Abstract
Mycorrhizas play an important role in plant growth and development. In mycorrhizal symbioses, fungi supply soil mineral nutrients, such as nitrogen and phosphorus, to their host plants in exchange for carbon resources. Plants gain as much as 80% of mineral nutrient requirements from mycorrhizal fungi, which form associations with the roots of over 90% of all plant species. Orchid seeds lack endosperms and contain very limited storage reserves. Therefore, the symbiosis with mycorrhizal fungi that form endomycorrhizas is essential for orchid seed germination and protocorm development under natural conditions. The rapid advancement of next-generation sequencing contributes to identifying the orchid and fungal genes involved in the orchid mycorrhizal symbiosis and unraveling the molecular mechanisms regulating the symbiosis. We aim to update and summarize the current understanding of the mechanisms on orchid-fungus symbiosis, and the main focus will be on the nutrient exchange between orchids and their fungal partners.
1. Introduction
All living organisms essentially interact and communicate with their surrounding organisms for their survival. In the case when all of the implied partners benefit from their interactions, the interaction is called symbiosis [1]. Over 90% of plants form a symbiotic relationship with a variety of environmental microorganisms and the associations, especially with fungi, occur in mycorrhizas, which are the parts located in plant roots [1,2,3]. One of the well-known benefits of mycorrhizas to both plants and fungi is nutrient acquisition. For example, fungi mainly obtain carbon (C) sources from their coexisting plants, and in return, enhance a plant’s ability to take up minerals or water, subsequently leading to improvement of plant growth and reproduction [1]. Furthermore, fungi are also known to contribute to the tolerance of their symbiotic plants to abiotic stresses including drought, salinity, too high and low temperature, and biotic stresses including pathogens and insects [4,5]. In nature, more than 6,000 fungal species are known to be capable of establishing mycorrhizal associations with about 240,000 plant species [6]. Mycorrhizas are commonly classified into the following groups based on the anatomical aspects, such as the presence of internal hyphae of mycorrhizal fungi in the host plant cells: (1) Ectomycorrhiza: about 5,000 species of fungi do not enter the cells, but create a mycelial sheath, which functions as a plant-fungus interface, surrounding the epidermal and outer cortical cells. The interface is used for the bidirectional nutrient transfer between a plant and its fungal partner [7]. (2) Endomycorrhiza: the fungi possess specific types of hyphae such as arbuscules and coils to penetrate and grow inside of the root cells of host plants [3]. Endomycorrhizas can be further classified as ectendomycorrhizas, ericoid, arbutoid, monotropoid, orchid and arbuscular mycorrhizas based on the specificity of plant families and the type of internal hyphae [3,8]. Ectomycorrhizas are also alternatively represented as arbuscular mycorrhizas because the class is the most prevalent type of mycorrhizae, as shown in approximately 71% of all vascular plant species [1,9].
The Orchidaceae is the largest family of flowering plants including over 700 genera and about 30,000–35,000 species and still hundreds of new species displaying variable floral features, lifestyles, habitat distributions and trophic patterns are being discovered and developed every year [10,11]. Mycorrhizas in orchid species are highly important throughout their whole life including germination and further development. In this article, we provide a brief review of the roles of mycorrhizas in orchid growth and development and the current understanding of the mechanisms in nutrient exchange between orchids and their fungal partners.
2. Orchid Mycorrhiza and Its Role in Orchid Seed Germination
The early stages of orchid reproduction and development, especially the germination of orchid seeds, are highly influenced by the relationship with their appropriate mycorrhizal fungi as well as their surrounding environments. Like dust particles, orchid seeds are incredibly tiny, ranging in weigh from 0.3 to 14 ug (micrograms), and each seed capsule may contain 1300 to 4 million seeds that are adapted for wind dispersal [12,13]. These characteristics are correlated to their reduced size of embryo and the absence of endosperm which causes difficulty in germination. Therefore, various attempts have been made to initiate the germination of orchid seeds. Currently, the symbiotic germination using mycorrhizal fungi is still considered for inducing a higher germination rate even though other methods by modulating the factors for germination including nutrients, photoperiod, temperature and plant hormone need to be combined [14]. The most important contribution of mycorrhizal fungi to orchids is supplying nutrients for growth and development. Orchid seeds under natural conditions are unable to germinate without a supply of C, mineral nutrients as well as vitamins from their symbiotic partners. Additionally, mycorrhizal fungi also contribute to supplying and retaining water for orchid species during germination. This is also a highly essential role of mycorrhizal fungi because orchid seeds commonly have a hydrophobic testa to avoid imbibition in unfavorable environments [12,14,15,16].
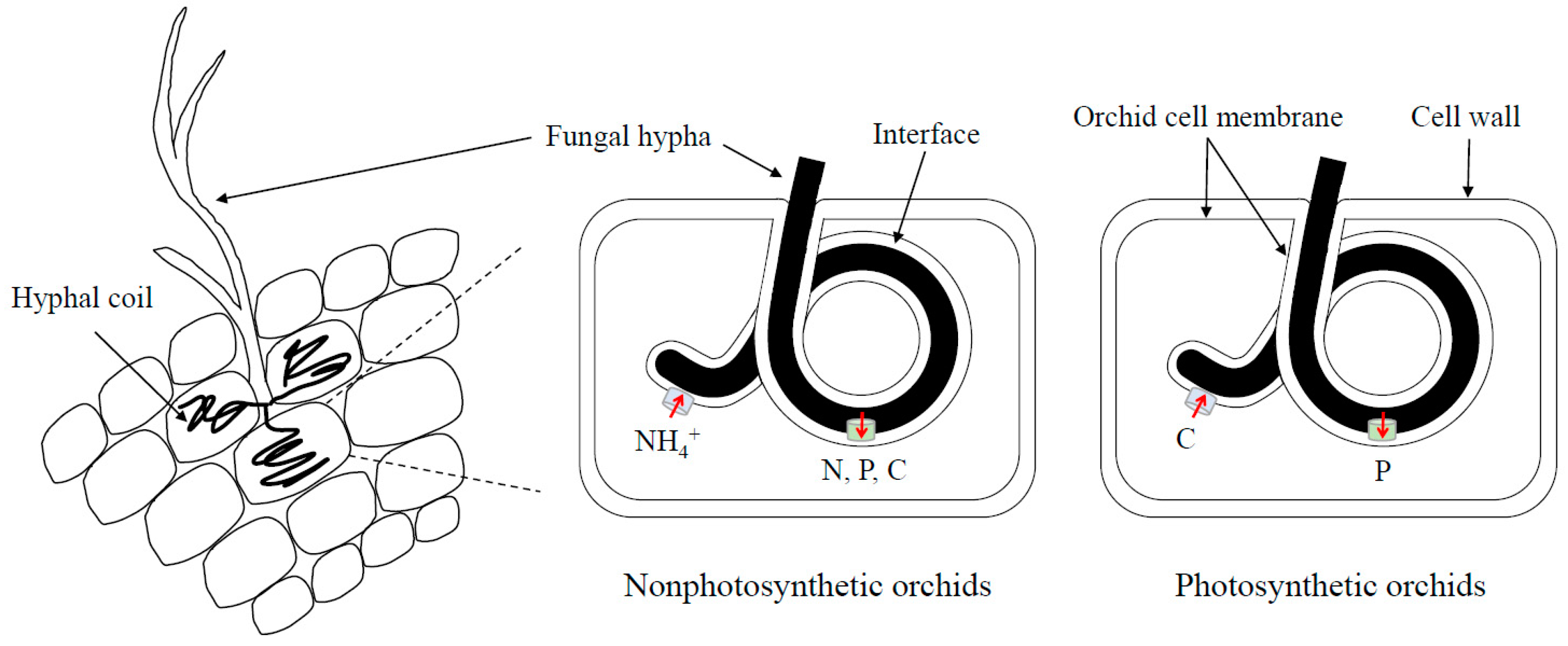
Under the co-existence with mycorrhizal fungi, orchid seeds can germinate and develop into unique seedling structures called protocorms that consist of parenchyma cells [17]. Then, further development of the protocorm into plant is also supported by their mycorrhizal fungi. In protocorms of orchids, the entered fungal hyphae form coiled complexes called pelotons. Orchids can obtain nutrients by digesting them [18]. It is likely that the site of hyphal entry can be the determinant factor for the success in symbiotic associations [17]. Although many fungal hyphae prefer suspensors for their entry, it has been shown that only the fungal hyphae which enter through rhizoids and not suspensors can establish successful mycorrhizal association in Dactyloriza majalis [19]. Therefore, the presence or absence of suspensors during orchid development and the preferred entry sites of mycorrhizal fungi might be the determinants of the specificity between orchids and their fungi.
Many environmental factors including abiotic factors, such as light, soil structure, moisture, and biotic factors, such as microorganisms, are required to be considered for the successful germination of orchid seeds and further development. These factors affect not only orchid seeds but also the success in symbiotic association between orchids and mycorrhizal fungi [12,16,20]. For example, it has been shown that organ development of symbiotic orchid seedlings and development of photosynthetic structures are affected by light intensity, whereas development of mycotrophic tissues is suppressed by light [20]. Furthermore, non-symbiotic microorganisms can act as competitors of the symbiotic partners and also influence microenvironments in soil [21,22]. Therefore, future studies on mechanisms of orchid seed germination should also pay attention on the effects of abiotic and biotic factors other than mycorrhizal fungi.
3. Nutrient Exchange between Orchids and Mycorrhizal Fungi
Mycorrhizal symbioses are important for the growth and survival of many plant species and are responsible for up to 80% of nitrogen (N) and phosphorus (P) uptake by plants [23]. In general, a mutualistic relationship for nutrient exchanges is established between plants and mycorrhizal fungi in that plants provide photosynthetically fixed C to the symbiotic fungi and they benefit from fungi with regard to absorption of mineral nutrients, such as N and P [24,25,26]. However, recent evidence suggests that not only carbohydrates but also lipids can be transferred from host plants to fungi as C resources [27,28]. The mechanisms of how ectomycorrhizal and arbuscular mycorrhizal fungi take up and transfer mineral nutrients to their host plants for exchanging C resources are extensively studied; however, much less attention has been paid to ericoid and orchid mycorrhizal fungi [3,23,29]. Several detailed reviews on the mutualistic interactions between plants and fungi that form ectomycorrhizal or arbuscular mycorrhizal symbiosis have been published [3,23,24,25,26,30]. Here, only the current understanding of mechanisms of nutrient exchange in orchid mycorrhizas is summarized.
Compared to the other types of mycorrhizas, the orchid-fungus symbiosis is considered to be unique since the nutrient movement is not bi-directional in the early developmental stages and even in adults of some orchid species [29]. As the orchid seeds are very tiny with very little storage, colonization by fungi is required for their germination and/or early protocorm development [15]. Therefore, the mycorrhizal fungi were thought to provide both mineral nutrients and C to the non-photosynthetic orchid seedlings and to receive no reward [29]. However, similar to all other mycorrhizal symbioses, the mycorrhizal fungi receive C as a reward for exchanging mineral nutrients after orchids develop green leaves for photosynthesis [31]. Although the majority of orchids are autotrophic with photosynthetic ability, more than 100 species are myco-heterotrophic which are completely achlorophyllous and nutritionally dependent on their symbiotic fungi throughout the whole life cycle [15,32]. In addition, mixotrophy, also called partial myco-heterotrophy, which consists of both nutritional strategies of autotrophy and myco-heterotrophy, has been reported [33,34]. Mixotrophic orchids, meeting their C demand via mycorrhizal fungi even when they are photosynthetic at the adult stage, may be the evolutionary intermediates between achlorophyllous and autotrophic orchids [15,31]. This partial myco-heterotrophy may enable orchids to adapt to the shaded environment of forests and the conditions when the photosynthetic abilities are reduced [35,36].
The well-known non-mutualistic interaction between non-photosynthetic orchids and their symbiotic fungi was challenged by a recent study using mycorrhizas formed between Serapias vomeracea (orchid) and Tulasnella calospora (fungus) to investigate expression of orchid and fungal N transport and assimilation genes [29,37]. It was demonstrated that the fungal low-affinity ammonium transporter gene, TcAMT2, is significantly upregulated in symbiosis and the transcripts corresponding to TcAMT2 are detected in the orchid protocorm cells containing fungal pelotons by laser microdissection analysis. In addition, TcGS1 and TcGOGAT, the fungal genes encoding glutamine synthetase and glutamate synthase in the N assimilation pathway, are also up-regulated in mycorrhizal protocorms. A commentary of this study by Dearnaley and Cameron (2017) concluded that the fungus, T. calospora, may receive ammonium from its orchid host, S. vomeracea, during seed germination and protocorm development. Therefore, a true mutualism may be also present in the initially mycoheterotrophic stage of autotrophic orchids or myco-heterotrophic orchids [29]. The finding that orchid seeds and protocorms may also pay for exchanging nutrients from their fungal partners could explain how a compatible mycorrhizal fungus recognizes and subsequently colonizes the seeds or protocorms of the host orchid in the soil. Although the orchid seeds have little storage, the mature embryo cells contain protein bodies, a protein storage organelle, which could potentially produce ammonium through a proper catalysis process after seed imbibition [38,39]. The ammonium produced from protein bodies may be used to attract fungi for colonization [29,37].
Growth of T. calospora on the media with different N sources indicated the fungus prefers utilizing ammonium and organic N including glutamic acid and glutamine. Poor growth on the nitrate-containing medium is consistent with the molecular evidence that the genome of T. calospora does not have the genes encoding nitrate uptake and assimilation. In addition, the research conducted by Fochi et al. (2017) also suggests that S. vomeracea may receive organic N from T. calospora because the fungal arginase and urease genes involved in amino acid breakdown are not significantly induced or lowly expressed, but the fungal Argininosuccinate lyase gene for arginine biosynthesis and the orchid amino acid transporter genes are significantly upregulated in symbiotic protocorms [29,37]. This finding is consistent with the previous study that Ceratobasidium cornigerum transfers amino acids to its orchid hosts [40]. However, Kuga et al. (2014) found that orchids obtain ammonium from mycorrhizal fungi by using isotope tracing experiments [41]. According to the above research, a novel model could be proposed that non-photosynthetic and photosynthetic orchids export ammonium and sugars, respectively, to their fungal partners for exchanging the required nutrients (Figure 1). Furthermore, orchids may be able to receive both ammonium and amino acids as N sources from mycorrhizal fungi.
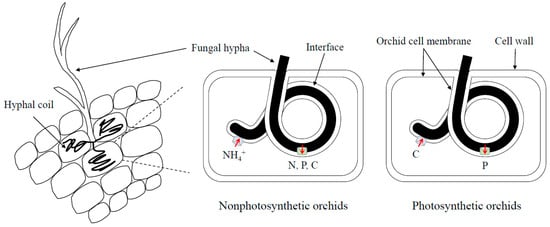
Figure 1.
The possible model of nutrient exchange in orchid mycorrhizas. In natural conditions, orchid seeds form symbiosis with orchid mycorrhizal fungi to receive nutrients for germination and further protocorm development. The hyphae of orchid mycorrhizal fungi can enter orchid cells and form coiled complexes called pelotons. At nonphotosynthetic stages, orchid cells export ammonium (NH4+) to their fungal partners and receive nitrogen (N), phosphorus (P) and carbon (C) for germination. In addition, the senescent pelotons can also release N, P and C to orchid cells when hyphal coils are digested. After orchids develop green leaves, mycorrhizal fungi can obtain photosynthetically fixed C as a reward. However, photosynthetic orchids may not further receive N from fungal cells and only P is exchanged to orchid cells [29].
The acquisition and movement of nutrients between orchids and their fungal partners could be more complex than the above single-directional or mutualistic mechanisms. In nature, many fully myco-heterotrophic orchids or orchids that have a myco-heterotrophic juvenile phase have been found to grow exclusively with specific autotrophic trees [42]. Therefore, it has been speculated that these orchids may obtain C from their symbiotic fungi which can also form ectomycorrhizas with those trees [43]. Campbell (1970) firstly found that the rhizomes of three Corallorhiza orchids are often very close to tree roots, and the hyphae connecting the tree roots and the Corallorhiza orchids are structures typical of many ectomycorrhizal basidiomycetes [44]. The fungi associated with both Corallorhiza orchids and their adjacent trees were indeed confirmed as the ectomycorrhizal fungi [45,46]. By autoradiographical and tissue oxidation analyses, McKendrick et al. (2000) demonstrated that 14C can move from the autotrophic shrubs (Betula pendula or Salix repens) to the mycoheterotrophic orchid (C. trifida) through the mycorrhizal fungus (Ceratobasidium cornigerum) [42,43]. This tripartite relationship among orchid, fungus and autotrophic shrub was further confirmed by investigation of Rhizanthella gardneri, a unique underground orchid that is entirely subterranean even during flowering, using isotopically labelled tracers, 13CO2 and double-labelled [13C-15N]glycine [47]. Bougoure et al. (2010) reported that the mycorrhizal fungus Ceratobasidium sp. associated with R. gardneri can simultaneously form ectomycorrhizas with autotrophic Melaleuca scalene, and C in R. gardneri was originally from photosynthetic carbohydrates in M. scalene. In addition, Ceratobasidium sp. is able to acquire C from M. scalene and access soil nutrients saprophytically, and subsequently provide both C and N to R. gardneri and N to M. scalene [47]. Increasing evidence indicates that different mycorrhizal types can be developed by the same mycorrhizal fungi in different host plants, especially by those fungi which can form ectomycorrhizas. Therefore, a tripartite system of nutrient exchange among orchids, mycorrhizal fungi and the adjacent trees may be more common than the relationship established by only orchids and their symbiotic fungi [32,47,48,49,50].
4. Molecular Studies on Orchid Mycorrhizal Symbiosis by Next-Generation Sequencing and Proteomics
In the past decade, the next-generation sequencing (NGS) tools were widely applied in whole genome sequencing and RNA sequencing (RNA-seq). The first whole genome sequence of the orchid Phalaenopsis equestris was published in 2015 [51] and the number of orchid genomes has continuously increased [52,53,54,55,56]. The whole genome sequencing of Orchidaceae provides a gateway into orchid research, including flower morphology, sexual deception, environmental stress responses and symbiosis. The platforms of 454-pyrosequencing and Illumina were adopted to generate the transcriptomes from Cymbidium hybridum [57], Serapias vomeraceae [37,58], Anoectochilus roxburghii [59], Gastrodia elata [60], Dendrobium officinale [61], Epipactis helleborine [62], and Bletilla striata [63] to identify the molecular processes involved in the symbiosis between orchids and orchid mycorrhizal fungi. Owing to the NGS technologies, the orchid-fungus symbiosis for facilitating seed germination and protocorm development is now better and comprehensively understood.
Recent studies have focused on comparative transcriptomes, nutrient transfer, and regulatory module between the symbiotic and asymbiotic orchids during seed germination and protocorm development stages. Transcriptome analysis of S. vomeracea by 454-pyrosequencing indicated that no significant induction of defense genes, such as pathogenesis-related genes, was found in the mycorrhizal protocorms (Table 1) [58]. Similarly, two wound response marker genes, SvWound2 and SvWound3, which code enzymes involved in the biosynthesis of many signaling compounds and the cellular processes against pathogen infection and oxidative stress, were not up-regulated in the mycorrhizal tissues. In contrast, several nodulin-like genes, such as SvNod1 and SvNod9, were apparently up-regulated. SvNod9 encodes a putative bidirectional sugar transporter belonging to the SWEET family which possesses activity for sugar export from cells and can be controlled by pathogens to acquire carbohydrates for C and energy [58,64]. Their preliminary data using laser microdissection analysis indicates that SvNod1 could be used as a marker gene of orchid symbiosis because it only accumulates in the fungi-colonized cells. The up-regulation of nodulin genes and the lack of induction of defense genes in mycorrhizal protocorms of S. vomeracea suggest a high level of orchid-fungus compatibility [58].

Table 1.
Overview of the current studies on the orchid-fungus symbiosis by using transcriptomic and proteomic analysis.
However, an inconsistent result was found in the deep sequencing-based comparative root transcriptomes of the C. hybridum co-cultured with Epulorhiza repens ML01 (mycorrhizal) and Umbelopsis nana ZH3A-3 (non-mycorrhizal) [57]. The transcriptome indicated that the mycorrhizal and non-mycorrhizal fungi or the combination of both fungi induced the general defense responses in the symbiotic orchid roots. The differentially expressed defense genes are involved in cell wall modification (probable xyloglucan endo-transglucosylase, beta-D-xylosidase and cellulose synthase), detoxification of reactive oxygen species (glutathione peroxidase, probable glutathione S-transferase and germin-like protein), plant hormone signaling and synthesis (ethylene receptor homolog 2, ethylene-responsive element binding factor 4, lipoxygenase, ABA and gibberellin biosynthesis) and secondary metabolism (Table 1). As found in other orchid mycorrhizal systems, two inorganic phosphate transporter genes were induced in the symbiotic roots of C. hybridum. In addition, several other nutrient transport genes including major facilitator family protein and oligopeptide transporter 1-like genes and root morphogenesis genes were also up-regulated. Although the non-mycorrhizal fungal strain, ZH3A-3, induced common stress responses with E. repens ML01, the mycorrhizal symbiosis specifically activates genes related to signaling (LysM domain receptor-like kinase 3-like), defense (chitinase 4-like, mannose specific lectin), transport (equilibrative nucleoside transporter, ATPase 7, plasma membrane-type-like, peptide transporter PTR3-A-like), auxin response and protein metabolism and processing. Furthermore, the three fungi treatments also induced genes encoding transcription factors including AP2 domain, ERF, WRKY, MYB, bHLH and zinc-finger families which are generally involved pathogen defense responses and root developmental process. In addition to identification of the orchid transcripts, the putative fungal genes related to remodeling of fungal cell wall, degradation of plant cell wall and nutrient transport were identified in the symbiotic roots. The authors previously reported that ZH3A-3 and another orchid mycorrhizal fungi isolate, CL01, induce a similar mineral nutrient uptake in C. hybridum root [67]. Here, they showed that the effect of ZH3A-3 on the growth of C. hybridum is comparable to that of ML01. Therefore, the ZH3A-3 isolate was designated as a mycorrhizal-associated fungus [57].
The fully achlorophyllous variants are sometimes found in the partially myco-heterotrophic orchids which gain C from both photosynthesis and their mycorrhizal fungi. Therefore, it may help to understand the myco-heterotrophy through investigating and comparing the chlorophyllous and achlorophyllous partially myco-heterotrophic orchids [62]. Suetsugu et al. (2017) demonstrated that the albino Epipactis helleborine is fully mycoheterotrophic by stable isotope analysis and the green and albino individuals of E. helleborine can form mycorrhizas with same fungi which are mainly belong to the Wilcoxina genus. Gene expression analyses according to RNA-seq and cDNA de novo assembly revealed that the albino variants may suffer greater oxidative stress than the green ones due to the higher expression of the genes involved in antioxidant metabolism, such as glutathione peroxidase and catalase-peroxidase thioredoxin genes, in the albino individuals (Table 1). In addition, the fungal DNA level was found to be lower in the albino plants. Blakeman et al. (1976) reported that activation of antioxidant enzymes is coincident with digestion of pelotons in the mycorrhizal protocorms of orchids [68]. Therefore, the authors speculated that the upregulation of antioxidant enzymes in the albino E. helleborine may cause more frequent peloton digestion, leading to the promotion of the C flux from the fungus to the orchid [62]. As shown in other orchid mycorrhizas, the gene ontology (GO) analysis represents an enrichment in transport genes in the albino variants and the fungal partner. In the albino E. helleborine, similar to the finding in the mycorrhizal protocorms of S. vomeracea, an upregulation of a putative SWEET gene was found [58,62]. As for the fungal genes, the major facilitator superfamily (MFS) genes with transporter activities for sugars, nucleosides, N and P were significantly upregulated in the albino variants [62,69], suggesting that the fully mycoheterotrophic E. helleborine may also transport some nutrients to its fungal partner. The results are somewhat consistent with the finding for T. calospora-S. vomeracea symbiosis [29,37]. It is worth noting that a glycerol-3-phosphate acyl transferase (GPAT) gene, RAM2, and a subtilisin-like protease gene required for arbuscular mycorrhizal symbiosis are up-regulated in the albino orchids. In contrast, two genes encoding gibberellin 20 oxidase and cytokinin dehydrogenase precursor which can reduce arbuscular mycorrhizal colonization are down-regulated. Furthermore, up-regulation of auxin efflux carrier component genes important for arbuscular mycorrhizal colonization is also detected in the albino variants (Table 1) [62]. This indicates that a common mechanism may be involved in both orchid mycorrhizal and arbuscular mycorrhizal symbioses.
In addition to isotope tracking analysis, the transcriptome-wide identification of transporter genes in orchid mycorrhizas has now been widely applied to investigate nutrient exchange between orchids and orchid mycorrhizal fungi. As we have summarized from the studies done in C. hybridum and E. hellebore, various transporter genes involved in the uptake of N, P, sugars and nucleosides have been identified [57,62]. In the T. calospora-S. vomeracea symbiosis, several genes putatively encoding transporters for ammonium and amino acids were isolated by RNA-seq using the Illumina HiSeq2000 system including TcAMT1 and TcAMT2 in the fungus T. calospora and SvAMT1, SvAMT2, SvAAP1, SvAAP2 and SvLHT in the orchid S. vomeracea [37]. The expression levels of these transporter genes were further confirmed by reverse transcription-quantitative PCR analysis and the ammonium transporter activities of TcAMT1 and TcAMT2 were verified in a yeast mep mutant with defects in all the three endogenous Mep ammonium transporters [37,70].
Not only transcriptome analysis but also proteomic tools have been applied to investigate orchid mycorrhizas. The studies on the orchid-fungus symbiosis mainly focused on terrestrial species. The epiphytic orchid Oncidium sphacelatum was firstly investigated by the 2D-LC-MS/MS and 2D-MALDI-TOF MS analysis for the symbiotic relationships with the mycorrhizal fungi Ceratobasidium sp. isolate and Thanatephorus sp. strain RG26, respectively [65,66]. The 88 differentially expressed proteins identified at different protocorm development stages (achlorophyllous or green) by the 2D-LC-MS/MS coupled to isobaric tagging are involved in molecular signaling, cell rescue and defense, energy and secondary metabolism (Table 1) [65]. Within these proteins, the fructose-bisphosphate aldolase and enolase in the glycolytic pathway and the isocitrate lyase in the Krebs cycle were accumulated in the achlorophyllous protocorms, suggesting the catabolism of organic C from the fungus may be induced. In contrast, the RuBisCO and Photosystem I P700 chlorophyll a apoprotein A1 were most abundant in the green protocorms. The result may imply the transition of metabolism from fully myco-heterotrophic to partially myco-heterotrophic or fully autotrophic. In the interaction between O. sphacelatum Lindl. and Thanatephorus sp., eleven orchid proteins related to cell cycle, energy, purine metabolism, ribosome biogenesis and vesicle secretion were identified, and the fungal proteins involved in protein-protein interaction, stress response and protein and saccharides biosynthesis were found under the symbiosis condition (Table 1) [66].
To better understand orchid seed germination and protocorm development, Chen et al. (2017) performed RNA-seq and proteomic analysis using iTRAQ (isobaric tags for relative and absolute quantification) at three different stages (germination, protocorm and seedling) to identify the differentially expressed genes/proteins involved in these processes in Dendrobium officinale [61]. For both symbiotic and asymbiotic germination, 308 out of the 2256 identified proteins were differentially expressed across the three stages. The results indicated that symbiotic and asymbiotic germination may share a common signaling pathway during the early stage. In addition, 229 proteins were identified to be differentially expressed in only symbiotic germination. Among them, 32 candidates were upregulated in both transcriptomic and proteomic experiments including those encode for symbiotic signal transduction, defense reaction, storage protein utilization and metabolisms of N, carbohydrates and lipids (Table 1). Interestingly, several genes or proteins identified in this study, such as acyl-CoA-binding proteins, acetyl-CoA carboxylase, β-oxidation multifunctional protein, may promote the utilization of the stored reserves in the embryo cells due to their key roles in lipid metabolism. By using isotopolog profiling, Keymer et al. (2017) demonstrated that lipids can be directly transferred from host plants to fungal cells in arbuscular mycorrhizal symbioses [27]. It would be also interesting to know whether the stored lipid in orchid embryos can be transported to the fungal partners for establishing orchid-fungus symbioses in further studies. Taken together, transcriptomic and proteomic analyses lead to a comprehensive understanding of the molecular processes activated in the orchid mycorrhizas. In addition to nutrient transporter genes, a set of genes regulating plant defense responses and secondary metabolism is also induced by symbiosis with mycorrhizal fungi. To fully understand the molecular mechanisms of the orchid-fungus symbiosis, further characterization of the differentially expressed genes is required.
5. Conclusions and Future Perspectives
Under natural conditions, orchid seeds cannot germinate in the absence of symbiosis with mycorrhizal fungi. It has been long believed that orchids receive not only mineral nutrients, such as N and P, but also organic C from orchid mycorrhizal fungi without rewards at the germination and the early stages in protocorm development in autotrophic orchids and the entire life cycle in myco-heterotrophic orchids. The mutualistic interaction between orchids and fungi has been focused on the flow of C without considering other potential benefits required for fungi, such as offering of vitamins and other nutrients or protection of fungal hyphae in orchid roots [71]. The recent molecular evidence in the T. calospora-S. vomeracea symbiosis implies that orchids in non-photosynthetic stages may export ammonium produced from protein bodies in the embryo cells to attract mycorrhizal fungi for symbiosis. In turn, the symbiotic fungi supply N, P and C to the host orchids. However, in the case of adult green orchids, the host plants export photosynthetically fixed C to the fungal partners for exchanging mineral nutrients. Therefore, the nutrient flow in orchids could also represent mutualism as in other types of mycorrhizas. This means the tiny orchid seeds without endosperm still have to pay in order to get nutrients in return. In addition, the partially myco-heterotrophic orchids (mixotrophs) have been shown to keep receiving C from mycorrhizal fungi even after initiation of photosynthesis and their distribution may be much wider among orchids than presumed previously [72]. Several orchid and fungal importer genes have been identified; however, how fungi export the nutrients to their host orchids is still not clear. Continued study of nutrient transport system in orchid mycorrhizas would help to understand the establishment and maintenance of the interaction and assist orchid conservation in the future. Furthermore, the efforts may also contribute to better understand other types of mycorrhizas due to the easier manipulation of in vitro study of orchid mycorrhizas under axenic condition [29].
Recent advances in NGS technologies and functional genomics methodologies provide an opportunity for a comprehensive understanding of orchid biology and offer new insights into the symbiotic relationship between orchids and orchid mycorrhizal fungi. However, the orchid-fungus symbiosis appears to be much more complex than previously thought, leading to difficulties in unravelling the mechanisms. For example, wild orchids often form symbioses with multiple fungi species within a single root [57] and the tripartite relationship among orchids, mycorrhizal fungi and the adjacent trees for nutrient exchange has been proved to be common in nature. Therefore, the future research directions in orchid mycorrhizas should consider how these three kinds of organisms interact with each other and the environment.
Author Contributions
Conceptualization, C.-M.Y. and W.-C.T.; literature search, C.-M.Y., K.C. and C.-K.L.; writing-original draft, C.-M.Y., K.C. and C.-K.L.; writing-review & editing, C.-M.Y., K.C., C.-K.L. and W.-C.T.; supervision, C.-M.Y. and W.-C.T.; funding acquisition, W.-C.T.
Funding
This research was funded by Ministry of Science and Technology, Taiwan, grant number 107-2313-B-006-002-MY3.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mohammadi, K.; Khalesro, S.; Sohrabi, Y.; Heidari, G. A Review: Beneficial Effects of the Mycorrhizal Fungi for Plant Growth. J. Appl. Environ. Biol. Sci. 2011, 1, 310–319. [Google Scholar]
- Pérez-Brocal, V.; Latorre, A.; Moya, A. Symbionts and Pathogens: What is the Difference? Curr. Top. Microbiol. Immunol. 2013, 358, 215–243. [Google Scholar] [PubMed]
- Bonfante, P.; Genre, A. Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat. Commun. 2010, 1, 48. [Google Scholar] [CrossRef] [PubMed]
- Latef, A.A.H.A.; Hashem, A.; Rasool, S.; Abd_Allah, E.F.; Alqarawi, A.A.; Egamberdieva, D.; Jan, S.; Anjum, N.A.; Ahmad, P. Arbuscular mycorrhizal symbiosis and abiotic stress in plants: A review. J. Plant Biol. 2016, 59, 407–426. [Google Scholar]
- Singh, L.P.; Gill, S.S.; Tuteja, N. Unravelling the role of fungal symbionts in plant abiotic stress tolerance. Plant Signal. Behav. 2011, 6, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Singh, A. Molecular basis of plant-symbiotic fungi interaction: An overview. SCI WORLD J. 2007, 5, 115–131. [Google Scholar] [CrossRef]
- Behie, S.W.; Bidochka, M.J. Nutrient transfer in plant-fungal symbioses. Trends Plant Sci. 2014, 19, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Peterson, R.L.; Massicotte, H.B.; Melville, L.H. Mycorrhizas: Anatomy and Cell Biology; National Research Council Research Press: Ottawa, ON, Canada, 2004. [Google Scholar]
- Cosme, M.; Fernández, I.; Van der Heijden, M.G.A.; Pieterse, C.M.J. Non-mycorrhizal plants: The exceptions that prove the rule. Trends Plant Sci. 2018, 23, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Gardes, M. An orchid-fungus marriage-physical promiscuity, conflict and cheating. New Phytol. 2003, 154, 46. [Google Scholar] [CrossRef]
- Herrera, H.; Valadares, R.; Contreras, D.; Bashan, Y.; Arriagada, C. Mycorrhizal compatibility and symbiotic seed germination of orchids from the Coastal Range and Andes in south central Chile. Mycorrhiza 2017, 27, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Arditti, J. Factors affecting the germination of orchid seeds. Bot. Rev. 1967, 33, 1–97. [Google Scholar] [CrossRef]
- Kauth, P.J.; Dutra, D.; Johnson, T.R.; Stewart, S.L.; Kane, M.E.; Vendram, W. Floriculture, Ornamental and Plant Biotechnology: Advances and Topical Issues, 5, Global Science Books, Techniques and Applications of In Vitro Orchid Seed Germination; Global Science Books, Ltd.: Ikenobe, Japan, 2008. [Google Scholar]
- Yamazaki, J.; Miyoshi, K. In vitro asymbiotic germination of immature seed and formation of protocorm by Cephalanthera falcata (Orchidaceae). Ann. Bot. 2006, 98, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Dearnaley, J.D.W. Further advances in orchid mycorrhizal research. Mycorrhiza 2007, 17, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, H.N.; Rasmussen, F.N. Climactic and seasonal regulation of seed plant establishment in Dactylorhiza majalis inferred from symbiotic experiments in vitro. Lindleyana 1991, 6, 221–227. [Google Scholar]
- Yeung, E.C. A perspective on orchid seed and protocorm development. Bot. Stud. 2017, 33, 1–14. [Google Scholar] [CrossRef]
- Peterson, R.L.; Farquhar, M.L. Mycorrhizas: Integrated development between roots and fungi. Mycologia 1994, 86, 311–326. [Google Scholar]
- Rasmussen, H.N. Cell differentiation and mycorrhizal infection in Dactylorhiza majalis (Rchb. F.) Hunt & Summerh. (Orchidaceae) during germination in vitro. New Phytol. 1990, 116, 137–143. [Google Scholar]
- Rasmussen, H.N.; Dixon, K.W.; Jersáková, J.; Těšitelová, T. Germination and seedling establishment in orchids: A complex of requirements. Ann. Bot. 2015, 116, 391–402. [Google Scholar] [CrossRef]
- Oehl, F.; Laczko, E.; Bogenrieder, A.; Stahr, K.; Bosch, R.; van der Hejiden, M.; Sieverding, E. Soil type and land use intensity determine the composition of arbuscular mycorrhizal fungal communities. Soil Biol. Biochem. 2010, 42, 724–738. [Google Scholar] [CrossRef]
- Xu, X.; Chen, C.; Zhang, Z.; Sun, S.; Chen, Y.; Jiang, J.; Shen, Z. The influence of environmental factors on communities of arbuscular mycorrhizal fungi associated with Chenopodium ambrosioides revealed by MiSeq sequencing investigation. Sci. Rep. 2017, 7, 45134. [Google Scholar] [CrossRef] [PubMed]
- Van der Heijden, M.G.; Martin, F.M.; Selosse, M.A.; Sanders, I.R. Mycorrhizal ecology and evolution: The past, the present, and the future. New Phytol. 2015, 205, 1406–1423. [Google Scholar] [CrossRef] [PubMed]
- Bücking, H.; Kafle, A. Role of Arbuscular mycorrhizal fungi in the nitrogen uptake of plants: Current knowledge and research gaps. Agronomy 2015, 5, 587–612. [Google Scholar] [CrossRef]
- Wang, W.; Shi, J.; Xie, Q.; Jiang, Y.; Yu, N.; Wang, E. Nutrient Exchange and Regulation in arbuscular mycorrhizal symbiosis. Mol. Plant. 2017, 10, 1147–1158. [Google Scholar] [CrossRef] [PubMed]
- Jacquemyn, H.; Brys, R.; Waud, M.; Busschaert, P.; Lievens, B. Mycorrhizal networks and coexistence in species-rich orchid communities. New Phytol. 2015, 206, 1127–1134. [Google Scholar] [CrossRef]
- Keymer, A.; Pimprikar, P.; Wewer, V.; Huber, C.; Brands, M.; Bucerius, S.L.; Delaux, P.M.; Klingl, V.; Röpenack-Lahaye, E.V.; Wang, T.L.; et al. Lipid transfer from plants to arbuscular mycorrhiza fungi. Elife 2017, 20, 6. [Google Scholar] [CrossRef]
- Keymer, A.; Gutjahr, C. Cross-kingdom lipid transfer in arbuscular mycorrhiza symbiosis and beyond. Curr. Opin. Plant Biol. 2018, 44, 137–144. [Google Scholar] [CrossRef]
- Dearnaley, J.D.W.; Cameron, D.D. Nitrogen transport in the orchid mycorrhizal symbiosis—Further evidence for a mutualistic association. New Phytol. 2017, 213, 10–12. [Google Scholar] [CrossRef]
- Chen, A.; Gu, M.; Wang, S.; Chen, J.; Xu, G. Transport properties and regulatory roles of nitrogen in arbuscular mycorrhizal symbiosis. Semin. Cell Dev. Biol. 2018, 74, 80–88. [Google Scholar] [CrossRef]
- Selosse, M.A.; Martos, F. Do chlorophyllous orchids heterotrophically use mycorrhizal fungal carbon? Trends Plant Sci. 2014, 19, 683–685. [Google Scholar] [CrossRef] [PubMed]
- Leake, J.R. Plants parasitic on fungi: Unearthing the fungi in myco-heterotrophs and debunking the ‘saprophytic’ plant myth. Mycologist 2005, 19, 113–122. [Google Scholar] [CrossRef]
- Julou, T.; Burghardt, B.; Gebauer, G.; Berveiller, D.; Damesin, C.; Selosse, M.-A. Mixotrophy in orchids: Insights from a comparative study of green individuals and nonphotosynthetic individuals of Cephalanthera damasonium. New Phytol. 2005, 166, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, G.; Preiss, K.; Gebauer, A.C. Partial mycoheterotrophy is more widespread among orchids than previously assumed. New Phytol. 2016, 211, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Girlanda, M.; Selosse, M.-A.; Cafasso, D.; Brilli, F.; Delfine, S.; Fabian, R.; Ghigone, P.; Pinelli, P.; Segreto, R.; Loreto, F.; et al. Inefficient photosynthesis in the Mediterranean orchid Limodorum abortivum is mirrored by specific association to ectomycorrhizal Russulaceae. Mol. Ecol. 2006, 15, 491–504. [Google Scholar] [CrossRef] [PubMed]
- Lallemand, F.; Figura, T.; Damesin, C.; Fresneau, C.; Griveau, C.; Fontaine, N.; Zeller, B.; Selosse, M.A. Mixotrophic orchids do not use photosynthates for perennial underground organs. New Phytol. 2019, 221, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Fochi, V.; Chitarra, W.; Kohler, A.; Voyron, S.; Singan, V.R.; Lindquist, E.A.; Barry, K.W.; Girlanda, M.; Grigoriev, I.V.; Martin, F.; et al. Fungal and plant gene expression in the Tulasnella calospora-Serapias vomeracea symbiosis provides clues about N pathways in orchid mycorrhizas. New Phytol. 2017, 213, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Richardson, K.A.; Peterson, R.L.; Currah, R.S. Seed reserves and early symbiotic protocorm development of Platanthera hyperborea (Orchidaceae). Can. J. Bot. 1992, 70, 291–300. [Google Scholar] [CrossRef]
- Yeung, E.C.; Zee, S.Y.; Ye, X.L. Embryology of Cymbidium sinense: Embryo development. Ann. Bot. 1996, 78, 105–110. [Google Scholar] [CrossRef]
- Cameron, D.D.; Leake, J.R.; Read, D.J. Mutualistic mycorrhiza in orchids: Evidence from plant-fungus carbon and nitrogen transfers in the green-leaved terrestrial orchid Goodyera repens. New Phytol. 2006, 171, 405–416. [Google Scholar] [CrossRef]
- Kuga, U.; Sakamoto, N.; Yurimoto, H. Stable isotope imaging reveals that both live and degenerating fungal pelotons transfer carbon and nitrogen to orchid protocorms. New Phytol. 2014, 202, 594–605. [Google Scholar] [CrossRef] [PubMed]
- McKendrick, S.L.; Leake, J.R.; Taylor, D.L.; Read, D.J. Symbiotic germination and development of myco-heterotrophic plants in nature: Ontogeny of Corallorhiza trifida and characterisation of its mycorrhizal fungi. New Phytol. 2000, 145, 523–537. [Google Scholar] [CrossRef]
- McKendrick, S.L.; Leake, J.R.; Read, D.J. Symbiotic germination and development of myco-heterotrophic plants in nature: Transfer of carbon from ectomycorrhizal Salix repens and Betula pendula to the orchid Corallorhiza trifida through shared hyphal connections. New Phytol. 2000, 145, 539–548. [Google Scholar] [CrossRef]
- Campbell, E.O. Morphology of the fungal association in three species of Corallorhiza in Michigan. Mich. Bot. 1970, 9, 108–113. [Google Scholar]
- Zelmer, C.D.; Currah, R.S. Evidence for a fungal liaison between Corallorhiza trifida (Orchidaceae) and Pinus contorta (Pinaceae). Can. J. Bot. 1995, 73, 862–866. [Google Scholar] [CrossRef]
- Taylor, D.L.; Bruns, T.D. Independent, specialized invasions of ectomycorrhizal mutualism by two nonphotosynthetic orchids. Proc. Natl. Acad. Sci. USA 1997, 94, 4510–4515. [Google Scholar] [CrossRef] [PubMed]
- Bougoure, J.J.; Brundrett, M.C.; Grierson, P.F. Carbon and nitrogen supply to the underground orchid, Rhizanthella gardneri. New Phytol. 2010, 186, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Bidartondo, M.I.; Burghardt, B.; Gebauer, G.; Bruns, T.D.; Read, D.J. Changing partners in the dark: Isotopic and molecular evidence of ectomycorrhizal liaisons between forest orchids and trees. Proc. R. Soc. Lond. B 2004, 271, 1799–1806. [Google Scholar] [CrossRef]
- Weiss, M.; Selosse, M.A.; Rexer, K.H.; Urban, A.; Oberwinkler, F. Sebacinales: A hitherto overlooked cosm of heterobasidiomycetes with broad mycorrhizal potential. Mycol. Res. 2004, 108, 1003–1010. [Google Scholar] [CrossRef]
- Waterman, R.J.; Bidartondo, M.I. Deception above, deception below: Linking pollination and mycorrhizal biology of orchids. J. Exp. Bot. 2008, 59, 1085–1096. [Google Scholar] [CrossRef]
- Cai, J.; Liu, X.; Vanneste, K.; Proost, S.; Tsai, W.C.; Liu, K.W.; Chen, L.J.; He, Y.; Xu, Q.; Bian, C.; et al. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 2015, 47, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Wang, X.; Liu, H.; Tian, Y.; Lian, J.; Yang, R.; Hao, S.; Wang, X.; Yang, S.; Li, Q.; et al. The genome of Dendrobium officinale illuminates the biology of the important traditional Chinese orchid herb. Mol. Plant 2015, 8, 922–934. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.Q.; Xu, Q.; Bian, C.; Tsai, W.C.; Yeh, C.M.; Liu, K.W.; Yoshida, K.; Zhang, L.S.; Chang, S.B.; Chen, F.; et al. The Dendrobium catenatum lindl. Genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016, 6, 19029. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.Q.; Liu, K.W.; Li, Z.; Lohaus, R.; Hsiao, Y.Y.; Niu, S.C.; Wang, J.Y.; Lin, Y.C.; Xu, Q.; Chen, L.J.; et al. The Apostasia genome and the evolution of orchids. Nature 2017, 549, 379–383. [Google Scholar] [CrossRef]
- Yuan, Y.; Jin, X.; Liu, J.; Zhao, X.; Zhou, J.; Wang, X.; Wang, D.; Lai, C.; Xu, W.; Huang, J.; et al. The Gastrodia elata genome provides insights into plant adaptation to heterotrophy. Nat. Commun. 2018, 9, 1615. [Google Scholar] [CrossRef] [PubMed]
- Chao, Y.T.; Chen, W.C.; Chen, C.Y.; Ho, H.Y.; Yeh, C.H.; Kuo, Y.T.; Su, C.L.; Yen, S.H.; Hsueh, H.Y.; Yeh, J.H.; et al. Chromosome-level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol. J. 2018, 16, 2027–2041. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, J.; Chen, C.; Yang, J.; Zhu, H.; Liu, M.; Lv, F. Deep sequencing-based comparative transcriptional profiles of Cymbidium hybridum roots in response to mycorrhizal and non-mycorrhizal beneficial fungi. BMC Genom. 2014, 15, 747. [Google Scholar] [CrossRef] [PubMed]
- Perotto, S.; Rodda, M.; Benetti, A.; Sillo, F.; Ercole, E.; Rodda, M.; Girlanda, M.; Murat, C.; Balestrini, R. Gene expression in mycorrhizal orchid protocorms suggests a friendly plant-fungus relationship. Planta 2014, 239, 1337–1349. [Google Scholar] [CrossRef]
- Liu, S.S.; Chen, J.; Li, S.C.; Zeng, X.; Meng, Z.X.; Guo, S.X. Comparative transcriptome analysis of genes involved in GA-GID1-DELLA regulatory module in symbiotic and asymbiotic seed germination of Anoectochilus roxburghii (Wall.) Lindl. (Orchidaceae). Int. J. Mol. Sci. 2015, 16, 30190–30203. [Google Scholar] [CrossRef]
- Tsai, C.C.; Wu, K.M.; Chiang, T.Y.; Huang, C.Y.; Chou, C.H.; Li, S.J.; Chiang, Y.C. Comparative transcriptome analysis of Gastrodia elata (Orchidaceae) in response to fungus symbiosis to identify gastrodin biosynthesis-related genes. BMC Genom. 2016, 17, 212. [Google Scholar] [CrossRef]
- Chen, J.; Liu, S.S.; Kohler, A.; Yan, B.; Luo, H.M.; Chen, X.M.; Guo, S.X. iTRAQ and RNA-Seq analyses provide new insights into regulation mechanism of symbiotic germination of Dendrobium officinale seeds (Orchidaceae). J. Proteome Res. 2017, 16, 2174–2187. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu, K.; Yamato, M.; Miura, C.; Yamaguchi, K.; Takahashi, K.; Ida, Y.; Shigenobu, S.; Kaminaka, H. Comparison of green and albino individuals of the partially mycoheterotrophic orchid Epipactis helleborine on molecular identities of mycorrhizal fungi, nutritional modes and gene expression in mycorrhizal roots. Mol. Ecol. 2017, 26, 1652–1669. [Google Scholar] [CrossRef] [PubMed]
- Miura, C.; Yamaguchi, K.; Miyahara, R.; Yamamoto, T.; Fuji, M.; Yagame, T.; Imaizumi-Anraku, H.; Yamato, M.; Shigenobu, S.; Kaminaka, H. The mycoheterotrophic symbiosis between orchids and mycorrhizal fungi possesses major components shared with mutualistic plant-mycorrhizal symbioses. Mol. Plant Microbe Interact. 2018, 31, 1032–1047. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Valadares, R.B.; Perotto, S.; Santos, E.C.; Lambais, M.R. Proteome changes in Oncidium sphacelatum (Orchidaceae) at different trophic stages of symbiotic germination. Mycorrhiza 2014, 24, 349–360. [Google Scholar] [CrossRef] [PubMed]
- López-Chávez, M.Y.; Guillén-Navarro, K.; Bertolini, V.; Encarnación, S.; Hernández-Ortiz, M.; Sánchez-Moreno, I.; Damon, A. Proteomic and morphometric study of the in vitro interaction between Oncidium sphacelatum Lindl. (Orchidaceae) and Thanatephorus sp. Rg26 (Ceratobasidiaceae). Mycorrhiza 2016, 26, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.L.; Yang, J.Z.; Liu, S.; Chen, C.L.; Zhu, H.Y.; Cao, J.X. The colonization patterns of different fungi on roots of Cymbidium hybridum plantlets and their respective inoculation effects on growth and nutrient uptake of orchid plantlets. World J. Microbiol. Biotechnol. 2014, 30, 1993–2003. [Google Scholar] [CrossRef] [PubMed]
- Blakeman, J.P.; Mokahel, M.A.; Hadley, G. Effect of mycorrhizal infection on respiration and activity of some oxidase enzymes of orchid protocorms. New Phytol. 1976, 77, 697–704. [Google Scholar] [CrossRef]
- Pao, S.S.; Paulsen, I.T.; Saier, M.H. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998, 62, 1–34. [Google Scholar]
- Marini, A.M.; Soussi-Boudekou, S.; Vissers, S.; Andre, B. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell Biol. 1997, 17, 4282–4293. [Google Scholar] [CrossRef]
- Selosse, M.A. The latest news from biological interactions in orchids: In love, head to toe. New Phytol. 2014, 202, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Stöckel, M.; Těšitelová, T.; Jersáková, J.; Bidartondo, M.I.; Gebauer, G. Carbon and nitrogen gain during the growth of orchid seedlings in nature. New Phytol. 2014, 202, 606–615. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).