366. [Google Scholar] [CrossRef]
- Steinberg, M.H.; Sebastiani, P. Genetic modifiers of sickle cell disease. Am. J. Hematol. 2012, 87, 795–803. [Google Scholar] [CrossRef]
- Driss, A.; Asare, K.O.; Hibbert, J.M.; Gee, B.E.; Adamkiewicz, T.V.; Stiles, J.K. Sickle Cell Disease in the Post Genomic Era: A Monogenic Disease with a Polygenic Phenotype. Genom. Insights 2009, 2, 23–48. [Google Scholar] [CrossRef]
- Fertrin, K.Y.; Costa, F.F. Genomic polymorphisms in sickle cell disease: Implications for clinical diversity and treatment: Expert Review of Hematology: Vol 3, No 4. Expert Rev. Hematol. 2010, 3, 443–458. [Google Scholar] [CrossRef]
- Giardine, B.; Borg, J.; Viennas, E.; Pavlidis, C.; Moradkhani, K.; Joly, P.; Bartsakoulia, M.; Riemer, C.; Miller, W.; Tzimas, G.; et al. Updates of the HbVar database of human hemoglobin variants and thalassemia mutations. Nucleic Acids Res. 2014, 42, D1063–D1069. [Google Scholar] [CrossRef]
- Sun, J.; Jia, P.; Fanous, A.H.; Webb, B.T.; van den Oord, E.J.C.G.; Chen, X.; Bukszar, J.; Kendler, K.S.; Zhao, Z. A multi-dimensional evidence-based candidate gene prioritization approach for complex diseases—Schizophrenia as a case. Bioinformatics 2009, 25, 2595–6602. [Google Scholar] [CrossRef]
- Larsen, E.; Menashe, I.; Ziats, M.N.; Pereanu, W.; Packer, A.; Banerjee-Basu, S. A systematic variant annotation approach for ranking genes associated with autism spectrum disorders. Mol. Autism 2016, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Ran, X.; Li, J.; Shao, Q.; Chen, H.; Lin, Z.; Sun, Z.S.; Wu, J. EpilepsyGene: A genetic resource for genes and mutations related to epilepsy. Nucleic Acids Res. 2015, 43, D893–D899. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, J.; Rao, S.; Ritter, M.; Manor, L.C.; Backer, R.; Cao, H.; Cheng, Z.; Liu, S.; Liu, Y.; et al. An Integrative Computational Approach to Evaluate Genetic Markers for Bipolar Disorder. Sci. Rep. 2017, 7, 6745. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Kuo, P.-H.; Riley, B.P.; Kendler, K.S.; Zhao, Z. Candidate genes for schizophrenia: A survey of association studies and gene ranking. Am. J. Med. Genet. Part B 2008, 147, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.; Mantzaris, D.; Georgitsi, M.; Drineas, P.; Paschou, P. Variant Ranker: A web-tool to rank genomic data according to functional significance. BMC Bioinform. 2017, 18, 341. [Google Scholar] [CrossRef]
- Köhler, S.; Doelken, S.C.; Mungall, C.J.; Bauer, S.; Firth, H.V.; Bailleul-Forestier, I.; Black, G.C.M.; Brown, D.L.; Brudno, M.; Campbell, J.; et al. The Human Phenotype Ontology project: Linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014, 42, D966–D974. [Google Scholar] [CrossRef]
- Robinson, P.N.; Köhler, S.; Bauer, S.; Seelow, D.; Horn, D.; Mundlos, S. The Human Phenotype Ontology: A Tool for Annotating and Analyzing Human Hereditary Disease. Am. J. Hum. Genet. 2008, 83, 610–615. [Google Scholar] [CrossRef]
- Strande, N.T.; Riggs, E.R.; Buchanan, A.H.; Ceyhan-Birsoy, O.; DiStefano, M.; Dwight, S.S.; Goldstein, J.; Ghosh, R.; Seifert, B.A.; Sneddon, T.P.; et al. Evaluating the Clinical Validity of Gene-Disease Associations: An Evidence-Based Framework Developed by the Clinical Genome Resource. Am. J. Hum. Genet. 2017, 100, 895–906. [Google Scholar] [CrossRef]
- Abrahams, B.S.; Arking, D.E.; Campbell, D.B.; Mefford, H.C.; Morrow, E.M.; Weiss, L.A.; Menashe, I.; Wadkins, T.; Banerjee-Basu, S.; Packer, A. SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 2013, 4, 36. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Pomaznoy, M.; Ha, B.; Peters, B. GOnet: A tool for interactive Gene Ontology analysis. BMC Bioinform. 2018, 19, 470. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; McMorran, R.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. The Comparative Toxicogenomics Database: Update 2019. Nucleic Acids Res. 2019, 47, D948–D954. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological network exloration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8–13. [Google Scholar] [CrossRef]
- MacArthur, D.G.; Manolio, T.A.; Dimmock, D.P.; Rehm, H.L.; Shendure, J.; Abecasis, G.R.; Adams, D.R.; Altman, R.B.; Antonarakis, S.E.; Ashley, E.A.; et al. Guidelines for investigating causality of sequence variants in human disease. Nature 2014, 508, 469–476. [Google Scholar] [CrossRef]
- Ah, Y.-M.; Kim, Y.-M.; Kim, M.-J.; Choi, Y.H.; Park, K.-H.; Son, I.-J.; Kim, S.G. Drug-induced Hyperbilirubinemia and the Clinical Influencing Factors. Drug Metab. Rev. 2008, 40, 511–537. [Google Scholar] [CrossRef]
- Lettre, G.; Sankaran, V.G.; Bezerra, M.A.C.; Araújo, A.S.; Uda, M.; Sanna, S.; Cao, A.; Schlessinger, D.; Costa, F.F.; Hirschhorn, J.N.; et al. DNA polymorphisms at the BCL11A, HBS1L-MYB, and β-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc. Natl. Acad. Sci. USA 2008, 105, 11869–11874. [Google Scholar] [CrossRef]
- Borg, J.; Papadopoulos, P.; Georgitsi, M.; Gutiérrez, L.; Grech, G.; Fanis, P.; Phylactides, M.; Verkerk, A.J.M.H.; van der Spek, P.J.; Scerri, C.A.; et al. Haploinsufficiency for the erythroid transcription factor KLF1 causes Hereditary Persistence of Fetal Hemoglobin. Nat. Genet. 2010, 42, 801–805. [Google Scholar] [CrossRef]
- Menzel, S.; Thein, S.L. Genetic Modifiers of Fetal Haemoglobin in Sickle Cell Disease. Mol. Diagn. Ther. 2019, 23, 235–244. [Google Scholar] [CrossRef]
- Ware, R.E. How I use hydroxyurea to treat young patients with sickle cell anemia. Blood 2010, 115, 5300–5311. [Google Scholar] [CrossRef]
- Thein, S.L. Genetic Basis and Genetic Modifiers of β-Thalassemia and Sickle Cell Disease. In Gene and Cell Therapies for Beta-Globinopathies; Malik, P., Tisdale, J., Eds.; Advances in Experimental Medicine and Biology; Springer New York: New York, NY, USA, 2017; pp. 27–57. ISBN 978-1-4939-7299-9. [Google Scholar]
- Pule, G.D.; Mowla, S.; Novitzky, N.; Wiysonge, C.S.; Wonkam, A. A Systematic Review of Known Mechanisms of Hydroxyurea-induced Foetal Haemoglobin for Treatment of Sickle Cell Disease. Expert Rev. Hematol. 2015, 8, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Ribeil, J.-A.; Arlet, J.-B.; Dussiot, M.; Cruz Moura, I.; Courtois, G.; Hermine, O. Ineffective Erythropoiesis in β-Thalassemia. Sci. World J. 2013, 2013, 11. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Zhou, S.; Kihm, A.J.; Katein, A.M.; Yu, X.; Gell, D.A.; Mackay, J.P.; Adachi, K.; Foster-Brown, L.; Louden, C.S.; et al. Loss of α-hemoglobin–stabilizing protein impairs erythropoiesis and exacerbates β-thalassemia. J. Clin. Investig. 2004, 114, 1457–1466. [Google Scholar] [CrossRef] [PubMed]
- Sankaran, V.G.; Ludwig, L.S.; Sicinska, E.; Xu, J.; Bauer, D.E.; Eng, J.C.; Patterson, H.C.; Metcalf, R.A.; Natkunam, Y.; Orkin, S.H.; et al. Cyclin D3 coordinates the cell cycle during differentiation to regulate erythrocyte size and number. Genes Dev. 2012, 26, 2075–2087. [Google Scholar] [CrossRef] [PubMed]
- Manwani, D.; Frenette, P.S. Vaso-occlusion in sickle cell disease: Pathophysiology and novel targeted therapies. Blood 2013, 122, 3892–3898. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Bakshi, N.; Krishnamurti, L. Acute Chest Syndrome in Children with Sickle Cell Disease. Pediatr. Allergy Immunol. Pulmonol. 2017, 30, 191–201. [Google Scholar] [CrossRef]
- Desai, P.C.; Ataga, K.I. The acute chest syndrome of sickle cell disease. Expert Opin. Pharmacother. 2013, 14, 991–999. [Google Scholar] [CrossRef]
- Dichgans, M. Genetics of ischaemic stroke. Lancet Neurol. 2007, 6, 149–161. [Google Scholar] [CrossRef]
- Steinberg, M.H.; Lu, Z.-H.; Barton, F.B.; Terrin, M.L.; Charache, S.; Dover, G.J. Fetal Hemoglobin in Sickle Cell Anemia: Determinants of Response to Hydroxyurea. Blood 1997, 89, 1078–1088. [Google Scholar] [CrossRef]
- Lebensburger, J.D.; Pestina, T.I.; Ware, R.E.; Boyd, K.L.; Persons, D.A. Hydroxyurea therapy requires HbF induction for clinical benefit in a sickle cell mouse model. Haematologica 2010, 95, 1599–1603. [Google Scholar] [CrossRef]
- King, S.B. A role for nitric oxide in hydroxyurea-mediated fetal hemoglobin induction. J. Clin. Investig. 2003, 111, 171–172. [Google Scholar] [CrossRef] [PubMed]
- Cokic, V.P.; Smith, R.D.; Beleslin-Cokic, B.B.; Njoroge, J.M.; Miller, J.L.; Gladwin, M.T.; Schechter, A.N. Hydroxyurea induces fetal hemoglobin by the nitric oxide—dependent activation of soluble guanylyl cyclase. J. Clin. Investig. 2003, 111, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Ikuta, T.; Ausenda, S.; Cappellini, M.D. Mechanism for fetal globin gene expression: Role of the soluble guanylate cyclase–cGMP-dependent protein kinase pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 1847–1852. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Morris, S.M. Arginine metabolism: Nitric oxide and beyond. Biochem. J. 1998, 336, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Denninger, J.W.; Marletta, M.A. Guanylate cyclase and the ⋅NO/cGMP signaling pathway. Biochim. Biophys. Acta BBA - Bioenerg. 1999, 1411, 334–350. [Google Scholar] [CrossRef]
- Bhatta, S.S.; Wroblewski, K.E.; Agarwal, K.L.; Sit, L.; Cohen, E.E.W.; Seiwert, T.Y.; Karrison, T.; Bakris, G.L.; Ratain, M.J.; Vokes, E.E.; et al. Effects of Vascular Endothelial Growth Factor Signaling Inhibition on Human Erythropoiesis. Oncologist 2013, 18, 965–970. [Google Scholar] [CrossRef] [PubMed]
- Greenwald, A.C.; Licht, T.; Kumar, S.; Oladipupo, S.S.; Iyer, S.; Grunewald, M.; Keshet, E. VEGF expands erythropoiesis via hypoxia-independent induction of erythropoietin in noncanonical perivascular stromal cells. J. Exp. Med. 2019, 216, 215–230. [Google Scholar] [CrossRef]
- Drogat, B.; Kalucka, J.; Gutierrez, L.; Hammad, H.; Goossens, S.; Farhang Ghahremani, M.; Bartunkova, S.; Haigh, K.; Deswarte, K.; Nyabi, O.; et al. Vegf regulates embryonic erythroid development through Gata1 modulation. Blood 2010, 116, 2141–2151. [Google Scholar] [CrossRef]
- Fang, S.; Nurmi, H.; Heinolainen, K.; Chen, S.; Salminen, E.; Saharinen, P.; Mikkola, H.K.A.; Alitalo, K. Critical requirement of VEGF-C in transition to fetal erythropoiesis. Blood 2016, 128, 710–720. [Google Scholar] [CrossRef]
- Cao, A.; Moi, P.; Galanello, R. Recent advances in β-thalassemias. Pediatric Rep. 2011, 3, e17. [Google Scholar] [CrossRef]
- Galanello, R.; Campus, S.; Origa, R. Deferasirox: Pharmacokinetics and clinical experience. Expert Opin. Drug Metab. Toxicol. 2012, 8, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, N.F.; Brittenham, G.M.; McLaren, C.E.; Templeton, D.M.; Cameron, R.G.; McClelland, R.A.; Burt, A.D.; Fleming, K.A. Long-Term Safety and Effectiveness of Iron-Chelation Therapy with Deferiprone for Thalassemia Major. N. Engl. J. Med. 2009, 339, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Galanello, R. Deferiprone in the treatment of transfusion-dependent thalassemia: A review and perspective. Ther. Clin. Risk Manag. 2007, 3, 795–805. [Google Scholar] [PubMed]
- Rowland, A.; Miners, J.O.; Mackenzie, P.I. The UDP-glucuronosyltransferases: Their role in drug metabolism and detoxification. Int. J. Biochem. Cell Biol. 2013, 45, 1121–1132. [Google Scholar] [CrossRef]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef]
- Münzel, P.A.; Schmohl, S.; Heel, H.; Kälberer, K.; Bock-Hennig, B.S.; Bock, K.W. Induction of Human UDP Glucuronosyltransferases (UGT1A6, UGT1A9, and UGT2B7) by t-Butylhydroquinone and 2,3,7,8-Tetrachlorodibenzo-p-Dioxin in Caco-2 Cells. Drug Metab. Dispos. 1999, 27, 569–573. [Google Scholar]
- Nebert, D.W.; Dalton, T.P. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat. Rev. Cancer 2006, 6, 947–960. [Google Scholar] [CrossRef]
- Exjade-European Public Assessment Report; European Medicines Evaluation Agency: London, UK, 2006.
- Waldmeier, F.; Bruin, G.J.; Glaenzel, U.; Hazell, K.; Sechaud, R.; Warrington, S.; Porter, J.B. Pharmacokinetics, Metabolism, and Disposition of Deferasirox in β-Thalassemic Patients with Transfusion-Dependent Iron Overload Who Are at Pharmacokinetic Steady State. Drug Metab. Dispos. 2010, 38, 808–816. [Google Scholar] [CrossRef]
- Bruin, G.J.M.; Faller, T.; Wiegand, H.; Schweitzer, A.; Nick, H.; Schneider, J.; Boernsen, K.-O.; Waldmeier, F. Pharmacokinetics, Distribution, Metabolism, and Excretion of Deferasirox and Its Iron Complex in Rats. Drug Metab. Dispos. 2008, 36, 2523–2538. [Google Scholar] [CrossRef]
- Jemnitz, K.; Heredi-Szabo, K.; Janossy, J.; Ioja, E.; Vereczkey, L.; Krajcsi, P. ABCC2/Abcc2: A multispecific transporter with dominant excretory functions. Drug Metab. Rev. 2010, 42, 402–436. [Google Scholar] [CrossRef]
- Haverfield, E.V.; Weatherall, D.J.; Graber, A.Y.; Ramirez, J.; Ratain, M.J. Pharmacogenomics of Deferiprone Metabolism. Blood 2005, 106, 2703. [Google Scholar]
- Benoit-Biancamano, M.-O.; Connelly, J.; Villeneuve, L.; Caron, P.; Guillemette, C. Deferiprone Glucuronidation by Human Tissues and Recombinant UDP Glucuronosyltransferase 1A6: An in Vitro Investigation of Genetic and Splice Variants. Drug Metab. Dispos. 2009, 37, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Martignoni, M.; Groothuis, G.M.M.; de Kanter, R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin. Drug Metab. Toxicol. 2006, 2, 875–894. [Google Scholar] [CrossRef] [PubMed]
- Miyagi, S.J.; Collier, A.C. Pediatric Development of Glucuronidation: The Ontogeny of Hepatic UGT1A4. Drug Metab. Dispos. 2007, 35, 1587–1592. [Google Scholar] [CrossRef]
- Miyagi, S.J.; Milne, A.M.; Coughtrie, M.W.H.; Collier, A.C. Neonatal Development of Hepatic UGT1A9: Implications of Pediatric Pharmacokinetics. Drug Metab. Dispos. 2012, 40, 1321–1327. [Google Scholar] [CrossRef]
- Kassim, A.A.; Galadanci, N.A.; Pruthi, S.; DeBaun, M.R. How I treat and manage strokes in sickle cell disease. Blood 2015, 125, 3401–3410. [Google Scholar] [CrossRef]
- Ohene-Frempong, K.; Weiner, S.J.; Sleeper, L.A.; Miller, S.T.; Embury, S.; Moohr, J.W.; Wethers, D.L.; Pegelow, C.H.; Gill, F.M. Cerebrovascular Accidents in Sickle Cell Disease: Rates and Risk Factors. Blood 1998, 91, 288–294. [Google Scholar]
- Quinn, C.T. Sickle Cell Disease in Childhood. Pediatr. Clin. N. Am. 2013, 60, 1363–1381. [Google Scholar] [CrossRef]
- Driscoll, M.C.; Hurlet, A.; Styles, L.; McKie, V.; Files, B.; Olivieri, N.; Pegelow, C.; Berman, B.; Drachtman, R.; Patel, K.; et al. Stroke risk in siblings with sickle cell anemia. Blood 2003, 101, 2401–2404. [Google Scholar] [CrossRef]
- Martella, M.; Quaglia, N.; Frigo, A.C.; Basso, G.; Colombatti, R.; Sainati, L. Association between a combination of single nucleotide polymorphisms and large vessel cerebral vasculopathy in African children with sickle cell disease. Blood Cells. Mol. Dis. 2016, 61, 1–3. [Google Scholar] [CrossRef]
- Pan, W.; Kastin, A.J. Tumor necrosis factor and stroke: Role of the blood-brain barrier. Prog. Neurobiol. 2007, 83, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Wajant, H.; Henkler, F.; Scheurich, P. The TNF-receptor-associated factor family: Scaffold molecules for cytokine receptors, kinases and their regulators. Cell. Signal. 2001, 13, 389–400. [Google Scholar] [CrossRef]
- Wang, J.; Hu, Z.; Yang, S.; Liu, C.; Yang, H.; Wang, D.; Guo, F. Inflammatory cytokines and cells are potential markers for patients with cerebral apoplexy in intensive care unit. Exp. Ther. Med. 2018, 16, 1014–1020. [Google Scholar] [CrossRef] [PubMed]
- Lambertsen, K.L.; Finsen, B.; Clausen, B.H. Post-stroke inflammation-target or tool for therapy? Acta Neuropathol. 2019, 137, 693–714. [Google Scholar] [CrossRef] [PubMed]
- Jickling, G.C.; Sharp, F.R. Biomarker Panels in Ischemic Stroke. Stroke 2015, 46, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Moon, G.J.; Bang, O.Y. Biomarkers for Stroke. J. Stroke 2013, 15, 27–37. [Google Scholar] [CrossRef]
- Katan, M.; Elkind, M.S. The potential role of blood biomarkers in patients with ischemic stroke: An expert opinion. Clin. Transl. Neurosci. 2018, 2. [Google Scholar] [CrossRef]
- Fang, C.; Lou, B.; Zhou, J.; Zhong, R.; Wang, R.; Zang, X.; Shen, H.; Li, Y. Blood biomarkers in ischemic stroke: Role of biomarkers in differentiation of clinical phenotype. Eur. J. Inflamm. 2018, 16, 1–10. [Google Scholar] [CrossRef]
- Riordan, J.D.; Nadeau, J.H. From Peas to Disease: Modifier Genes, Network Resilience, and the Genetics of Health. Am. J. Hum. Genet. 2017, 101, 177–191. [Google Scholar] [CrossRef]
- McCarthy, M.I.; Abecasis, G.R.; Cardon, L.R.; Goldstein, D.B.; Little, J.; Ioannidis, J.P.A.; Hirschhorn, J.N. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat. Rev. Genet. 2008, 9, 356–369. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).