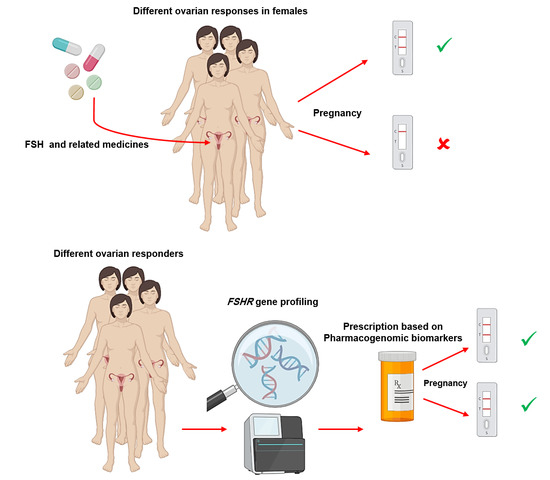
Pharmacogenomic Biomarkers of Follicle-Stimulating Hormone Receptor Malfunction in Females with Impaired Ovarian Response—A Genetic Survey
Abstract
1. Introduction: FSHR, Related Infertility Medicines, and the Role of Pharmacogenetics
2. Methods
3. A Quick Overview on Pharmacogenomics of FSHR in Females with Poor Ovarian Response
4. Genetic and Pharmacogenetic Variations in FSHR Gene
5. Advanced Genetic Screening Methods for FSHR Profiling
6. Genetic Variations in FSHR Related Cell Signaling Genes as Potential Players for Diverse Infertility Drug Response
7. Discussion
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ulloa-Aguirre, A.; Zariñán, T.; Pasapera, A.M.; Casas-González, P.; Dias, J.A. Multiple facets of follicle-stimulating hormone receptor function. Endocrine 2007, 32, 251–263. [Google Scholar] [CrossRef]
- Yan, K.; Gao, L.-N.; Cui, Y.-L.; Zhang, Y.; Zhou, X. The cyclic AMP signaling pathway: Exploring targets for successful drug discovery (Review). Mol. Med. Rep. 2016, 13, 3715–3723. [Google Scholar] [CrossRef]
- Thatcher, J.D. The Ras-MAPK Signal Transduction Pathway. Sci. Signal. 2010, 3. [Google Scholar] [CrossRef]
- Conti, M. Specificity of the cyclic adenosine 3′,5′-monophosphate signal in granulosa cell function. Biol. Reprod. 2002, 67, 4. [Google Scholar] [CrossRef] [PubMed]
- Borght, M.V.; Wyns, C. Fertility and infertility: Definition and epidemiology. Clin. Biochem. 2018, 62, 2–10. [Google Scholar] [CrossRef]
- Lindsay, T.J.; Vitrikas, K.R. Evaluation and treatment of infertility. Am. Fam. Physician 2015, 91, 308–314. [Google Scholar]
- Hrometz, S.L.; Gates, V. Review of available infertility treatments. Drugs Today 2009, 45, 275–291. [Google Scholar] [CrossRef]
- Derman, S.G.; Adashi, E.Y. Adverse Effects of Fertility Drugs. Drug Saf. 1994, 11, 408–421. [Google Scholar] [CrossRef]
- Samplaski, M.K.; Nangia, A.K. Adverse effects of common medications on male fertility. Nat. Rev. Urol. 2015, 12, 401–413. [Google Scholar] [CrossRef]
- Conforti, A.; Cariati, F.; Vallone, R.; Alviggi, C.; de Placido, G. Individualization of treatment in controlled ovarian stimulation: Myth or reality. Biochim. Clin. 2017, 41, 294–305. [Google Scholar]
- Momenimovahed, Z.; Taheri, S.; Tiznobaik, A.; Salehiniya, H. Do the Fertility Drugs Increase the Risk of Cancer? A Review Study. Front. Endocrinol. 2019, 10, 313. [Google Scholar] [CrossRef]
- Ferraretti, A.P.; La Marca, A.; Fauser, B.C.J.M.; Tarlatzis, B.; Nargund, G.; Gianaroli, L.; on behalf of the ESHRE working group on Poor Ovarian Response Definition. ESHRE consensus on the definition of ’poor response’ to ovarian stimulation for in vitro fertilization: The Bologna criteria. Hum. Reprod. 2011, 26, 1616–1624. [Google Scholar] [CrossRef]
- Esteves, S.C.; Roque, M.; Bedoschi, G.M.; Conforti, A.; Humaidan, P.; Alviggi, C. Defining Low Prognosis Patients Undergoing Assisted Reproductive Technology: POSEIDON Criteria—The Why. Front. Endocrinol. 2018, 9, 461. [Google Scholar] [CrossRef]
- Esteves, S.C.; Alviggi, C.; Humaidan, P.; Fischer, R.; Andersen, C.Y.; Conforti, A.; Bühler, K.; Sunkara, S.K.; Polyzos, N.P.; Galliano, D.; et al. The POSEIDON Criteria and Its Measure of Success through the Eyes of Clinicians and Embryologists. Front. Endocrinol. 2019, 10, 814. [Google Scholar] [CrossRef]
- Abrahams, E. Right drug—right patient—right time: Personalized Medicine Coalition. Clin. Transl. Sci. 2008, 1, 11. [Google Scholar] [CrossRef]
- Lemire, F. The right drug for the right patient: Caring for our patients while minimizing prescription drug misuse. Can. Fam. Physician 2013, 59, 708. [Google Scholar]
- Morón, F.J.; Ruiz, A. Pharmacogenetics of controlled ovarian hyperstimulation: Time to corroborate the clinical utility of FSH receptor genetic markers. Pharmacogenomics 2010, 11, 1613–1618. [Google Scholar] [CrossRef]
- Nenonen, H.; Lindgren, I.A.; Prahl, A.S.; Trzybulska, D.; Kharraziha, I.; Hultén, M.; Giwercman, Y.L.; Henic, E. The N680S variant in the follicle-stimulating hormone receptor gene identifies hyperresponders to controlled ovarian stimulation. Pharm. Genom. 2019, 29, 114–120. [Google Scholar] [CrossRef]
- Achrekar, S.K.; Modi, D.N.; Desai, S.K.; Mangoli, V.S.; Mangoli, R.V.; Mahale, S.D. Poor ovarian response to gonadotrophin stimulation is associated with FSH receptor polymorphism. Reprod. Biomed. Online 2009, 18, 509–515. [Google Scholar] [CrossRef]
- Riccetti, L.; De Pascali, F.; Gilioli, L.; Santi, D.; Brigante, G.; Simoni, M.; Casarini, L. Genetics of gonadotropins and their receptors as markers of ovarian reserve and response in controlled ovarian stimulation. Best Pr. Res. Clin. Obstet. Gynaecol. 2017, 44, 15–25. [Google Scholar] [CrossRef]
- Alviggi, C.; Conforti, A.; Santi, D.; Esteves, S.C.; Andersen, C.Y.; Humaidan, P.; Chiodini, P.; De Placido, G.; Simoni, M. Clinical relevance of genetic variants of gonadotrophins and their receptors in controlled ovarian stimulation: A systematic review and meta-analysis. Hum. Reprod. Updat. 2018, 24, 599–614. [Google Scholar] [CrossRef] [PubMed]
- Gerasimova, T.; Thanasoula, M.N.; Zattas, D.; Seli, E.; Sakkas, D.; Lalioti, M.D. Identification and in vitro characterization of follicle stimulating hormone (FSH) receptor variants associated with abnormal ovarian response to FSH. J. Clin. Endocrinol. Metab. 2010, 95, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Lalioti, M.; Gerasimova, T.; Zattas, D.; Anastasakis, D.; Seli, E.; Sakkas, D. A Deleted Form of FSH Receptor, Found in Women Undergoing Infertility Treatment, Impairs the Function of the Normal Receptor When Co-Expressed In Vitro. Biol. Reprod. 2010, 83, 193. [Google Scholar] [CrossRef]
- Kuechler, A.; Hauffa, B.P.; Köninger, A.; Kleinau, G.; Albrecht, B.; Horsthemke, B.; Gromoll, J. An unbalanced translocation unmasks a recessive mutation in the follicle-stimulating hormone receptor (FSHR) gene and causes FSH resistance. Eur. J. Hum. Genet. 2010, 18, 656–661. [Google Scholar] [CrossRef] [PubMed]
- Beau, I.; Touraine, P.; Meduri, G.; Gougeon, A.; Desroches, A.; Matuchansky, C.; Milgrom, E.; Kuttenn, F.; Misrahi, M. A novel phenotype related to partial loss of function mutations of the follicle stimulating hormone receptor. J. Clin. Investig. 1998, 102, 1352–1359. [Google Scholar] [CrossRef]
- Ramadhan, R.S. Molecular analysis of FSH receptor gene in Iraqi women with PCOS syndrome. Middle East Fertil. Soc. J. 2018, 23, 404–408. [Google Scholar] [CrossRef]
- Čuš, M.; Vlaisavljević, V.; Repnik, K.; Potočnik, U.; Kovačič, B. Could polymorphisms of some hormonal receptor genes, involved in folliculogenesis help in predicting patient response to controlled ovarian stimulation? J. Assist. Reprod. Genet. 2019, 36, 47–55. [Google Scholar] [CrossRef]
- De Castro, F.; Morón, F.J.; Montoro, L.; Galán, J.J.; Hernández, D.P.; Padilla, E.S.-C.; Ramírez-Lorca, R.; Real, L.M.; Ruiz, A. Human controlled ovarian hyperstimulation outcome is a polygenic trait. Pharmacogenetics 2004, 14, 285–293. [Google Scholar] [CrossRef]
- Loutradis, D.; Patsoula, E.; Minas, V.; Koussidis, G.A.; Antsaklis, A.; Michalas, S.; Makrigiannakis, A. FSH receptor gene polymorphisms have a role for different ovarian response to stimulation in patients entering IVF/ICSI-ET programs. J. Assist. Reprod. Genet. 2006, 23, 177–184. [Google Scholar] [CrossRef]
- Livshyts, G.B.; Podlesnaja, S.; Kravchenko, S.; Sudoma, I.; Livshits, L. A distribution of two SNPs in exon 10 of the FSHR gene among the women with a diminished ovarian reserve in Ukraine. J. Assist. Reprod. Genet. 2008, 26, 29–34. [Google Scholar] [CrossRef]
- Mayorga, M.P.; Gromoll, J.; Behre, H.M.; Gassner, C.; Nieschlag, E.; Simoni, M. Ovarian Response to Follicle-Stimulating Hormone (FSH) Stimulation Depends on the FSH Receptor Genotype*. J. Clin. Endocrinol. Metab. 2000, 85, 3365–3369. [Google Scholar] [CrossRef]
- Behre, H.M.; Greb, R.R.; Mempel, A.; Sonntag, B.; Kiesel, L.; Kaltwasser, P.; Seliger, E.; Röpke, F.; Gromoll, J.; Nieschlag, E.; et al. Significance of a common single nucleotide polymorphism in exon 10 of the follicle-stimulating hormone (FSH) receptor gene for the ovarian response to FSH: A pharmacogenetic approach to controlled ovarian hyperstimulation. Pharmacogenetics Genom. 2005, 15, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Ma, C.-H.; Tang, H.; Hu, Y.-F. Influence of follicle-stimulating hormone receptor (FSHR) Ser680Asn polymorphism on ovarian function and in-vitro fertilization outcome: A meta-analysis. Mol. Genet. Metab. 2011, 103, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Altmäe, S.; Hovatta, O.; Stavreus-Evers, A.; Salumets, A. Genetic predictors of controlled ovarian hyperstimulation: Where do we stand today? Hum. Reprod. Updat. 2011, 17, 813–828. [Google Scholar] [CrossRef] [PubMed]
- Lussiana, C.; Guani, B.; Mari, C.; Restagno, G.; Massobrio, M.; Revelli, A. Mutations and polymorphisms of the FSH receptor (FSHR) gene: Clinical implications in female fecundity and molecular biology of FSHR protein and gene. Obstet. Gynecol. Surv. 2008, 63, 785–795. [Google Scholar] [CrossRef]
- Ilgaz, N.S.; Aydos, O.S.E.; Karadag, A.; Taspinar, M.; Eryilmaz, O.G.; Sunguroglu, A. Impact of follicle-stimulating hormone receptor variants in female infertility. J. Assist. Reprod. Genet. 2015, 32, 1659–1668. [Google Scholar] [CrossRef]
- Desai, S.S.; Roy, B.S.; Mahale, S.D. Mutations and polymorphisms in FSH receptor: Functional implications in human reproduction. Reproduction 2013, 146, R235–R248. [Google Scholar] [CrossRef]
- Rizk, B. Genetics of ovarian hyperstimulation syndrome. Reprod. Biomed. Online 2009, 19, 14–27. [Google Scholar] [CrossRef]
- Jun, J.K.; Yoon, J.S.; Ku, S.-Y.; Choi, Y.M.; Hwang, K.R.; Park, S.Y.; Lee, G.H.; Lee, W.D.; Kim, S.H.; Kim, J.G.; et al. Follicle-stimulating hormone receptor gene polymorphism and ovarian responses to controlled ovarian hyperstimulation for IVF-ET. J. Hum. Genet. 2006, 51, 665–670. [Google Scholar] [CrossRef]
- Tranchant, T.; Durand, G.; Gauthier, C.; Crépieux, P.; Ulloa-Aguirre, A.; Royère, D.; Reiter, E. Preferential β-arrestin signalling at low receptor density revealed by functional characterization of the human FSH receptor A189 V mutation☆. Mol. Cell. Endocrinol. 2011, 331, 109–118. [Google Scholar] [CrossRef]
- Bogerd, J. Ligand-selective determinants in gonadotropin receptors. Mol. Cell. Endocrinol. 2007, 260, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Schofield, P.R.; Sprengel, R. Amino-terminal leucine-rich repeats in gonadotropin receptors determine hormone selectivity. EMBO J. 1991, 10, 1885–1890. [Google Scholar] [CrossRef] [PubMed]
- Lazaros, L.; Hatzi, E.; Xita, N.; Takenaka, A.; Sofikitis, N.; Zikopoulos, K.; Georgiou, I. Influence of FSHR diplotypes on ovarian response to standard gonadotropin stimulation for IVF/ICSI. J. Reprod. Med. 2013, 58, 395–401. [Google Scholar] [PubMed]
- Valkenburg, O. (Olivier); Uitterlinden, A.; Piersma, D.; Hofman, A.; Themmen, A.; De Jong, F.; Fauser, B.; Laven, J. Genetic polymorphisms of GnRH and gonadotrophic hormone receptors affect the phenotype of polycystic ovary syndrome. Hum. Reprod. 2009, 24, 2014–2022. [Google Scholar] [CrossRef] [PubMed]
- Casarini, L.; Santi, D.; Marino, M. Impact of gene polymorphisms of gonadotropins and their receptors on human reproductive success. Reproduction 2015, 150, R175–R184. [Google Scholar] [CrossRef] [PubMed]
- He, W.-B.; Du, J.; Yang, X.-W.; Li, W.; Tang, W.-L.; Dai, C.; Chen, Y.-Z.; Zhang, Y.-X.; Lu, G.-X.; Lin, G.; et al. Novel inactivating mutations in the FSH receptor cause premature ovarian insufficiency with resistant ovary syndrome. Reprod. Biomed. Online 2019, 38, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Zariñán, T.; Mayorga, J.; Jardón-Valadez, E.; Gutiérrez-Sagal, R.; Maravillas-Montero, J.L.; Mejía-Domínguez, N.R.; Martínez-Luis, I.; Yacini-Torres, O.G.; Cravioto, M.-D.-C.; Reiter, E.; et al. A Novel Mutation in the FSH Receptor (I423T) Affecting Receptor Activation and Leading to Primary Ovarian Failure. J. Clin. Endocrinol. Metab. 2020. [Google Scholar] [CrossRef]
- Patel, B.; Parets, S.; Akana, M.; Kellogg, G.; Jansen, M.; Chang, C.; Cai, Y.; Fox, R.; Niknazar, M.; Shraga, R.; et al. Comprehensive genetic testing for female and male infertility using next-generation sequencing. J. Assist. Reprod. Genet. 2018, 35, 1489–1496. [Google Scholar] [CrossRef]
- Friemel, C.; Ammerpohl, O.; Gutwein, J.; Schmutzler, A.G.; Caliebe, A.; Kautza, M.; Von Otte, S.; Siebert, R.; Bens, S. Array-based DNA methylation profiling in male infertility reveals allele-specific DNA methylation in PIWIL1 and PIWIL2. Fertil. Steril. 2014, 101, 1097–1103.e1. [Google Scholar] [CrossRef]
- Flugent. Available online: https://www.fulgentgenetics.com/Infertility-Female (accessed on 14 March 2020).
- Centogene. Available online: https://www.centogene.com/diagnostics/infertility-testing.html (accessed on 14 March 2020).
- CGCgenetics. Available online: https://www.cgcgenetics.com/en/by-test-a-z/5099 (accessed on 14 March 2020).
- França, M.M.; Lerário, A.M.; Funari, M.F.; Nishi, M.Y.; Narcizo, A.M.; De Mello, M.P.; Guerra-Junior, G.; Maciel-Guerra, A.T.; De Mendonça, B.B. A Novel Homozygous Missense FSHR Variant Associated with Hypergonadotropic Hypogonadism in Two Siblings from a Brazilian Family. Sex. Dev. 2017, 11, 137–142. [Google Scholar] [CrossRef]
- Liu, H.; Xu, X.; Han, T.; Yan, L.; Cheng, L.; Qin, Y.; Liu, W.; Zhao, S.; Chen, Z.-J. A novel homozygous mutation in the FSHR gene is causative for primary ovarian insufficiency. Fertil. Steril. 2017, 108, 1050–1055.e2. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Guo, T.; Gong, Z.; Yu, Y.; Zhang, Y.; Zhao, S.; Qin, Y. Novel FSHR mutations in Han Chinese women with sporadic premature ovarian insufficiency. Mol. Cell. Endocrinol. 2019, 492, 110446. [Google Scholar] [CrossRef] [PubMed]
- Lorenzi, D.; Fernández, C.; Bilinski, M.; Fabbro, M.; Galain, M.; Menazzi, S.; Miguens, M.; Perassi, P.N.; Fulco, M.F.; Kopelman, S.; et al. First custom next-generation sequencing infertility panel in Latin America: Design and first results. JBRA Assist. Reprod. 2020, 24, 104–114. [Google Scholar] [CrossRef]
- França, M.M.; Mendonca, B.B. Genetics of Primary Ovarian Insufficiency in the Next-Generation Sequencing Era. J. Endocr. Soc. 2020, 4, bvz037. [Google Scholar] [CrossRef] [PubMed]
- Reisberg, S.; Krebs, K.; Lepamets, M.; Kals, M.; Mägi, R.; Metsalu, K.; Lauschke, V.M.; Vilo, J.; Milani, L. Translating genotype data of 44,000 biobank participants into clinical pharmacogenetic recommendations: Challenges and solutions. Genet. Med. 2019, 21, 1345–1354. [Google Scholar] [CrossRef] [PubMed]
- Bramble, M.S.; Goldstein, E.H.; Lipson, A.; Ngun, T.; Eskin, A.; Gosschalk, J.E.; Roach, L.; Vashist, N.; Barseghyan, H.; Lee, E.; et al. A novel follicle-stimulating hormone receptor mutation causing primary ovarian failure: A fertility application of whole exome sequencing. Hum. Reprod. 2016, 31, 905–914. [Google Scholar] [CrossRef]
- Zhou, Y.; Fujikura, K.; Mkrtchian, S.; Lauschke, V.M. Computational Methods for the Pharmacogenetic Interpretation of Next Generation Sequencing Data. Front. Pharmacol. 2018, 9, 1437. [Google Scholar] [CrossRef] [PubMed]
- Ay, B.G.B.; May, J.V.; Davis, J.S.; Dias, J.A.; Kumar, T.R. In Vivo and In Vitro Impact of Carbohydrate Variation on Human Follicle-Stimulating Hormone Function. Front. Endocrinol. 2018, 9, 216. [Google Scholar] [CrossRef]
- Davis, D.; Liu, X.; Segaloff, D.L. Identification of the sites of N-linked glycosylation on the follicle-stimulating hormone (FSH) receptor and assessment of their role in FSH receptor function. Mol. Endocrinol. 1995, 9, 159–170. [Google Scholar] [CrossRef][Green Version]
- Sayers, N.; Hanyaloglu, A.C. Intracellular Follicle-Stimulating Hormone Receptor Trafficking and Signaling. Front. Endocrinol. 2018, 9, 653. [Google Scholar] [CrossRef]
- Landomiel, F.; Gallay, N.; Jégot, G.; Tranchant, T.; Durand, G.; Bourquard, T.; Crépieux, P.; Poupon, A.; Reiter, E. Biased signalling in follicle stimulating hormone action. Mol. Cell. Endocrinol. 2014, 382, 452–459. [Google Scholar] [CrossRef]
- Alviggi, C.; Conforti, A.; Cariati, F.; Alfano, S.; Huhtaniemi, I.; Santi, D.; De Placido, G.; Humaidan, P. Impact of polymorphisms of gonadotropins and their receptors on controlled ovarian stimulation: A prospective observational study. In Proceedings of the 32nd Annual Meeting of ESHRE, Helsinki, Finland, 3–6 July 2016. [Google Scholar]
- Badawy, A.; Wageah, A.; El Gharib, M.; Osman, E.E. Prediction and Diagnosis of Poor Ovarian Response: The Dilemma. J. Reprod. Infertil. 2011, 12, 241–248. [Google Scholar]
- Loutradis, D.; Drakakis, P.; Vomvolaki, E.; Antsaklis, A. Different ovarian stimulation protocols for women with diminished ovarian reserve. J. Assist. Reprod. Genet. 2007, 24, 597–611. [Google Scholar] [CrossRef]
- Conforti, A.; Esteves, S.C.; Cimadomo, D.; Vaiarelli, A.; Di Rella, F.; Ubaldi, F.M.; Zullo, F.; De Placido, G.; Alviggi, C. Management of Women With an Unexpected Low Ovarian Response to Gonadotropin. Front. Endocrinol. 2019, 10, 387. [Google Scholar] [CrossRef]
- García-Jiménez, G.; Zariñán, T.; Rodríguez-Valentín, R.; Mejía-Domínguez, N.R.; Gutiérrez-Sagal, R.; Hernández-Montes, G.; Tovar, A.R.; Arechavaleta-Velasco, F.; Canto, P.; Granados, J.; et al. Frequency of the T307A, N680S, and -29G>A single-nucleotide polymorphisms in the follicle-stimulating hormone receptor in Mexican subjects of Hispanic ancestry. Reprod. Biol. Endocrinol. 2018, 16, 100. [Google Scholar] [CrossRef]
- Paschalidou, C.; Anagnostou, E.; Mavrogianni, D.; Raouasnte, R.; Klimis, N.; Drakakis, P.; Loutradis, D. The effects of follicle-stimulating hormone receptor (FSHR) -29 and Ser680Asn polymorphisms in IVF/ICSI. Horm. Mol. Biol. Clin. Investig. 2020. [Google Scholar] [CrossRef]
- Ammar, R.; Paton, T.A.; Torti, D.; Shlien, A.; Bader, G.D. Long read nanopore sequencing for detection of HLA and CYP2D6 variants and haplotypes. F1000Research 2015, 4, 17. [Google Scholar] [CrossRef]
- Conforti, A.; Vaiarelli, A.; Cimadomo, D.; Bagnulo, F.; Peluso, S.; Carbone, L.; Di Rella, F.; De Placido, G.; Ubaldi, F.M.; Huhtaniemi, I.; et al. Pharmacogenetics of FSH Action in the Female. Front. Endocrinol. 2019, 10, 398. [Google Scholar] [CrossRef]

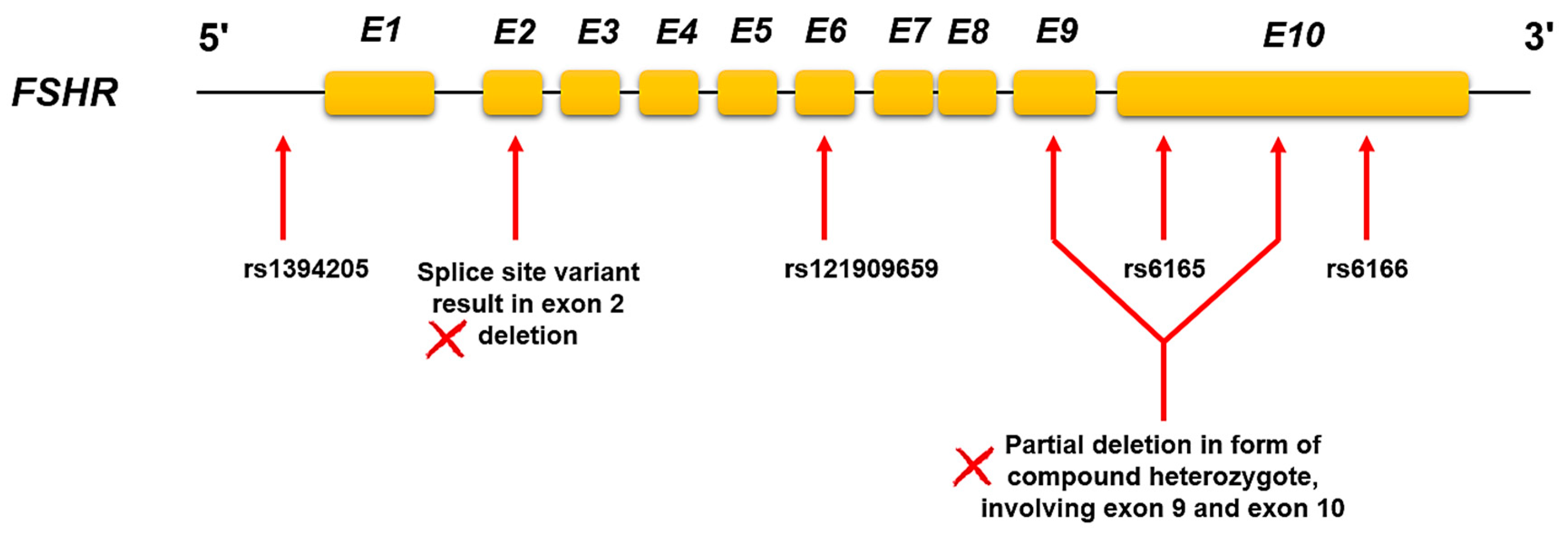

| Gene | Variants with PGx Effects | Nucleotide Changes | MAF* (%) | Variation Effects | Reference |
|---|---|---|---|---|---|
| FSHR | rs6166 | c.2038 G > A | 0.3–0.4 | Allele G: poor response to infertility drugs | [17,18] |
| FSHR | rs6165 | c.919 G > A | 0.3–0.4 | Allele G: poor response to ligands (decreased sensitivity) | [19] |
| FSHR | rs1394205 | -29 G > A | 0.2–0.3 | Allele A: reduced gene expression level leads to a decreased level of the response to drugs | [20,21] |
| FSHR | - | exon2 del | NA* | Reduced response of the receptor for infertility treatments due to impaired receptor transferring to the cell membrane | [22,23] |
| FSHR | - | exon9 and 10 del | NA | Loss of function of receptor lead to FSH resistance | [24] |
| FSHR | rs121909659 | c.479 A > G,T | 0.000025 | Partial loss of function and impaired cell surface expression of FSHR causes the reduced response in patients (Classified as pathogenic SNP) | [25,26] |
| Country | Study Population | FSHR Evaluated SNP(s) | Different Responders to the Treatments Due to FSHR Altered Function | Conclusion | Reference |
|---|---|---|---|---|---|
| Slovenia | 60 women undergoing ovarian stimulation were selected | −29 G > A c.2038 G > A | Poor Responders: 28.3% Normal Responders: 43.4% High Responders: 28.3% | The GG genotype in rs1394205 is associated with poor ovarian response to COH 1, and the related patients may require higher doses of rFSH for ovulation induction. | [27] |
| Spain | 170 women undergoing controlled ovarian stimulation included | c.2038 G > A | Poor responders: 58.4% Normal Responders: - 2 High responders: 27.7% | Discrete set of genes and polymorphisms, including rs6166 in the FSHR gene, may partially explain the poor response to FSH hormone during controlled ovarian stimulation treatments. | [28] |
| Greece | 79 sub-fertile women and 46 normo-ovulatory women with diverse respond to IVF were included | c.2038 G > A | Poor responders: 28% Normal Responders: 36.8% High responders: - | Good (normal) responder group had a statistically significant Asn/Ser heterozygous variant (rs6166) with more follicles and oocytes in patients. | [29] |
| Ukraine | 374 women, including ovary dysfunction patients and healthy individuals with different treatment responses, were selected | c.919 G > A c.2038 G > A | Poor responders: 10.42% Normal responders: 10.7% High responders: - (Study also included control groups: 51.6%) | Combined allelic distribution for rs6165 and rs6166 (Ala307-Ser680/Ala307-Ser680) genotype should have an impact on the delineation of stimulation protocols. | [30] |
| Germany | 161 ovulatory women below the age of 40 years with different FSH stimulation requirements were included | c.919 G > A c.2038 G > A | Poor responders: - Normal Responders: - High responders: - | The Asn/Ser heterozygote genotype for rs6166 was significantly more common in infertile patients with diverse ovarian response. Ovarian response to FSH stimulation depends on the FSHR genotype. | [31] |
| Germany | 93 women (homozygous for Asn/Asn or Ser/Ser) undergoing controlled ovarian hyperstimulation in IVF and ICSI | c.2038 G > A | Poor responders: 25.80% Normal Responders: 74.19% High responders: - | Lower FSH receptor sensitivity due to p.N680S sequence variation in FSHR (rs6166) resulted in lower estradiol levels following FSH stimulation, which cause the patients to need to receive higher FSH doses. | [32] |
| Sweden and China | Systematic review and meta-analysis on special FSHR variants and altered ovarian response in women undergoing IVF | c.2038 G > A | - | FSHR polymorphism Ser680Asn (rs6166), through the other pharmacogenomics variants, is the most optimal biomarker for implementing in routine clinical practice. | [33,34] |
| Armenia origin | Case report of a woman with secondary amenorrhea and very high plasma gonadotropin concentrations (especially FSH) | c.479 A > G,T | Poor responders: 100% Normal Responders: - High responders: - | rs121909659 causes partial loss of function, and impaired cell surface expression of FSHR resulted in reduced response in COH. The study reminds us of the population-specific assessments of FSHR. | [25] |
| United States | 35 women undergoing in vitro fertilization included | exon2 del exon6 del exon9 del intron 8 insertion | Poor responders: 8.5% Normal Responders: 68.57%% High responders: 22.85% | FSHR splicing variants, seen in women with a normal menstrual cycle that show an abnormal response to FSH stimulation described. Exon 2 deletion was associated with low ovarian response. | [22] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tafazoli, A.; Wołczyński, S.; Wawrusiewicz-Kurylonek, N.; Esmaeili, S.-A.; Miltyk, W. Pharmacogenomic Biomarkers of Follicle-Stimulating Hormone Receptor Malfunction in Females with Impaired Ovarian Response—A Genetic Survey. J. Clin. Med. 2021, 10, 170. https://doi.org/10.3390/jcm10020170
Tafazoli A, Wołczyński S, Wawrusiewicz-Kurylonek N, Esmaeili S-A, Miltyk W. Pharmacogenomic Biomarkers of Follicle-Stimulating Hormone Receptor Malfunction in Females with Impaired Ovarian Response—A Genetic Survey. Journal of Clinical Medicine. 2021; 10(2):170. https://doi.org/10.3390/jcm10020170
Chicago/Turabian StyleTafazoli, Alireza, Sławomir Wołczyński, Natalia Wawrusiewicz-Kurylonek, Seyed-Alireza Esmaeili, and Wojciech Miltyk. 2021. "Pharmacogenomic Biomarkers of Follicle-Stimulating Hormone Receptor Malfunction in Females with Impaired Ovarian Response—A Genetic Survey" Journal of Clinical Medicine 10, no. 2: 170. https://doi.org/10.3390/jcm10020170
APA StyleTafazoli, A., Wołczyński, S., Wawrusiewicz-Kurylonek, N., Esmaeili, S.-A., & Miltyk, W. (2021). Pharmacogenomic Biomarkers of Follicle-Stimulating Hormone Receptor Malfunction in Females with Impaired Ovarian Response—A Genetic Survey. Journal of Clinical Medicine, 10(2), 170. https://doi.org/10.3390/jcm10020170