Adaptation of the Vaccine Prophylaxis Strategy to Variants of the SARS-CoV-2 Virus
Abstract
1. Introduction
2. Pre-Omicron Era: The Race Between Vaccine Development and SARS-CoV-2 Evolution
| Variant | Date of Emergence | Key Mutation | Reference | Key Features |
|---|---|---|---|---|
| G614 | April 2020 | D614G * | [23,33] | Enhanced viral replication by increased infectivity and stability of virion-ACE2 interaction |
| Alpha (B.1.1.7) | October 2020 (UK) | D614G, N501Y, P681H, R203K, G204R, D1118H, ΔH69/V70, ∆Y144 (+IE), A570D | [26,27,34,35] | Higher viral load Increased infectivity and infectiousness Improved binding to ACE2 receptor Enhanced S protein cleavage |
| Beta (B.1.351) | September 2020 (South Africa) | D614G, D215G, K417N (+IE), E484K, N501Y | [36] | Enhanced transmissibility Resistance to neutralizing antibodies |
| Gamma (P.1) | November 2020 (Brazil) | D614G, N501Y, E484K, K417T | [37,38] | Increased RBD affinity to ACE2 Increased transmissibility and virulence Immune evasion |
| Delta (B.1.617.2) | October 2020 (India) | D614G, L452R, T478K, P681R | [37,39,40] | Increased fusogenicity and infectivity 60% more transmissible than the Alpha variant Improved furin site recognition by proteases |
| Omicron BA.1 (B.1.1.529.1) | November 2021 (South Africa) | D614G, E484A, N501Y, Q493K, Q498R, K417N, S477N, Y505H, G496S, T478K, L452R S375F, S371L | [39,41,42,43,44] | Diminished fusogenicity, pathogenicity and cleavage efficacy (relative to Delta variant) Heightened transmissibility and infectivity Higher reinfection possibility Immune evasion |
| BA.2 (B.1.1.529.2) | November 2021 (India) | BA.1+ T19I, ∆PPA25–27, G142D, V231G, S371F, T376A, D405N, R408S | [45,46,47] | Increased transmissibility Improved fusogenicity Better S protein cleavage and ACE2 affinity (compared to BA.1) |
| XBB | August 2022 (India) | ∆Y144 (+IE), P681H V83A (+IE), H146Q, Q183E, V213E, R346T (+IE), N460K G339H, R368I, V445P, G446S, F490S, F486S (+IE) | [48,49,50] | Increasing its fitness through recombination rather than substitutions. (recombination of BJ.1 and BM.1.1.1) Improved transmissibility and Enhanced immune evasion (relative to BA.1 Omicron) |
| XBB.1 | August 2022 (India) | XBB+ G252V | [49] | Greater fusogenicity (in comparison to BA.2.75) Profound resistance to antiviral humoral immunity induced by prior Omicron subvariants |
| XBB.1.5—Kraken | December 2022 | XBB.1+ S486P | [51] | Enhanced binding affinity to ACE2 receptor (compared to XBB.1) Increased transmissibility Immune evasion capabilities are the same as XBB.1 |
| BA.2.86 | July 2023 (Israel, Denmark) | ∆Y144 (+IE) F157S, P681R, G339H, N460K, F486P, A484K, L452W (+IE), A445H (+IE), N450D, ∆N211, A264D, S50L, L216F, K356T (+IE), R403K | [52,53] | Substantial antigenic drift Enhanced receptor affinity |
| JN.1 | September 2023 (India) | BA.2.86+ L455S | [54,55] | Significantly improved fusogenicity Increased infectivity |
| SLip | JN.1+ F456L (Flip mutation) | [56,57] | Decreased infectivity and membrane fusion Declined spike processing compared to JN.1 Immune evasion | |
| FLiRT | SLip+ R346T | [57] | Immune evasion Decreased infectivity, cell-cell fusion, and spike processing relative to JN.1 | |
| KP.2 | January 2024 | Flirt+ V1140L | [58] | Immune evasion Decreased infectivity, cell-cell fusion, and spike processing relative to JN.1 |
| HK.3 | January 2023 (East Asia) | EG.5.1+ L455F (Flip mutation) | [54] | Enhanced immune evasion Increased fusogenicity (compared with EG.5.1) |
| KP.3 | February 2024 | JN.1+ Q493E | [59,60,61] | Diminished infectivity and affinity to ACE2 Immune evasion |
| KP.3.1.1 | March 2024 | KP.3+ ΔS31 | [61] | KP.3, KP.3.1.1 and XEC showed a significant increase in ACE2-Spike binding affinity compared with JN.1; no significant changes in the receptor binding of KP.3.1.1 and XEC relative to KP.3 |
| XEC | June 2024 | KP.3+ F59S, T22N | [61,62] | Enhanced binding affinity to ACE2 receptor in comparison to JN.1, but not to KP.3 Reduced cell–cell fusion relative to its parental KP.3 |
3. Vaccine Efficiency in the Omicron Landscape
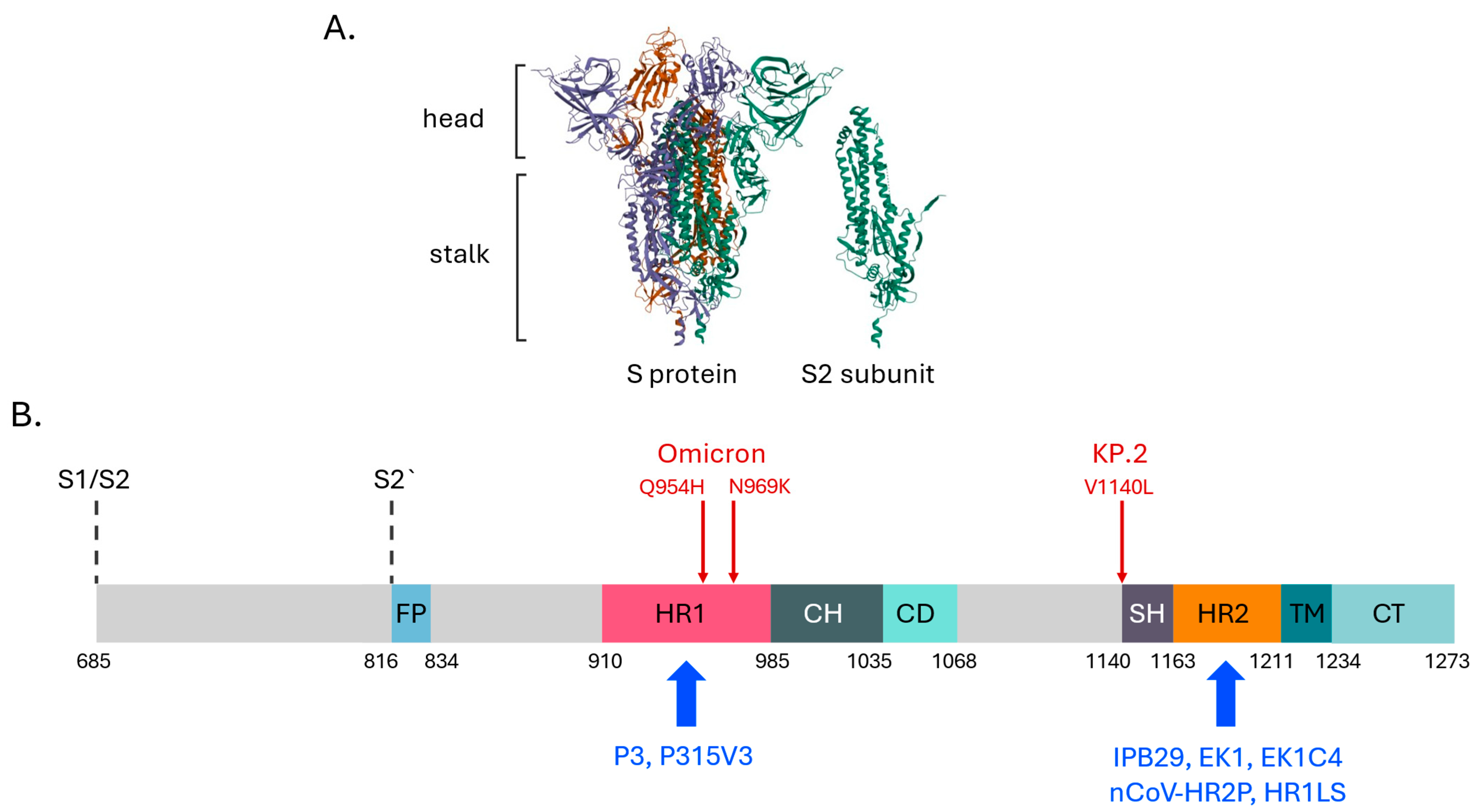
4. Prospects for Pan-Coronaviral Antivirals: The S2 Subunit in the Spotlight
5. Conclusions and Future Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-NCoV and Naming It SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- National Library for Medicine. Available online: https://www.ncbi.nlm.nih.gov/datasets/taxonomy/3418604/ (accessed on 8 July 2025).
- Soheili, M.; Khateri, S.; Moradpour, F.; Mohammadzedeh, P.; Zareie, M.; Mortazavi, S.M.M.; Manifar, S.; Kohan, H.G.; Moradi, Y. The Efficacy and Effectiveness of COVID-19 Vaccines around the World: A Mini-Review and Meta-Analysis. Ann. Clin. Microbiol. Antimicrob. 2023, 22, 42. [Google Scholar] [CrossRef] [PubMed]
- Sharif, N.; Alzahrani, K.J.; Ahmed, S.N.; Dey, S.K. Efficacy, Immunogenicity and Safety of COVID-19 Vaccines: A Systematic Review and Meta-Analysis. Front. Immunol. 2021, 12, 714170. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human Neutralizing Antibodies Elicited by SARS-CoV-2 Infection. Nature 2020, 584, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Papageorgiou, A.C.; Mohsin, I. The SARS-CoV-2 Spike Glycoprotein as a Drug and Vaccine Target: Structural Insights into Its Complexes with ACE2 and Antibodies. Cells 2020, 9, 2343. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Sette, A. From Alpha to Omicron: The Response of T Cells. Curr. Res. Immunol. 2022, 3, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Gushchin, V.A.; Pochtovyi, A.A.; Kustova, D.D.; Ogarkova, D.A.; Tarnovetskii, I.Y.; Belyaeva, E.D.; Divisenko, E.V.; Vasilchenko, L.A.; Shidlovskaya, E.V.; Kuznetsova, N.A.; et al. Dynamics of SARS-CoV-2 Major Genetic Lineages in Moscow in the Context of Vaccine Prophylaxis. Int. J. Mol. Sci. 2022, 23, 14670. [Google Scholar] [CrossRef] [PubMed]
- Malheiro, D.T.; Parreira, K.C.J.; Celeghini, P.D.; Callado, G.Y.; Cotia, A.L.F.; Cendoroglo Neto, M.; Bragatte, M.A.S.; Negretto Schrarstzhaupt, I.; Sampaio, V.; Kobayashi, T.; et al. COVID-19 Reinfections in the City of São Paulo, Brazil: Prevalence and Socioeconomic Factors. Open Forum Infect. Dis. 2025, 12, ofaf181. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Yang, J.; He, X.; Hong, W.; Lei, H.; Chen, Z.; Shen, G.; Yang, L.; Li, J.; Wang, Z.; et al. A Bivalent Recombinant Vaccine Targeting the S1 Protein Induces Neutralizing Antibodies against Both SARS-CoV-2 Variants and Wild-Type of the Virus. MedComm 2021, 2, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Heidary, M.; Kaviar, V.H.; Shirani, M.; Ghanavati, R.; Motahar, M.; Sholeh, M.; Ghahramanpour, H.; Khoshnood, S. A Comprehensive Review of the Protein Subunit Vaccines Against COVID-19. Front. Microbiol. 2022, 13, 927306. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.-W.; Fan, Y.; Tang, K.; Ong, C.P.; Luo, C.; Chung, H.-L.; Leong, T.-L.; Liang, R.; Lui, W.-Y.; Zhou, R.; et al. Cross-Variant Protection against SARS-CoV-2 Infection in Hamsters Immunized with Monovalent and Bivalent Inactivated Vaccines. Int. J. Biol. Sci. 2022, 18, 4781–4791. [Google Scholar] [CrossRef] [PubMed]
- Chalkias, S.; Harper, C.; Vrbicky, K.; Walsh, S.R.; Essink, B.; Brosz, A.; McGhee, N.; Tomassini, J.E.; Chen, X.; Chang, Y.; et al. A Bivalent Omicron-Containing Booster Vaccine against Covid-19. N. Engl. J. Med. 2022, 387, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wang, Z.; Lu, S.; Qi, Z.; Zhang, Z.; Luan, J.; Ba, J. Monitoring Reported SARS-CoV-2 Variants to Assess the Status of COVID-19 Epidemics in the Low Epidemic State. Sci. Rep. 2025, 15, 10169. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, S.; Dehkharghani, M.Z.; Fudolig, M. Emergence to Dominance: Estimating Time to Dominance of SARS-CoV-2 Variants Using Nonlinear Statistical Models. PLoS ONE 2025, 20, e0311459. [Google Scholar] [CrossRef] [PubMed]
- Abavisani, M.; Rahimian, K.; Mahdavi, B.; Tokhanbigli, S.; Mollapour Siasakht, M.; Farhadi, A.; Kodori, M.; Mahmanzar, M.; Meshkat, Z. Mutations in SARS-CoV-2 Structural Proteins: A Global Analysis. Virol. J. 2022, 19, 220. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.; Akter, S.; Rashid, A.A.; Khair, S.; Alam, A.S.M.R.U. Unique Mutations in SARS-CoV-2 Omicron Subvariants’ Non-Spike Proteins: Potential Impacts on Viral Pathogenesis and Host Immune Evasion. Microb. Pathog. 2022, 170, 105699. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Weeraratne, D.; Snowdon, J.L.; Parida, L. Emergence of Drift Variants That May Affect COVID-19 Vaccine Development and Antibody Treatment. Pathogens 2020, 9, 324. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Platt, D.; Parida, L. Variant Analysis of SARS-CoV-2 Genomes. Bull. World Health Organ. 2020, 98, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike Mutation D614G Alters SARS-CoV-2 Fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.J.; Chiba, S.; Halfmann, P.; Ehre, C.; Kuroda, M.; Dinnon, K.H., 3rd; Leist, S.R.; Schäfer, A.; Nakajima, N.; Takahashi, K.; et al. SARS-CoV-2 D614G Variant Exhibits Efficient Replication Ex Vivo and Transmission in Vivo. Science 2020, 370, 1464–1468. [Google Scholar] [CrossRef] [PubMed]
- Morbidity and Mortality Weekly Report (MMWR). Available online: https://www.cdc.gov/mmwr/volumes/70/wr/mm7003e2.htm?utm_source=mp-fotoscapes#suggestedcitation (accessed on 25 April 2025).
- Rajah, M.M.; Hubert, M.; Bishop, E.; Saunders, N.; Robinot, R.; Grzelak, L.; Planas, D.; Dufloo, J.; Gellenoncourt, S.; Bongers, A.; et al. SARS-CoV-2 Alpha, Beta, and Delta Variants Display Enhanced Spike-mediated Syncytia Formation. EMBO J. 2021, 40, e108944. [Google Scholar] [CrossRef] [PubMed]
- Telenti, A.; Hodcroft, E.B.; Robertson, D.L. The Evolution and Biology of SARS-CoV-2 Variants. Cold Spring Harb. Perspect. Med. 2022, 12, a041390. [Google Scholar] [CrossRef] [PubMed]
- Ikegame, S.; Siddiquey, M.N.A.; Hung, C.-T.; Haas, G.; Brambilla, L.; Oguntuyo, K.Y.; Kowdle, S.; Chiu, H.-P.; Stevens, C.S.; Vilardo, A.E.; et al. Neutralizing Activity of Sputnik V Vaccine Sera against SARS-CoV-2 Variants. Nat. Commun. 2021, 12, 4598. [Google Scholar] [CrossRef] [PubMed]
- Muik, A.; Wallisch, A.-K.; Sänger, B.; Swanson, K.A.; Mühl, J.; Chen, W.; Cai, H.; Maurus, D.; Sarkar, R.; Türeci, Ö.; et al. Neutralization of SARS-CoV-2 Lineage B.1.1.7 Pseudovirus by BNT162b2 Vaccine–Elicited Human Sera. Science 2021, 371, 1152–1153. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Tang, H.; McDanal, C.; Wagh, K.; Fischer, W.; Theiler, J.; Yoon, H.; Li, D.; Haynes, B.F.; Sanders, K.O.; et al. SARS-CoV-2 Variant B.1.1.7 Is Susceptible to Neutralizing Antibodies Elicited by Ancestral Spike Vaccines. Cell Host Microbe 2021, 29, 529–539.e3. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shen, H.; Huang, R.; Tong, X.; Wu, C. Serum Neutralising Activity against SARS-CoV-2 Variants Elicited by CoronaVac. Lancet Infect. Dis. 2021, 21, 1071–1072. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Shayestehpour, M.; Mirzaei, H. The Impact of Spike Mutated Variants of SARS-CoV2 [Alpha, Beta, Gamma, Delta, and Lambda] on the Efficacy of Subunit Recombinant Vaccines. Brazilian J. Infect. Dis. 2021, 25, 101606. [Google Scholar] [CrossRef] [PubMed]
- Plante, J.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.; Lokugamage, K.; Zhang, X.; Muruato, A.; Zou, J.; Fontes-Garfias, C.; et al. Spike Mutation D614G AltersSARS-CoV-2 Fitness and Neutralization Susceptibility. Nature 2020, 592, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The Evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, C.; Wang, Y.; Hong, Q.; Zhang, C.; Li, Z.; Xu, S.; Zuo, Q.; Liu, C.; Huang, Z.; et al. Conformational Dynamics of the Beta and Kappa SARS-CoV-2 Spike Proteins and Their Complexes with ACE2 Receptor Revealed by Cryo-EM. Nat. Commun. 2021, 12, 7345. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.H.; Krieger, J.M.; Banerjee, A.; Xiang, Y.; Kaynak, B.; Shi, Y.; Arditi, M.; Bahar, I. Impact of New Variants on SARS-CoV-2 Infectivity and Neutralization: A Molecular Assessment of the Alterations in the Spike-Host Protein Interactions. iScience 2022, 25, 103939. [Google Scholar] [CrossRef] [PubMed]
- Padilha, D.A.; Benetti Filho, V.; Moreira, R.S.; Soratto, T.A.; Maia, G.A.; Christoff, A.P.; Barazzetti, F.H.; Schörner, M.A.; Ferrari, F.L.; Martins, C.L.; et al. Emergence of Two Distinct SARS-CoV-2 Gamma Variants and the Rapid Spread of P.1-like-II SARS-CoV-2 during the Second Wave of COVID-19 in Santa Catarina, Southern Brazil. Viruses 2022, 14, 695. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, T.; Fang, Y.; Liu, J.; Ye, Q.; Ding, L. SARS-CoV-2 Spike L452R Mutation Increases Omicron Variant Fusogenicity and Infectivity as Well as Host Glycolysis. Signal Transduct. Target. Ther. 2022, 7, 76. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced Sensitivity of SARS-CoV-2 Variant Delta to Antibody Neutralization. Nature 2021, 596, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Syed, A.M.; Ciling, A.; Taha, T.Y.; Chen, I.P.; Khalid, M.M.; Sreekumar, B.; Chen, P.-Y.; Kumar, G.R.; Suryawanshi, R.; Silva, I.; et al. Omicron Mutations Enhance Infectivity and Reduce Antibody Neutralization of SARS-CoV-2 Virus-like Particles. Proc. Natl. Acad. Sci. USA 2022, 119, e2200592119. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Bhattacharya, M.; Sharma, A.R.; Mallik, B. Omicron (B.1.1.529)—A New Heavily Mutated Variant: Mapped Location and Probable Properties of Its Mutations with an Emphasis on S-Glycoprotein. Int. J. Biol. Macromol. 2022, 219, 980–997. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Li, T.; Lai, K.K.; Johnson, R.F.; Yewdell, J.W.; Compton, A.A. Omicron Spike Confers Enhanced Infectivity and Interferon Resistance to SARS-CoV-2 in Human Nasal Tissue. Nat. Commun. 2024, 15, 889. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, L.B.; Foster, C.; Rawlinson, W.; Tedla, N.; Bull, R.A. Evolution of the SARS-CoV-2 Omicron Variants BA.1 to BA.5: Implications for Immune Escape and Transmission. Rev. Med. Virol. 2022, 32, e2381. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liao, H.; Meng, Y.; Li, W.; Han, P.; Liu, K.; Wang, Q.; Li, D.; Zhang, Y.; Wang, L.; et al. Structural Basis of Human ACE2 Higher Binding Affinity to Currently Circulating Omicron SARS-CoV-2 Sub-Variants BA.2 and BA.1.1. Cell 2022, 185, 2952–2960.e10. [Google Scholar] [CrossRef] [PubMed]
- Izumi, K.; Daichi, Y.; Hesham, N.; Hayato, I.; Jiri, Z.; Jiaqi, W.; Shigeru, F.; Keiya, U.; Jiei, S.; Tomokazu, T.; et al. Multiple Mutations of SARS-CoV-2 Omicron BA.2 Variant Orchestrate Its Virological Characteristics. J. Virol. 2023, 97, e01011-23. [Google Scholar] [CrossRef]
- Kannan, S.R.; Spratt, A.N.; Sharma, K.; Goyal, R.; Sönnerborg, A.; Apparsundaram, S.; Lorson, C.L.; Byrareddy, S.N.; Singh, K. Complex Mutation Pattern of Omicron BA.2: Evading Antibodies without Losing Receptor Interactions. Int. J. Mol. Sci. 2022, 23, 5534. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Bhattacharya, M.; Chopra, H.; Islam, M.A.; Saikumar, G.; Dhama, K. The SARS-CoV-2 Omicron Recombinant Subvariants XBB, XBB.1, and XBB.1.5 Are Expanding Rapidly with Unique Mutations, Antibody Evasion, and Immune Escape Properties—An Alarming Global Threat of a Surge in COVID-19 Cases Again? Int. J. Surg. 2023, 109, 1041–1043. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Ito, J.; Uriu, K.; Zahradnik, J.; Kida, I.; Anraku, Y.; Nasser, H.; Shofa, M.; Oda, Y.; Lytras, S.; et al. Virological Characteristics of the SARS-CoV-2 XBB Variant Derived from Recombination of Two Omicron Subvariants. Nat. Commun. 2023, 14, 2800. [Google Scholar] [CrossRef] [PubMed]
- Giancotti, R.; Lomoio, U.; Puccio, B.; Tradigo, G.; Vizza, P.; Torti, C.; Veltri, P.; Guzzi, P.H. The Omicron XBB.1 Variant and Its Descendants: Genomic Mutations, Rapid Dissemination and Notable Characteristics. Biology 2024, 13, 90. [Google Scholar] [CrossRef] [PubMed]
- Criscuolo, E.; Giuliani, B.; Castelli, M.; Cavallaro, M.; Sisti, S.; Burioni, R.; Ferrari, D.; Mancini, N.; Locatelli, M.; Clementi, N. Single Spike Mutation Differentiating XBB.1 and XBB.1.5 Enhances SARS-CoV-2 Cell-to-Cell Transmission and Facilitates Serum-Mediated Enhancement. Front. Immunol. 2024, 15, 1501200. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, L.; Jiang, S. SARS-CoV-2 Omicron Subvariant BA.2.86: Limited Potential for Global Spread. Signal Transduct. Target. Ther. 2023, 8, 439. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Mizuma, K.; Nasser, H.; Deguchi, S.; Padilla-Blanco, M.; Oda, Y.; Uriu, K.; Tolentino, J.E.M.; Tsujino, S.; Suzuki, R.; et al. Virological Characteristics of the SARS-CoV-2 BA.2.86 Variant. Cell Host Microbe 2024, 32, 170–180.e12. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhao, X.; Shi, J.; Wang, Y.; Liu, H.; Hu, Y.-F.; Hu, B.; Shuai, H.; Yuen, T.T.-T.; Chai, Y.; et al. Lineage-Specific Pathogenicity, Immune Evasion, and Virological Features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3. Nat. Commun. 2024, 15, 8728. [Google Scholar] [CrossRef] [PubMed]
- Selvavinayagam, S.T.; Sankar, S.; Yong, Y.K.; Murugesan, A.; Suvaithenamudhan, S.; Hemashree, K.; Rajeshkumar, M.; Kumaresan, A.; Pandey, R.P.; Shanmugam, S.; et al. Emergence of SARS-CoV-2 Omicron Variant JN.1 in Tamil Nadu, India—Clinical Characteristics and Novel Mutations. Sci. Rep. 2024, 14, 17476. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; Spezia, P.G.; Gueli, F.; Maggi, F. The Era of the FLips: How Spike Mutations L455F and F456L (and A475V) Are Shaping SARS-CoV-2 Evolution. Viruses 2024, 16, 3. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Faraone, J.N.; Hsu, C.C.; Chamblee, M.; Zheng, Y.; Carlin, C.; Bednash, J.S.; Horowitz, J.C.; Mallampalli, R.K.; Saif, J.; et al. Neutralization Escape, Infectivity, and Membrane Fusion of JN.1-Derived SARS-CoV-2 SLip, FLiRT, and KP.2 Variants. Cell Rep. 2024, 44, 114520. [Google Scholar] [CrossRef]
- Li, P.; Faraone, J.N.; Hsu, C.C.; Chamblee, M.; Zheng, Y.-M.; Carlin, C.; Bednash, J.S.; Horowitz, J.C.; Mallampalli, R.K.; Saif, L.J.; et al. Characteristics of JN.1-Derived SARS-CoV-2 Subvariants SLip, FLiRT, and KP.2 in Neutralization Escape, Infectivity and Membrane Fusion. bioRxiv Prepr. Serv. Biol. 2024. preprint. [Google Scholar] [CrossRef]
- Xu, K.; An, Y.; Liu, X.; Xie, H.; Li, D.; Yang, T.; Duan, M.; Wang, Y.; Zhao, X.; Dai, L.; et al. Neutralization of SARS-CoV-2 KP.1, KP.1.1, KP.2 and KP.3 by Human and Murine Sera. Npj Vaccines 2024, 9, 215. [Google Scholar] [CrossRef] [PubMed]
- Kaku, Y.; Uriu, K.; Kosugi, Y.; Okumura, K.; Yamasoba, D.; Uwamino, Y.; Kuramochi, J.; Sadamasu, K.; Yoshimura, K.; Asakura, H.; et al. Virological Characteristics of the SARS-CoV-2 KP.2 Variant. Lancet Infect. Dis. 2024, 24, e416. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yu, Y.; Jian, F.; Yang, S.; Song, W.; Wang, P.; Yu, L.; Shao, F.; Cao, Y. Enhanced Immune Evasion of SARS-CoV-2 Variants KP.3.1.1 and XEC through N-Terminal Domain Mutations. Lancet Infect. Dis. 2025, 25, e6–e7. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Faraone, J.N.; Hsu, C.C.; Chamblee, M.; Liu, Y.; Zheng, Y.-M.; Xu, Y.; Carlin, C.; Horowitz, J.C.; Mallampalli, R.K.; et al. Role of Glycosylation Mutations at the N-Terminal Domain of SARS-CoV-2 XEC Variant in Immune Evasion, Cell-Cell Fusion, and Spike Stability. J. Virol. 2025, 99, e00242-25. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 Variant of Concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Widera, M.; Wilhelm, A.; Hoehl, S.; Pallas, C.; Kohmer, N.; Wolf, T.; Rabenau, H.F.; Corman, V.M.; Drosten, C.; Vehreschild, M.J.G.T.; et al. Limited Neutralization of Authentic Severe Acute Respiratory Syndrome Coronavirus 2 Variants Carrying E484K In Vitro. J. Infect. Dis. 2021, 224, 1109–1114. [Google Scholar] [CrossRef] [PubMed]
- Chemaitelly, H.; Yassine, H.M.; Benslimane, F.M.; Al Khatib, H.A.; Tang, P.; Hasan, M.R.; Malek, J.A.; Coyle, P.; Ayoub, H.H.; Al Kanaani, Z.; et al. MRNA-1273 COVID-19 Vaccine Effectiveness against the B.1.1.7 and B.1.351 Variants and Severe COVID-19 Disease in Qatar. Nat. Med. 2021, 27, 1614–1621. [Google Scholar] [CrossRef] [PubMed]
- Singer, S.R.; Angulo, F.J.; Swerdlow, D.L.; McLaughlin, J.M.; Hazan, I.; Ginish, N.; Anis, E.; Mendelson, E.; Mor, O.; Zuckerman, N.S.; et al. Effectiveness of BNT162b2 MRNA COVID-19 Vaccine against SARS-CoV-2 Variant Beta (B.1.351) among Persons Identified through Contact Tracing in Israel: A Prospective Cohort Study. eClinicalMedicine 2021, 42, 101190. [Google Scholar] [CrossRef] [PubMed]
- Madhi, S.A.; Baillie, V.; Cutland, C.L.; Voysey, M.; Koen, A.L.; Fairlie, L.; Padayachee, S.D.; Dheda, K.; Barnabas, S.L.; Bhorat, Q.E.; et al. Efficacy of the ChAdOx1 NCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021, 384, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Dejnirattisai, W.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.E.; Tuekprakhon, A.; Nutalai, R.; et al. Evidence of Escape of SARS-CoV-2 Variant B.1.351 from Natural and Vaccine-Induced Sera. Cell 2021, 184, 2348–2361.e6. [Google Scholar] [CrossRef] [PubMed]
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; da S Candido, D.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.S.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and Epidemiology of the P.1 SARS-CoV-2 Lineage in Manaus, Brazil. Science 2021, 821, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Luna-Muschi, A.; Borges, I.C.; de Faria, E.; Barboza, A.S.; Maia, F.L.; Leme, M.D.; Guedes, A.R.; Mendes-Correa, M.C.; Kallas, E.G.; Segurado, A.C.; et al. Clinical features of COVID-19 by SARS-CoV-2 Gamma variant: A prospective cohort study of vaccinated and unvaccinated healthcare workers. J. Infect. 2022, 84, 248–288. [Google Scholar] [CrossRef]
- Banho, C.A.; Sacchetto, L.; Rodrigues, G.; Campos, F.; Bittar, C.; Possebon, F.S.; Ullmann, L.S.; Marques, B.D.C.; Ceslestino, G.; Moraes, M.M.; et al. Impact of SARS-CoV-2 Gamma Lineage Introduction and COVID-19 Vaccination on the epidemiological landscape of a Brazilian city. Commun. Med. 2022, 2, 41. [Google Scholar] [CrossRef] [PubMed]
- Hitchings, M.D.T.; Ranzani, O.T.; Dorion, M.; D’aGostini, T.L.; de Paula, R.C.; de Paula, O.F.P.; Villela, E.F.d.M.; Torres, M.S.S.; de Oliveira, S.B.; Schulz, W.; et al. Effectiveness of ChAdOx1 Vaccine in Older Adults. Nat. Commun. 2021, 12, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Redd, A.D.; Nardin, A.; Kared, H.; Bloch, E.M.; Pekosz, A.; Laeyendecker, O.; Abel, B.; Fehlings, M.; Quinn, T.C.; Tobian, A.A.R. CD8+ T-Cell Responses in COVID-19 Convalescent Individuals Target Conserved Epitopes From Multiple Prominent SARS-CoV-2 Circulating Variants. Open Forum Infect. Dis. 2021, 8, ofab143. [Google Scholar] [CrossRef] [PubMed]
- Hart, W.S.; Miller, E.; Andrews, N.J.; Waight, P.; Maini, P.K.; Funk, S.; Thompson, R.N. Generation Time of the Alpha and Delta SARS-CoV-2 Variants: An Epidemiological Analysis. Lancet Infect. Dis. 2022, 22, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Johnson, B.A.; Xia, H.; Ku, Z.; Schindewolf, C.; Widen, S.G.; An, Z.; Weaver, S.C.; Menachery, V.D.; et al. Delta Spike P681R Mutation Enhances SARS-CoV-2 Fitness over Alpha Variant. Cell Rep. 2022, 39, 110829. [Google Scholar] [CrossRef] [PubMed]
- Hattab, D.; Amer, M.F.A.; Al-Alami, Z.M.; Bakhtiar, A. SARS-CoV-2 Journey: From Alpha Variant to Omicron and Its Sub-Variants. Infection 2024, 52, 767–786. [Google Scholar] [CrossRef] [PubMed]
- Pouwels, K.B.; Pritchard, E.; Matthews, P.C.; Stoesser, N.; Eyre, D.W.; Vihta, K.D.; House, T.; Hay, J.; Bell, J.I.; Newton, J.N.; et al. Effect of Delta Variant on Viral Burden and Vaccine Effectiveness against New SARS-CoV-2 Infections in the UK. Nat. Med. 2021, 27, 2127–2135. [Google Scholar] [CrossRef] [PubMed]
- Eyre, D.W.; Taylor, D.; Purver, M.; Chapman, D.; Fowler, T.; Pouwels, K.B.; Walker, A.S.; Peto, T.E.A. Effect of Covid-19 Vaccination on Transmission of Alpha and Delta Variants. N. Engl. J. Med. 2022, 386, 744–756. [Google Scholar] [CrossRef] [PubMed]
- Vikkurthi, R.; Ansari, A.; Pai, A.R.; Jha, S.N.; Sachan, S.; Pandit, S.; Nikam, B.; Kalia, A.; Jit, B.P.; Parray, H.A.; et al. Inactivated Whole-Virion Vaccine BBV152/Covaxin Elicits Robust Cellular Immune Memory to SARS-CoV-2 and Variants of Concern. Nat. Microbiol. 2022, 7, 974–985. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Zhang, X.; Gong, T.; Ma, J.; Zhang, P.; Cai, Z.; Ren, D.; Zhang, C. A Systematic Mutation Analysis of 13 Major SARS-CoV-2 Variants. Virus Res. 2024, 345, 199392. [Google Scholar] [CrossRef] [PubMed]
- Chemaitelly, H.; Ayoub, H.H.; AlMukdad, S.; Coyle, P.; Tang, P.; Yassine, H.M.; Al-Khatib, H.A.; Smatti, M.K.; Hasan, M.R.; Al-Kanaani, Z.; et al. Duration of MRNA Vaccine Protection against SARS-CoV-2 Omicron BA.1 and BA.2 Subvariants in Qatar. Nat. Commun. 2022, 13, 3082. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.; Wang, X.; He, X.; Zhao, X.; Zhang, Y.; Jiang, Y.; Li, M.; Cui, Y.; Chen, Y.; Qiao, R.; et al. Antibody Evasion of SARS-CoV-2 Omicron BA.1, BA.1.1, BA.2, and BA.3 Sub-Lineages. Cell Host Microbe 2022, 30, 1077–1083.e4. [Google Scholar] [CrossRef] [PubMed]
- Seki, Y.; Yoshihara, Y.; Nojima, K.; Momose, H.; Fukushi, S.; Moriyama, S.; Wagatsuma, A.; Numata, N.; Sasaki, K.; Kuzuoka, T.; et al. Safety and Immunogenicity of the Pfizer/BioNTech SARS-CoV-2 MRNA Third Booster Vaccine Dose against the BA.1 and BA.2 Omicron Variants. Med 2022, 3, 406–421.e4. [Google Scholar] [CrossRef] [PubMed]
- Petrelli, F.; Luciani, A.; Borgonovo, K.; Ghilardi, M.; Parati, M.C.; Petrò, D.; Lonati, V.; Pesenti, A.; Cabiddu, M. Third Dose of SARS-CoV-2 Vaccine: A Systematic Review of 30 Published Studies. J. Med. Virol. 2022, 94, 2837–2844. [Google Scholar] [CrossRef] [PubMed]
- Vasin, A.V.; Stukova, M.A. Bivalent Omicron (BA.1) Booster Vaccination against SARS-CoV-2. Lancet Infect. Dis. 2023, 23, 880–881. [Google Scholar] [CrossRef] [PubMed]
- Bhiman, J.N.; Richardson, S.I.; Lambson, B.E.; Kgagudi, P.; Mzindle, N.; Kaldine, H.; Crowther, C.; Gray, G.; Bekker, L.-G.; Koen, A.; et al. Novavax NVX-COV2373 Triggers Neutralization of Omicron Sub-Lineages. Sci. Rep. 2023, 13, 1222. [Google Scholar] [CrossRef] [PubMed]
- Qu, P.; Xu, K.; Faraone, J.N.; Goodarzi, N.; Zheng, Y.M.; Carlin, C.; Bednash, J.S.; Horowitz, J.C.; Mallampalli, R.K.; Saif, L.J.; et al. Immune Evasion, Infectivity, and Fusogenicity of SARS-CoV-2 BA.2.86 and FLip Variants. Cell 2024, 187, 585–595.e6. [Google Scholar] [CrossRef] [PubMed]
- Qu, P.; Faraone, J.N.; Evans, J.P.; Zheng, Y.M.; Carlin, C.; Anghelina, M.; Stevens, P.; Fernandez, S.; Jones, D.; Panchal, A.R.; et al. Enhanced Evasion of Neutralizing Antibody Response by Omicron XBB.1.5, CH.1.1, and CA.3.1 Variants. Cell Rep. 2023, 42, 112443. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jiang, S.; Ma, W.; Li, X.; Wei, K.; Xie, F.; Zhao, C.; Zhao, X.; Wang, S.; Li, C.; et al. Enhanced Neutralization of SARS-CoV-2 Variant BA.2.86 and XBB Sub-Lineages by a Tetravalent COVID-19 Vaccine Booster. Cell Host Microbe 2024, 32, 25–34.e5. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Hong, W.; Shi, H.; He, C.; Lei, H.; Zhou, Y.; Yang, H.; Alu, A.; Chen, Z.; Yang, Y.; et al. Trivalent Recombinant Protein Vaccine Induces Cross-Neutralization against XBB Lineage and JN.1 Subvariants: Preclinical and Phase 1 Clinical Trials. Nat. Commun. 2024, 15, 10778. [Google Scholar] [CrossRef] [PubMed]
- Klotz, S.A.; Yates, S.; Smith, S.L.; Dudley, S.; Schmidt, J.O.; Shirazi, F.M. Neutralization of BA.4–BA.5, BA.4.6, BA.2.75.2, BQ.1.1, and XBB.1 with Bivalent Vaccine. N. Engl. J. Med. 2023, 388, 853–854. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhao, J.; Zhu, X.; Guan, Q.; Liu, S.; Li, M.; Gao, J.; Tan, J.; Cao, F.; Gan, B.; et al. Efficacy of the Tetravalent Protein COVID-19 Vaccine, SCTV01E: A Phase 3 Double-Blind, Randomized, Placebo-Controlled Trial. Nat. Commun. 2024, 15, 6255. [Google Scholar] [CrossRef] [PubMed]
- Chenchula, S.; Karunakaran, P.; Sharma, S.; Chavan, M. Current Evidence on Efficacy of COVID-19 Booster Dose Vaccination against the Omicron Variant: A Systematic Review. J. Med. Virol. 2022, 94, 2969–2976. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, Y.; Bowen, A.; Mellis, I.A.; Valdez, R.; Gherasim, C.; Gordon, A.; Liu, L.; Ho, D.D. XBB.1.5 Monovalent MRNA Vaccine Booster Elicits Robust Neutralizing Antibodies against XBB Subvariants and JN.1. Cell Host Microbe 2024, 32, 315–321.e3. [Google Scholar] [CrossRef] [PubMed]
- Alves, K.; Kotloff, K.; McClelland, R.S.; Kouassi, A.; Plested, J.S.; Kalkeri, R.; Zhu, M.; Cloney-Clark, S.; Cai, Z.; Smith, K.; et al. Immunogenicity and Safety of a Monovalent Omicron XBB.1.5 SARS-CoV-2 Recombinant Spike Protein Vaccine as a Heterologous Booster Dose in US Adults: Interim Analysis of a Single-Arm Phase 2/3 Study. Lancet Infect. Dis. 2025, 25, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Nesamari, R.; Omondi, M.A.; Baguma, R.; Höft, M.A.; Ngomti, A.; Nkayi, A.A.; Besethi, A.S.; Magugu, S.F.J.; Mosala, P.; Walters, A.; et al. Post-Pandemic Memory T Cell Response to SARS-CoV-2 Is Durable, Broadly Targeted, and Cross-Reactive to the Hypermutated BA.2.86 Variant. Cell Host Microbe 2024, 32, 162–169.e3. [Google Scholar] [CrossRef] [PubMed]
- Jian, F.; Wang, J.; Yisimayi, A.; Song, W.; Xu, Y.; Chen, X.; Niu, X.; Yang, S.; Yu, Y.; Wang, P.; et al. Evolving Antibody Response to SARS-CoV-2 Antigenic Shift from XBB to JN.1. Nature 2025, 637, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Kaku, Y.; Okumura, K.; Padilla-Blanco, M.; Kosugi, Y.; Uriu, K.; Hinay, A.A.; Chen, L.; Plianchaisuk, A.; Kobiyama, K.; Ishii, K.J.; et al. Virological Characteristics of the SARS-CoV-2 JN.1 Variant. Lancet Infect. Dis. 2024, 24, e82. [Google Scholar] [CrossRef] [PubMed]
- CDC COVID Data Tracker. Available online: https://covid.cdc.gov/covid-data-tracker/#variant-summary (accessed on 23 March 2025).
- Kosugi, Y.; Plianchaisuk, A.; Putri, O.; Uriu, K.; Kaku, Y.; Hinay, A.A.; Chen, L.; Kuramochi, J.; Sadamasu, K.; Yoshimura, K.; et al. Characteristics of the SARS-CoV-2 Omicron HK.3 Variant Harbouring the FLip Substitution. Lancet Microbe 2024, 5, e313. [Google Scholar] [CrossRef] [PubMed]
- Happle, C.; Hoffmann, M.; Kempf, A.; Nehlmeier, I.; Stankov, M.V.; Calderon Hampel, N.; Witte, T.; Pöhlmann, S.; Behrens, G.M.N.; Dopfer-Jablonka, A. Humoral Immunity after MRNA SARS-CoV-2 Omicron JN.1 Vaccination. Lancet Infect. Dis. 2024, 24, 674–676. [Google Scholar] [CrossRef] [PubMed]
- Uriu, K.; Kaku, Y.; Uwamino, Y.; Fujiwara, H.; Saito, F.; Sato, K. Antiviral Humoral Immunity Induced by JN.1 Monovalent MRNA Vaccines against SARS-CoV-2 Omicron Subvariants Including JN.1, KP.3.1.1, and XEC. Lancet Infect. Dis. 2025, 25, e61. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Then, E.; Miric, M.; Qian, H.-Z.; Chen, Y.Q.; Wang, Y.; Vallejo, V.; Quezada, W.; Flaquer, M.; Olivo, J.; Castillo, J.; et al. Population-Level Effectiveness of an Inactivated Whole-Virion COVID-19 Vaccine: A Test Negative Case-Control Study in the Dominican Republic. Open Forum Infect. Dis. 2023, 10, ofad075. [Google Scholar] [CrossRef] [PubMed]
- Hitchings, M.D.T.; Ranzani, O.T.; Torres, M.S.S.; de Oliveira, S.B.; Almiron, M.; Said, R.; Borg, R.; Schulz, W.L.; de Oliveira, R.D.; da Silva, P.V.; et al. Effectiveness of CoronaVac among Healthcare Workers in the Setting of High SARS-CoV-2 Gamma Variant Transmission in Manaus, Brazil: A Test-Negative Case-Control Study. Lancet Reg. Health Am. 2021, 1, 100025. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-N.; Yong, H.; Wen, W.; Qin-Long, J.; Chun-Huan, Z.; Peng-Zhe, Q.; Wei-Jie, G.; Lin, G.; Yi-Lan, L.; Wen-Hui, L.; et al. Effectiveness of Inactivated SARS-CoV-2 Vaccines against the Delta Variant Infection in Guangzhou: A Test-Negative Case–Control Real-World Study. Emerg. Microbes Infect. 2021, 10, 1751–1759. [Google Scholar] [CrossRef] [PubMed]
- Jeewandara, C.; Aberathna, I.S.; Danasekara, S.; Gomes, L.; Fernando, S.; Guruge, D.; Ranasinghe, T.; Gunasekera, B.; Kamaladasa, A.; Kuruppu, H.; et al. Comparison of the Immunogenicity of Five COVID-19 Vaccines in Sri Lanka. Immunology 2022, 167, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Law, M.; Ho, S.S.H.; Tsang, G.K.C.; Ho, C.M.Y.; Kwan, C.M.; Yan, V.K.C.; Yiu, H.H.E.; Lai, F.T.T.; Wong, I.C.K.; Chan, E.W.Y. Efficacy and Effectiveness of Inactivated Vaccines against Symptomatic COVID-19, Severe COVID-19, and COVID-19 Clinical Outcomes in the General Population: A Systematic Review and Meta-Analysis. Lancet Reg. Health West. Pacific 2023, 37, 100788. [Google Scholar] [CrossRef] [PubMed]
- Jara, A.; Undurraga, E.A.; González, C.; Paredes, F.; Fontecilla, T.; Jara, G.; Pizarro, A.; Acevedo, J.; Leo, K.; Leon, F.; et al. Effectiveness of an Inactivated SARS-CoV-2 Vaccine in Chile. N. Engl. J. Med. 2021, 385, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Svetlova, J.; Gustin, D.; Manuvera, V.; Shirokov, D.; Shokina, V.; Prusakov, K.; Aldarov, K.; Kharlampieva, D.; Matyushkina, D.; Bespyatykh, J.; et al. Microarray Profiling of Vaccination-Induced Antibody Responses to SARS-CoV-2 Variants of Interest and Concern. Int. J. Mol. Sci. 2022, 23, 13220. [Google Scholar] [CrossRef] [PubMed]
- Leung, D.; Mu, X.; Duque, J.S.R.; Cheng, S.M.S.; Wang, M.; Zhang, W.; Zhang, Y.; Tam, I.Y.S.; Lee, T.S.S.; Lam, J.H.Y.; et al. Safety and Immunogenicity of 3 Doses of BNT162b2 and CoronaVac in Children and Adults with Inborn Errors of Immunity. Front. Immunol. 2022, 13, 982155. [Google Scholar] [CrossRef] [PubMed]
- Rakshit, S.; Babji, S.; Parthiban, C.; Madhavan, R.; Adiga, V.; J, S.E.; Kumar, N.C.; Ahmed, A.; Shivalingaiah, S.; Shashikumar, N.; et al. Polyfunctional CD4 T-Cells Correlating with Neutralising Antibody Is a Hallmark of COVISHIELDTM and COVAXIN® Induced Immunity in COVID-19 Exposed Indians. Npj Vaccines 2023, 8, 134. [Google Scholar] [CrossRef] [PubMed]
- Kruglov, A.A.; Bondareva, M.A.; Gogoleva, V.S.; Semin, I.K.; Astrakhantseva, I.V.; Zvartsev, R.; Lunin, A.S.; Apolokhov, V.D.; Shustova, E.Y.; Volok, V.P.; et al. Inactivated Whole Virion Vaccine Protects K18-HACE2 Tg Mice against the Omicron SARS-CoV-2 Variant via Cross-Reactive T Cells and Nonneutralizing Antibody Responses. Eur. J. Immunol. 2024, 54, e2350664. [Google Scholar] [CrossRef] [PubMed]
- Morens, D.M.; Taubenberger, J.K.; Fauci, A.S. Universal Coronavirus Vaccines—An Urgent Need. N. Engl. J. Med. 2022, 386, 297–299. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Sauer, M.M.; Czudnochowski, N.; Low, J.S.; Alejandra Tortorici, M.; Housley, M.P.; Noack, J.; Walls, A.C.; Bowen, J.E.; Guarino, B.; et al. Broad Betacoronavirus Neutralization by a Stem Helix–Specific Human Antibody. Science 2021, 373, 1109–1116. [Google Scholar] [CrossRef] [PubMed]
- Dacon, C.; Tucker, C.; Peng, L.; Lee, C.C.D.; Lin, T.H.; Yuan, M.; Cong, Y.; Wang, L.; Purser, L.; Williams, J.K.; et al. Broadly Neutralizing Antibodies Target the Coronavirus Fusion Peptide. Science 2022, 377, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.; Klumpp-Thomas, C.; Kalish, H.; Shunmugavel, A.; Mehalko, J.; Denson, J.P.; Snead, K.R.; Drew, M.; Corbett, K.S.; Graham, B.S.; et al. Serologic Cross-Reactivity of SARS-CoV-2 with Endemic and Seasonal Betacoronaviruses. J. Clin. Immunol. 2021, 41, 906–913. [Google Scholar] [CrossRef] [PubMed]
- Ladner, J.T.; Henson, S.N.; Boyle, A.S.; Engelbrektson, A.L.; Fink, Z.W.; Rahee, F.; D’ambrozio, J.; Schaecher, K.E.; Stone, M.; Dong, W.; et al. Epitope-Resolved Profiling of the SARS-CoV-2 Antibody Response Identifies Cross-Reactivity with Endemic Human Coronaviruses. Cell Reports Med. 2021, 2, 100189. [Google Scholar] [CrossRef] [PubMed]
- Takeda, M. Proteolytic Activation of SARS-CoV-2 Spike Protein. Microbiol. Immunol. 2022, 66, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Lin, S.; Chen, Z.; Cao, Y.; He, B.; Lu, G. Targetable Elements in SARS-CoV-2 S2 Subunit for the Design of Pan-Coronavirus Fusion Inhibitors and Vaccines. Signal Transduct. Target. Ther. 2023, 8, 197. [Google Scholar] [CrossRef] [PubMed]
- Hatmal, M.M.; Alshaer, W.; Al-Hatamleh, M.A.I.; Hatmal, M.; Smadi, O.; Taha, M.O.; Oweida, A.J.; Boer, J.C.; Mohamud, R.; Plebanski, M. Comprehensive Structural and Molecular Comparison of Spike Proteins of SARS-CoV-2, SARS-CoV and MERS-CoV, and Their Interactions with ACE2. Cells 2020, 9, 2638. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Canziani, G.A.; Carter, E.P.; Chaiken, I. The Case for S2: The Potential Benefits of the S2 Subunit of the SARS-CoV-2 Spike Protein as an Immunogen in Fighting the COVID-19 Pandemic. Front. Immunol. 2021, 12, 637651. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zheng, A.; Tang, Y.; Chai, Y.; Chen, J.; Cheng, L.; Hu, Y.; Qu, J.; Lei, W.; Liu, W.J.; et al. A Pan-Coronavirus Peptide Inhibitor Prevents SARS-CoV-2 Infection in Mice by Intranasal Delivery. Sci. China Life Sci. 2023, 66, 2201–2213. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Jiao, F.; Wang, L.; Yu, X.; Lu, T.; Fu, Y.; Huang, Z.; Li, X.; Huang, J.; Wang, Q.; et al. SARS-CoV-2 Omicron XBB Subvariants Exhibit Enhanced Fusogenicity and Substantial Immune Evasion in Elderly Population, but High Sensitivity to Pan-Coronavirus Fusion Inhibitors. J. Med. Virol. 2023, 95, e28641. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Gao, Z.; Feng, X.; Cheng, L.; Liu, N.; Liu, C.; Han, S.; Yang, Q.; Zou, Q.; Chong, H.; et al. Development of Potent Pan-Coronavirus Fusion Inhibitors with a New Design Strategy. MedComm 2024, 5, e666. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Zhu, Y.; Liu, M.; Lan, Q.; Xu, W.; Wu, Y.; Ying, T.; Liu, S.; Shi, Z.; Jiang, S.; et al. Fusion Mechanism of 2019-NCoV and Fusion Inhibitors Targeting HR1 Domain in Spike Protein. Cell. Mol. Immunol. 2020, 17, 765–767. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, L.; Liu, Z.; Xing, L.; Zhu, Y.; Xu, W.; Xia, S.; Lu, L.; Jiang, S. An Engineered Recombinant Protein Containing Three Structural Domains in SARS-CoV-2 S2 Protein Has Potential to Act as a Pan-Human Coronavirus Entry Inhibitor or Vaccine Antigen. Emerg. Microbes Infect. 2023, 12, 2244084. [Google Scholar] [CrossRef] [PubMed]
- Shiakolas, A.R.; Kramer, K.J.; Wrapp, D.; Richardson, S.I.; Schäfer, A.; Wall, S.; Wang, N.; Janowska, K.; Pilewski, K.A.; Venkat, R.; et al. Cross-Reactive Coronavirus Antibodies with Diverse Epitope Specificities and Fc Effector Functions. Cell Reports Med. 2021, 2, 100313. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Contant, P.; Embong, A.K.; Kanagaiah, P.; Chaves, F.A.; Yang, H.; Branche, A.R.; Topham, D.J.; Sangster, M.Y.; Ellebedy, A.; Schultz-Cherry, S. S Protein-Reactive IgG and Memory B Cell Production after Human SARS-CoV-2 Infection Includes Broad Reactivity to the S2 Subunit. MBio 2020, 11, e01991-20. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.; Loyal, L.; Frentsch, M.; Wendisch, D.; Georg, P.; Kurth, F.; Hippenstiel, S.; Dingeldey, M.; Kruse, B.; Fauchere, F.; et al. SARS-CoV-2-Reactive T Cells in Healthy Donors and Patients with COVID-19. Nature 2020, 587, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.W.; Faulkner, N.; Cornish, G.H.; Rosa, A.; Harvey, R.; Hussain, S.; Ulferts, R.; Earl, C.; Wrobel, A.G.; Benton, D.J.; et al. Preexisting and de Novo Humoral Immunity to SARS-CoV-2 in Humans. Science 2020, 370, 1339–1343. [Google Scholar] [CrossRef] [PubMed]
- Grobben, M.; Van Der Straten, K.; Brouwer, P.J.; Brinkkemper, M.; Maisonnasse, P.; Dereuddre-Bosquet, N.; Appelman, B.; Ayesha Lavell, A.H.; Van Vught, L.A.; Burger, J.A.; et al. Cross-Reactive Antibodies after Sars-Cov-2 Infection and Vaccination. Elife 2021, 10, e70330. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, Y.; Furukawa, K.; Ise, T.; Takayama, M.; Ota, T.; Kuroda, T.; Shano, S.; Hashimoto, T.; Konishi, H.; Ishihara, T.; et al. Discovery of Anti-SARS-CoV-2 S2 Protein Antibody CV804 with Broad-Spectrum Reactivity with Various Beta Coronaviruses and Analysis of Its Pharmacological Properties in Vitro and in Vivo. PLoS ONE 2024, 19, e0300297. [Google Scholar] [CrossRef] [PubMed]
- Felbinger, N.; Trudil, D.; Loomis, L.; Ascione, R.; Siragusa, G.; Haba, S.; Rastogi, S.; Mucci, A.; Claycomb, M.; Snowberger, S.; et al. Epitope Mapping of SARS-CoV-2 Spike Protein Differentiates the Antibody Binding Activity in Vaccinated and Infected Individuals. Front. Virol. 2023, 3, 988109. [Google Scholar] [CrossRef]
- Forni, D.; Filippi, G.; Cagliani, R.; De Gioia, L.; Pozzoli, U.; Al-Daghri, N.; Clerici, M.; Sironi, M. The Heptad Repeat Region Is a Major Selection Target in MERS-CoV and Related Coronaviruses. Sci. Rep. 2015, 5, srep14480. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xiao, G.; Chen, Y.; He, Y.; Niu, J.; Escalante, C.R.; Xiong, H.; Farmar, J.; Debnath, A.K.; Tien, P.; et al. Interaction between Heptad Repeat 1 and 2 Regions in Spike Protein of SARS-Associated Coronavirus: Implications for Virus Fusogenic Mechanism and Identification of Fusion Inhibitors. Lancet 2004, 363, 938–947. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, P.J.; Patel, R.S.; Loeffler, K.; Yasuhara, A.; Van De Velde, L.A.; Yang, J.E.; Chervin, J.; Troxell, C.; Huang, M.; Zheng, N.; et al. Multivalent S2 Subunit Vaccines Provide Broad Protection against Clade 1 Sarbecoviruses in Female Mice. Nat. Commun. 2025, 16, 462. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, P.J.; Frey, S.J.; Loeffler, K.; Kuroda, M.; Maemura, T.; Armbrust, T.; Yang, J.E.; Hou, Y.J.; Baric, R.; Wright, E.R.; et al. Multivalent S2-Based Vaccines Provide Broad Protection against SARS-CoV-2 Variants of Concern and Pangolin Coronaviruses. eBioMedicine 2022, 86, 104341. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.V.; Wall, S.C.; Kramer, K.J.; Holt, C.M.; Periasamy, S.; Richardson, S.I.; Manamela, N.P.; Suryadevara, N.; Andreano, E.; Paciello, I.; et al. Discovery and Characterization of a Pan-Betacoronavirus S2-Binding Antibody. Structure 2024, 32, 1893–1909.e11. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zou, F.; Yu, F.; Li, R.; Yuan, Y.; Zhang, Y.; Zhang, X.; Deng, J.; Chen, T.; Song, Z.; et al. Nanoparticle Vaccines Based on the Receptor Binding Domain (RBD) and Heptad Repeat (HR) of SARS-CoV-2 Elicit Robust Protective Immune Responses. Immunity 2020, 53, 1315–1330.e9. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Shen, F.; He, W.-Q.; Li, A.-Q.; Li, M.-H.; Feng, X.-L.; Zheng, Y.-T.; Pang, W. HR121 Targeting HR2 Domain in S2 Subunit of Spike Protein Can Serve as a Broad-Spectrum SARS-CoV-2 Inhibitor via Intranasal Administration. Acta Pharm. Sin. B 2023, 13, 3339–3351. [Google Scholar] [CrossRef] [PubMed]
- Pang, W.; Lu, Y.; Zhao, Y.-B.; Shen, F.; Fan, C.-F.; Wang, Q.; He, W.-Q.; He, X.-Y.; Li, Z.-K.; Chen, T.-T.; et al. A Variant-Proof SARS-CoV-2 Vaccine Targeting HR1 Domain in S2 Subunit of Spike Protein. Cell Res. 2022, 32, 1068–1085. [Google Scholar] [CrossRef] [PubMed]
- Evtushenko, E.; Ryabchevskaya, E.; Kovalenko, A.; Granovskiy, D.; Arkhipenko, M.; Vasiliev, Y.; Nikitin, N.; Karpova, O. Wuhan Sequence-Based Recombinant Antigens Expressed in E. Coli Elicit Antibodies Capable of Binding with Omicron S-Protein. Int. J. Mol. Sci. 2024, 25, 9016. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, A.O.; Ryabchevskaya, E.M.; Evtushenko, E.A.; Kondakova, O.A.; Ivanov, P.A.; Arkhipenko, M.V.; Nikitin, N.A.; Karpova, O. V Dataset on Safety and Protective Efficacy Studies of COVID-19 Vaccine Candidates Based on Structurally Modified Plant Virus in Female Hamsters. Data Br. 2023, 48, 109158. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-C.; Sayedahmed, E.E.; Sambhara, S.; Mittal, S.K. Progress towards the Development of a Universal Influenza Vaccine. Viruses 2022, 14, 1684. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Ni, W.; Li, W.; Wu, Z.; Yao, X.; Zheng, Y.; Zhao, Y.; Yuan, W. SARS-CoV-2 S Protein Disrupts the Formation of ISGF3 Complex through Conserved S2 Subunit to Antagonize Type I Interferon Response. J. Virol. 2025, 99, e0151624. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Law, A.H.T.; Cheung, J.K.; Lin, Y.; Wu, P.; Li, Z.; Cowling, B.J.; Yang, W.; Wong, J.Y. Comparison of Excess Deaths and Laboratory-Confirmed COVID-19 Deaths during a Large Omicron Epidemic in 2022 in Hong Kong. J. Glob. Health 2025, 15, 4105. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Wu, C.; Wu, X.; Ma, X.; Shu, C.; Chen, Q.; Zheng, A.; Yang, H.; Lu, J.; Du, P.; et al. Classification of Five SARS-CoV-2 Serotypes Based on RBD Antigenicities. Sci. Bull. 2023, 68, 3003–3012. [Google Scholar] [CrossRef] [PubMed]
- Simon-Loriere, E.; Schwartz, O. Towards SARS-CoV-2 Serotypes? Nat. Rev. Microbiol. 2022, 20, 187–188. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.; Shi, J.; Gong, Z.; Wen, H.; Lan, Y.; Deng, X.; Fan, Q.; Li, J.; Jiang, M.; Tang, X.; et al. Intra- vs. Interhost Evolution of SARS-CoV-2 Driven by Uncorrelated Selection—The Evolution Thwarted. Mol. Biol. Evol. 2023, 40, msad204. [Google Scholar] [CrossRef] [PubMed]
- Hamelin, D.J.; Scicluna, M.; Saadie, I.; Mostefai, F.; Grenier, J.C.; Baron, C.; Caron, E.; Hussin, J.G. Predicting Pathogen Evolution and Immune Evasion in the Age of Artificial Intelligence. Comput. Struct. Biotechnol. J. 2025, 27, 1370–1382. [Google Scholar] [CrossRef] [PubMed]
- Tai, J.-H.; Lee, D.-C.; Lin, H.-F.; Chao, T.-L.; Ruan, Y.; Cheng, Y.-W.; Chou, Y.-C.; Lin, Y.-Y.; Chang, S.-Y.; Chen, P.-J.; et al. Tradeoffs between Proliferation and Transmission in Virus Evolution– Insights from Evolutionary and Functional Analyses of SARS-CoV-2. Virol. J. 2025, 22, 107. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, B.F.; Saad-Roy, C.M.; Li, Y.; Sneppen, K.; Simonsen, L.; Viboud, C.; Levin, S.A.; Grenfell, B.T. Host Heterogeneity and Epistasis Explain Punctuated Evolution of SARS-CoV-2. PLOS Comput. Biol. 2023, 19, e1010896. [Google Scholar] [CrossRef] [PubMed]
- Hammer, A.S.; Quaade, M.L.; Rasmussen, T.B.; Fonager, J.; Rasmussen, M.; Mundbjerg, K.; Lohse, L.; Strandbygaard, B.; Jørgensen, C.S.; Alfaro-Núñez, A.; et al. SARS-CoV-2 Transmission between Mink (Neovison Vison) and Humans, Denmark. Emerg. Infect. Dis. 2021, 27, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Yang, X.; Guo, Y.; Peng, B.; Dong, R.; Li, S.; Xu, S. SARS-CoV-2 Infection in Animals: Patterns, Transmission Routes, and Drivers. Eco-Environ. Health 2024, 3, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Raglow, Z.; Surie, D.; Chappell, J.D.; Zhu, Y.; Martin, E.T.; Kwon, J.H.; Frosch, A.E.; Mohamed, A.; Gilbert, J.; Bendall, E.E.; et al. SARS-CoV-2 Shedding and Evolution in Patients Who Were Immunocompromised during the Omicron Period: A Multicentre, Prospective Analysis. Lancet Microbe 2024, 5, e235–e246. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.D.; Graham-Wooten, J.; Fitzgerald, A.S.; Leonard, A.S.; Cook, E.J.; Everett, J.K.; Rodino, K.G.; Moncla, L.H.; Kelly, B.J.; Collman, R.G.; et al. SARS-CoV-2 Evolution during Prolonged Infection in Immunocompromised Patients. MBio 2024, 15, e00110-24. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Li, N.; Xiong, X.; Tang, S.; Dai, Q.; Liu, Z.; Wang, T.; Gu, X.; Zhou, Z. A Bivalent Vaccine Containing D614G and BA.1 Spike Trimer Proteins or a BA.1 Spike Trimer Protein Booster Shows Broad Neutralizing Immunity. J. Med. Virol. 2022, 94, 4287–4293. [Google Scholar] [CrossRef] [PubMed]
- Launay, O.; Gupta, R.; Machabert, T.; Konate, E.; Rousseau, A.; Vigne, C.; Beckers, F.; Devlin, L.; Botelho-Nevers, E.; Cachanado, M.; et al. Beta-Variant Recombinant SARS CoV-2 Vaccine Induces Durable Cross-Reactive Antibodies against Omicron BA Variants. Commun. Med. 2025, 5, 21. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-S.; Matsuoka, Y.; Santos, C.; Luongo, C.; Liu, X.; Yang, L.; Kaiser, J.A.; Duncan, E.F.; Johnson, R.F.; Teng, I.-T.; et al. Intranasal Parainfluenza Virus-Vectored Vaccine Expressing SARS-CoV-2 Spike Protein of Delta or Omicron B.1.1.529 Induces Mucosal and Systemic Immunity and Protects Hamsters against Homologous and Heterologous Challenge. PLoS Pathog. 2025, 21, e1012585. [Google Scholar] [CrossRef] [PubMed]
- Perez Marc, G.; Coria, L.M.; Ceballos, A.; Rodriguez, J.M.; Lombardo, M.E.; Bruno, L.; Páez Córdoba, F.; Fascetto Cassero, C.G.; Salvatori, M.; Rios Medrano, M.; et al. Immunogenicity and Safety of Monovalent and Bivalent SARS-CoV-2 Variant Adapted RBD-Based Protein Booster Vaccines in Adults Previously Immunized with Different Vaccine Platforms: A Phase II/III, Randomized Clinical Trial. Vaccine 2025, 54, 127045. [Google Scholar] [CrossRef] [PubMed]
- Ao, D.; Peng, D.; He, C.; Ye, C.; Hong, W.; Huang, X.; Lu, Y.; Shi, J.; Zhang, Y.; Liu, J.; et al. Correction: A Promising MRNA Vaccine Derived from the JN.1 Spike Protein Confers Protective Immunity against Multiple Emerged Omicron Variants. Mol. Biomed. 2025, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Piano Mortari, E.; Ferrucci, F.; Zografaki, I.; Carsetti, R.; Pacelli, L. T and B Cell Responses in Different Immunization Scenarios for COVID-19: A Narrative Review. Front. Immunol. 2025, 16, 1535014. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Kobayashi, N.; Deguchi, K.; Omori, S.; Ichinohe, T. SARS-CoV-2 Infection Primes Cross-Protective Respiratory IgA in a MyD88- and MAVS-Dependent Manner. Npj Vaccines 2025, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Ghosh Roy, S.; Dwivedi, R.; Tripathi, P.; Kumar, K.; Nambiar, S.M.; Pathak, R. Beyond the Pandemic Era: Recent Advances and Efficacy of SARS-CoV-2 Vaccines Against Emerging Variants of Concern. Vaccines 2025, 13, 424. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.D.; Bentley, K.; Fielding, C.A.; Hatfield, K.M.; Ings, D.P.; Harnum, D.; Wang, E.C.; Stanton, R.J.; Holder, K.A. Combined Anti-S1 and Anti-S2 Antibodies from Hybrid Immunity Elicit Potent Cross-Variant ADCC against SARS-CoV-2. JCI Insight 2023, 8, e170681. [Google Scholar] [CrossRef] [PubMed]
- Astrakhantseva, I.V.; Krut’, V.G.; Chuvpilo, S.A.; Shevyrev, D.V.; Shumeev, A.N.; Rybtsov, S.A.; Nedospasov, S.A. On Immunological Studies at Sirius University of Science and Technology. Mol. Biol. 2023, 57, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.-L.; Goldsmith, J.A.; Schaub, J.M.; DiVenere, A.M.; Kuo, H.-C.; Javanmardi, K.; Le, K.C.; Wrapp, D.; Lee, A.G.; Liu, Y.; et al. Structure-Based Design of Prefusion-Stabilized SARS-CoV-2 Spikes. Science 2020, 369, 1501–1505. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Mendonça, L.; Allen, E.R.; Howe, A.; Lee, M.; Allen, J.D.; Chawla, H.; Pulido, D.; Donnellan, F.; Davies, H.; et al. Native-like SARS-CoV-2 Spike Glycoprotein Expressed by ChAdOx1 NCoV-19/AZD1222 Vaccine. ACS Cent. Sci. 2021, 7, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Ke, Z.; Oton, J.; Qu, K.; Cortese, M.; Zila, V.; McKeane, L.; Nakane, T.; Zivanov, J.; Neufeldt, C.J.; Cerikan, B.; et al. Structures and Distributions of SARS-CoV-2 Spike Proteins on Intact Virions. Nature 2020, 588, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Kordyukova, L.V.; Moiseenko, A.V.; Serebryakova, M.V.; Shuklina, M.A.; Sergeeva, M.V.; Lioznov, D.A.; Shanko, A. V Structural and Immunoreactivity Properties of the SARS-CoV-2 Spike Protein upon the Development of an Inactivated Vaccine. Viruses 2023, 15, 480. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wu, Y.; Gao, G.F. A Structural Voyage Toward the Landscape of Humoral and Cellular Immune Escapes of SARS-CoV-2. Immunol. Rev. 2025, 330, e70000. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gulova, S.M.; Veselkina, U.S.; Astrakhantseva, I.V. Adaptation of the Vaccine Prophylaxis Strategy to Variants of the SARS-CoV-2 Virus. Vaccines 2025, 13, 761. https://doi.org/10.3390/vaccines13070761
Gulova SM, Veselkina US, Astrakhantseva IV. Adaptation of the Vaccine Prophylaxis Strategy to Variants of the SARS-CoV-2 Virus. Vaccines. 2025; 13(7):761. https://doi.org/10.3390/vaccines13070761
Chicago/Turabian StyleGulova, Sofia M., Uliana S. Veselkina, and Irina V. Astrakhantseva. 2025. "Adaptation of the Vaccine Prophylaxis Strategy to Variants of the SARS-CoV-2 Virus" Vaccines 13, no. 7: 761. https://doi.org/10.3390/vaccines13070761
APA StyleGulova, S. M., Veselkina, U. S., & Astrakhantseva, I. V. (2025). Adaptation of the Vaccine Prophylaxis Strategy to Variants of the SARS-CoV-2 Virus. Vaccines, 13(7), 761. https://doi.org/10.3390/vaccines13070761




