Featured Application
This study provides the validation of a DIVA qPCR assay to differentiate a vaccine strain from wild Salmonella sv. Typhimurium strains, thereby improving the efficiency and accuracy of microbiological control in poultry farms and ultimately supporting public health.
Abstract
Salmonella enterica subsp. enterica serovar Typhimurium is an important foodborne pathogen, and poultry products are a major source of human infection. Live attenuated vaccines for poultry are an effective tool for reducing the prevalence of infection, but vaccine strains must be differentiated from wild strains to ensure effective disease surveillance and control. This study reports the validation of the SalTypm&PriSal-T qPCR Duplex kit, a DIVA qPCR assay for the differentiation of the Primun Salmonella T vaccine from wild strains using DNA extracted from isolated colonies. Analytical specificity and sensitivity, as well as diagnostic specificity and sensitivity, were evaluated with optimal results. This qPCR assay significantly reduces the time required to obtain a diagnostic result compared to reference methods based on antibiogram differentiation. Notably, this is the first qPCR test available worldwide for distinguishing this vaccine from wild strains, providing a valuable tool for improving the efficiency and accuracy of Salmonella surveillance programs in poultry production systems.
1. Introduction
Salmonella is one of the most important human pathogens responsible for foodborne diseases worldwide. Non-typhoidal Salmonella enterica serotypes can colonize the intestinal tract and cause salmonellosis, which is characterised by self-limiting gastrointestinal symptoms, and can rarely spread through the bloodstream, causing sepsis. It is mainly acquired through the consumption of contaminated food or water. There are over 2600 serovars, each exhibiting different levels of virulence and the ability to infect specific animal hosts, with only a few infecting humans [1]. In 2023, salmonellosis was the second most frequently reported foodborne disease in the EU, with S. enterica subsp. enterica sv. Typhimurium being the second most common serovar implicated in human infections [2]. While Enteritidis is the most frequently reported serovar in industrialized countries, Typhimurium is more prevalent in developing countries [3]. Effective monitoring and control measures are essential to reduce the prevalence of Salmonella in food sources and hence the number of human cases of salmonellosis [4].
Eggs and poultry meat are the main sources of Salmonella infections in humans [5,6]. Control of Salmonella in poultry is therefore a priority for the industry and public health authorities. However, Salmonella infections in poultry caused by serotypes with a broad host range, such as Typhimurium, Enteritidis or Indiana, are usually asymptomatic and are therefore difficult to detect [7]. Nonetheless, they can transmit salmonellosis to humans via the food chain. Strategies to reduce its prevalence include biosecurity measures, regular testing, and vaccination programs [8,9].
The use of live attenuated vaccines is an effective approach to controlling S. enterica subsp. enterica sv. Typhimurium infections in poultry [10]. Primun Salmonella T vaccine strain ST CAL16 Str+/Rif+/Enr- (Calier, León, Spain) is a live attenuated vaccine that can be used in breeding hens, laying hens, and broilers to reduce the prevalence of S. enterica subsp. enterica sv. Typhimurium in eggs and poultry meat. While live attenuated vaccines are effective in reducing disease incidence, they can complicate the diagnostic process because they can be misidentified as wild virulent strains. Consequently, detection methods that can distinguish the vaccine strain from wild strains are required for the safe use of a live vaccine in the field.
Wild and Primun Salmonella T vaccine strains can be distinguished by their antimicrobial resistance profiles. Unlike the field strains, the vaccine strain is sensitive to enrofloxacin and resistant to streptomycin and rifampicin. Therefore, the initial method for differentiating Primun Salmonella T vaccine strain from wild strains relied on antibiograms. However, this approach presents significant limitations as it depends on the stability of resistance patterns among circulating wild strains. Wild strains may eventually show the same antibiotic resistance profile as the ST CAL16 Str+/Rif+/Enr- strain [11]. In fact, antibiotic overuse has led to an increase in antimicrobial resistance [12]. Consequently, the antibiogram method could fail to differentiate the vaccine strain from wild strains.
The development of a more accurate and easy-to-use technique for differentiating infected from vaccinated animals (DIVA) is essential to ensure a rapid and appropriate response to detected infections and the effectiveness of vaccination programs. Indeed, incorrect characterisation of wild strains as vaccine strains could lead to a salmonellosis outbreak, while incorrect characterisation of vaccine strains as wild strains could lead to inappropriate slaughtering of animals and important economic losses. DIVA assays typically rely on the differentiation of specific antigens, genetic markers, or phenotypic traits unique to the vaccine strain. Molecular approaches, particularly those based on polymerase chain reaction (PCR) technologies, have emerged as the most promising tools due to their superior sensitivity, specificity, and turnaround time compared to conventional microbiological methods. A duplex qPCR assay was specifically designed for the accurate and reliable differentiation of wild S. enterica subsp. enterica sv. Typhimurium strains (SalTypm assay) and the Primun Salmonella T vaccine (PriSal-T assay) from isolated colonies. This qPCR-based DIVA assay has the advantage of being significantly faster than antibiogram-based techniques; however, it is important to ensure its sensitivity and specificity.
In this study, the SalTypm&PriSal-T qPCR Duplex kit was validated by assessing its analytical and diagnostic performance.
2. Materials and Methods
2.1. Bacterial Strains
To assess the analytical specificity of the kit, the Primun Salmonella T vaccine strain and six Salmonella strains obtained from the Colección Española de Cultivos Tipo (CECT, Valencia, Spain) were used: Salmonella enterica subsp. enterica sv. Typhi CECT 409T, Salmonella enterica subsp. enterica sv. Typhimurium CECT 443, Salmonella enterica subsp. enterica sv. Paratyphi A CECT 825, Salmonella enterica subsp. enterica sv. Enteritidis CECT 4155, Salmonella enterica subsp. arizonae CECT 4395, and Salmonella enterica subsp. salamae CECT 4000T.
Diagnostic validation of the kit was performed using 41 wild S. enterica subsp. enterica sv. Typhimurium strains and 10 Primun Salmonella T vaccine strains. Thirty-seven wild strains and two vaccine strains isolated by three laboratories of the Spanish National Salmonella Control Program and four additional wild strains and two vaccine strains were used. All strains were previously characterised as S. enterica subsp. enterica sv. Typhimurium by plate culture and serotyping (according to ISO 6887-6:2013 and ISO 6579-1:2017 [13,14]) and wild and vaccine strains were differentiated by antibiogram. The vaccine strain is sensitive to enrofloxacin and resistant to streptomycin and rifampicin. Of the 41 wild strains (10 from stool samples and 31 from environmental samples), 21 were isolated from laying hens, 11 from broilers, 2 from breeding hens, and 7 were of undetermined origin. Of the 10 vaccine strains, 2 were isolated from stool samples of laying hens and 8 were vaccine samples from different stages of production.
2.2. DNA Extraction
DNA was extracted from isolated colonies using the GPStraction DNA colonies rapid extraction kit (GPS™, Orihuela, Spain). Briefly, the biomass of a single colony was resuspended in 200 µL of Colony Extraction Buffer by vigorous vortexing and kept at −20 °C for at least 5 min. The extracted DNA can be used directly for qPCR analysis.
2.3. qPCR Tests
For qPCR analysis using the SalTypm&PriSal-T qPCR Duplex kit (GPS™, Orihuela, Spain), reaction mixtures were prepared according to the manufacturer’s instructions using 5 µL of extracted DNA per reaction. Experiments were performed on a QuantStudio5 (Applied Biosystems, Waltham, MA, USA) or a CFX96 (Bio-Rad, Hercules, CA, USA) thermocycler. The thermal protocol consisted of an initial activation step at 95 °C for 2 min, followed by 40 amplification cycles organised as follows: denaturation at 95 °C for 5 s, annealing/extension at 60 °C for 20 s, and data acquisition. Positive control, negative control (nuclease-free water), and internal control were included in all assays. The internal control included in the kit is directed against an external target, the concentration of which is minimised to detect the presence of possible qPCR inhibitors in the case of negative samples without affecting the sensitivity of the main targets. Fluorogenic signals were detected using the FAM channel for the PriSal-T, the Cy5 channel for SalTypm, and the HEX channel for the internal control. Calibration of the qPCR was performed using a standard curve prepared from 10-fold serial dilutions from 106 to 10 copies of a standard template.
2.4. Analytical Specificity
Analytical specificity, as defined by the WOAH, is the ability of a qPCR assay to recognize only the target sequence but no other homologous sequences belonging to different related strains [15]. To evaluate the analytical specificity of the duplex qPCR assay, several strains were tested in vitro. The assay was performed with 5 µL of extracted DNA from Primun Salmonella T vaccine strain, which should give a positive result for both assays, Salmonella enterica subsp. enterica serovar Typhimurium, which should be positive for SalTypm assay and negative for PriSal-T assay, and Salmonella enterica subsp. enterica sv. Enteritidis, Salmonella enterica subsp. enterica serovar Typhi, Salmonella enterica subsp. enterica serovar Paratyphi A, Salmonella enterica subsp. arizonae, and Salmonella enterica subsp. salamae, which should be negative for both assays.
2.5. Analytical Sensitivity
Analytical sensitivity is defined as the lowest quantity of target nucleic acid that can be reliably detected and quantified by the assay. The SalTypm&PriSal-T qPCR Duplex assay in MONODOSE format was validated according to the recommendations outlined in the international standard ISO/IEC 17025:2017 and following internal quality protocols [16,17,18,19]. Standard curves were generated with 10-fold serial dilutions from 106 to 10 copies of the standard template, which is a synthetic DNA fragment containing the target of the assay. One technician (Technician A) ran 10 standard curves with one template decimal dilution and another technician (Technician B) ran 5 standard curves with 5 different template decimal dilutions. To obtain statistically relevant results, all of the parameters were evaluated with at least 10 replicates. The slope, coefficient of determination (R2), and efficiency (e) of the mean of the 15 standard curves were calculated for each assay (SalTypm and PriSal-T). The validation of the linear model was performed with a Fisher test with a 95% confidence interval using the efficiency of 5 standard curves generated by Technician A and 5 standard curves generated by Technician B. Reliability (repeatability and reproducibility), limit of detection (LOD), and limit of quantification (LOQ) were determined for each qPCR assay. Repeatability was assessed by calculating the coefficient of variation of each dilution for the 10 standard curves generated by Technician A. Reproducibility was assessed by calculating the coefficient of variation of each dilution between the 10 standard curves generated by Technician A and the 5 standard curves generated by Technician B. The LOD was determined by assessing the reproducibility of the detection of the lowest dilution (10 copies). The LOQ was determined by performing a Student’s t-test using the Cts obtained for the 10-copies dilutions of the 10 standard curves generated by Technician A and the 5 standard curves generated by Technician B.
2.6. Diagnostic Performance
Diagnostic performance refers to the accuracy of a test in identifying the presence or absence of a disease, typically measured by diagnostic specificity (proportion of negatives correctly identified) and diagnostic sensitivity (proportion of positives correctly identified) [15]. These parameters were assessed by testing 5 µL of extracted DNA from 41 wild S. enterica subsp. enterica sv. Typhimurium strains and 10 Primun Salmonella T vaccine strains from the Spanish National Salmonella Control Program with the SalTypm&PriSal-T qPCR Duplex assay.
3. Results
The SalTypm&PriSal-T qPCR Duplex kit, previously developed by GPS™ and commercially available, consists of a duplex assay for the simultaneous detection of Salmonella enterica subsp. enterica serovar Typhimurium and the Primun Salmonella T vaccine strain. The analytical specificity of the duplex assay was validated in vitro using genetic material from the Primun Salmonella T vaccine strain ST CAL16 Str+/Rif+/Enr-, S. enterica subsp. enterica sv. Typhimurium, S. enterica subsp. enterica sv. Enteritidis, S. enterica subsp. enterica sv. Typhi, S. enterica subsp. enterica sv. Paratyphi A, S. enterica subsp. arizonae, and S. enterica subsp. salamae. The internal control of the assay amplified in all samples, which indicates that there is no PCR inhibition (Figure 1). The PriSal-T assay gave positive results only for the Primun Salmonella T vaccine strain, while the SalTypm assay yielded positive results for both the Primun Salmonella T vaccine strain and the S. enterica subsp. enterica sv. Typhimurium reference strain (Figure 1). The other S. enterica subspecies and serovars tested were negative for both assays. Therefore, the PriSal-T assay was specific for the Primun Salmonella T strain, and the SalTypm assay was inclusive for wild and vaccine strains of S. enterica subsp. enterica sv. Typhimurium and exclusive for the other S. enterica subspecies and serovars tested.
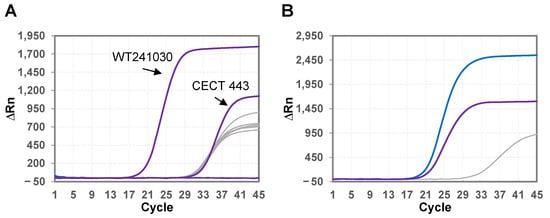
Figure 1.
Amplification plots of (A) S. enterica subsp. enterica sv. Typhimurium strains WT241030 and CECT 443, S. enterica subsp. enterica sv. Typhi CECT 409T, S. enterica subsp. enterica sv. Paratyphi A CECT 825, S. enterica subsp. enterica sv. Enteritidis CECT 4155, S. enterica subsp. arizonae CECT 4395, and S. enterica subsp. salamae CECT 4000T and (B) Primun Salmonella T vaccine (ST0116-MSB). The figure shows amplification with the SalTypm assay in purple (Cy5), the PriSal-T assay in blue (FAM), and the internal control in grey (HEX).
Several analytical sensitivity parameters of the duplex assay were evaluated in relation to established acceptance criteria. Each parameter was tested at least 10 times to ensure reproducibility. The study included qPCR standard curves generated using multiple template dilutions across six decimal ranges (from 106 copies to 10 copies). The template used for these assays is included in the kit as a positive control and can be used to generate a standard curve from 104 to 10 copies. For each individual assay, the slope (a) and coefficient of determination (R2) were calculated for the mean of 15 standard curves to assess the linear regression. The SalTypm and PriSal-T assays had slopes of −3.322 and −3.321, respectively, and both achieved an R2 value of 1.000, meeting the criteria for acceptability. A Fisher test with a 95% confidence interval and n = 10 was performed to validate the linear model. The Ffisher value was 5.318 for both assays, and the Fassay values were 0.572 and 0.145 for the SalTypm and PriSal-T assays, respectively. Since the Fassay values were lower than the Ffisher value, the linear model was validated for both assays. The efficiency was 100.0% for the mean of 15 standard curves for both assays, falling within the acceptability range (75–125%). Repeatability is the ability of a test to produce the same results under the same conditions, while reproducibility is the ability of a measurement or test to produce consistent results under different conditions. Standard curves were performed by the same technician with one template decimal dilution to evaluate the repeatability of the assays and by two technicians with different template decimal dilutions to evaluate reproducibility. The coefficient of variation (CV) of the Ct values ranged from 0.37% to 1.39% for repeatability, and from 0.92% to 2.43% for reproducibility in the SalTypm assay and from 0.28% to 1.30% for repeatability, and from 1.08% to 1.83% for reproducibility in the PriSal-T assay. All CV values were below 10% and therefore the repeatability and the reproducibility results were acceptable. Both assays were thus repeatable and reproducible. The limit of detection (LOD) and the limit of quantification (LOQ) are the lowest number of copies that can be detected and quantified. In these experiments, the lowest number of copies analysed was 10 copies. The LOD for 10 genomic copies was 100% reproducible for both assays (all 10 copies dilutions were detected) and the LOQ for 10 genomic copies was accepted, as the t-values for the SalTypm and PriSal-T assays (1.500 and 0.581 respectively) were lower than the theoretical value from a Student’s t-table (tstudent = 2.145 for both assays). All these parameters met the acceptance criteria and the corresponding values are summarised in Table 1.

Table 1.
Obtained values, criteria for acceptance, and results of the SalTypm&PriSal-T qPCR Duplex kit (n: number of replicates for each parameter).
The SalTypm&PriSal-T qPCR Duplex kit was subjected to diagnostic validation, and the results were compared with those previously obtained with the reference diagnostic method of the Spanish National Salmonella Control Program. Diagnostic specificity, defined as the ability of the test to correctly identify positive cases, and diagnostic sensitivity, defined as the ability of a test to correctly identify negative cases, were evaluated with 51 isolated colonies (41 wild S. enterica subsp. enterica sv. Typhimurium strains and 10 vaccine strains). DNA from each colony was extracted using the GPStraction DNA colonies kit (GPS™) and analysed by the qPCR assay. All samples were positive for the SalTypm assay as expected, since all samples are S. enterica subsp. enterica sv. Typhimurium strains (vaccine or wild strains). The results of the SalTypm&PriSal-T qPCR Duplex kit were in full agreement with the reference method, yielding a total of 10 positive samples and 41 negative samples for the PriSal-T assay (Table 2). Therefore, diagnostic specificity and diagnostic sensitivity were both 100%, resulting in an overall diagnostic efficiency of 100%.

Table 2.
Diagnostic specificity and sensitivity results for the SalTypm&PriSal-T qPCR Duplex kit.
4. Discussion
The comprehensive results obtained from the validation of the SalTypm&PriSal-T qPCR Duplex kit demonstrate its high specificity, sensitivity, and reliability in distinguishing between the vaccine and wild strains. These results provide a strong basis for discussing the implications of using this assay in the effective management of Salmonella infections within poultry production systems. The accurate differentiation between vaccinated and infected animals is crucial for ensuring the success of vaccination programs while maintaining robust surveillance and outbreak prevention strategies.
Bacteria of the genus Salmonella spp. are one of the most relevant foodborne pathogens worldwide, with S. enterica subsp. enterica sv. Typhimurium being one of the predominant serotypes associated with human infections. Consumption of poultry products is the primary source of transmission to humans. The use of live attenuated S. enterica subsp. enterica sv. Typhimurium vaccines, such as Primun Salmonella T, is considered an effective tool for reducing its prevalence in poultry flocks, thereby decreasing the risk of transmission along the food chain. Therefore, a rapid and reliable method to distinguish the vaccine strain from wild strains of S. enterica subsp. enterica sv. Typhimurium is essential to ensure a diagnosis that can distinguish infected from vaccinated animals. GPS™ has previously developed a qPCR DIVA assay that allows effective discrimination of Primun Salmonella T vaccine and wild strains from S. enterica subsp. enterica sv. Typhimurium colonies isolated following current normalised culturing. This qPCR assay, including a rapid DNA extraction step, provided accurate results in less than 40 min, whereas an antibiogram to identify strain ST CAL16 Str+/Rif+/Enr- may take 24 h or longer. This remarkable improvement in turnaround time facilitates timely decision-making for infection control measures.
The specificity of the SalTypm and PriSal-T assays was evaluated in vitro using reference genomic material. The SalTypm assay was specific for the detection of S. enterica subsp. enterica sv. Typhimurium, including the vaccine strain. Conversely, the PriSal-T assay specifically detected the vaccine strain but not wild S. enterica subsp. enterica sv. Typhimurium strains. The SalTypm&PriSal-T qPCR Duplex kit was subjected to analytical validation following the guidelines recommended by the international standard ISO/IEC 17025:2017 and the results obtained were optimal according to the ranges established as acceptance criteria (Table 1). These acceptance thresholds are those previously employed for the validation of other qPCR assays [17,18,19]. In summary, the linearity of the calibration curve was validated, with a high efficiency that ensures an optimal yield of PCR amplification. The method was considered repeatable and reproducible, and the limits of detection and quantification were established at 10 copies. The diagnostic performance was evaluated using 51 culture samples isolated from a diverse range of samples, including stool and environmental samples from laying hens, broilers, and breeding hens, all identified with the reference diagnostic method. All strains were correctly identified by the qPCR assay, showing optimal diagnostic specificity and sensitivity.
In conclusion, the results of the parameters analysed met the acceptance criteria established for validation and were in agreement with the reference diagnostic method, making the assay suitable to distinguish the vaccine strain from wild S. enterica subsp. enterica sv. Typhimurium obtained from isolated colonies. The ability to accurately distinguish between vaccinated and infected animals ensures the integrity of surveillance programs and prevents unnecessary culling of vaccinated animals. The availability of this DIVA assay is essential for the safe use of the Primum Salmonella T vaccine in the field. To the best of our knowledge, this is the first available qPCR test for this purpose. DIVA assays based on qPCR, ELISA, or chromogenic media are available for other Salmonella vaccines [20,21,22]. qPCR-based DIVA assays for other organisms have been validated with optimal diagnostic results [23,24]. However, validations of Salmonella qPCR DIVA assays have not been published, to the best of our knowledge. This product was registered in the Registry of Entities and Animal Health Products under the Ministry of Agriculture, Fisheries and Food of Spain (MAPA) with the following references: entity authorization number R-10046 and SalTypm&PriSal-T qPCR Duplex registration number 12186-RD.
The SalTypm&PriSal-T qPCR Duplex assay has been conceived as an identification method for the analysis of S. enterica subsp. enterica sv. Typhimurium colonies obtained from pure cultures following ISO 6887-6:2013 and ISO 6579-1:2017 standards. While the current assay targets isolated colonies, the authors of the present work have suggested undertaking future validations using the SalTypm&PriSal-T qPCR Duplex kit as a detection method directly on DNA extracted from stool or environmental samples, with simultaneous detection of S. enterica subsp. enterica sv. Typhimurium and discrimination of the Primun Salmonella T vaccine from wild strains, which would significantly reduce the time needed to obtain a diagnosis. A significant part of the proposed work would focus on the method of sampling and DNA extraction/purification in order to increase the sensitivity of the method.
In conclusion, the SalTypm&PriSal-T qPCR Duplex kit represents a significant step forward in the use of Primun Salmonella T vaccine, providing a rapid and reliable method for differentiating vaccine and wild strains, ultimately contributing to safer food production and public health protection.
Author Contributions
Conceptualization, A.M.-M., A.N., A.G.-S., P.R., C.M.-P., J.M.R. and A.B.; methodology, A.M.-M., A.N., A.G.-S., P.R. and C.M.-P.; formal analysis, A.M.-M., A.N., A.G.-S., P.R. and C.M.-P.; writing—original draft, A.M.-M., A.N., A.G.-S., P.R. and C.M.-P.; writing—review and editing, A.M.-M., A.N., A.G.-S., P.R. and C.M.-P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
A.M.-M. is a professor at the University Miguel Hernández and a scientific advisor at Genetic PCR Solutions™. A.N., A.G.-S., P.R. and C.M.-P. are employees of Genetic PCR Solutions™. J.-M.R. and A.B. are employees of Laboratorios Calier S.A.
References
- Wallis, T.S. Host-Specificity of Salmonella Infections in Animal Species. In Salmonella Infections: Clinical, Immunological and Molecular Aspects; Maskell, D., Mastroeni, P., Eds.; Advances in Molecular and Cellular Microbiology; Cambridge University Press: Cambridge, UK, 2006; pp. 57–88. ISBN 978-0-521-83504-6. [Google Scholar]
- EFSA; ECDC. The European Union One Health 2023 Zoonoses Report. EFSA J. 2024, 22, e9106. [Google Scholar] [CrossRef]
- Nazir, J.; Manzoor, T.; Saleem, A.; Gani, U.; Bhat, S.S.; Khan, S.; Haq, Z.; Jha, P.; Ahmad, S.M. Combatting Salmonella: A Focus on Antimicrobial Resistance and the Need for Effective Vaccination. BMC Infect. Dis. 2025, 25, 84. [Google Scholar] [CrossRef] [PubMed]
- Ricke, S.C.; Kim, S.A.; Shi, Z.; Park, S.H. Molecular-Based Identification and Detection of Salmonella in Food Production Systems: Current Perspectives. J. Appl. Microbiol. 2018, 125, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Antunes, P.; Mourão, J.; Campos, J.; Peixe, L. Salmonellosis: The Role of Poultry Meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Shaji, S.; Selvaraj, R.K.; Shanmugasundaram, R. Salmonella Infection in Poultry: A Review on the Pathogen and Control Strategies. Microorganisms 2023, 11, 2814. [Google Scholar] [CrossRef] [PubMed]
- Rakov, A.V.; Mastriani, E.; Liu, S.-L.; Schifferli, D.M. Association of Salmonella Virulence Factor Alleles with Intestinal and Invasive Serovars. BMC Genom. 2019, 20, 429. [Google Scholar] [CrossRef] [PubMed]
- Sylejmani, D.; Musliu, A.; Ramadani, N.; Sparagano, O.; Hamidi, A. Associations Between the Level of Biosecurity and Occurrence of Dermanyssus gallinae and Salmonella spp. in Layer Farms. Avian Dis. 2016, 60, 454–459. [Google Scholar] [CrossRef] [PubMed]
- Ruvalcaba-Gómez, J.M.; Villagrán, Z.; Valdez-Alarcón, J.J.; Martínez-Núñez, M.; Gomez-Godínez, L.J.; Ruesga-Gutiérrez, E.; Anaya-Esparza, L.M.; Arteaga-Garibay, R.I.; Villarruel-López, A. Non-Antibiotics Strategies to Control Salmonella Infection in Poultry. Animals 2022, 12, 102. [Google Scholar] [CrossRef] [PubMed]
- Barrow, P.A. Salmonella Infections: Immune and Non-Immune Protection with Vaccines. Avian Pathol. 2007, 36, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.H.; Paul, N.C.; Sischo, W.C.; Crespo, R.; Guard, J. Population Dynamics and Antimicrobial Resistance of the Most Prevalent Poultry-Associated Salmonella Serotypes. Poult. Sci. 2017, 96, 687–702. [Google Scholar] [CrossRef] [PubMed]
- Crump, J.A.; Medalla, F.M.; Joyce, K.W.; Krueger, A.L.; Hoekstra, R.M.; Whichard, J.M.; Barzilay, E.J. Antimicrobial Resistance among Invasive Nontyphoidal Salmonella enterica Isolates in the United States: National Antimicrobial Resistance Monitoring System, 1996 to 2007. Antimicrob. Agents Chemother. 2011, 55, 1148–1154. [Google Scholar] [CrossRef] [PubMed]
- ISO 6887-6:2013; Microbiology of Food and Animal Feed—Preparation of Test Samples, Initial Suspension and Decimal Dilutions for Microbiological Examination. ISO: Geneva, Switzerland, 2013. Available online: https://www.iso.org/standard/53664.html (accessed on 19 February 2025).
- ISO 6579-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella. ISO: Geneva, Switzerland, 2017. Available online: https://www.iso.org/standard/56712.html (accessed on 19 February 2025).
- World Organization for Animal Health. Terrestrial Manual, Chapter 2.2.3. Development and Optimisation of Nucleic Acid Detection Assays. Online Access. 2024. Available online: https://www.woah.org/en/what-we-do/standards/codes-and-manuals/terrestrial-manual-online-access/ (accessed on 14 February 2025).
- ISO/IEC 17025:2017; General Requirements for the Competence of Testing and Calibration Laboratories. ISO/IEC: Geneva, Switzerland, 2017. Available online: https://www.iso.org/standard/66912.html (accessed on 19 February 2025).
- Martínez-Murcia, A.; Navarro, A.; Garcia-Sirera, A.; Pérez, L.; Bru, G. Internal Validation of a Real-Time qPCR Kit Following the UNE/EN ISO/IEC 17025:2005 for Detection of the Re-Emerging Monkeypox Virus. Diagnostics 2023, 13, 1560. [Google Scholar] [CrossRef] [PubMed]
- Bru, G.; Martínez-Candela, M.; Romero, P.; Navarro, A.; Martínez-Murcia, A. Internal Validation of the ASFV MONODOSE Dtec-qPCR Kit for African Swine Fever Virus Detection under the UNE-EN ISO/IEC 17025:2005 Criteria. Vet. Sci. 2023, 10, 564. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Murcia, A.; Navarro, A.; Bru, G.; Chowdhary, A.; Hagen, F.; Meis, J.F. Internal Validation of GPSTM MONODOSE CanAur Dtec-qPCR Kit Following the UNE/EN ISO/IEC 17025:2005 for Detection of the Emerging Yeast Candida Auris. Mycoses 2018, 61, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Llorens, J.; Garcia, C.; Paulet, P.; Le-Tallec, B.; Dauphin, G.; Comte, S.; Catalá-Gregori, P.; Simon, F.; Sevilla-Navarro, S.; Sarabia, J. Research Note: Validation of a New Differentiation Approach Using the Commercial ASAPTM Media to Detect the Salmonella 441/014 Vaccine Strain. Poult. Sci. 2024, 103, 103679. [Google Scholar] [CrossRef] [PubMed]
- Aribam, S.D.; Nakayama, M.; Ichimura, S.; Tokuyama, K.; Hara, Y.; Ogawa, Y.; Shimoji, Y.; Eguchi, M. Differentiation of Salmonella Vaccinated and Infected Animals by Serological Detection of Antibody to T3SS Effector SsaK in an Indirect ELISA. J. Microbiol. Methods 2023, 209, 106729. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, J.; Kudlackova, H.; Kosina, M.; Kovarcik, K.; Tesarik, R.; Osvaldova, A.; Faldyna, M.; Matiasovic, J. A Proteomic Approach to the Development of DIVA ELISA Distinguishing Pigs Infected with Salmonella Typhimurium and Pigs Vaccinated with a Salmonella Typhimurium-Based Inactivated Vaccine. BMC Vet. Res. 2016, 12, 252. [Google Scholar] [CrossRef] [PubMed]
- Dijkman, R.; Feberwee, A.; Landman, W.J.M. Development, Validation and Field Evaluation of a Quantitative Real-Time PCR Able to Differentiate Between Field Mycoplasma synoviae and the MS-H-Live Vaccine Strain. Avian Pathol. 2017, 46, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Vidanović, D.; Tešović, B.; Šekler, M.; Debeljak, Z.; Vasković, N.; Matović, K.; Koltsov, A.; Krstevski, K.; Petrović, T.; De Leeuw, I.; et al. Validation of TaqMan-Based Assays for Specific Detection and Differentiation of Wild-Type and Neethling Vaccine Strains of LSDV. Microorganisms 2021, 9, 1234. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).