Abstract
A growing body of work has linked key biological activities to the mechanical properties of cellular membranes, and as a means of identification. Here, we present a computational approach to simulate and compare the vibrational spectra in the low-THz region for mammalian and bacterial membranes, investigating the effect of membrane asymmetry and composition, as well as the conserved frequencies of a specific cell. We find that asymmetry does not impact the vibrational spectra, and the impact of sterols depends on the mobility of the components of the membrane. We demonstrate that vibrational spectra can be used to distinguish between membranes and, therefore, could be used in identification of different organisms. The method presented, here, can be immediately extended to other biological structures (e.g., amyloid fibers, polysaccharides, and protein-ligand structures) in order to fingerprint and understand vibrations of numerous biologically-relevant nanoscale structures.
1. Introduction
The internal motions of biological membranes have increasingly been the focus of biological research, as they provide a connection between membrane composition and many biological processes [1]. For example, membranes’ vibrations and density fluctuations have been linked to the transport of small molecules across membranes [2,3,4,5]. These processes and the membrane mechanical properties are influenced not only by the presence of transmembrane proteins and membrane composition [6,7,8,9,10], but also emerging molecular structures and their distributions play a critical role. For example, lipid asymmetry across bacterial membranes has been linked to varying susceptibility to antibiotics [11,12,13], and lipid rafts—the non-homogeneous distribution of lipids into localized regions—play a role in several biological processes [1,7,8,9].
First proposed in 1911 [14], the idea that mechanical vibrations are an identifying feature of various compounds has been extensively studied in the past century [15,16,17,18,19,20,21]. Likewise, there exists a clear link between the mechanical properties of a membrane and its properties, and in turn its functionality, which has motivated research investigating the relationship between membrane vibration and cellular activity. Recently, this line of thinking has been used to show that vibrations can be used as a means of distinguishing among microorganisms [22,23], and to study the interactions between membranes and anchored or adjacent external structures, like in bacterial biofilms [24,25]. Moreover, an increasing number of studies have identified modes of membrane-adjacent structures. For example, functional amyloid fibers, proteinaceous fibers that grow in biofilm and anchor to the bacterial membranes, have been suggested to mechanically vibrate and deliver a damped vibrational signal to an adjacent bacterial cell [24,25,26]. Electromagnetic signals on the order of kHz of bacterial DNA that match DNA extracted from Alzheimer’s and other amyloid-induced diseased patients [27] suggest that bacterial infections are present in such illnesses. THz vibrations have also been observed in protein-ligand binding [28] and other biological polymers [29,30], suggesting that protein-ligand interactions trigger unique changes in vibration that can be used in detection and diagnoses.
Despite these promising results, work in this direction has been hindered by several factors. Experimentally, membranes’ mechanical and structural characterization, as well as cellular identification, have been expensive and time-consuming [1,7,8,9,21,22,23]. Indeed, many of the early works that discussed the use of mechanical vibrations as an identification tool speculated that computation would eventually dominate the field [16,18,19]. Nonetheless, while computationally probing the vibrational modes of biological structures, like membranes, is simpler, it is computationally demanding, which has led to less accurate approaches (e.g., coarse-graining, continuous models) and assumptions (e.g., membrane composition and structure) [31,32,33,34,35,36,37,38] of limited usefulness or reproducibility. Finally, even when data is available, an unbiased method for the identification and comparison of the vibrational spectra has long been a complex challenge [18,20,39,40,41,42].
To fill this gap, we propose an approach that combines atomistic molecular dynamics simulations, to gather information about the low-THz vibration of disparate membranes, with signal processing, to identify and compare their vibrational spectra. Using this approach, we discuss the effect of membrane asymmetry and lipid composition (with and without sterols) on the vibrations, as well as some hidden pitfalls that are potentially introduced by the use of atomistic simulations. Moreover, by employing a nonparametric test, our comparisons allow us to test the variability among samples obtained of the same system, and quantify spectral uncertainty.
2. Materials and Methods
The approach used in this work is detailed below and illustrated in Figure 1 and Figure 2. All abbreviations are listed at the end of the manuscript. Briefly, we used Molecular Dynamics (MD) to simulate different cellular membranes, and used the time evolution of the atomic positions to compute the infrared absorption in the THz region. The resulting spectra were then filtered to help with peak identification and compared to obtain a simple measure of the difference between spectra.
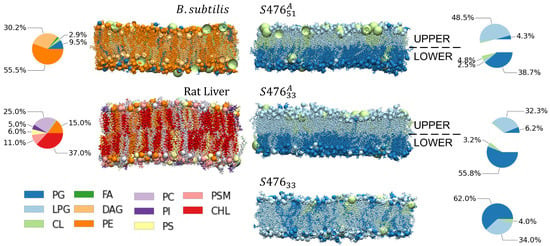
Figure 1.
Systems and Nomenclature Summary. Composition and distribution of five types of plasma membranes modeled in this work: one for B. Subtilis, one for rat liver cell, and three for S. aureus (S476) membranes. S. aureus membrane are labeled according to symmetry (A for asymmetric) and percent concentration of LPG (33% or 51%). Only composition fractions greater than 2% are shown here. Only non-sterol containing membranes are shown. All abbreviations are listed at the end of the manuscript.
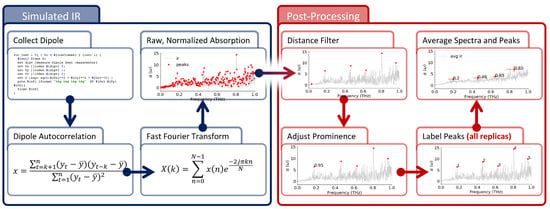
Figure 2.
Spectra Calculation and Signal Analysis. Infrared spectra are computed from MD trajectories (blue panel) and, then, post-processed to find local maxima. Spectra are computed from MD trajectories as Fourier transform of the membrane total dipole autocorrelation; then, the spectra are normalized before peaks are identified using a distance and prominence filter (Figures S1–S4). For more details, see Section 2.
2.1. Systems
In this work, instead of taking the standard approach of exploring the effect of each possible parameter (e.g., the concentration of each possible lipid), we focused on three complex membranes using realistic compositions, as discussed in the following. We chose this approach because looking into the effect of all the possible parameters of a membrane is a generally daunting task, given the number of possible lipids and their concentrations (e.g., B. Subtilis has at least 127 different lipids), and their often nonlinear relations. The latter is a critical consideration, as it can make the effort of decomposing the problem in simpler tasks very challenging; the addition of a single type of lipid can markedly change certain observed properties, like the spectra in the range of interest here. We show such an example at the end of this manuscript, when discussing the effect of sterols and LPG.
With this in mind, instead of trying to create a general model, we aimed to (1) determine if we could detect any difference between real compositions and (2) discuss the problems simulations would encounter in sampling more realistic and therefore complicated systems. We studied the plasma membranes of three types of cells, two of bacterial (Staphylococcus aureus and Bacillus subtilis) and one of mammalian (rat liver plasma [43]) origin. We chose S. aureus because of its high pathogenicity and prevalence in hospital-acquired infections [44,45], B. Subtilis thanks to its ubiquity and innocuousness in healthy individuals [46], and rat liver cell membrane as an example of mammalian plasma membranes. For S. aureus, we considered two asymmetric membranes with different composition (S476, S476), observed at two different values of pH (i.e., 5.5 and 7.4), as well as a symmetric membrane (S476) as close as possible to S476, to study the effect of lipid distribution between leaflets [11,31,47,48,49,50,51,52]. For B. Subtilis and rat liver cells, however, we could only find information about the total composition of the plasma membranes and therefore, we simulated homogenous bilayers. These five systems are illustrated in Figure 1. Additionally, we simulated four membranes, derived from S476 and S476 by adding to each one of them 1.3% molar [53] of either ergosterol or cholesterol. For all the membranes, 3 to 9 independent replicas were generated and simulated.
Except when noted otherwise, the S. aureus membranes consist of a periodic bilayer of 15 × 15 in size (approximately 700 lipids, exact number depends on composition), the B. Subtilis membrane 16 × 16 in size (840 lipids), and the rat liver membrane, 12 × 12 in size (600 lipids). These sizes were chosen based on the estimated lowest frequency mode () that could be observed,
where is the speed of sound in the membrane and the longest distance between two points on the x-y dimension (membrane plan) of the periodic box. Since is hard to estimate accurately, we conservatively used the speed of sound of water (1550 m s−1), which is greater than alcohols and alkenes with long aliphatic chains (1150–1250 m s−1), which places an upper limit to of approximately .
2.2. Molecular Dynamics Simulations
All the systems were prepared using the Membrane Builder in CHARMM GUI [54] and, then, post-processed when cropped systems were needed. Nanoscale Molecular Dynamics [55] software was used to perform Molecular Dynamics simulations, a time step of 2 was employed to integrate the equations of motion, while hydrogen atoms were kept rigid via the SHAKE algorithm. Membranes were fully solvated in a NaCl solution using TIP3P for water [56] and CHARMM, version 36 [57], to model atomic interactions. Non-bonded short-range interactions smoothly approached 0 using an X-PLOR switching function between 1 and , in conjunction with the particle mesh Ewald algorithm, to evaluate long-range Coulomb forces.
The systems were equilibrated using constrained canonical simulations, followed by isothermal-isobaric ensemble simulations with vanishing restraint, as per CHARMM GUI protocol. This preliminary equilibration was followed by 100 simulation in an NPsT ensemble, an isothermal-isobaric ensemble where changes in dimensions in the direction of the membrane’s plane are coupled. Starting from the equilibrated systems, spectra were computed from 50 microcanonical ensemble simulations, while Langmuir isotherms were computed from canonical simulations 20 long. In all cases, temperature was kept constant at 310 K by using a Langevin thermostat with a period of 1 , and pressure was imposed by using a Nosé-Hoover Langevin piston method, with a period of 200 and 50 decay. Processing and preparation of trajectories and structures were performed with the help of Visual Molecular Dynamics (VMD) software [58], as well as the MDAnalysis and Scipy Python libraries [59,60,61].
2.3. Spectra
Absorption cross-section as a function of the frequency were calculated from trajectories, using the relation:
where ℏ is the reduced Plank constant, c is the speed of light in vacuum, the vacuum permittivity, the refractive index of the solution, is the inverse of the Boltzmann constant times the temperature, is the harmonic quantum correction [62],
and is the classical spectral density,
where angular brackets indicate ensemble averaging and is the membrane’s total dipole moment. For practical reason, we assumed = 1 as, in this range, the correction is almost linear [63]. Finally, numerical noise was reduced by using Blackman windowing when computing the Fourier transform and by applying a Savitsky-Golay filter (polynomial order of 6 over 21 windows, Figure S5) on .
Since different membranes are represented by periodic systems of different size, to compare different spectra and obtain size-independent quantities, we divided each spectra by the average value of the signal in our interval
and used in all the comparisons.
2.4. Signal Analysis
Peaks were identified from the spectra by tuning the (1) prominence (vertical distance between the peak and its lowest contour line) and the (2) minimum distance between peaks. Prominence, alone, is not used as a peak-finding metric because doing so would only select the highest intensity peaks, which are located on the right side of the spectra in this case. Instead, after setting the minimum peak-to-peak distance ( 125 for the rat liver cells, and 142 for bacterial membranes), we computed the prominence distribution for different prominence thresholds. A demonstration of how prominence and distance between peaks are tuned is available in the Supplementary Materials (Figures S1–S4). We then identified the range of values that were closer to the average number of peaks (computer over all the prominence value) and selected the highest value. For each membrane, we compute the average frequency and intensity of each peak using the peaks computed over replicas (3 replicas for S476, S476, B. Subtilis, and rat liver; 4 replicas for S476, see a more detailed discussion in the Results section).
To quantify the similarity between two spectra, we used a two sample Kolmogorov-Smirnov (KS) test [64]. As replicas of the same membrane are (or should be, see the discussion in the results) statistically equivalent, variation between replicas of the same system can be assumed to originate from the computational method. As such, when comparing different membranes, we used the mean KS (and the corresponding standard error) of the replicas of a membrane as a baseline for the comparison with other systems. Of note, all the comparisons were performed by computing the KS value for all the possible pairs of replicas of one system with another, and not by comparing the average spectra.
2.5. Root-Mean-Squared Fluctuations (RMSF)
RMSF, that is the mean deviation from the average position of an atom, was calculated from the microcanonical simulations, using the Python MDAnalysis module [60,61]. Due to a systematic numerical bias in the RMSF value for molecules that are close to the periodic boundaries, all the contributions from these molecules were removed. To allow a meaningful comparison with other membranes, we only computed the RMSF of the atoms that are not part of sterol molecules.
3. Results
Before analyzing the differences among replicas of different membranes, we tested the force field and relaxation protocol. As atomistic molecular dynamics has been extensively proven in the literature to be suitable to model bilayer dynamics, we performed a minimal validation by computing the Langmuir isotherms in the between and for S476, S476, and S476. The comparison with experimental data (see Supplementary Material, Figure S6) shows that the difference of our estimates is well below the uncertainty (standard deviation).
As a second step, we tested the potential bias introduced by using periodic boundary conditions (i.e., size effect). To this end, we compared the spectra of membranes with identical composition (S476) but having four different periodic system sizes (15 × 15 , 4 × 15 , 3 × 15 , and 3 × 12 ). The results show (see Supplementary Material, Figure S7) that peak location is unaffected by the size of the bilayer patch, but the normalized intensity is marginally weaker for the smallest membranes. In the following, however, we will only use square periodic patches (15 × 15 for S. aureus systems, 16 × 16 for B. Subtilis, and 12 × 12 for rat liver, see Section 2), to avoid introducing any anisotropy in the systems.
3.1. Membrane Asymmetry
The asymmetry in the membrane composition is suggested to play a key role in many cellular processes. At the same time, accurate information about the distribution of species between leaflets is generally scarce, due to the difficulty of experimental measurements, as well as the dynamic nature of the cellular membrane make-up. As a larger number of average compositions are available in the literature, we compared the differences in the vibrational spectra between symmetric and asymmetric S. aureus systems (S476 and S476). The initial comparison (see Figure 3), while showing a statistical equivalence between the peaks of the two systems, was surprisingly affected by large uncertainty, despite the number of replicas used for each system (9 for S476 and 6 for S476).
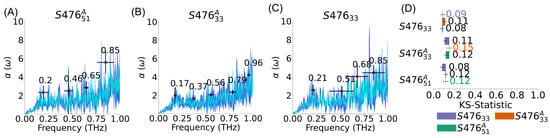
Figure 3.
Effect of composition and distribution on S. aureus membrane’s spectra. (A–C) S. aureus membrane average spectra (cyan), standard deviation of spectra (dark blue), and peaks (black); error bars represent standard deviations. For S476 and S476 the spectra are computed from the largest cluster of replicas (see Supplementary Material, Figure S8). (D) Kolmogorov-Smirnov (KS) statistics; error bars represent the standard error of the mean. Self comparison (e.g., S476/ S476) indicates the average difference between replicas of the same system. KS statistic shows that this part of the absorption spectra for the three systems are not distinct.
To investigate the rationale behind this observation, we looked into the similarity among replicas of a given membrane. To make sense of all these comparisons, we built an undirected weighted graph (see Supplementary Material, Figure S8a), where each node is a replica and the weight of each edge is equal to 1-KS (i.e., similar spectra are connected by an edge with higher value). Different types of clustering analyses can be performed to obtain such a graph, but the consistent result is that the replicas are not separated, as expected, in two groups based on their leaflet symmetry, but rather in three groups, where both types of systems are somewhat mixed. Given the microcanonical nature of the simulations used to generate the spectra, the reason for this clustering is a dependence on the initial conditions, likely resulting in some violation of the ergodic hypothesis. To narrow down the source of this difference, we considered the effects of the membrane thickness (see Supplementary Material, Figure S8a), periodic system size, and surface tension (see Supplementary Material, Figure S8b) on the spectra, but we found no strong correlation in all cases. Thus, we hypothesize that the differences are related to slightly different stability in the vibrational modes that are sampled, due to slightly different initial velocity distribution. This hypothesis is corroborated by the analysis of the peaks, which shows that, with the notable exception of the lowest frequency (∼ ), the peaks display variability in intensity and location between groups (see Supplementary Material, Figure S8c).
This analysis leads to three main conclusions. First, it shows that the leaflet symmetry, for equivalent total composition, does not have a statistically significant effect on the spectra, whether we consider KS statistic of the average of all replicas or we restrict ourselves to one of the replica clusters, like in Figure 3. Of note, the membrane symmetry can still affect other processes, as well as other mechanical properties. Second, despite the differences and the complexity in comparing the spectra of each replica, the lowest frequency peak (between 0.17–0.21 THz) is conserved in all S. aureus systems and replicas. Finally, the spectra obtained from simulations should be carefully tested for internal consistently. Even though we observed statistical variability only among replicas of two S. aureus systems in this work, this issue should be tested to avoid adding systematic uncertainty to the results, especially as we will show, below, for more rigid membranes.
3.2. Cell Type
After establishing the effect of composition and lipid distribution for S. aureus membrane, we compared the spectra of the plasma membrane of different cells, namely we compared S. aureus with one other common bacterium as well as a mammalian cell.
The results (Figure 4) show that the S. aureus absorption spectra in the low-THz region is rather distinct from the other two, with a characteristic peak, just below . Of note, this distinction holds regardless of the S. aureus replicas or the symmetry chosen (see Supplementary Material, Figure S9). B. Subtilis and rat liver cell are also statistically distinct, although notably more similar than S. aureus, despite the remarkably different composition. Notably, these differences are the results of the interplay between lipids and not a simple inertial behavior due to the difference in the membranes’ density: the mass per unit area of S476 (approximately 2.4 kDa nm−2) falls between the one for B. Subtilis (approximately 2.2 kDa nm−2) and the mammalian cell (approximately 2.6 kDa nm−2).
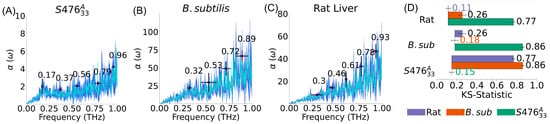
Figure 4.
Plasma membranes absorption spectra of different species. (A–C) Average spectra are shown in cyan, standard deviation in spectra (dark blue), and peaks are labeled by black points; error bars represent standard deviations in peak location and intensity. The S. aureus membrane is the only one among the three that has a peak in the (0.17–0.2 THz) region. (D) Kolmogorov-Smirnov (KS) statistics; error bars represent the standard error of the mean. Self comparisons (e.g., Rat/Rat) indicate the average difference between replicas of the same system. KS statistic shows that S. aureus spectra is very distinct from the other two, even though B. Subtilis and Rat spectra are still statistically discernible.
3.3. Sterols
Finally, we looked in the effect of the presence of sterols on the S. aureus membrane. Bacterial cells do not typically synthesize sterols, as the bacterial cell wall occupies the same function fulfilled by sterol-containing plasma membranes in eukaryotic cells by maintaining structural integrity and fluidity. However, cell-wall-deficient forms of S. aureus, called L-forms, do exist [65]. As staphylococcal L-forms lack cell walls, sterols provide a means of maintaining structural integrity and fluidity [66]. More importantly, the presence of sterols has been linked to increased resistance to antimicrobial peptides [67] and lipid raft formation, demonstrating their importance in membrane function and biological processes. For this reason, we analyzed the effect of sterols on the absorption spectra by comparing the spectra of the asymmetric S. aureus membranes (without sterols), with identical membranes to which we added either 1.3% cholesterol or ergosterol [53].
The presence of sterols in the membranes (see Figure 5) had a greater effect on the spectra of S476 than that of S476. This difference can be related to the different content of LPG between the two membranes, as higher levels of LPG decrease the membrane fluidity [12,68,69,70,71], causing a slight change in the absorption spectra. These results agree with a study in which mutant bacteria, producing less LPG, have membranes with a reduced rigidity [72]. Indeed, the presence of high LPG concentration, much like low concentration of sterols, has been shown to have a stabilizing effect on membrane fluidity [68,69]. While these effects do not compound (S476), a membrane with a lower concentration of LPG (S476) would be more susceptible to changes caused by sterols. It is interesting that sterols in the membrane with more LPG cause an increase in mobility, as measured by the average atomic RMSF, while sterols in the membrane with less LPG cause an increase in rigidity (Figure 5C). Finally, cholesterol typically affects the membrane mechanical properties more than ergosterol (Figure 5B), which agrees with the experimental observation that ergosterol has a smaller effect on membrane mobility than cholesterol [67].
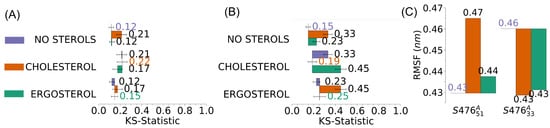
Figure 5.
Difference in the spectra due to the presence of sterols in S. aureus membranes. Plots show the results of two-sample Kolmogorov-Smirnov (KS) tests for the average spectra of (A) S476 and (B) S476, without any additional sterol, and with 1.3% cholesterol or ergosterol. Error bars represent the standard error of the mean. (C) Average mobility measured as root-mean-squared fluctuations (RMSF) of the positions of non-sterol atoms in S476 and S476 membranes, without sterols (baseline) and with cholesterol (orange) or ergosterol (green).
4. Discussion
The unique absorption spectra of biological membranes are a promising metric for species differentiation and bio-process identification. In this work, we show how to estimate, analyze, and compare the absorption spectra of bacterial and mammalian membranes, by combining molecular dynamics simulations and signal processing techniques (i.e., peak detection and KS statistics). The analysis of S. aureus, B. Subtilis, and rat liver cells, shows that distinct peaks can be identified for different species in the low-THz region and that certain peaks, for S. aureus around , are present even for different compositions and symmetries, making them good identifiers under a variety of conditions.
The ability to rigorously compare noisy and complex spectra, like the one studies here, opens the door to finding unexpected correlations. For example, S. aureus membranes (at 7.4 pH) have a lower LPG content, which we found is associated with both increased variability among replicas and higher susceptibility to changes in mobility in the presence of sterols. This change could speak to higher rigidity in membranes with more LPG, as LPG tends to maintain membrane fluidity [12,68,69,70,71]. While higher fluidity may have biological advantages, it seems inversely correlated to antibiotic resistance as, generally, an increase in resistance of membranes to antimicrobial peptides is associated with higher concentrations of LPG [68,73] and sterols [67].
The comparison of spectra of different samples of the same system also provides a way to find similarities, even for complex signals like the one presented here. Different samples can be clustered and analyzed by building fully connected graphs, where edges are weighted based on the values of a two-sample KS test. This representation, allows visualizing and finding similarities on high dimensional spaces, like the 105th-dimensional space of the comparisons between 15 S. aureus samples in this work. While we found that the existence of clusters among the samples affected only the most rigid membrane (see the previous discussion about LPG content), it is nevertheless allowed extracting data from samples that would have been otherwise affected by a high uncertainty.
Finally, while this work was designed around the low-THz absorption spectra of plasma membranes, it can immediately generalize to other structures (e.g., fibers present in biofilm matrix), other regions of the vibrational spectra, as well as to data obtained from experimental techniques. Our work reinforces the idea that vibrations can be used to successfully differentiate between different biological complexes and how they are affected by specific changes, which, then, can potentially be related to biological functions. This link, if present, could inform a route by which these vibrations could be manipulated, for very targeted effects [27].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/membranes13020139/s1, Figure S1: Prominence Filter Tuning; Figure S2: Distance Filter Selection for S476; Figure S3: Distance Filter Selection for B. Subtilis Membrane; Figure S4: Distance Filter Selection for Rat Liver Plasma Membrane; Figure S5: Savitsky-Golay Filter Selection; Figure S6: Langmuir Isotherms; Figure S7: Effect of S476 Periodic Boundary Size on Spectra after Normalization; Figure S8: Clustering Analysis for KS Statistics of and Replicas; Figure S9: KS Statistics for S476 and S476 Clusters; Figure S10: Area per Lipid Equilibration; Table S1: Asymmetric S. aureus Membrane Compositions per Leaflet; Table S2: Detailed Membrane Compositions; Table S3: Complete KS Statistics; TCL Script Dipole Calculation.
Author Contributions
A.V. conceived and designed the study, reviewed the paper; C.L. designed, ran, and analyzed the simulations, implemented post-processing code for simulations, and wrote the paper; P.E. conceived and designed the study, designed and analyzed the simulations, implemented post-processing code for membrane vibrations; and wrote the paper; J.V. ran simulations and wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This project was funded by the Department of Defense Advanced Research Projects Agency (DARPA), grant n. HR00111720067 and by the BlueSky project, University of Michigan.
Data Availability Statement
The data presented in this study are openly available in University of Michigan DeepBlue Documents at 10.7302/6571.
Acknowledgments
We acknowledge University of Michigan’s ARC-TS for use of their high performance computing resources, particularly Great Lakes. We also acknowledge the use of BioRender in the creation of the Graphical Abstract.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| CL | cardiolipin |
| DAG | diacylglycerol |
| FA | free fatty acid |
| KS | Kolmogorov-Smirnov |
| LPG | lysophosphatidylglycerol |
| MD | Molecular Dynamics |
| PC | phosphatidylcholine |
| PE | phosphatidylethanolamine |
| PG | phosphatidylglycerol |
| PI | phosphatidylinositol |
| PS | phosphatidylserine |
| PSM | sphingomyelin |
| S476 | Asymmetric S. aureus membrane with 33% LPG content |
| S476 | Symmetric S. aureus membrane with 33% LPG content |
| S476 | Asymmetric S. aureus membrane with 51% LPG content |
References
- Leonov, D.V.; Dzuba, S.A.; Surovtsev, N.V. Normal vibrations of ternary DOPC/DPPC/cholesterol lipid bilayers by low-frequency Raman spectroscopy. RSC Adv. 2019, 9, 34451–34456. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Elvati, P.; Majumder, S.; Wang, Y.; Liu, A.P.; Violi, A. Predicting the Time of Entry of Nanoparticles in Lipid Membranes. ACS Nano 2019, 13, 10221–10232. [Google Scholar] [CrossRef] [PubMed]
- Paula, S.; Volkov, A.; Van Hoek, A.; Haines, T.; Deamer, D.W. Permeation of protons, potassium ions, and small polar molecules through phospholipid bilayers as a function of membrane thickness. Biophys. J. 1996, 70, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Zhernenkov, M.; Bolmatov, D.; Soloviov, D.; Zhernenkov, K.; Toperverg, B.P.; Cunsolo, A.; Bosak, A.; Cai, Y.Q. Revealing the mechanism of passive transport in lipid bilayers via phonon-mediated nanometre-scale density fluctuations. Nat. Commun. 2016, 7, 12292. [Google Scholar] [CrossRef]
- Chen, S.; Liao, C.; Huang, H.; Weiss, T.; Bellisent-Funel, M.; Sette, F. Collective dynamics in fully hydrated phospholipid bilayers studied by inelastic X-ray scattering. Phys. Rev. Lett. 2001, 86, 740. [Google Scholar] [CrossRef]
- Liu, C.; Elvati, P.; Violi, A. On Drug-Membrane Permeability of Antivirals for SARS-CoV-2. J. Phys. Chem. Lett. 2021, 12, 1384–1389. [Google Scholar] [CrossRef]
- Rheinstädter, M.; Ollinger, C.; Fragneto, G.; Demmel, F.; Salditt, T. Collective dynamics of lipid membranes studied by inelastic neutron scattering. Phys. Rev. Lett. 2004, 93, 108107. [Google Scholar] [CrossRef]
- Armstrong, C.L.; Barrett, M.A.; Hiess, A.; Salditt, T.; Katsaras, J.; Shi, A.C.; Rheinstädter, M.C. Effect of cholesterol on the lateral nanoscale dynamics of fluid membranes. Eur. Biophys. J. 2012, 41, 901–913. [Google Scholar] [CrossRef]
- Surovtsev, N.; Adichtchev, S. Dynamic response on a nanometer scale of binary phospholipid-cholesterol vesicles: Low-frequency Raman scattering insight. Phys. Rev. E 2021, 104, 054406. [Google Scholar] [CrossRef]
- Foley, S.L.; Hossein, A.; Deserno, M. Fluid-gel coexistence in lipid membranes under differential stress. Biophys. J. 2022, 121, 2997–3009. [Google Scholar] [CrossRef]
- Rosado, H.; Turner, R.D.; Foster, S.J.; Taylor, P.W. Impact of the β-Lactam resistance modifier (-)-epicatechin gallate on the non-random distribution of phospholipids across the cytoplasmic membrane of staphylococcus aureus. Int. J. Mol. Sci. 2015, 16, 16710–16727. [Google Scholar] [CrossRef]
- Jones, T.; Yeaman, M.R.; Sakoulas, G.; Yang, S.J.; Proctor, R.A.; Sahl, H.G.; Schrenzel, J.; Xiong, Y.Q.; Bayer, A.S. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 2008, 52, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Shireen, T.; Singh, M.; Das, T.; Mukhopadhyay, K. Differential adaptive responses of Staphylococcus aureus to in vitro selection with different antimicrobial peptides. Antimicrob. Agents Chemother. 2013, 57, 5134–5137. [Google Scholar] [CrossRef] [PubMed]
- Coblentz, W.W. Radiometric Investigation of Water of Crystallization, Light Filters, and Standard Absorption Bands; Number 168; US Government Printing Office: Washington, DC, USA, 1911. [Google Scholar]
- Horbach, I.; Naumann, D.; Fehrenbach, F.J. Simultaneous infections with different serogroups of Legionella pneumophila investigated by routine methods and Fourier transform infrared spectroscopy. J. Clin. Microbiol. 1988, 26, 1106–1110. [Google Scholar] [CrossRef] [PubMed]
- Naumann, D.; Helm, D.; Labischinski, H. Microbiological characterizations by FT-IR spectroscopy. Nature 1991, 351, 81–82. [Google Scholar] [CrossRef] [PubMed]
- Curk, M.; Peledan, F.; Hubert, J. Fourier transform infrared (FTIR) spectroscopy for identifying Lactobacillus species. FEMS Microbiol. Lett. 1994, 123, 241–248. [Google Scholar] [CrossRef]
- Helm, D.; Labischinski, H.; Schallehn, G.; Naumann, D. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. Microbiology 1991, 137, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Naumann, D.; Fijala, V.; Labischinski, H.; Giesbrecht, P. The rapid differentiation and identification of pathogenic bacteria using Fourier transform infrared spectroscopic and multivariate statistical analysis. J. Mol. Struct. 1988, 174, 165–170. [Google Scholar] [CrossRef]
- Thomas, L.; Greenstreet, J. The identification of micro-organisms by infrared spectrophotometry. Spectrochim. Acta 1954, 6, 302–319. [Google Scholar] [CrossRef]
- Kenner, B.A.; Riddle, J.W.; Rockwood, S.W.; Bordner, R.H. Bacterial Identification by Infrared Spectrophotometry II: Effect of Instrumental and Environmental Variables. J. Bacteriol. 1958, 75, 16–20. [Google Scholar] [CrossRef]
- Berrier, A.; Schaafsma, M.C.; Nonglaton, G.; Bergquist, J.; Rivas, J.G. Selective detection of bacterial layers with terahertz plasmonic antennas. Biomed. Opt. Express 2012, 3, 2937–2949. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Cha, S.; Jun, S.; Park, S.; Park, J.Y.; Lee, S.; Kim, H.; Ahn, Y. Identifying different types of microorganisms with terahertz spectroscopy. Biomed. Opt. Express 2020, 11, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Barani, N.; Sarabandi, K. Electromagnetic Signaling and Quorum Sensing within Biofilms: Which Mechanism Is the Most Probable Means of Communication? In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; IEEE: New York, NY, USA, 2020; pp. 2459–2462. [Google Scholar]
- Barani, N.; Sarabandi, K.; Kotov, N.A.; Vanepps, J.S.; Elvati, P.; Wang, Y.; Violi, A. A Multiphysics Modeling of Electromagnetic Signaling Phenomena at kHz-GHz Frequencies in Bacterial Biofilms. IEEE Access 2022, 10, 39344–39361. [Google Scholar] [CrossRef]
- Barani, N.; Kashanianfard, M.; Sarabandi, K. A Mechanical Antenna with Frequency Multiplication and Phase Modulation Capability. IEEE Trans. Antennas Propag. 2020, 69, 3726–3739. [Google Scholar] [CrossRef]
- Montagnier, L.; Aissa, J.; Ferris, S.; Montagnier, J.L.; Lavalléee, C. Electromagnetic signals are produced by aqueous nanostructures derived from bacterial DNA sequences. Interdiscip. Sci. Comput. Life Sci. 2009, 1, 81–90. [Google Scholar] [CrossRef]
- Turton, D.A.; Senn, H.M.; Harwood, T.; Lapthorn, A.J.; Ellis, E.M.; Wynne, K. Terahertz underdamped vibrational motion governs protein-ligand binding in solution. Nat. Commun. 2014, 5, 3999. [Google Scholar] [CrossRef]
- Xu, J.; Plaxco, K.W.; Allen, S.J. Probing the collective vibrational dynamics of a protein in liquid water by terahertz absorption spectroscopy. Protein Sci. 2006, 15, 1175–1181. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.J.; Cheng, G.; Huang, Z.; Zhang, S.; Norris, T.B.; Kotov, N.A. Terahertz circular dichroism spectroscopy of biomaterials enabled by kirigami polarization modulators. Nat. Mater. 2019, 18, 820–826. [Google Scholar] [CrossRef]
- Piggot, T.J.; Holdbrook, D.A.; Khalid, S. Electroporation of the E. coli and S. aureus membranes: Molecular dynamics simulations of complex bacterial membranes. J. Phys. Chem. B 2011, 115, 13381–13388. [Google Scholar] [CrossRef]
- Brandt, E.G.; Braun, A.R.; Sachs, J.N.; Nagle, J.F.; Edholm, O. Interpretation of fluctuation spectra in lipid bilayer simulations. Biophys. J. 2011, 100, 2104–2111. [Google Scholar] [CrossRef]
- Braun, A.R.; Brandt, E.G.; Edholm, O.; Nagle, J.F.; Sachs, J.N. Determination of electron density profiles and area from simulations of undulating membranes. Biophys. J. 2011, 100, 2112–2120. [Google Scholar] [CrossRef] [PubMed]
- Boek, E.; Padding, J.; den Otter, W.K.; Briels, W.J. Mechanical properties of surfactant bilayer membranes from atomistic and coarse-grained molecular dynamics simulations. J. Phys. Chem. B 2005, 109, 19851–19858. [Google Scholar] [CrossRef] [PubMed]
- Stecki, J. Variation of lateral tension and a new transition in model bilayers made of chain molecules. J. Chem. Phys. 2005, 122, 111102. [Google Scholar] [CrossRef]
- Stecki, J. Note: On the power spectrum of undulations of simulated bilayers. J. Chem. Phys. 2012, 137, 116102. [Google Scholar] [CrossRef] [PubMed]
- Stecki, J. Correlations in simulated model bilayers. J. Chem. Phys. 2004, 120, 3508–3516. [Google Scholar] [CrossRef]
- Różycki, B.; Lipowsky, R. Spontaneous curvature of bilayer membranes from molecular simulations: Asymmetric lipid densities and asymmetric adsorption. J. Chem. Phys. 2015, 142, 02B601_1. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; He, Z.; Yu, W. Comparison of public peak detection algorithms for MALDI mass spectrometry data analysis. BMC Bioinform. 2009, 10, 1–13. [Google Scholar] [CrossRef]
- Lipkus, A.H.; Chittur, K.K.; Vesper, S.J.; Robinson, J.B.; Pierce, G.E. Evaluation of infrared spectroscopy as a bacterial identification method. J. Ind. Microbiol. 1990, 6, 71–75. [Google Scholar] [CrossRef]
- Helm, D.; Labischinski, H.; Naumann, D. Elaboration of a procedure for identification of bacteria using Fourier-transform IR spectral libraries: A stepwise correlation approach. J. Microbiol. Methods 1991, 14, 127–142. [Google Scholar] [CrossRef]
- Naumann, D. The ultra rapid differentiation and identification of pathogenic bacteria using FT-IR techniques. In Fourier and computerized Infrared Spectroscopy; SPIE: Bellingham, WA, USA, 1985; Volume 553, pp. 268–269. [Google Scholar]
- Horvath, S.E.; Daum, G. Lipids of mitochondria. Prog. Lipid Res. 2013, 52, 590–614. [Google Scholar] [CrossRef]
- Van Hal, S.J.; Jensen, S.O.; Vaska, V.L.; Espedido, B.A.; Paterson, D.L.; Gosbell, I.B. Predictors of mortality in Staphylococcus aureus bacteremia. Clin. Microbiol. Rev. 2012, 25, 362–386. [Google Scholar] [CrossRef]
- Kourtis, A.; Hatfield, K.; Baggs, J.; Mu, Y.; See, I.; Epson, E.; Nadle, J.; Kainer, M.; Dumyati, G.; Petit, S.; et al. Emerging Infections Program MRSA Author Group: Vital signs: Epidemiology and recent trends in methicillin-resistant and in methicillin-susceptible Staphylococcus aureus bloodstream infections-United States. MMWR Morb. Mortal Wkly. Rep. 2019, 68, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, D.J.; Haste, N.M.; Hollands, A.; Fleming, T.C.; Hamby, M.; Pogliano, K.; Nizet, V.; Dorrestein, P.C. Microbial competition between Bacillus subtilis and Staphylococcus aureus monitored by imaging mass spectrometry. Microbiology 2011, 157, 2485. [Google Scholar] [CrossRef] [PubMed]
- Rehal, R.P.; Marbach, H.; Hubbard, A.T.; Sacranie, A.A.; Sebastiani, F.; Fragneto, G.; Harvey, R.D. The influence of mild acidity on lysyl-phosphatidylglycerol biosynthesis and lipid membrane physico-chemical properties in methicillin-resistant Staphylococcus aureus. Chem. Phys. Lipids 2017, 206, 60–70. [Google Scholar] [CrossRef]
- Drew Bennett, W.; Fox, S.J.; Sun, D.; Maupin, C.M. Bacterial Membranes Are More Perturbed by the Asymmetric Versus Symmetric Loading of Amphiphilic Molecules. Membranes 2022, 12, 350. [Google Scholar] [CrossRef]
- White, D.C.; Frerman, F.E. Fatty acid composition of the complex lipids of Staphylococcus aureus during the formation of the membrane-bound electron transport system. J. Bacteriol. 1968, 95, 2198–2209. [Google Scholar] [CrossRef] [PubMed]
- Haest, C.; De Gier, J.; Den Kamp, J.O.; Bartels, P.; Van Deenen, L. Changes in permeability of Staphylococcus aureus and derived liposomes with varying lipid composition. Biochim. Biophys. Acta BBA-Biomembr. 1972, 255, 720–733. [Google Scholar] [CrossRef]
- Gould, R.M.; Lennarz, W. Metabolism of phosphatidylglycerol and lysyl phosphatidylglycerol in Staphylococcus aureus. J. Bacteriol. 1970, 104, 1135–1144. [Google Scholar] [CrossRef]
- Witzke, S.; Petersen, M.; Carpenter, T.S.; Khalid, S. Molecular dynamics simulations reveal the conformational flexibility of lipid II and its loose association with the defensin plectasin in the Staphylococcus aureus membrane. Biochemistry 2016, 55, 3303–3314. [Google Scholar] [CrossRef]
- Hayami, M.; Okabe, A.; Kariyama, R.; Abe, M.; Kanemasa, Y. Lipid composition of Staphylococcus aureus and its derived L-forms. Microbiol. Immunol. 1979, 23, 435–442. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Michaud-Agrawal, N.; Denning, E.J.; Woolf, T.B.; Beckstein, O. MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011, 32, 2319–2327.61. [Google Scholar] [CrossRef]
- Gowers, R.J.; Linke, M.; Barnoud, J.; Reddy, T.J.; Melo, M.N.; Seyler, S.L.; Domanski, J.; Dotson, D.L.; Buchoux, S.; Kenney, I.M.; et al. MDAnalysis: A Python package for the rapid analysis of molecular dynamics simulations. In Proceedings of the 15th Python in Science Conference (SciPy), Austin, TX, USA, 11–17 July 2016; Volume 98, p. 105. [Google Scholar]
- Ramírez, R.; López-Ciudad, T.; Kumar, P.P.; Marx, D. Quantum corrections to classical time-correlation functions: Hydrogen bonding and anharmonic floppy modes. J. Chem. Phys. 2004, 121, 3973–3983. [Google Scholar] [CrossRef]
- Wilmink, G.J.; Ibey, B.L.; Rivest, B.D.; Grundt, J.E.; Roach, W.P.; Tongue, T.D.; Schulkin, B.J.; Laman, N.; Peralta, X.G.; Roth, C.C.; et al. Development of a compact terahertz time-domain spectrometer for the measurement of the optical properties of biological tissues. J. Biomed. Opt. 2011, 16, 047006. [Google Scholar] [CrossRef]
- Hodges, J.L. The significance probability of the Smirnov two-sample test. Ark. Mat. 1958, 3, 469–486. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, B.; Wang, L.; Jing, T.; Chen, J.; Xu, X.; Zhang, W.; Zhang, Y.; Han, J. Unusual features and molecular pathways of Staphylococcus aureus L-form bacteria. Microb. Pathog. 2020, 140, 103970. [Google Scholar] [CrossRef]
- Nishiyama, Y.; Yamaguchi, H. Morphological Detection of Filipin-Sterol Complexes in the Cytoplasmic Membrane of Staphylococcal L-Form. Microbiol. Immunol. 1990, 34, 25–34. [Google Scholar] [CrossRef]
- Dufourc, E.J. Sterols and membrane dynamics. J. Chem. Biol. 2008, 1, 63–77. [Google Scholar] [CrossRef]
- Kilelee, E.; Pokorny, A.; Yeaman, M.R.; Bayer, A.S. Lysyl-phosphatidylglycerol attenuates membrane perturbation rather than surface association of the cationic antimicrobial peptide 6W-RP-1 in a model membrane system: Implications for daptomycin resistance. Antimicrob. Agents Chemother. 2010, 54, 4476–4479. [Google Scholar] [CrossRef] [PubMed]
- Wepy, J.A.; Galligan, J.J.; Kingsley, P.J.; Xu, S.; Goodman, M.C.; Tallman, K.A.; Rouzer, C.A.; Marnett, L.J. Lysophospholipases cooperate to mediate lipid homeostasis and lysophospholipid signaling [S]. J. Lipid Res. 2019, 60, 360–374. [Google Scholar] [CrossRef] [PubMed]
- Brown, W.J.; Chambers, K.; Doody, A. Phospholipase A2 (PLA2) enzymes in membrane trafficking: Mediators of membrane shape and function. Traffic 2003, 4, 214–221. [Google Scholar] [CrossRef]
- Grzelczyk, A.; Gendaszewska-Darmach, E. Novel bioactive glycerol-based lysophospholipids: New data–new insight into their function. Biochimie 2013, 95, 667–679. [Google Scholar] [CrossRef]
- Xu, W.; Hsu, F.F.; Baykal, E.; Huang, J.; Zhang, K. Sterol biosynthesis is required for heat resistance but not extracellular survival in Leishmania. PLoS Pathog. 2014, 10, e1004427. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, A.T.; Barker, R.; Rehal, R.; Vandera, K.K.A.; Harvey, R.D.; Coates, A.R. Mechanism of Action of a Membrane-Active Quinoline-Based Antimicrobial on Natural and Model Bacterial Membranes. Biochemistry 2017, 56, 1163–1174. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).