Abstract
The accurate identification of pills is essential for their safe administration in the medical field. Despite technological advancements, pill classification encounters hurdles such as ambiguous images, pattern similarities, mixed pills, and variations in pill shapes. A significant factor is the inability of 2D imaging to capture a pill’s 3D structure efficiently. Additionally, the scarcity of diverse datasets reflecting various pill shapes and colors hampers accurate prediction. Our experimental investigation shows that while color-based classification obtains a 95% accuracy rate, shape-based classification only reaches 66%, underscoring the inherent difficulty distinguishing between pills with similar patterns. In response to these challenges, we propose a novel system integrating Multi Combination Pattern Labeling (MCPL), a new method designed to accurately extract feature points and pill patterns. MCPL extracts feature points invariant to rotation and scale and effectively identifies unique edges, thereby emphasizing pills’ contour and structural features. This innovative approach enables the robust extraction of information regarding various shapes, sizes, and complex pill patterns, considering even the 3D structure of the pills. Experimental results show that the proposed method improves the existing recognition performance by about 1.2 times. By improving the accuracy and reliability of pill classification and recognition, MCPL can significantly enhance patient safety and medical efficiency. By overcoming the limitations inherent in existing classification methods, MCPL provides high-accuracy pill classification, even with constrained datasets. It substantially enhances the reliability of pill classification and recognition, contributing to improved patient safety and medical efficiency.
1. Introduction
Medication safety is a critical concern in healthcare due to its direct impact on patient health. The accurate identification of medications is essential, as errors can lead to serious adverse effects, including injuries and complications. Oral pills, widely used because of their stability and ease of administration, pose significant challenges, particularly for patients with memory problems. Reports from the Organization for Economic Co-operation and Development (OECD) indicate that medication errors contribute to 18.3% of adverse patient events [1], with a significant portion resulting from confusion over pill shapes and colors, as highlighted by studies in the British Medical Journal (BMJ) [2] and by the World Health Organization (WHO) [3]. This problem is exacerbated during the medication cycle—prescribing, dispensing, and administration—where physical characteristics of pills often lead to errors [4,5].
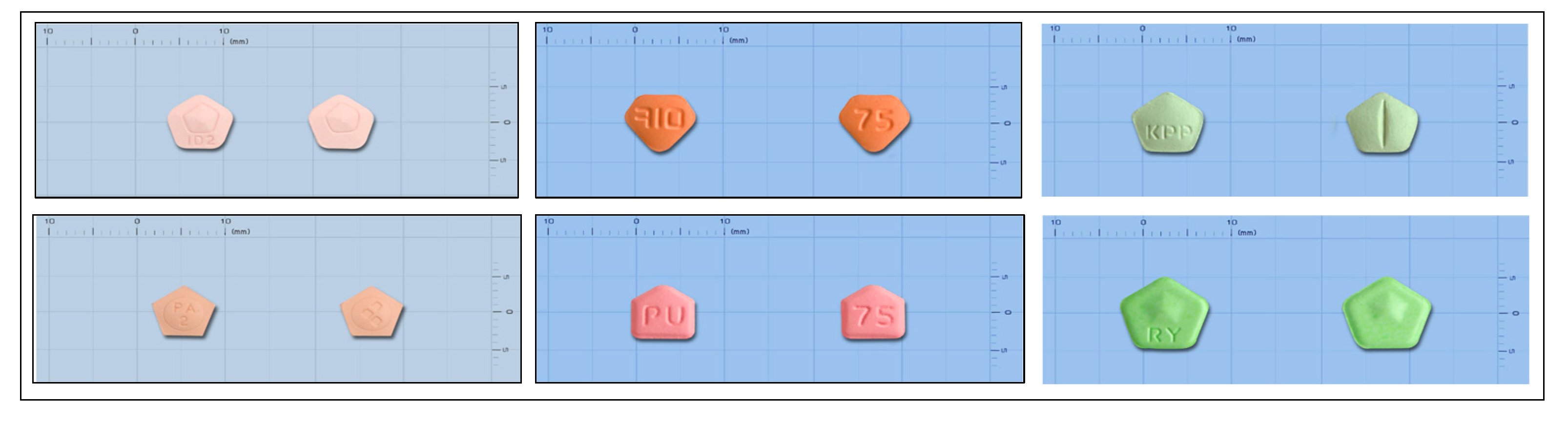
The variability in pill shapes, sizes, and colors, which may change over time due to factors like wear and discoloration, complicates reliable identification. Traditional machine learning models used for pill detection struggle to keep pace with the rapid introduction of new drugs, often failing to adapt to these changes, as evidenced by studies showing that nearly 25% of medications go unidentified in university hospitals [6,7]. Pentagonal pills present unique challenges among the various pill shapes due to their complex geometry, including pointed edges and a mix of curves and straight lines, as illustrated in Figure 1.

Figure 1.
Pentagonal pill shapes with subtle differences.
These identification challenges are compounded by rotation invariance and perspective distortion, where a pill’s orientation in an image can drastically alter its perceived shape. Variable lighting conditions that distort visual features further exacerbate such issues, underscoring the need for more sophisticated algorithms that can effectively navigate these complexities.
To address these issues, numerous studies have explored the potential of computer vision technology in pill detection. A widely used analytical technique involves comparing similarities between input and reference images based on shape, color, and imprinting features. These methods are broadly categorized into pixel-based and shape-based matching. Pixel-based methods calculate differences between corresponding pixels and compute similarity metrics such as the sum of squared differences or normalized cross-correlation [8,9]. Although effective against image distortion, they cannot detect changes in size or rotation. In contrast, shape-based matching extracts the subject’s region of interest (ROI) from a reference image, better accommodating object size and orientation variations, and excels in variable illumination settings [10,11,12].
Recent advancements in deep learning-based object-detection algorithms have been pivotal in developing self-learning and decision-making capabilities for pill-recognition solutions [13]. These neural networks excel in tasks such as object detection, biomedical diagnosis, and variant detection thanks to their ability to process and learn from large datasets [14,15,16,17]. Popular methods include You Only Look Once (YOLO), convolutional neural networks (CNNs), and recurrent neural networks (RNNs). Achieving high detection performance, however, necessitates extensive data due to the nuanced variability in pill shapes, textures, and colors. This variability demands a comprehensive approach that accounts for these subtle differences, making pill identification a complex visual classification challenge that requires precise differentiation between pill fragments.
To address these challenges, we propose the Multi-Combination Pattern Labeling (MCPL) method, a novel approach designed to enhance the accuracy and reliability of pill classification in challenging image conditions. The MCPL method extracts unique and representative features from pill images, providing a rich representation of the pill’s characteristics to improve classification performance. To accomplish this, MCPL combines features obtained from multiple established image feature-extraction algorithms: KAZE, Oriented FAST and Rotated BRIEF (ORB), Shi-Tomasi, and Harris. KAZE offers scale and rotation-invariant feature-detection robustness, capturing intricate details and broader patterns within images. ORB combines the FAST key point detector with the BRIEF descriptor for fast and efficient feature matching. Shi-Tomasi and Harris focus on corner detection, with Harris excelling under changes in rotation and illumination.
By combining these algorithms, MCPL comprehensively represents pill images, capturing a wide array of features, including shapes, patterns, and details that may be missed by using a single extraction method.
To effectively tackle these identification challenges, we employ the YOLO version 5 object-detection model to initially identify pills within images. This step is crucial as it sets the stage for more detailed analysis. Following this initial detection, our convolutional networks undertake a thorough analysis of the pills’ shape, color, and form, focusing particularly on those with complex geometries. This targeted approach is vital for the MCPL method to refine the feature-extraction processes, significantly enhancing classification accuracy and adapting to the diverse challenges presented by different pill types. The contributions of this study are as follows.
- Proposal of a new approach, Multi-Combination Pattern Labeling method, designed to enhance the accuracy and reliability of pill classification in challenging image conditions. The proposed method extracts unique and representative features from pill images, providing a rich representation of the pill’s characteristics to improve classification performance.
- Proposal of a pill-detection model-based MCPL to extract pills’ shape, color, and form, focusing particularly on those with complex geometries.
- Verification of the proposed methods through the experimental results.
This paper is organized as follows: Section 2 introduces the proposed method and summarizes related studies, providing the necessary context and background for our approach. Section 3 details the deep learning-based pill-classification system developed in this study, elucidating technical details and integration of the MCPL method. Section 4 elaborates further on the MCPL method, describing how it utilizes various image feature-extraction algorithms to surmount the identified challenges. Section 5 examines the experimental results, assessing the efficacy of the MCPL method across various scenarios and pill types. Finally, Section 6 presents our concluding remarks, discussing the implications of our findings and suggesting potential avenues for future research.
2. Related Studies
This section introduces the proposed method and presents a review of related works. In the present study, we employed a deep learning-based object-recognition model to identify pills with complex shapes. Object recognition, primarily a branch of computer vision and image processing, is concerned with the simultaneous classification and localization of objects within an image. DetectorNet [18] is a pioneering algorithm in deep learning-based object detection, marking this task’s first implementation of a Convolutional Neural Network (CNN) model. One of the landmark developments in object-detection technology is the region-based CNN (R-CNN), which has laid the foundation for several subsequent models. For instance, the Spatial Pyramid Pooling Network (SPPNet) [19], Fast R-CNN [20,21], and YOLO [22].
CNNs are composed of convolutional and pooling layers that are used to efficiently process input images. It performs tasks such as classification and object detection by recognizing and learning local patterns in images [23]. Ref. [24] proposed a CNN-based classification model and conducted a comparative experiment for classifying images trained in ImageNet. In [25], a fine-grained pill identification based on images captured by handheld devices was conducted using a deep convolutional network (DCN). Ref. [26] proposed a CNN for classifying images taken under various conditions in the ImageNet Large-Scale Visual Recognition Challenge 2014 (ILSVRC14). Ref. [27] focused on a content-based image-retrieval (CBIR) system that retrieves similar images from an extensive database when provided with a query image. The R-CNN offers an ideal structure for accurately localizing objects. An R-CNN splits input images into candidate areas and performs a pooling process to generate images of the same size for those areas. These images are then input to the CNN model for the extraction of feature values. Classification is performed using a support vector machine (SVM) for each class. The R-CNN series include representative algorithms such as Fast R-CNN [20], Faster R-CNN [21], and Mask R-CNN [28]. Although R-CNNs yield satisfactory performance in object-detection tasks, they present disadvantages including a slow processing speed, complex structure, and high computational cost, as the CNN is individually trained in each candidate area. YOLO is a popular object-detection algorithm that processes entire images with one forward pass and predicts the bounding box coordinates and class probabilities for multiple objects [29,30]. This algorithm is capable of faster learning than R-CNN using the same quantity of training data, as well as detecting objects in real time at a rate of up to 65 fps. YOLO divides input images into grids, places multiple bounding boxes in each grid cell, and then predicts the detection probabilities associated with these boxes. YOLOv5, the latest algorithm of the YOLO series, exhibits a higher accuracy and smaller model size with significantly enhanced object-detection speed and performance. The YOLOv5, similar to the YOLO algorithm, predicts the locations and classes of bounding boxes; in addition, it can simultaneously process all the images and detect objects of various sizes more accurately, [31,32]. However, previous object-detection studies have focused on real-time detection, and the accuracy of pill identification has not yet been considered.
In pill-identification research, extracting distinctive features for classification primarily focuses on characteristics such as color, shape, texture, and imprinted characters on the pill. A study by [33] used Local Feature Regions (LFR), a method derived from color histograms, to obtain effective image-retrieval results. Another research initiative by [34] developed an automated pill-recognition system based on color histogram approaches, incorporating three attributes—shape, color, and texture of the pill imprint. Their empirical outcomes displayed a matching accuracy of 73% against a dataset of 13,000 legitimate pill images. In a separate study [35], Support Vector Machines (SVM) were utilized to investigate color characteristics, achieving a classification accuracy of 97.90%. However, this precision was constrained by specific factors such as lighting conditions, camera resolution, and the contrast between the pill and background colors.
Despite the substantial results of most existing research, these studies predominantly identify pills based on a subset of color and imprint features. There is a significant research gap in extracting distinctive features from complex pill shapes for classification purposes. We propose that leveraging such shape-based features could considerably enhance pill-classification performance. Hence, this paper specifically focuses on identifying unique feature points from intricately shaped pills. We employ a combination of four distinct algorithms—KAZE, ORB, Shi-Tomasi, and Harris—to fortify the robustness and accuracy of pill classification.
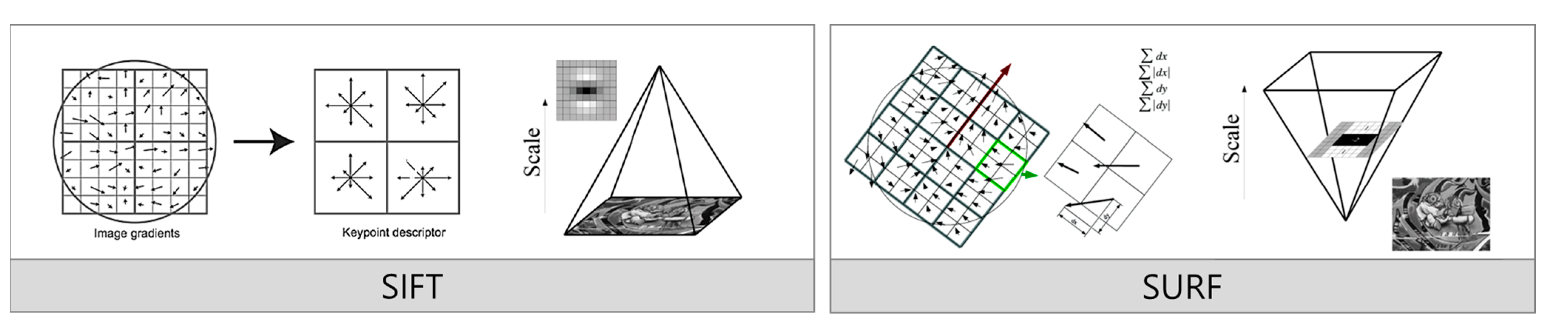
The tasks of tracking or recognizing objects in images or videos are primarily performed via feature extraction. KAZE and ORB are representative feature-extraction algorithms in computer vision. The KAZE algorithm [36] was developed to improve the shortcomings of the conventional Scale-Invariant Feature Transform (SIFT) and Speeded-Up Robust Features (SURF) algorithms [37]. The SIFT algorithm detects features by generating image pyramids via Gaussian smoothing and Difference-of-Gaussian (DOG) operations. SIFT incurs a high computation cost because it generates images at variable scales by adjusting the size of the kernel used in Gaussian smoothing. The SURF algorithm detects features by generating images at various scales using the box filter rather than Gaussian smoothing.
Figure 2 compares the feature extraction processes of the SIFT and SURF algorithms. Each algorithm offers a distinct method for detecting essential image features, and the diagram visually represents their respective approaches. In SIFT, the process begins by sampling gradients around a point of interest (POI). Figure 2 (left side) illustrates this with a circular region representing where gradients are computed. This step calculates changes in direction and magnitude at specific points to identify key features. After sampling the gradients, SIFT divides the area into 4 × 4 blocks. Inside each block, eight orientations shown as arrows are defined to capture local image features with represent different gradient directions. SIFT extracts strong key points that are robust to transformations like scaling and rotation by assigning higher weights to pixels closer to the edges. The right side of the SIFT diagram encodes these local gradient orientations into a vector representation. This feature vector later matches features across different images, maintaining robustness against image transformations. On the right side of the figure, the SURF algorithm uses integral images created by calculating cumulative pixel values at each point. This approach enables rapid image scaling without iterative downscaling, reducing the overall computational cost. SURF detects features by recording neighborhood statistics within 4 × 4 blocks. The diagram depicts these blocks with green arrows highlighting the regions where the neighborhood statistics are calculated. Unlike SIFT, which uses eight directional histograms to capture gradients, SURF simplifies the process by employing only four neighborhood statistics, significantly reducing computation time while extracting critical features. Although SURF uses a box filter to increase computational speed, this filter has limitations, such as weaker orientation handling and lower rotational invariability.
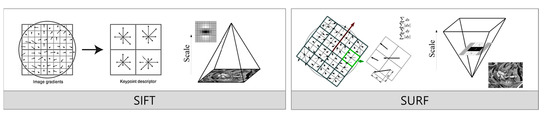
Figure 2.
Comparison of feature-extraction method for SIFT and SURF.
To mitigate these disadvantages, KAZE was designed to respond to the size, direction, rotation, and transformation of images using a nonlinear scale space in place of Gaussian smoothing. Consequently, the KAZE algorithm can extract more robust features [38,39]. Additionally, during feature detection, KAZE utilizes a Hessian reaction instead of the pixel value difference, which enables the detection of more powerful features and stabilizes performance under varying lighting conditions. In contrast, the ORB algorithm combines the Features from Accelerated Segment Test (FAST) and Binary Robust Independent Elementary Features (BRIEF) algorithms to extract image features. ORB detects features and standardizes their directions irrespective of rotation using FAST, and it calculates the corresponding feature vectors using BRIEF. Furthermore, ORB offers the advantage of enabling fast calculation and matching using binary descriptors. Therefore, when used in conjunction, the KAZE and ORB algorithms can complement each other by extracting image features in different ways.
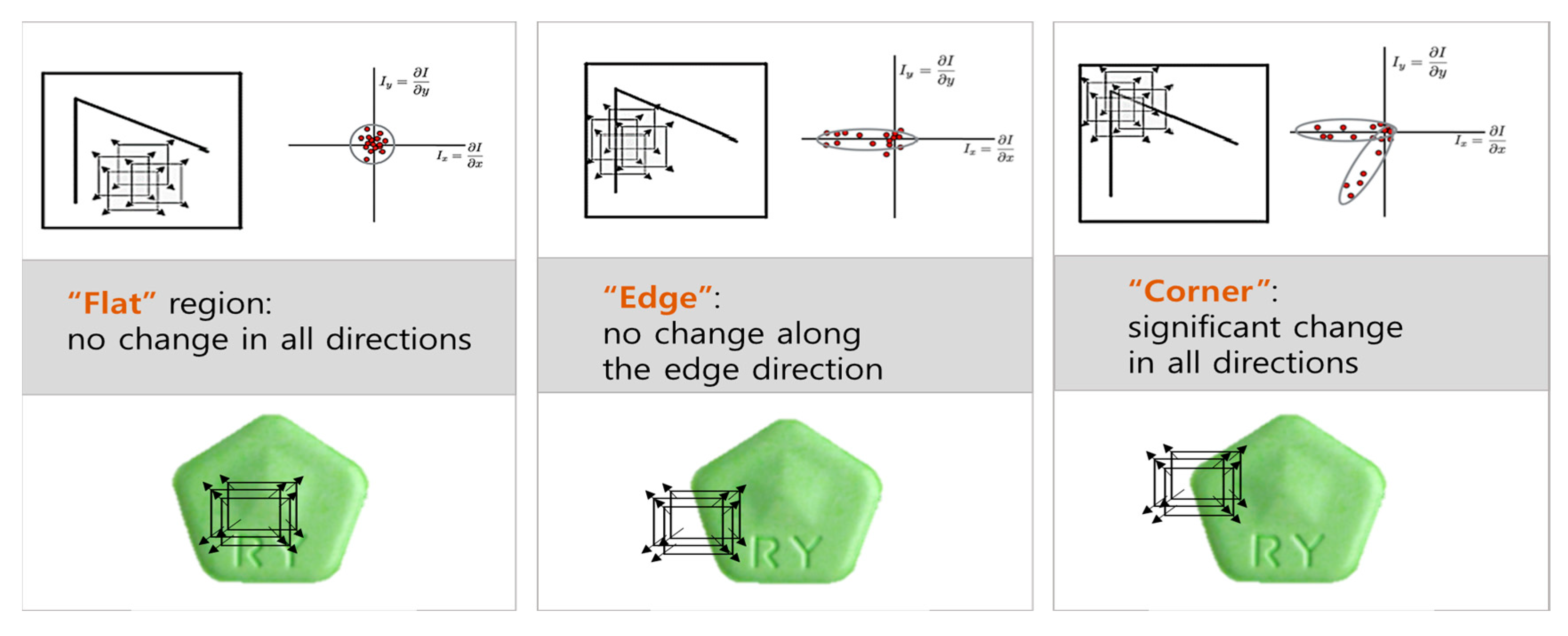
The Shi-Tomasi and Harris algorithms are suitable for the identification of corner features in images [40]. These corner features represent the intersections of edges within objects, which can be utilized to extract the outlines of objects, thereby facilitating the matching process. Although both algorithms use similar principles to detect the POI in images, they have slight differences [41]. The Harris algorithm applies a window to a specific location in an image, and then calculates the gradient change rate in the window alongside the differences between surrounding and central pixels in the window. It computes the eigenvalue that determines the feature strength for the gradient change rate, and then considers the point with the largest eigenvalue as the corner. In contrast, the Shi-Tomasi algorithm considers the smaller of the two eigenvalues for determining whether a point is a corner. Figure 3 illustrates the behavior of the Harris algorithm as the window moves across different regions of the image: flat, edge, and corner regions. The red dots in the figure represent vital points within the image where the gradient change is being evaluated. In the case of flat regions, the red dots show minimal or no change in gradient across all directions. In edge regions, the red dots demonstrate changes perpendicular to the edge, while there is little to no change along the edge itself. This indicates the presence of an edge but not a corner. In corner regions, significant changes occur in all directions, and the Harris algorithm detects a corner due to the high values in both eigenvalues. This characteristic allows the algorithm to confidently identify the corner where the most complex features (i.e., sharp changes in pixel intensity in all directions) are located.
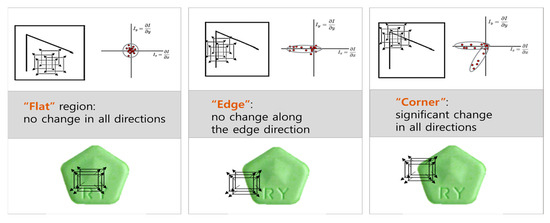
Figure 3.
Window movement in Harris algorithm.
3. The Proposed Pill-Classification System
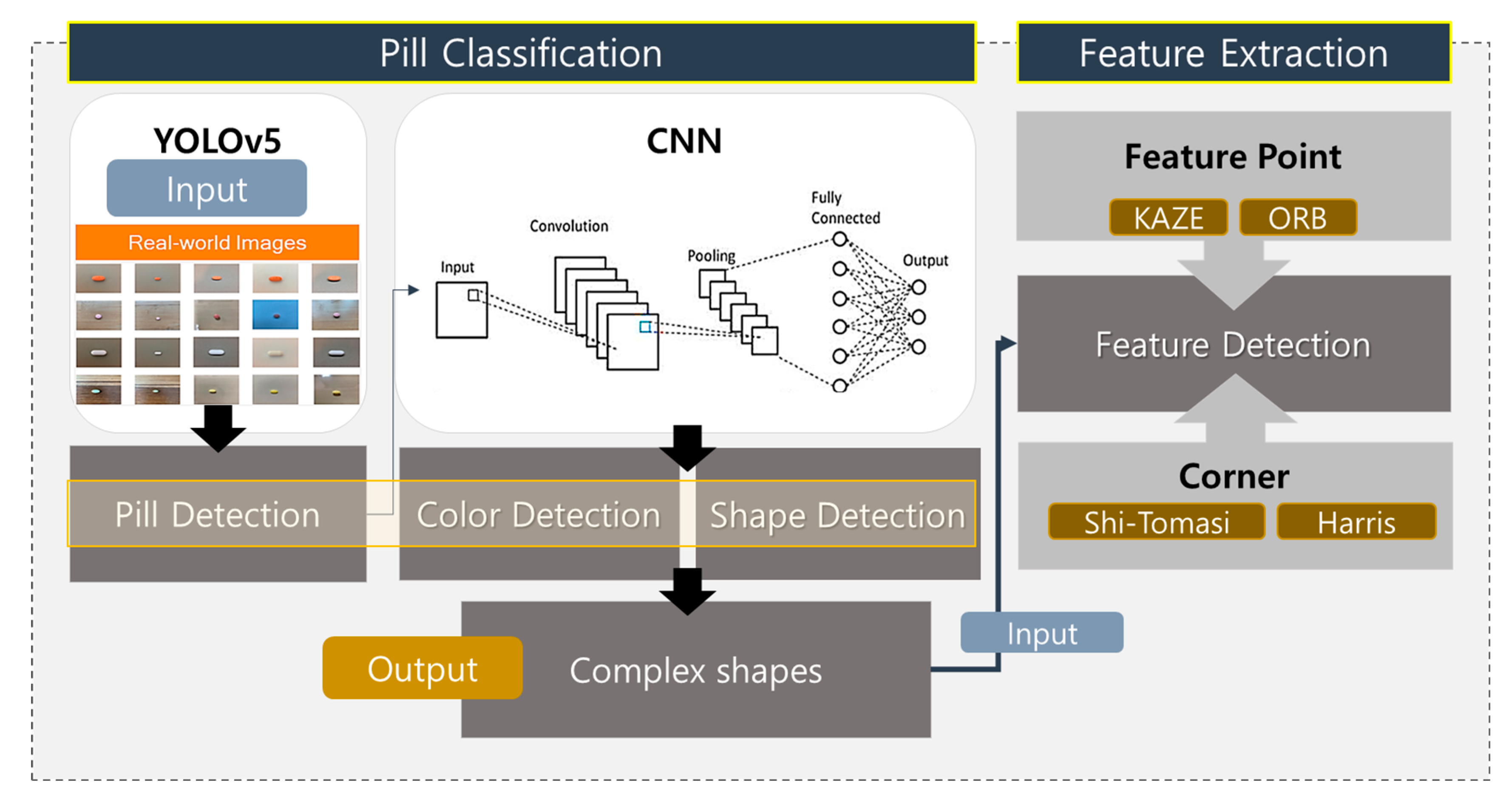
The following sections elaborate on the deep learning-based pill-classification system developed in this research, incorporating feedback to enhance the clarity and depth of the methodologies employed. The MCPL algorithm is pivotal in our approach, aiming to improve identification accuracy by extracting discernible features from pills, particularly those that are challenging to classify based on limited training data. We first collected around 25,000 pill images for building the dataset, either by directly downloading them or by crawling various sites that provide pill images. As illustrated in Figure 4, the system architecture is structured first to detect and analyze the colors and shapes of pills—fundamental attributes for initial sorting and identification.
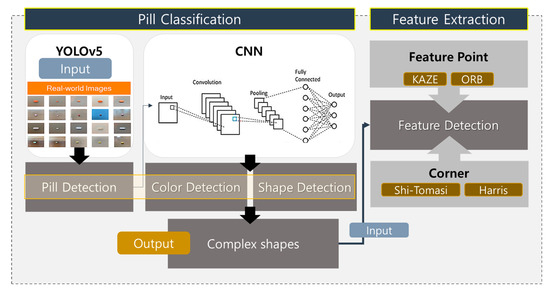
Figure 4.
Overview of proposed pill-classification system.
3.1. Pill Detection
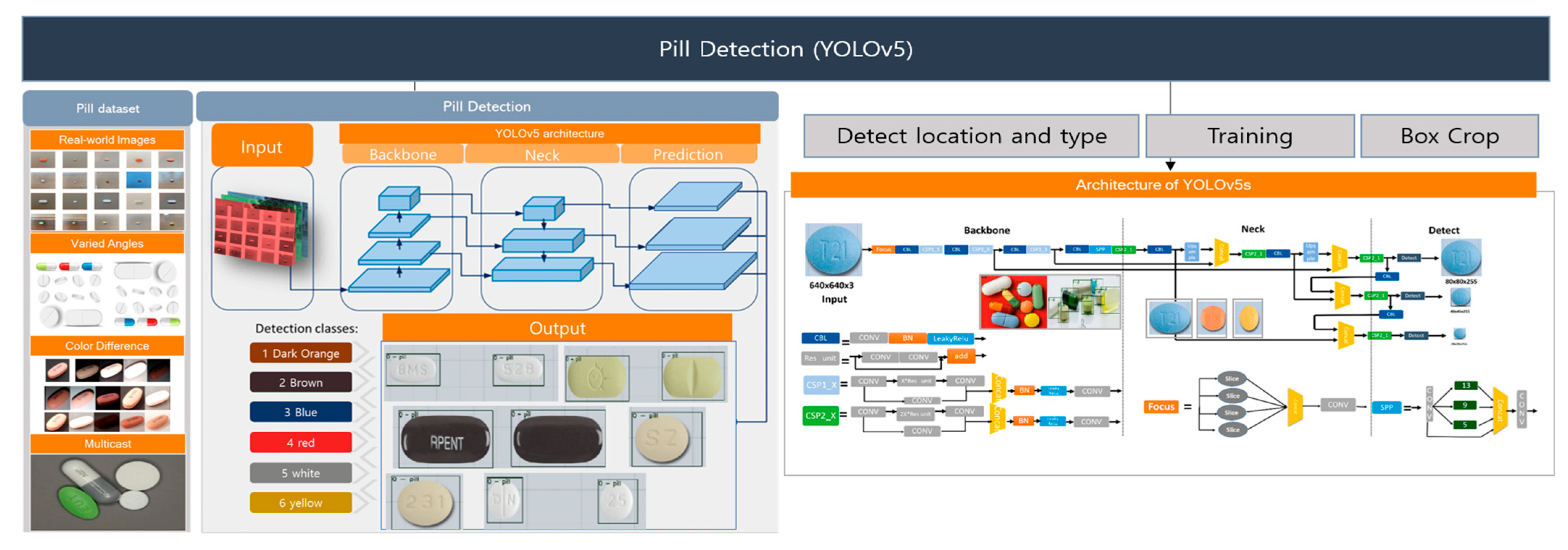
We utilize the YOLOv5 [S] architecture for pill detection, composed of four distinct subnetworks: the backbone, neck, head, and detect layer. YOLOv5 models—available in sizes S, M, L, and X—differ in capacity and computational efficiency, each tailored to specific operational requirements. Our implementation in PyTorch version 1. 7 leverages the generality of the trained YOLOv5 model, making it highly adaptable across varied pill-identification scenarios. As depicted in Figure 5, the pill-detection workflow entails three primary steps:
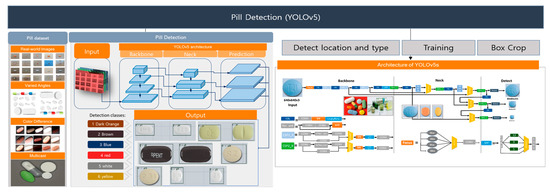
Figure 5.
YOLOv5-based pill-detection step for pill classification.
- -
- Step1: Localization and Configuration: Initial detection of pill locations and configurations crucial for setting up the training dataset.
- -
- Step2: Feature Extraction: The backbone network, utilizing a Darknet architecture enhanced by a Cross Stage Partial Network (CSPNet), extracts a broad spectrum of features from pill images, including edges, textures, and distinctive patterns.
- -
- Step3: Detection and Classification: The combined efforts of the neck and head networks, which employ spatial pyramid pooling (SPP) and anchor boxes, refine the detection and classification processes. This stage culminates with applying non-maximum suppression (NMS) to accurately identify and categorize pill types based on the extracted features.
3.2. Color and Shape Detection
The identification of pills also heavily relies on distinguishing features such as color and shape. For example, Amaryl pills, used in diabetes management, share the same shape and size but are differentiated by color based on dosage. We employ a CNN model to detect these features, focusing mainly on color because of its significance in the identification of the pill. The CNN’s ability to process and classify images based on learned features allows it to effectively separate pills that appear similar but differ in critical attributes like color. This section details how the CNN model is specifically trained to recognize and categorize pills based on these attributes, providing a foundational step in the classification process.
The CNN model utilized for color detection has also been adapted for shape identification, leveraging the robust feature-detection capabilities of CNNs. This approach is supported by extensive research in the field. References [42,43] applied neural-network-based methods to detect pills in images. References [44,45] focused on identifying pills in images portraying multiple different pills. Reference [46] aimed to detect the presence of pills in blister images for pill tracking, as well as identify pills in images of the mouth. Reference [47] attempted to learn potential associations between pills using information extracted from prescriptions. Although the problems covered in previous studies differ from the present study’s objective, our work aims to complement the results of these studies by presenting an accurate and efficient pill-identification method.
In this study, two primary methods are implemented to classify pills using the CNN framework. The first method processes entire images as input, dividing them into multiple layers and applying a specific filter to each layer. This technique demonstrates high accuracy but requires extensive processing time and significant data. The second method focuses on extracting key features from pill images and using these as direct inputs to the CNN. This method significantly reduces processing time and increases accuracy by concentrating on essential information extracted directly from the pills.
After verifying the accuracy of both methods, we introduced an additional feature-extraction process tailored for pills that are difficult to classify based on standard attributes. This enhancement improves the robustness of our pill-classification system, allowing for more precise identification across a broader range of pill types and conditions.
3.3. Feature Detection
In the proposed MCPL-based pill-classification system, feature detection plays a crucial role in enhancing the accuracy and reliability of pill identification. The feature-detection process is designed to capture and analyze the overt and subtle characteristics of pills that are significant for their classification.
First, we detect color and texture as specific features of the pill. These are essential for distinguishing pills with similar shapes but different therapeutic uses. The system identifies subtle variations in color gradients and texture patterns characteristic of different pill coatings and materials. We also detect the shape and size of the pill, as contour analysis techniques detect the geometric features of pills, including round, oval, rectangular, and other irregular shapes. Size measurements help differentiate between dosage forms.
For feature-extraction techniques, we first perform initial image processing, then utilize advanced algorithms for feature detection, and finally apply deep learning models. High-resolution images are pre-processed using techniques such as denoising, normalization, and color correction to enhance the quality of the input data for feature extraction. Next, the proposed method employs a combination of edge detection, corner detection, and blob-detection algorithms to identify and outline critical features. Algorithms like the Shi-Tomasi and Harris corner detectors identify key points on the pill images that are invariant to rotation and scale. CNNs are used for more complex feature detection, such as identifying textural patterns and classifying pills based on learned image features from extensive datasets. This approach enhances the pill-classification system’s ability to handle variability in pill appearances and ensures high identification accuracy.
4. Multi-Combination Pattern Labeling
In this section, we propose the MCPL method to achieve accurate pill classification based on extracted features. In pill-classification systems, the feature extraction of hard-to-classify pills is indispensable in maximizing accuracy and reliability by minimizing the risks of drug misuse and adverse effects. To this end, we implemented appropriate feature-extraction procedures to supplement the information needed to identify hard-to-classify pills.
The MCPL model detects features and utilizes them as criteria for the classification of complex pill shapes. To achieve this, the KAZE algorithm, which responds to various changes in image size, rotation, and orientation, is used in conjunction with the ORB algorithm, which performs fast computation and matching. Furthermore, kernel features are found using the Shi-Tomasi and Harris algorithms to ensure more accurate feature extraction and matching.
4.1. KAZE
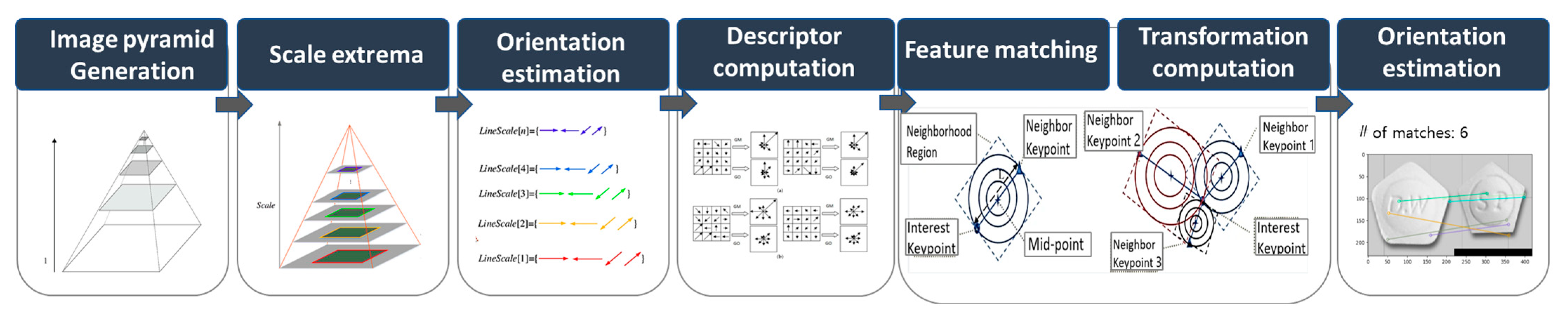
The KAZE algorithm employs a nonlinear scale space, which utilizes nonlinear scale filtering to make image feature blurring locally adaptive. This approach helps in reducing noise while preserving image boundaries. The KAZE detector is configured based on a scale-normalized determinant of the Hessian matrix computed at multiple scale levels. The maxima of the detector response are selected as features using a moving window. Feature description introduces the rotation invariance property by finding the dominant orientation in the circular neighborhood around each detected feature. Although KAZE features are invariant to rotation, scale, and limited affine transformations and have more distinct characteristics at different scales, they incur a longer computational time. Figure 6 illustrates the structure of KAZE-based feature detection. The process begins with image pyramid generation, creating multiple image scales. Scale extrema are identified across these scales, followed by orientation estimation represented by the colored lines, each indicating different gradient directions at specific points. Next, descriptor computation generates feature vectors that capture key point information, while feature matching compares these descriptors across different images. In the transformation computation phase, matching key points determine geometric transformations between the images. The final step, orientation estimation, refines the key points matches, ensuring accurate alignment and orientation, visualized by the connected key points and transformation overlays.
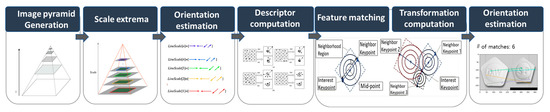
Figure 6.
Structure of KAZE-based feature detection.
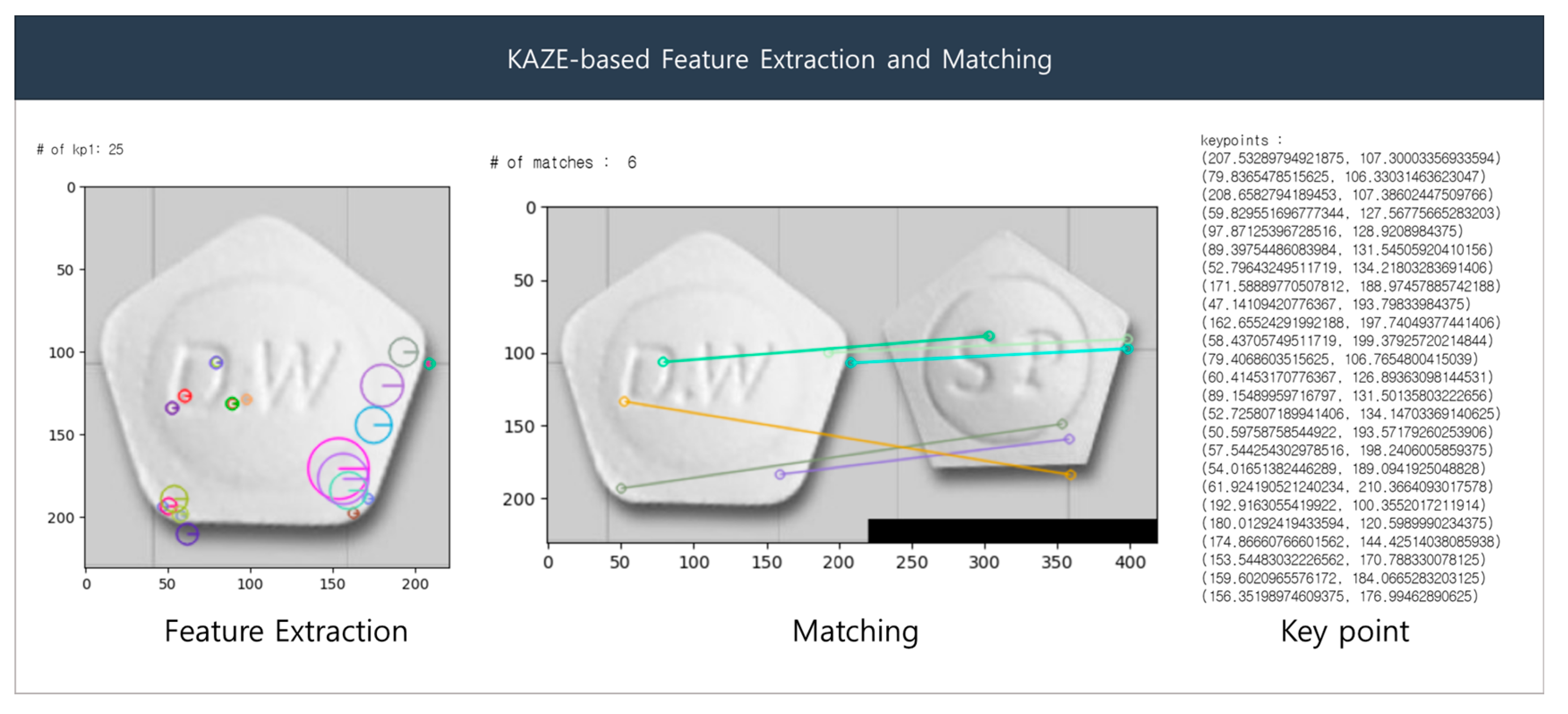
The KAZE detector initially generates an image pyramid, wherein the input image is repeatedly down-sampled to detect features at different scales. The scale extrema step detects the maximum and minimum points in the scale space of each image based on the LoG filter. Orientation estimation is performed using the gradient orientation of the image to approximate the orientation at each key point. The descriptor computation step obtains the path of the region around the key point to extract local features from that key point. The rotation is corrected using the orientation information from the path and computed based on the SIFT descriptor. In the feature-matching step, distances are calculated between descriptors to match key points between two images via Nearest-Neighbor (NN) and the Nearest-Neighbor-Ratio (NNR) matching. To compute the transformation, the NNR matching results are used to calculate the affine transformation matrix, which is used to align features between the two images and subsequently identify the key points between them. Figure 7 presents sample results of the KAZE algorithm. The left side of the Figure 7 shows the extracted features as colored circles, and the right side shows the matching result based on the features.
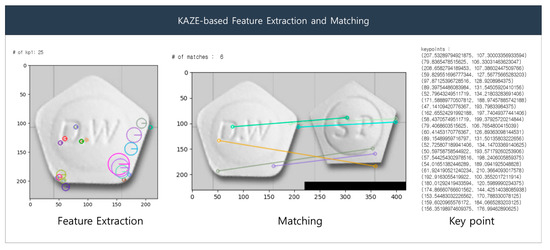
Figure 7.
KAZE-based feature extraction and matching.
4.2. ORB
The ORB algorithm is a combination of the modified FAST detection and orientation-normalized BRIEF techniques. FAST detects possible corner points at each layer of the scale pyramid, and each point is then evaluated using the Harris corner score to filter points with the highest quality. The features of ORB are points that are invariant to scalar, rotational, and limited affine changes.
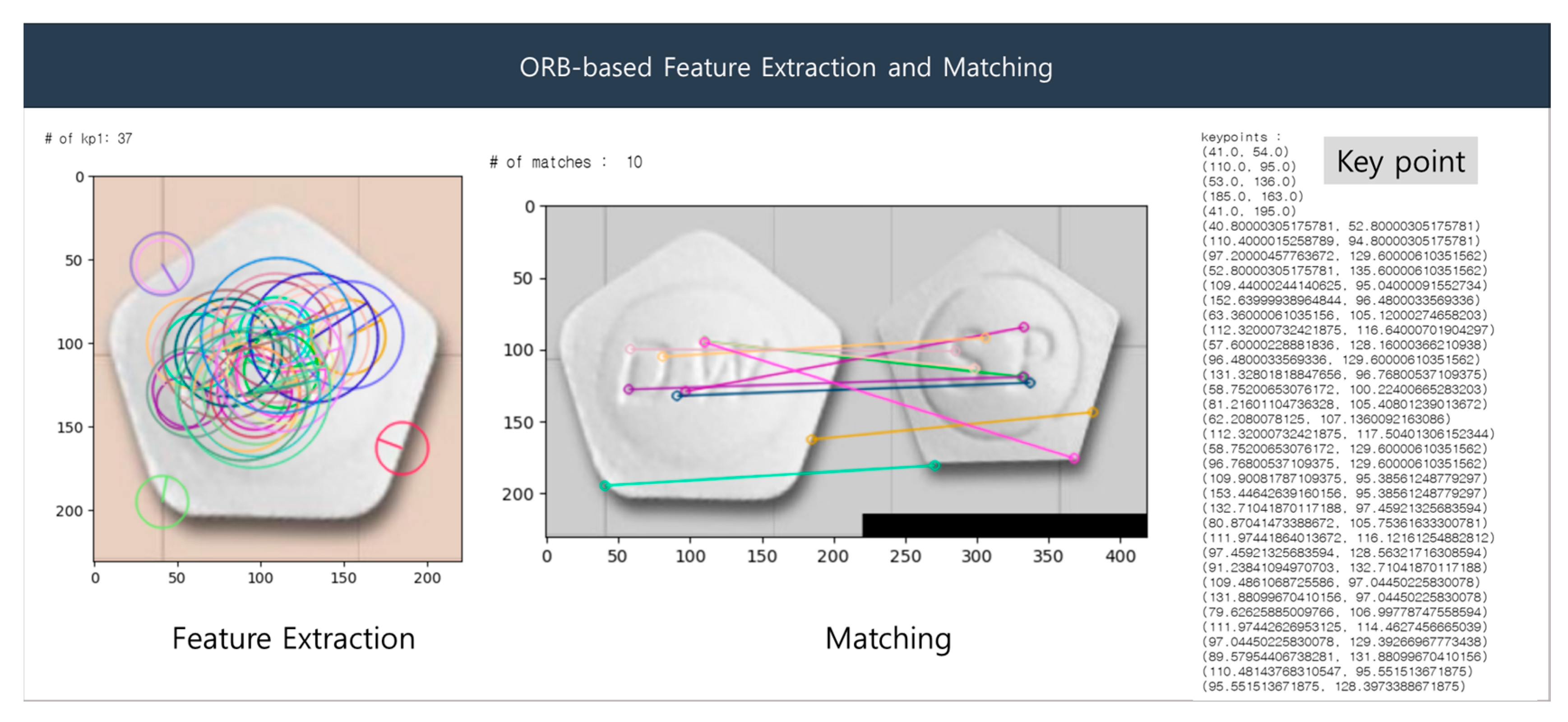
The ORB algorithm comprises the following procedure: (1) configure a multi-resolution pyramid of the image; (2) perform FAST corner detection at each resolution; (3) evaluate the ‘cornerness’ value by calculating the Harris corner score from the detected corners; (4) remove redundant corners by selecting only the corners with the highest Harris corner scores; (5) estimate the orientation from the selected corners; (6) generate a BRIEF descriptor based on the orientation to ensure invariance to rotation; (7) display the features using the descriptors. Figure 8 presents a sample result of ORB-based feature detection.
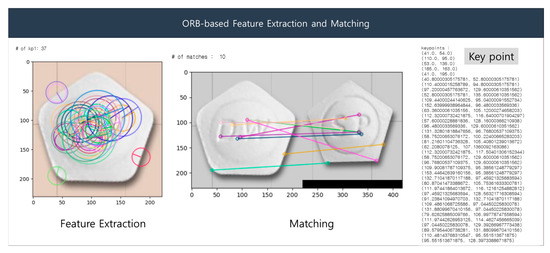
Figure 8.
ORB-based feature extraction and matching.
4.3. Shi-Tomasi and Harris Algorithms
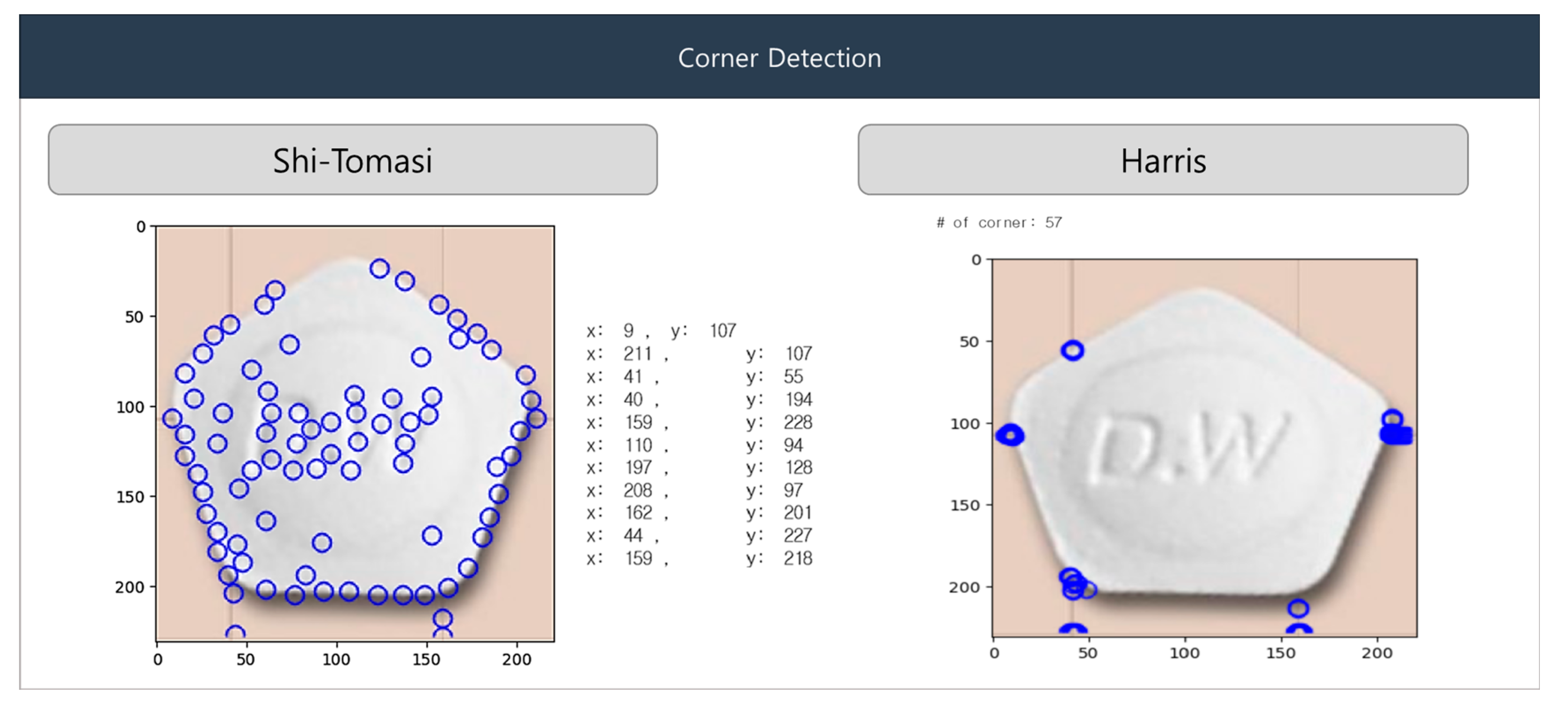
The Shi-Tomasi and Harris algorithms are among the most popular corner-detection algorithms. To identify the corners of pills in an input image, the Shi-Tomasi algorithm creates a grayscale version of the image, selects a window in the image, and calculates the gradient at every pixel inside the window. It then applies the derivative at each pixel to compute a feature value from the magnitude and direction of the gradient. The feature value is the smallest of the gradient magnitudes of the pixels inside the window. The top N features are selected by comparing the magnitudes of feature values over the entire image.
Although the Harris algorithm is similar to the Shi-Tomasi algorithm, it uses a corner-response function to detect corners. In Figure 9, Shi-Tomasi (left), the blue circles show corner points detected based on the smallest eigenvalue of the gradient matrix. This method’s sensitivity to subtle image variations results in more scattered corners. In Harris (right), the blue circles indicate fewer but stronger corner detections. The algorithm focuses on areas with significant intensity changes in multiple directions, offering more stable corner identification.
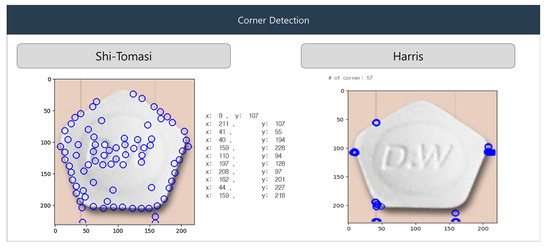
Figure 9.
Shi-Tomasi and Harris algorithm for corner detection.
5. Experimental Results
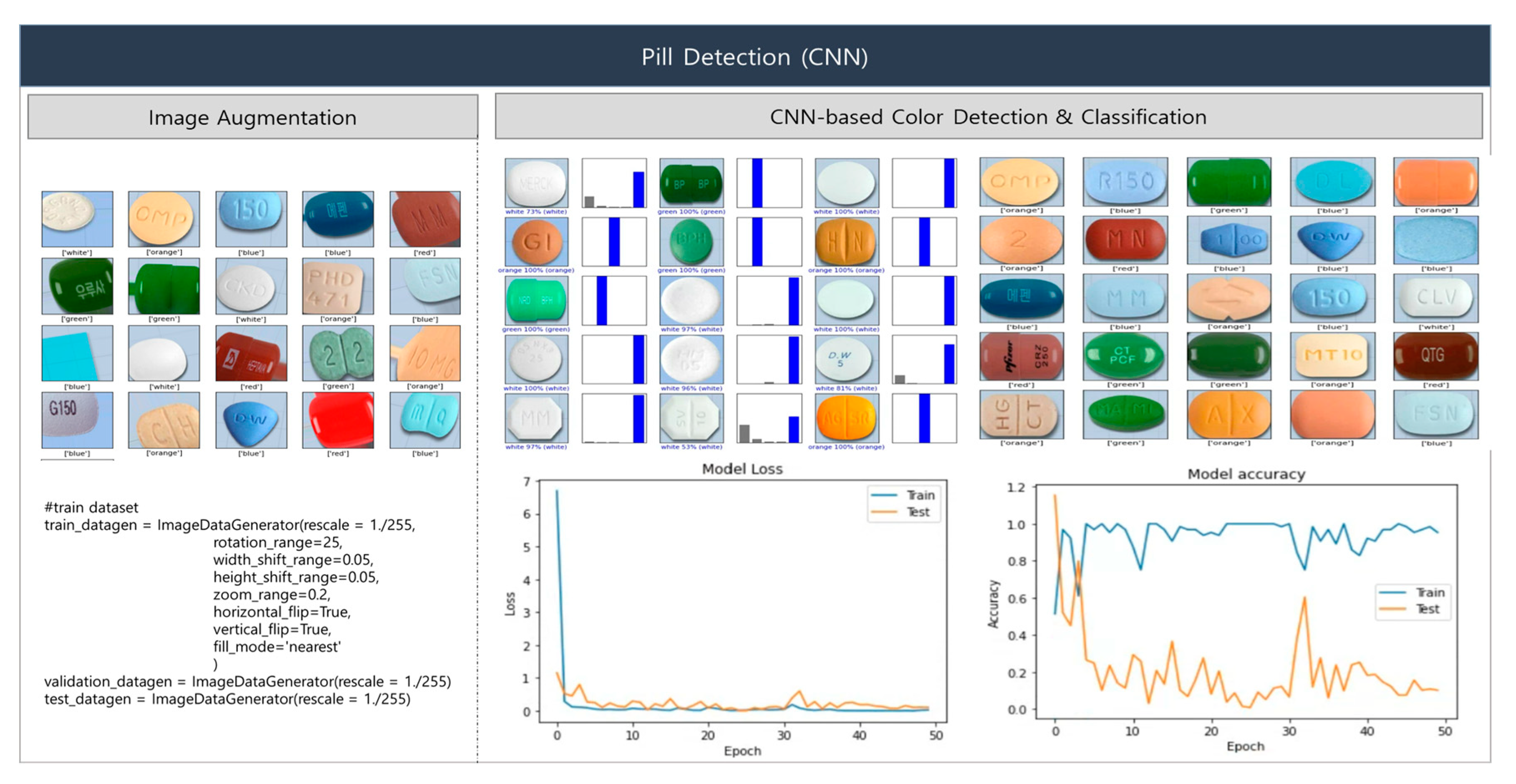
This section presents and interprets the experimental results of our proposed deep learning-based pill-classification system. First, we describe the results of the proposed base pill-recognition system, and then the experimental results based on MCPL. The proposed system uses a CNN to recognize the colors and shapes of pills. Although CNNs have exhibited high performance in tasks including image classification, object detection, and segmentation, they require large quantities of data for training. Accordingly, we applied the image-augmentation technique to generate new training data from existing images via transformations, allowing the CNN model to learn more diverse patterns. To ensure a sufficiently large and diverse dataset, we rotated the image data by random angles between 0 and 25 degrees, shifted the orientation in horizontal and vertical directions by random values within a range of 0.05, performed scaling by random values between 0.8 and 1.2, and inverted the original images. Figure 10 illustrates the CNN-facilitated processes of image augmentation and color detection. Five color classes were used for classification: class = [‘red’, ‘green’, ‘blue’, ‘orange’, ‘white’]. The experimental results show a pill color detection accuracy of 95%.
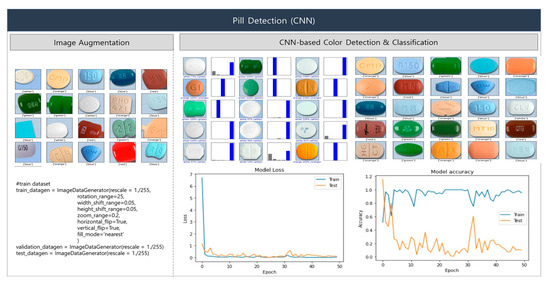
Figure 10.
Image augmentation and color detection using CNN.
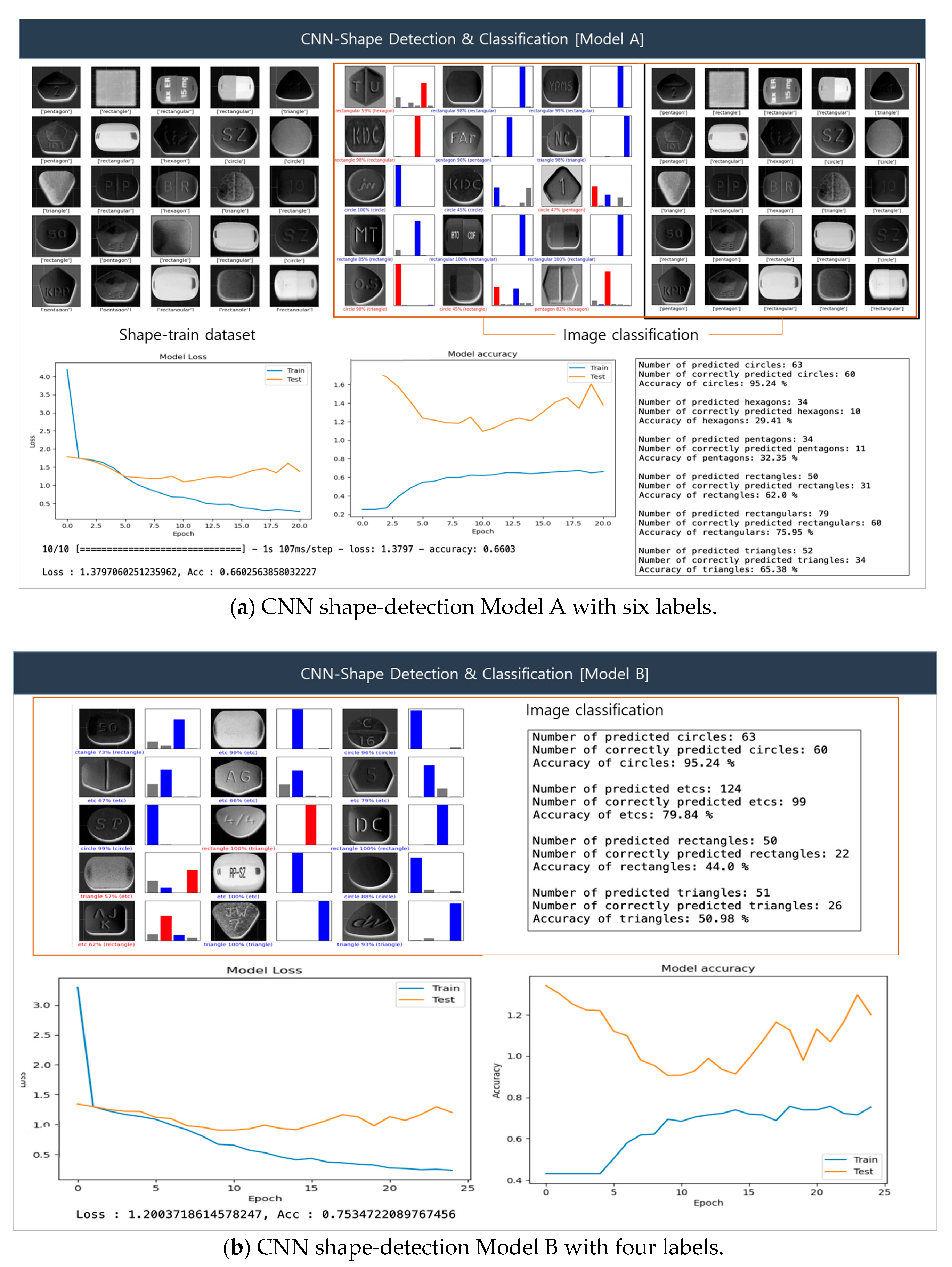
Figure 11 illustrates the CNN-based shape-detection procedure. In our experimental setup, we designated Models A and B to compare detection accuracy according to class. In the classification results of the figure, the blue graph indicates correct classifications, while the red graph signifies incorrect classifications. Figure 11a presents the pill shape-detection results obtained by Model A. For shape detection, six classes were initially defined—class = [‘circle’, ‘hexagon’, ‘pentagon’, ‘rectangle’, ‘capsular’, ‘triangle’]—yielding an accuracy of 66%. Specifically, the detection accuracies of circle, triangle, rectangle, and capsular shapes were 95%, 65%, 62%, and 75%, respectively, whereas those of hexagon and pentagon shapes were 29% and 32%, respectively.
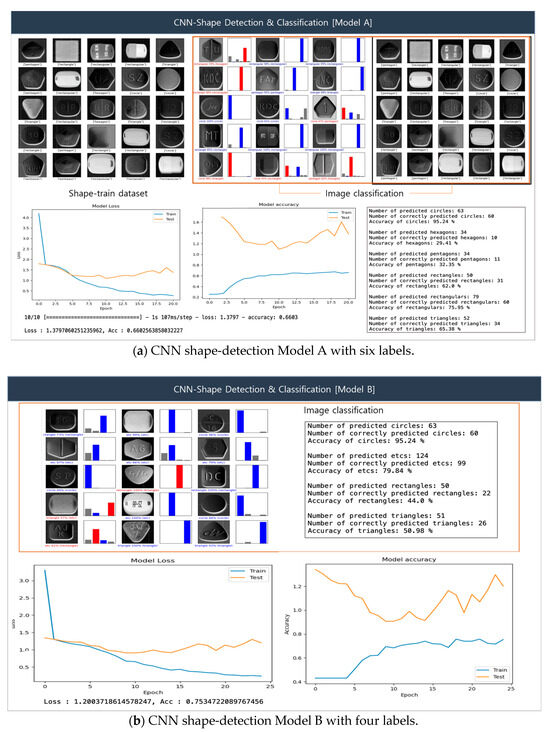
Figure 11.
CNN-facilitated shape detection.
Figure 11b presents the pill shape-detection results obtained by Model B, where in four classes were defined: class = [‘circle’, ‘rectangle’, ‘triangle’, ‘etc’]. The number of classes was reduced to make the pill classification simpler and more intuitive. The etc class was added to allow shapes that were not correctly classified by Model A to be organized within a single category. The resulting detection accuracy was 75%, which is an improvement over Model A. The experimental results once again confirm the importance of identifying features for accurate classification, as complex shapes may negatively impact overall model performance. Accordingly, features identified as etc require further detection based on complex shapes.
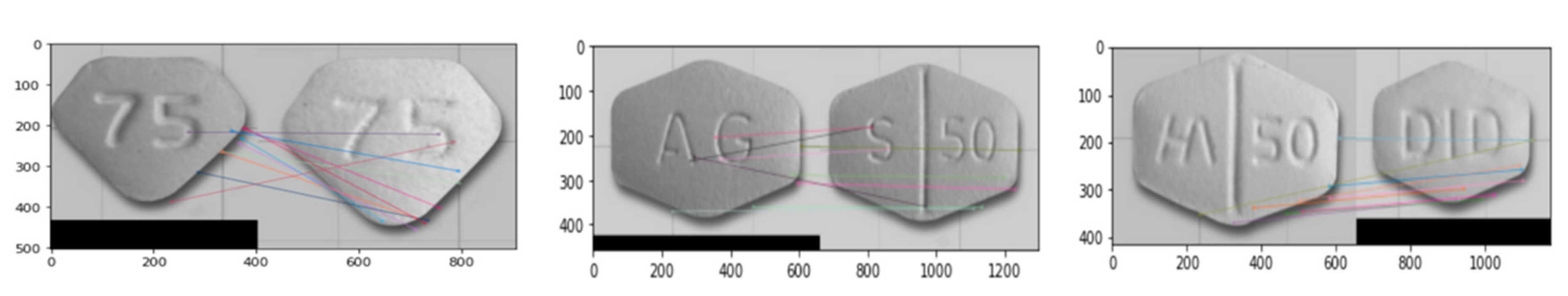
We detect shapes of pill using MCPL for the hexagon and pentagon classes, which are not well classified in the proposed base system, as shown in Figure 11a. We experimented with KAZE, ORB, Shi-Tomasi and Harris for feature extraction. Figure 12 shows the experimental results of MCPL-based pill detection and the colored lines represent similar features.
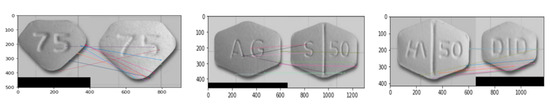
Figure 12.
Experimental results of MCPL-based pill detection.
The experimental results comparing the combination of feature-extraction algorithms and the proposed system are shown in Table 1. In response to the lower accuracy in detecting complex shapes like hexagons and pentagons, we implemented the MCPL method, incorporating advanced feature-extraction algorithms (KAZE, ORB, Shi-Tomasi, Harris). This corresponds to a relative performance enhancement of approximately 1.2 times the base accuracy, providing a clearer and more concrete demonstration of the MCPL method’s effectiveness.

Table 1.
Comparative experimental results of the combination of feature-extraction algorithm and proposed method.
6. Conclusions
In this study, we introduced a deep learning-based pill-classification system leveraging Multi-Combination Pattern Labeling (MCPL) to enhance the identification of complex-shaped pills such as hexagons and pentagons. While our approach improved classification accuracy significantly, achieving about 35% accuracy for these shapes, there is room for further enhancement. The MCPL system, integrating algorithms like KAZE, ORB, Shi-Tomasi, and Harris, proved effective in refining feature detection, thus improving the system’s accuracy and potentially reducing medication errors due to misclassification. Moreover, image quality variations such as blur and noise were recognized as crucial factors affecting performance, suggesting the need for robust preprocessing techniques to standardize input data and enhance the system’s reliability. This study not only pushed the boundaries of current pill-classification systems by employing advanced deep learning methods but also highlighted several areas for further investigation to optimize performance and expand usability.
In future research, we aim to integrate the latest advancements in deep learning architectures to enhance the classification accuracy of complex-shaped pills. We plan to experiment with newer object-detection models, such as improved versions of YOLO and EfficientDet, comparing these against our current MCPL-based system to quantify any performance improvements. Additionally, recognizing the critical impact of image quality on classification success, we will conduct a systematic study to evaluate how variations in image quality—such as blur and noise—affect the system’s performance. This will include testing advanced preprocessing techniques to standardize input data quality. To clarify and expand upon the dataset used, we will provide a detailed subsection in our study that describes the dataset’s publicly available or proprietary sources and analyze the distribution of different pill types better to understand their impact on our system’s effectiveness. Moreover, we will undertake an ablation study to distinctly showcase the contribution of each algorithmic component to the overall system efficacy. Finally, our method will be benchmarked against other state-of-the-art pill-classification systems to place our findings in a broader context and outline our system’s specific limitations and challenges, setting the stage for ongoing improvements in automated pill identification.
Author Contributions
Conceptualization, S.K. and S.-Y.I.; methodology, S.-Y.I.; software, J.-S.K.; validation, S.K., E.-Y.P. and S.-Y.I.; formal analysis, S.-Y.I.; resources, E.-Y.P.; data curation, J.-S.K. and S.-Y.I.; writing—original draft preparation, S.K.; writing—review and editing, S.-Y.I.; visualization, S.-Y.I.; supervision, S.-Y.I.; project administration, S.-Y.I.; funding acquisition, S.-Y.I. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2021R1C1C2011105). This work was supported by Innovative Human Resource Development for Local Intellectualization program through the Institute of Information & Communications Technology Planning & Evaluation(IITP) grant funded by the Korea government(MSIT)(IITP-2024-RS-2022-00156334).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Slawomirski, L.; Auraaen, A.; Klazinga, N. The Economics of Patient Safety: Strengthening a Value-Based Approach to Reducing Patient Harm at National Level; OECD Health Working Papers, no.96; OECD: Paris, France, 2017. [Google Scholar]
- Alrabadi, N.; Shawagfeh, S.; Haddad, R.; Mukattash, T.; Abuhammad, S.; Al-rabadi, D.; Farha, R.A.; AlRabadi, S.; Al-Faouri, I. Medication errors: A focus on nursing practice. J. Pharm. Health Serv. Res. 2021, 12, 78–86. [Google Scholar] [CrossRef]
- Isaacs, A.N.; Ch’ng, K.; Delhiwale, N.; Taylor, K.; Kent, B.; Raymond, A. Hospital medication errors: A cross-sectional study. Int. J. Qual. Health Care 2021, 33, mzaa136. [Google Scholar] [CrossRef] [PubMed]
- Naseralallah, L.; Stewart, D.; Price, M.; Paudyal, V. Prevalence, contributing factors, and interventions to reduce medication errors in outpatient and ambulatory settings: A systematic review. Int. J. Clin. Pharm. 2023, 45, 1359–1377. [Google Scholar] [CrossRef] [PubMed]
- Keers, R.N.; Williams, S.D.; Cooke, J.; Ashcroft, D.M. Causes of Medication Administration Errors in Hospitals: A Systematic Review of Quantitative and Qualitative Evidence. Drug Saf. 2013, 36, 1045–1067. [Google Scholar] [CrossRef]
- Tefera, Y.G.; Gebresillassie, B.M.; Ayele, A.A.; Belay, A.B.; Emiru, A.K. The characteristics of drug information inquiries in an Ethiopian university hospital: A two-year observational study. Sci. Rep. 2019, 9, 13835. [Google Scholar] [CrossRef]
- Almazrou, D.A.; Ali, S.; Alzhrani, J.A. Assessment of queries received by the drug information center at King Saud Medical City. J. Pharm. BioAllied Sci. 2017, 9, 246–250. [Google Scholar]
- Chantara, W.; Mun, J.H.; Shin, D.W.; Ho, Y.S. Object tracking using adaptive template matching. IEIE Trans. Smart Process. Comput. 2015, 4, 1–9. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, Y.; Xiao, C.; Zhu, Q.; Lu, X.; Zhang, H.; Ge, J.; Zhao, H. Automated visual inspection of glass bottle bottom with saliency detection and template matching. IEEE Trans. Instrum. Meas. 2019, 68, 4253–4267. [Google Scholar] [CrossRef]
- Ushizima, D.; Carneiro, A.; Souza, M.; Medeiros, F. Investigating pill recognition methods for a new national library of medicine image dataset. In Proceedings of the ISVC 2015, Las Vegas, NV, USA, 14–16 December 2015; pp. 410–419. [Google Scholar]
- Ling, S.; Pastor, A.; Li, J.; Che, Z.; Wang, J.; Kim, J.; Callet, P.L. Few-Shot Pill Recognition. In Proceedings of the CVPR 2020, Seattle, WA, USA, 13–19 June 2020; pp. 9789–9798. [Google Scholar]
- Yaniv, Z.; Faruque, J.; Howe, S.; Dunn, K.; Sharlip, D.; Bond, A.; Perillan, P.; Bodenreider, O.; Ackerman, M.J.; Yoo, T.S. The national library of medicine pill image recognition challenge: An initial report. In Proceedings of the IEEE Applied Imagery Pattern Recognition Workshop, Washington, DC, USA, 18–20 October 2016; pp. 1–9. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Sahraeian, S.M.E.; Liu, R.; Lau, B.; Podesta, K.; Mohiyuddin, M.; Lam, H.Y.K. Deep convolutional neural networks for accurate somatic mutation detection. Nat. Commun. 2019, 10, 1041. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018, 172, 1122–1131. [Google Scholar] [CrossRef] [PubMed]
- Eraslan, G.; Avsec, Z.; Gagneur, J.; Theis, F.J. Deep learning: New computational modelling techniques for genomics. Nat. Rev. Genet. 2019, 20, 389–403. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 37, 1904–1916. [Google Scholar] [CrossRef]
- Girshick, R. Fast R-CNN. In Proceedings of the CVPR 2015, Santiago, Chile, 7–13 December 2015; pp. 1440–1448. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the CVPR 2016, Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Delgado, N.L.; Usuyama, N.; Hall, A.K.; Hazen, R.J.; Ma, M.; Sahu, S.; Lundin, J. Fast and accurate medication identification. Npj Digit. Med. 2019, 2, 10. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Wong, Y.F.; Ng, H.T.; Leung, K.Y.; Chan, K.Y.; Chan, S.Y.; Loy, C.C. Development of fine-grained pill identification algorithm using deep convolutional network. J. Biomed. Inf. 2017, 74, 130–136. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the CVPR 2015, Boston, MA, USA, 8–10 June 2015; pp. 1–9. [Google Scholar]
- Maji, S.; Bose, S. CBIR using features derived by deep learning. ACM/IMS Trans. Data Sci. 2021, 2, 1–24. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollar, P.; Girshick, R. Mask R-CNN. In Proceedings of the ICCV 2017, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Yao, J.; Qi, J.; Zhang, J.; Shao, H.; Yang, J.; Li, X. A real-time detection algorithm for kiwifruit defects based on YOLOv5. Electronics 2021, 10, 1711. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, X.; Yan, J.; Qiu, X.; Yao, X.; Tian, Y.; Zhu, Y.; Cao, W. A wheat spike detection method in UAV images based on improved YOLOv5. Remote Sens. 2021, 13, 3095. [Google Scholar] [CrossRef]
- Darma, I.W.A.S.; Suciati, N.; Siahaan, D. A Performance Comparison of Balinese Carving Motif Detection and Recognition using YOLOv5 and Mask R-CNN. In Proceedings of the ICICoS 2021, Semarang, Indonesia, 24–25 November 2021; pp. 52–57. [Google Scholar]
- Zhou, F.; Zhao, H.; Nie, Z. Safety helmet detection based on YOLOv5. In Proceedings of the ICPECA 2021, Shenyang, China, 22–24 January 2021; pp. 6–11. [Google Scholar]
- Wang, X.Y.; Wu, J.F.; Yang, H.Y. Robust image retrieval based on color histogram of local feature regions. Multimed. Tools Appl. 2010, 49, 323–345. [Google Scholar] [CrossRef]
- Lee, Y.B.; Park, U.; Jain, A.K.; Lee, S.W. Pill-ID: Matching and retrieval of drug pill images. Pattern Recognit. Lett. 2012, 33, 904–910. [Google Scholar] [CrossRef]
- Guo, P.; Stanley, R.J.; Cole, J.G.; Hagerty, J.; Stoecker, W.V. Color Feature-based Pillbox Image Color Recognition. In Proceedings of the VISIGRAPP 2017, Porto, Portugal, 27 February–1 March 2017; pp. 188–194. [Google Scholar]
- Alcantarilla, P.F.; Bartoli, A.; Davison, A.J. KAZE features. In Proceedings of the ECCV 2012, Florence, Italy, 7–13 October 2012; pp. 214–227. [Google Scholar]
- Bay, H.; Tuytelaars, T.; Gool, L.V. SURF: Speeded up Robust Features. In Proceedings of the ECCV 2006, Graz, Austria, 7–13 May 2006; pp. 404–417. [Google Scholar]
- Viswanathan, D.B. Features from accelerated segment test (fast). In Proceedings of the WIAMIS 2009, London, UK, 6–8 May 2009; pp. 6–8. [Google Scholar]
- Lowe, D.G. Distinctive image features from scale-invariant key points. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Szeliski, R. Computer Vision: Algorithms and Applications Second Edition; Springer Nature: Berlin/Heidelberg, Germany, 2022. [Google Scholar]
- Harris, C.; Stephens, M. A combined corner and edge detector. In Proceedings of the Alvey Vision Conference 1988, Manchester, UK, 31 August–2 September 1988; pp. 147–151. [Google Scholar]
- Kwon, H.J.; Kim, H.G.; Lee, S.H. Pill Detection Model for Medicine Inspection Based on Deep Learning. Chemosensors 2022, 10, 4. [Google Scholar] [CrossRef]
- Holtkötter, J.; Amaral, R.; Almeida, R.; Jácome, C.; Cardoso, R.; Pereira, A.; Pereira, M.; Chon, K.H.; Fonseca, J.A. Development and Validation of a Digital Image Processing–Based Pill Detection Tool for an Oral Medication Self-Monitoring System. Sensors 2022, 22, 2958. [Google Scholar] [CrossRef]
- Nguyen, A.D.; Pham, H.H.; Trung, H.T.; Nguyen, Q.V.H.; Truong, T.N.; Nguyen, P.L. Highly Accurate and Explainable Multi-Pill Detection Framework with Graph Neural Network–Assisted Multimodal Data Fusion. arXiv 2023, arXiv:2303.09782. [Google Scholar] [CrossRef]
- Chang, W.J.; Chen, L.B.; Hsu, C.H.; Lin, C.P.; Yang, T.C. A Deep Learning–Based Intelligent Medicine Recognition System for Chronic Patients. IEEE Access 2019, 7, 44441–44458. [Google Scholar] [CrossRef]
- Ting, H.W.; Chung, S.L.; Chen, C.F.; Chiu, H.Y.; Hsieh, Y.W. A drug identification model developed using deep learning technologies: Experience of a medical center in Taiwan. BMC Health Serv. Res. 2019, 20, 1–9. [Google Scholar] [CrossRef]
- Nguyen, A.D.; Nguyen, T.D.; Pham, H.H.; Nguyen, T.H.; Nguyen, P.L. Image-based Contextual Pill Recognition with Medical Knowledge Graph Assistance. In Proceedings of the ACIIDS 2022, Ho Chi Minh City, Vietnam, 28–30 November 2022; pp. 354–369. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).