Abstract
Transition metal complexes with dppz-type ligands (dppz = dipyrido[3,2-a:2′,3′-c]phenazine) are extensively studied and attract a considerable amount of attention, becoming, from the very beginning and increasingly over time, a powerful tool for investigating the structure of the DNA helix. In particular, [Ru(bpy)2(dppz)]2+ and [Ru(phen)2(dppz)]2+ and their derivatives were extensively investigated as DNA light-switches. The purpose of this mini-review, which is not and could not be exhaustive, was to first introduce DNA and its importance at a biological level and research in the field of small molecules that are capable of interacting with it, in all its forms. A brief overview is given of the results obtained on the Ru-dppz complexes that bind to DNA. The mechanism of the light-switch active in this type of species is also briefly introduced along with its effects on structural modifications on both the dppz ligand and the ancillary ligands. Finally, a brief mention is made of biological applications and the developments obtained due to new spectroscopic techniques, both for understanding the mechanism of action and for cellular imaging applications.
1. Introduction
DNA has a vital role in life—cells use DNA to store and communicate all information necessary for the evolution of life. Its replication and transcription are the key mechanism underlying all biological functioning.
DNA is essentially polymorphic, although its most common structure is the anti-parallel double helix (B-DNA) [1]. However, its existence in several conformations, which are indispensable for its functioning and for the regulation of its functions, makes DNA a difficult target for small molecules. Therefore, research aimed at developing small molecules capable of binding to and reacting with DNA in all its forms is in continuous development. In fact, over the last 30 years, research led to the synthesis and study of molecules that bind to DNA as both diagnostic probes and therapeutic agents [2,3,4].
Among many others, inspired by the discovery and pharmaceutical activity of the effective anticancer agent cisplatin [5,6,7], the search for metal complexes capable of interacting with DNA is increasingly flourishing. In particular, attention is often directed towards a large family of transition metal complexes, as these species are able to combine structural elements, such as defined and modulable coordination geometries with appropriate ligand stability/lability, and distinctive electrochemical and photophysical properties.
On the one hand, the negative charge of the DNA surface implicitly allows one to consider electrostatic attraction to all cationic complexes as a possible interaction; on the other hand, one can also imagine that appropriately designed ligands (with extensive planar surfaces) might allow the metal complex to fit between the base pairs of double-stranded DNA in the B form (i.e., bind by intercalation).
This type of reasoning makes it possible to justify, for example, the different types of interaction with DNA offered by [Ru(bpy)3]2+ and [Ru(phen)3]2+ [8,9]. In fact, while the former binds only weakly to DNA, the interaction of [Ru(phen)3]2+ is much stronger, thanks to the presence of the phenanthroline ligand that allows for an at least partial intercalation into DNA base pairs, called semi-intercalation [10,11,12].
The initial interest was amplified by Barton′s research into the use of enantiopure forms of complexes like possible chiral probes for DNA [13,14,15]. Over time, it was realized that these were the photophysical and electrochemical properties of a given complex, that was achieved by appropriate ligand substitution, which actually expressed the great potential of these metal complexes as luminescent probes and photochemical reagents.
Furthermore, as well as allowing modulation of redox and implicitly photophysical properties, the design of appropriate ligands also provided a means of varying the character and strength of binding to nucleic acids.
In this review, rather than presenting a complete history of the development of metal complexes used as DNA binders, we focused on certain aspects of this class of compounds and, in particular, on the light-switch effect offered by the Ru(II) dppz derivatives [16].
2. DNA Structure
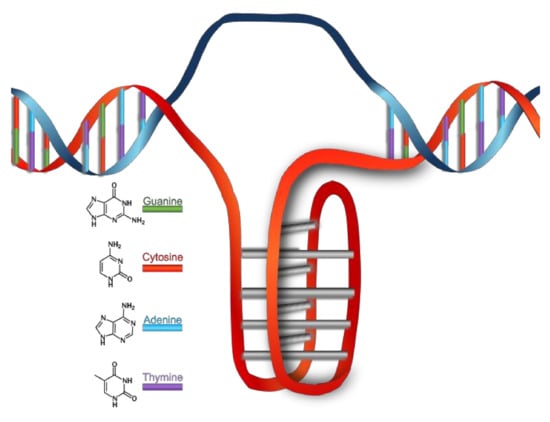
The classic 3D-model, proposed by Watson and Crick in 1953, describes the DNA (in B conformation) as two polynucleotide chains that coil around each other to form a right-handed double helix [17]. The two filaments of the DNA run in opposite directions to each other, so they are antiparallel, and according to the model, this is the reason such winding is present. In the center of the double helix there are nitrogenous bases, which keep the two chains bound by hydrophobic π-π interactions (which reduce the interaction with water molecules) and by the hydrogen bond between complementary bases, each of them belonging to a different chain. It is exclusively the interaction between a purine and pyrimidine derived base that grants the necessary distance so that a hydrogen bond can form—this can happen only by the A-T and G-C pairs.
Since hydrogen bonds are weak, it is relatively easy to break and form them again, like a zip, for example, due to the high temperature or mechanic interactions. As a consequence, in addition to the complementary bases, DNA molecules might be replicated such that all biological information contained within might be duplicated—such is the DNA replication [18].
It is quite well-known that G-C stability (three hydrogen bonds) is higher than that of A-T (two hydrogen bonds), therefore, the global stability of a DNA helix is related to the amount of G-C pairs, as well as the chain length.
The B form is the most common DNA conformation found within cells [19]. B-DNA winds up in a right-handed manner, see Scheme 1. There are approximately ten nitrogenous bases per turn, paired on the same plane and arranged perpendicularly to the DNA axis. The glycosidic bond between the 2-deoxyribose and the nucleobases shows anti-conformation. The stacking of the nucleobases leads to the formation of a major and a minor groove. In B-DNA, the major groove is 11.6 Å wide and the minor one is 6.0 Å wide—this difference leads to a different accessibility to the nitrogenous bases, depending on their position. Since on these grooves the nitrogenous bases are exposed to solvent, they might specifically interact with other species, without implicating modifications to the double helix. The majority of DNA-specific proteins interact with it right in one of the two grooves, especially into the major one because the sequence-specific recognition is greater in this groove.
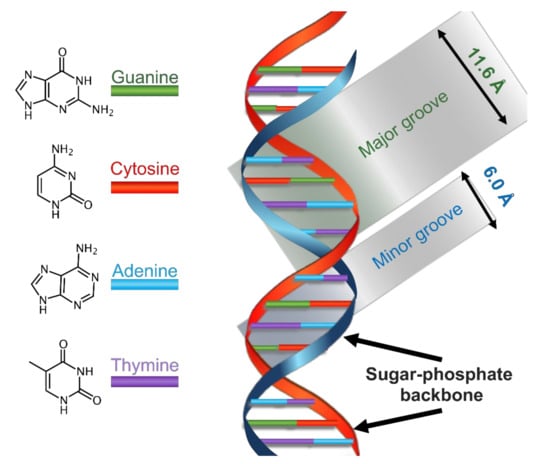
Scheme 1.
Schematization of the main physical features of the structure of B-DNA.
3. DNA Binding
3.1. Interaction of Small Molecules with DNA
It is possible to distinguish two kinds of interactions with DNA—reversible and irreversible ones [20,21]. Irreversible interactions involve the formation of a covalent or coordination bond between a species and an atom site of the helix; such a linkage implicates the total inhibition of DNA processes (and the cell death as a consequence), if DNA repair systems, such as the NER (nucleotide excision repair) fail. Very often, the steric hindrance of species bound irreversibly might cause a distortion of the phosphate groups, which leads to negative effects on DNA transcription and replication, such as preventing the protein-complex identification [2,3,4].
Reversible interactions are usually less toxic than the irreversible ones. These can be classified into four types (see Scheme 2)—(i) external electrostatic interaction, (ii) groove binding, (iii) external self-aggregation, and (iv) intercalation.
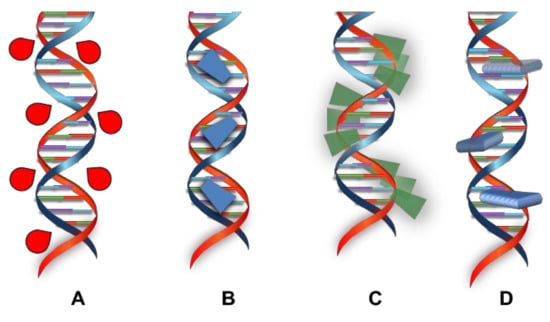
Scheme 2.
Schematization of the main reversible interactions between small molecules and DNA—(A) external interaction; (B) interaction in grooves; (C) external self-aggregation; and (D) intercalation.
The external electrostatic interaction (i) is due to the presence of the phosphate groups in the chain, because their negative charge might attractively interact above any positive ion. In particular, simple cations like alkaline ones associate with nucleic acids in the base of the density charge generated by the phosphate groups, resulting in a stabilization of the DNA itself. This association is called ‘condensation’, because the ions are associated to the overall charge of the biomolecule and not to specific sites, which remain capable of moving along the double helix. The typical DNA counterions are Na+ ions, which compensate most of the phosphate groups’ negative charge. The remaining charges are neutralized by other cations through the classic Debye-Huckel mechanism.
The groove binding (ii) occurs for species that have the right dimension and conformation to “lay down” in one of the grooves, which interact via electrostatic interactions, Van der Waals forces, and especially, hydrogen bonds [22,23]. The typical molecules that are able to make this kind of interaction have an appropriate length and flexibility, which might assume an optimal conformation. Usually, these molecules have several aromatic rings (like pyrroles, furans, or benzenes) that are bound to substituents that are capable of rotating around the bonds, letting them adapt to the double helix shape without significantly perturbing the DNA structure.
External self-aggregation (iii) occurs when molecules, especially aromatic planar species, stack along the external part of the double helix, forming dimers or even huge aggregates through a non-specific bond called external stacking. It is known that interactions of porphyrin and metal-porphyrin with DNA, lead to the formation of chiral supramolecular structures [24,25,26,27,28], Pt(II) organometal complexes coordinated by terpyridine [29,30] or Pt(II) complexes with extended aromatic ligands like dipyrido[3,2-a:2′,3′-c]phenazine or benzodipyrido[b:3,2-h:2′,3′-j]phenazine, in the case of low [DNA]/[complex] ratio, and consequently, the presence of few intercalative sites [31]. Pasternack et al. highlighted that the structure of intercalators influences the interaction type with DNA and might lead an external interaction to be preferred to an intercalation [32,33].
Intercalation (iv) is essentially a stacking interaction resulting from the insertion of a planar aromatic system between the base pairs of the DNA double helix, and is permitted by a significant overlap between π orbitals of the intercalative species and those of nitrogenous bases, forming a system stabilized by Van der Waals forces, hydrophobic interaction, and especially, π-π stacking [34,35].
This type of interaction was first proposed by Lerman [34] in order to rationalize his results on the high affinity for DNA of certain acridine derivatives. The combination of these structural modifications results in functional changes, and frequently, in the inhibition of DNA transcription and replication and repair processes, thus making the intercalators mutagens.
To permit this intercalation process, it is necessary that a “cavity” forms between the nucleobases, so they have to distance each other; this process causes the elongation of DNA [36,37,38,39]. Vertical separation between nucleobases (about 3.4 Å in the B-DNA) might increase from 1.8 to 4.5 Å per intercalated molecule when a huge aromatic molecule intercalates. The angle of rotation might vary from 11° to 48°, depending on the intercalator structure and the type of nucleotides involved [40]. The process causes distortions in the DNA structure that can be used to study DNA-molecule interactions by, for example, viscosity measurements [41] sedimentation studies [42], circular dichroism (CD) variations [36,37,38,39,40,43], and electrophoresis studies on circular DNA supercoil [44].
The intercalation model proposed by Lerman, however, did not take into account the possibility of the species to insert itself between nucleobases pairs, until a double helix saturation. Crothers introduced the neighbor exclusion model: [45,46] in case a molecule intercalates into a site, it distorts the vicinal ones, so that they would not be available to permit the intercalation of other molecules. Therefore the intercalation is anti-cooperative at adjacent sites, and therefore, only 50% of DNA sites can undergo an intercalative interaction. Moreover, it is also (highly) possible that the molecules do not occupy every other site, obstructing more than two of them; also, huge molecules might prevent this kind of interaction due their steric hindrance.
Several studies were conducted to understand whether there exists a specificity in the intercalative interactions related to the type or the sequence of the nucleobases involved. Some intercalators showed preferential interactions, but almost all of them interact aspecifically with the nucleobases.
At first it was believed that only small molecules, with a planar structure, could intercalate, but after several studies during the years a distinction between ‘classical’ and ‘non-classical’ intercalators was made. Classical intercalators are characterised by aromatic condensed rings and have positive charge(s) on the rings themselves or on the side chains.
Among non-classical intercalators there are compounds that have huge or polar (or charged) substituents on the opposite sites of the intercalating aromatic system [47]. These compounds need to insert at least one of the substituents between the nucleobases pairs to intercalate. Among these species there are known compounds like naphthalene bisimide and cationic porphyrins, which, although they are not supposed to intercalate, might interact efficiently with DNA through such an interaction [48]. Even if thermodynamically favorable, this interaction is kinetically unfavorable, and needs opening of the double helix (causing distortion or breaking of hydrogen bonds); such effect makes the DNA–molecule interaction slow.
The initial studies of the intercalative process involved simple organic molecules, but they were followed by studies focused on the interaction between DNA and metal complexes. The presence of a transition metal indeed provides several advantages, as described below [49,50,51,52,53,54].
- The presence of d orbitals grants more ways (bonds and geometries) such that the species might interact with DNA other than those of organic molecules.
- Steric and electronic properties can be modulated choosing opportune ancillary ligands.
- Bridging ligands of certain dimensions might modify the size of the intercalator.
- It is possible to take advantages from their spectroscopic, magnetic, redox, photophysical, and photochemical properties.
Since the antitumoral properties of cisplatin, square planar Pt(II) complexes were the first inorganic species whose interaction with DNA was studied. Lippard, in the ‘70s, demonstrated that Pt(II) complexes with heterocyclic aromatic ligands (like 2,2′:6′,2″-terpyridine) intercalate into the DNA [55]. It is worth noting that the geometry of Pt(II) complexes is compatible with the steric requisites for the intercalation process, and by modifying the bound ligands, it is possible for them to modulate their steric and electronic properties [56,57,58,59,60].
3.2. Ru(II) Complexes as Intercalators
In the last decades, more attention was given to metal complexes with an octahedral geometry, to verify if the species with high steric hindrance would intercalate and how this influences the process. In fact, ancillary ligands could prevent the insertion of small molecules between DNA adjacent base pairs.
Ru(II) complexes containing rigid aromatic rings (stable, inert, and soluble in water) were among the first classes of octahedral complexes studied [61,62].
One of these was [Ru(phen)3]2+ species (phen = 1,10-phenanthroline) [63,64,65]. In this complex, each ligand has an aromatic planar structure that might interact with DNA; moreover, the three ligands give a chiral effect to the species. Chirality might have a relevant effect on the interaction process—according to Barton4, the intercalation is favored for Δ isomers of these kinds of complexes. This is due to the orientation of ligands in the Δ isomers, which interfere sterically less than those in Λ isomers, with the skeleton of DNA. Later studies by Chaires and Nordèn clarified that rather than intercalation, it is more correct to refer to this interaction as semi-intercalation, since there is only a partial insertion of a phenanthroline between the nucleobases, without the complete separation of the latter [10,11,12].
Taking enantioselectivity of Ru(II) tris-phenanthroline complexes as a starting point for designing probes that are able to recognize the double helix chirality, there were synthesized complexes that could selectively bind to DNA. For example, Λ isomers of metal complexes based on 4,7-diphenylphenanthroline (DIP) are efficient probes for Z-DNA [66,67]. [Ru(TMP)3]2+ (TMP = 3,4,7,8-tetramethylphenanthroline) is, instead, a selective probe for A-DNA [68]. Methyl substituents, in fact, do not allow the complex to intercalate between nucleobases; it interacts with DNA through electrostatic and hydrophobic interactions. This complex is too huge to insert into a B-DNA minor groove, but can insert into the major one of A-DNA and, through photoactivation and subsequent generation of 1O2, can break the double helix.
However, the real research success in this field, and in fact, one of the most studied complexes is [Ru(bpy)2(dppz)]2+ (dppz = dipyrido[3,2-a:2′,3′-c phenazine]); see Figure 1.
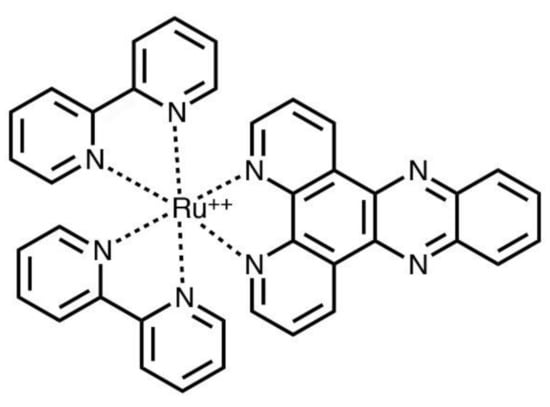
Figure 1.
Molecular structure of [Ru(bpy)2(dppz)]2+.
The dppz ligand possesses a large aromatic surface area, allowing extensive intercalation in the DNA base pairs. Most Ru polypyridine complexes based on this ligand bind strongly to the DNA duplex via the intercalation mechanism. One measure of the affinity of these interactions is the log K value (K= DNA binding constant). For the classic DNA intercalator ethidium bromide (EB), this value is >6, while for [Ru(bpy)2dppz]2+, under the same experimental conditions, this value is >7 [69], demonstrating the high affinity of this ligand for DNA.
In 1990, Barton first reported that [Ru(bpy)2(dppz)]2+ could serve as a luminescent DNA probe [70]. This species does not exhibit luminescence in aqueous solution at room temperature, but shows intense emission in the presence of DNA, due to the radiative decay of an MLCT excited state, once the complex intercalates into the base pairs [71]. In water solution, instead, the luminescence is quenched due to proton-transfer from water molecules to phenazine’s nitrogen atoms. Intercalation does not permit the interaction between water molecules and the Ru(II) complex, so the latter becomes luminescent.
This phenomenon is known as the ‘light-switch’ effect of DNA and is widely used to study the interaction of polypyridyl metal complexes with DNA.
Interaction of Ru(II) complexes with DNA usually generates an increase in luminescence intensity due to the reduced probability of:
- non-radiative decay, because the intercalated luminophore is surrounded by the DNA hydrophobic environment and is therefore protected from water-induced deactivation;
- intersystem crossing to the 3MC excited state, as the complex is caged into the double helix and its rigidity result is enhanced;
- O2 luminescence quenching.
On the other hand, if there are photoinduced reactions between the metal complex and nucleobases, luminescence is quenched.
Similar to the [Ru(bpy)2(dppz)]2+ species, the two Ru(II) complexes reported in Figure 2 behave as luminescent DNA probes; they are based on TAP (1,4,5,8-tetra-azaphenanthrene) and PHEHAT (1,10-phenanthroline[5,6-b]-1,4,5,8,9,12-hexa-azatriphenylene) ligands. Interestingly, in the presence of DNA, the behavior of these two complexes is different—the first complex acts as an ON–OFF molecular switch; the second one acts, instead, as an OFF–ON molecular switch [72].
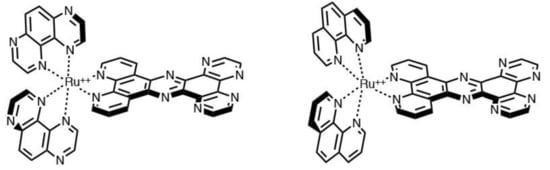
Figure 2.
Representation of [Ru(TAP)2(PHEHAT)]2+ (left) and [Ru(phen)2(PHEHAT)]2+ (right).
[Ru(TAP)2(PHEHAT)]2+ is a photo-oxidant complex (the two TAP ligands give it a high reduction potential) and shows a great affinity for the DNA, thanks to the PHEHAT ligand, which can intercalate. DNA efficaciously quenches its luminescence due to the ‘electron transfer’ process with guanine; which is a ‘light-switch’ON–OFF’.
[Ru(phen)2(PHEHAT)]2+ in water solution at room temperature does not exhibit any luminescence; the lone pairs present on nitrogen atoms in the PHEHAT ligand, interacting with water molecules, stabilize a non-emissive excited state. When the complex intercalates into DNA, the PHEHAT ligand is surrounded by the DNA hydrophobic environment and the stabilization by water molecules cannot happen. Therefore, the non-emissive excited state is not stabilized and the complex exhibits luminescence; which is called ‘light-switch OFF–ON’.
A wide variety of dppz-based complexes were designed, synthesized, and studied by modifying the bipyridine ancillary ligand or the dppz ligand [73,74,75,76,77,78], or changing the metal center to Re [79,80,81,82], Co [83,84], Cu [85,86], Rh [87,88,89,90], Os [91,92], Pt [31,93,94,95], and Ir [96,97,98] cations.
Many experiments were conducted in an attempt to rationalize and “observe” the light-switch mechanism. In the case of [Ru(phen)2(dppz)]2+ [69], see Figure 3, the mechanism seems to occur in two distinct phases, the first involving intersystem crossing between the state called MLCT′ (bright state) and the one called MLCT″ and subsequently by non-radiative deactivation from the latter. The MLCT′ state represents the dominant state of [Ru(phen)2(dppz)]2+ in aprotic organic solvent, and is the state that is initially populated in other solvents as well; see Scheme 3.
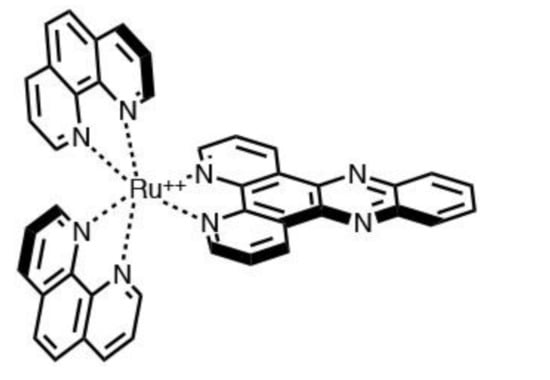
Figure 3.
Molecular structure of [Ru(phen)2(dppz)]2+.
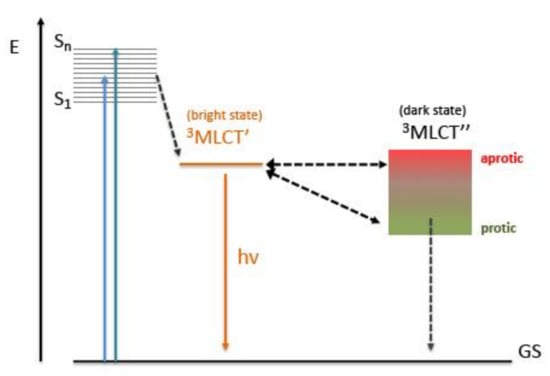
Scheme 3.
Jablonski diagram describing the excited states and transitions of [Ru(bpy/phen)2(dppz)]2+complexes and the solvent influence on the dark state (GS: Ground State).
The MLCT” state (dark state), on the other hand, is a state involving the phenazine portion of dppz and its energy is a function of the solvent. In fact, by rationalizing the photophysical properties of [Ru(phen)2(dppz)]2+ in a wide range of solvent environments, it can be inferred that both the polarity of the latter and the ability to form hydrogen bonds with the N atoms of the phenazine fragment govern the population of this dark state [48].
The light-switch behavior is also a function of temperature, which governs the extent of equilibration between the two states and thus the relative population. It was shown that the light-switch process results from a competition between energetic factors that promote the population of the dark state (MLCT″) and entropic factors that tend to favor the population of the bright state (MLCT″) [99,100,101,102,103,104].
3.3. Solvent Effects on the Photophysical Properties of the Ru-Dppz Complexes
The effect of the solvent on the spectroscopic and photophysical properties of the [Ru(phen)2dppz]2+ complex was systematically studied for the first time by Murphy [105]. The studies carried out showed that the absorption spectra in water and in various non-aqueous solvents were very similar, while the emission energies, as well as the lifetime and quantum yield of luminescence, were extremely solvent-dependent. It was shown that there was no correlation between the maximum luminescence and the polarity of the solvent. In particular, as already pointed out, no emission was observed in water and trifluoroethanol. On the other hand, quantum yield and luminescence lifetime were particularly sensitive to solvent polarity.
Both of these quantities were reduced in high-polarity solvents, and this trend was correlated with the empirical quantity ET, which reflected, to some extent, both a measure of the polarity of the solvent and its ability to make h-bond. Furthermore, studies carried out on [Ru(phen)2dppz]2+ showed that knr (rate constants for non-radiative processes) was almost independent of solvent polarity, while all non-radiative constants were, and somehow became greater, as polarity increased.
The energy gap law that correlated the non-radiative constants with the difference in energy between the fundamental and excited luminescent states did not appear to be respected in this case, unlike what happened for example for the parent compound [Ru(bpy)3]2+.
Contrary to the assumption that H-bonding to phenazine nitrogen atoms in the excited state was capable of activating non-radiative channels and thus quenching the emission, the improved correlation observed between ln(knr) and ET, compared to the H-bond-donating capacity of the solvent alone [106], made it possible to highlight that the forces at play, and the mechanism, were subtler (or involve concomitant events) than predicted by early studies.
The light-switch phenomenon for this family of compounds was indeed closely related to the effect of the solvent on the two possible states, involving the metal and the bipyridine or phenazine moiety of the dppz. In two fundamental studies, it was shown that this effect resulted from a thermal equilibrium between these two excited states, one enthalpically favored (the state involving the phenazine fragment) and the other entropically favored. This behavior could be attributed to the fact that charge separation induced a rearrangement (and thus greater order) of the surrounding solvent [107].
The competition between the two different thermodynamic contributions results in a non-obvious inversion of the populations of excited states, as a function of solvent—the photoluminescent 3MLCT state essentially dominates in aprotic solvents (or in the presence of double-stranded DNA), while the CS state population becomes prominent in protic solvents [106].
3.4. Effects of Tuning the Photophyisical and Redox Properties
Modifications on complexes based on the dppz ligand are aimed at two main effects—the light-switch effect and DNA affinity [108].
The coordination geometry in the case of Ru(II) is that of an octahedral d6 and, therefore, in addition to the bidentate dppz ligand, four other coordination sites are available for the so-called ancillary ligands [109,110,111,112].
Modifications made to these ligands can result in important electronic effects that lead to changes in the photophysical and redox properties of these types of systems; see Table 1.

Table 1.
Absorption and photophysical properties, in MeCN and H2O of the representative Ru-dppz complexes.
An example of that is the use of primary amines or acetylacetonates that leads to complete non-emissive species, as in the case of [Ru(NH3)4(dppz)]2+ and [Ru(acac)2(dppz)] [118,119], while the use of polypyridine ligands with ligand-centered (LC) states (which are much more stabilized species than phenanthroline or bipyridine and which become acceptors of the MLCT transition) leads to luminescence in both the presence and absence of DNA [120].
The affinity of these species for DNA can also be modulated by peripheral ligands. Polypyridine ligands modified with carboxylic acids, as in the case of [Ru(bpy-COOH)2(dppz)]2+ lead to a greater selectivity of duplex DNA to single-stranded DNA than the corresponding esterified ligands ([Ru(bpy-COOMe)2(dppz)]2+) [128]. This type of behavior is due to the fact that deprotonation of the acidic groups results in the formation of a formally neutral species that induces greater repulsion between the ancillary ligands and the DNA surface and leads to duplex selectivity.
The electronic effect of the ancillary ligands leads to a modulation of the redox potentials of these species, and as in the case of [Ru(tap)2(dppz)]2+ [117], a photoinduced electron transfer process can occur with guanine, leading to the formation of a covalent bond between the species and the polynucleotide [109,110,111,112].
Substitution on the ancillary ligands is also important to manage the selectivity of the light-switch. Indeed, by incorporating methyl groups on the phenanthrolines of the [Ru(phen)2(dppz)]2+ system, a highly specific probe for a DNA mismatch was obtained. The selectivity was shown to be due to a higher binding affinity towards the mismatched DNA, which results in a much higher luminescence lifetime when bound to a mismatch [129].
It is very interesting to point out that the electron transfer processes involving DNA and metal complexes also depend on the type of interaction and thus on whether the reagents are metallo-intercalators or groove-binding agents. It was shown that photoinduced electron transfer and charge recombination (back electron transfer) becomes more efficient across the DNA base stack. The efficiency and yield of these processes are also a function of the base sequence into which the metal-binding agent fits, due to the different redox potentials of the transition metal complexes and the different bases [130,131,132].
A particularly interesting effect of substitution on the photophysical, and in particular, the photochemical properties of the polypyridyl ruthenium complexes is obtained by using a dppz analogue, i.e., 1,4,5,8-tetraazafenanthrene (TAP) or 1,4,5,8,9,12-hexaazatriphenylene (HAT), see Figure 4 [133]. In the case of the homoleptic [Ru(TAP)3]2+ complex, it was shown that this complex is capable of forming covalent photoadducts with DNA. The ability of this type of complexes to give photochemistry with DNA is a function of the oxidizing power of the complex and becomes scarce in the case of the complex containing two TAP or HAT ligands, to the point of being absent in the case of the mono-substituted [Ru(bpy)2L]2+ complex (L = TAP or HAT) [134,135,136]. The interest in this type of research is fundamentally based on the selectivity of the photochemical action with adduct formation, which is attributed to the reaction of the radical products formed by the initial electron transfer and the subsequent proton transfer.
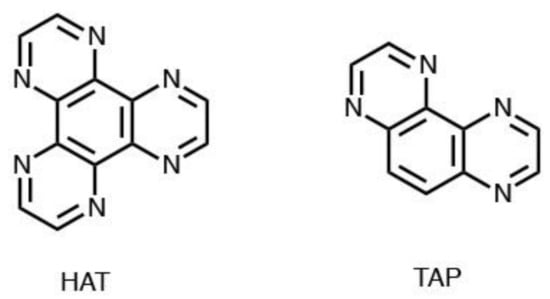
Figure 4.
Molecular structure of HAT and TAP ligands.
Both luminescence lifetime and time-resolved absorption spectroscopy confirmed that the excited states of these species were able to oxidize guanine, either as a free nucleotide (5-GMP) or in DNA, but not the other nucleobases.
It was also shown that the photo-oxidation of guanine by [Ru(TAP)2(dppz)]2+, and in particular its isomer Λ, was a function of the nucleobase sequence. In fact, if bound to (CCGGATCCGG)2 or (CCGGTACCGG)2, its behavior was different. In the case of the TA sequence, the electronic transfer was not very efficient [137]. However, after irradiation with visible light, the TAP containing complexes, exhibited both photocleavage and formation of mono- and bi-adducts with DNA [138].
In particular, the photochemical product of [Ru(TAP)3]2+ with 5-GMP was isolated and identified by demonstrating that the adduct was formed by a bond between the TAP ligand and the external NH2 group of guanine. In the case of the [Ru(HAT)2(phen)]2+ species, however, the photochemical reaction with 5-GMP resulted in the formation of a bond between HAT and the 6-O atom of guanine [139].
Similar behavior was observed with the [Ru(bpy)2(tpphz)]2+ complex, see Figure 5 (tpphz = tetrapyrido[3,2-a:2′,3′-c:3″,2″-h:2‴,3‴-j]-phenazine). In particular, this species demonstrated that the light-switch ON effect could be cycled OFF and ON through the successive introduction of Co2+ and EDTA. The presence of a further chelating site on the dppz-like ligand results in the coordination of cobalt ions that affect the energy of the two 3MLCT state, promoting the population of the non-emissive state [126,127].
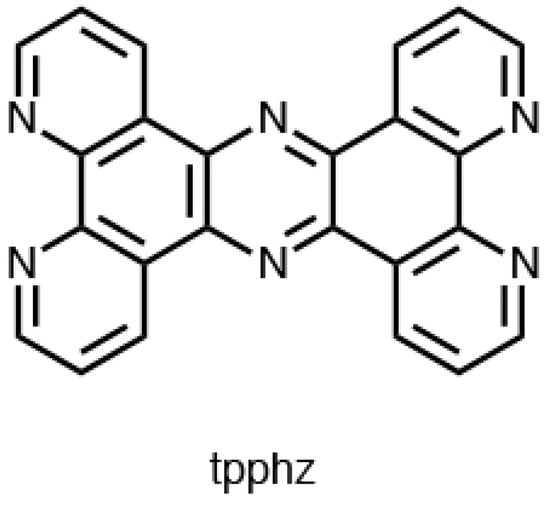
Figure 5.
Molecular structure of the tpphz ligand.
The interaction of these molecules with the particular structure of DNA was also studied.
Both [Ru(bpy)2(dppz)]2+ and [Ru(phen)2(dppz)]2+ were shown to induce the human single-stranded telomeric repeat 5′-AGGGTTAGGTTAGGG-3′(22AG) to fold into an intramolecular quadruplex [140,141]. In contrast, a cytosine-rich filament oligomer 5′-CCCTAACCCTAACTAACCCT-3′(22CT) could self-associate into an i-motif structure; see Scheme 4. In general, Ru-dppz-type complexes only weakly interact at the i-motif, making them somewhat selective for G-quadruplexes. This type of behavior was palpably attributable to the interaction of [Ru(phen)2(dppz)]2+, which stacked on the ends of G-quadruplexes, while it might bind to the i-motif only by non-specific interaction [142]. Similar to the G-quadruplex DNA structures (containing essential guanine residues), i-motifs (intercalated motifs) consist of a fragment of oligodeoxynucleotides strands, mostly based on cytosine residues.
Scheme 4.
Schematic representation of DNA i-motifs found in human cells.
I-motif forming DNA was recently identified as a key step in the recognition and regulation of transcriptional proteins in cells. It was also present in many promoter regions of oncogenes [143,144].
Although the interaction seemed too weak to be studied in detail, recently, [Ru(phen)2(dppz)]2+ was used to collect information on the binding sites to i-motif structures. IR transient spectroscopy made it possible to distinguish the presence of different bases in loops (as was shown for the human telomeric sequence), demonstrating that this archetype species, together with advanced spectroscopic techniques, has a great potential for identifying binding for biologically-relevant sequences [145,146].
The electronic effect becomes even more pronounced by making changes to the dppz ligand. Conjugation with an active redox fragment was able to activate photoinduced intramolecular electron transfer processes that led to the quenching of luminescence (e.g., Ru-qdppz and Ru-qdpq, see Figure 6). An external redox input (chemical or electrochemical) could modify the active redox fragment and restore the light-switch effect. This was the case, for example, with the quinone/hydroquinone pair. The former was, in fact, able to switch off the luminescence through a process of oxidative electron transfer involving the 3MLCT state characteristic of Ru-dppz. Electrochemical or chemical reduction of the quinone fragment restores luminescence, thus allowing dual-input operation of the molecular probe [121,123].
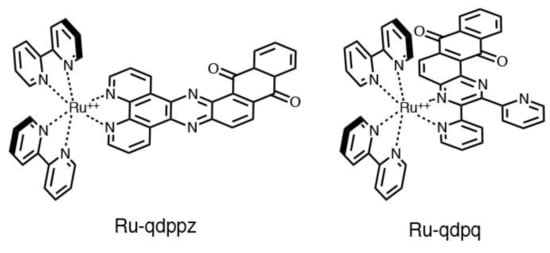
Figure 6.
Molecular structure of Ru-qdppz and Ru-qdpq.
In general, the low luminescence quantum yield and short excited-state lifetimes led to less interest in ruthenium dppz complexes carrying the tridentate ligand tpy (tpy = 2,2′:6′,2″-terpyridyl) than in complexes based on bipyridine and phenanthroline ligands, despite the fact that the absence of a metal stereocenter eliminated the need to separate racemic mixtures typical of tris-bidentate systems.
However, it is important to emphasize that, when designing a system capable of targeting DNA in vivo, not only is the efficiency of the luminescence processes to be important, but also the range of emission, which must overlap with the optical therapeutic window, as much as possible.
The [Ru(tpy)2]2+ complex (tpy = 2,2′:6′,2″-terpyridine), for example, is characterized by a short excited state and a low quantum yield, making it, in principle, unsuitable for photobiological applications [147,148].
However, it is possible to modulate the properties of this family of complexes by operating substitutions on the terpyridine ligands [149].
Turro et al. reported that the [Ru(tpy)(pydppz)]2+ species, based on a Ru(II)-terpyridine fragment and a tridentate dppz derivative known as pydppz (3-(pyrid-2′-yl)dipyrido-[3,2-a:2′,3′-c]phenazine), is capable of intercalating into DNA and behaving as a light-switch [149]. The photophysical properties of this complex, characterized by a weak emission centered at 698 nm in acetonitrile (τ = 5.4 ns and φ = 2.1 × 10−4), although more interesting than the model species [Ru(tpy)2]2+, are not such as to warrant its use for photobiological applications.
Starting with these bases, and since it is known that (i) with the same ligands, Os(II) polypyridine complexes exhibit excited states with significantly lower energy than that of the analogous Ru(II) compounds, and that (ii) osmium compounds with tridentate ligands such as terpyridine possess excited states with a longer lifetime, the complex [Os(tpy)(pydppz)]2+, shown in Figure 7, was synthesized. This new species of Os(II) exhibits an absorption spectrum extending to 800 nm. Its emission characterized in aprotic organic solvent, by relatively high quantum yields and lifetimes in the order of hundreds of ns, is centered at 800 nm, and the complex represents the first example of an [Os(tpy)2]-like species capable of intercalating into the DNA [91]. Moreover, it is also interesting to note that, despite the structural and chemical variation, it behaves as a true DNA OFF–ON light switch, like the analogous [Os(phen)2(dppz)]2+ [150,151,152,153,154].
Figure 7.
Molecular structure of [M(tpy)(pydppz)]2+.
3.5. Few Examples of Applications: Cellular Imaging, Photodynamic Therapy (PDT), and Sensing
Studies carried out on cells, including cytoetric and imaging studies [155], showed that cellular uptake is generally a function of the integrity of the cell membrane and the nuclear membrane [156].
Species based on dppz were studied from a photophysical point of view in a variety of solvents, in the presence and absence of DNA [157,158]. Studies were also carried out on systems embedded in phospholipid vesicles [159,160], and in general, it was confirmed in the biological environment that these properties of Ru(II)-based complexes were highly dependent on the environment [161]. Recently, [Ru(bpy)2(dppz)]2+ was studied for the first time in cells, using localized pump-probe spectroscopy, showing that the ultrafast dynamics of this species is a function of the position where the complex was internalized. The results obtained on a sample of fixed HepG2 cells clearly showed that it was necessary to have a good understanding of the dynamics of photoactive complexes in the actual target environment of the drug, so that the properties of the photodrug could be unambiguously correlated with its in vitro activity.
The DNA light-switching effect of the Ru-dppz complexes offers potential for use in fluorescent cellular imaging, particularly for nuclear staining with high-density DNA.
One of the key parameters for the use of these species at the cellular level is to optimize the cellular uptake process, especially, the crossing of the nuclear membrane [162].
In particular, research by Professor Barton′s group showed that a key parameter was the level of lipophilicity of the Ru-dppz photo-switch derivative obtained using different accessory ligands (bpy, phen, CO2Et-bpy, …) [163,164,165]. However, this did not constrain absorption in the nuclear zone.
For example, in the case of the [Ru(DIP)2(dppz)]2+ complex, confocal microscopy images of HeLa cells showed that this species is able to localize in the mitochondria or in the endoplasmic reticulum region, but not in the nuclear one [149].
When the lipophilicity of Ru-dppz complexes was modulated by the use of aliphatic chains on the ancillary ligands, cell imaging of fixed CHO-K1 cells revealed that the less lipophilic complex was preferentially localized within the nucleus, whereas, when the degree of lipophilicity became more pronounced, the light-switch essentially localised in the cell membrane. It was clear therefore that in a complex system such as the cellular one, it was difficult to find a univocal behavior, but the structural properties of these complexes must be dosed appropriately in order to convey them univocally [166,167,168].
One way to achieve this was to conjugate Ru-dppz like complexes with a nuclear targeting fragment such as the peptide D-octaarginine (D-R8) or the tetrapeptide RrRK, in which case nuclear localization was facilitated [124,125].
It is known that one of the key steps to increase the efficiency of the photosensitizers in DNA photocleavage is to bind, more or less strongly, to the DNA. Therefore, the use of intercalators was particularly studied in the field of PDT. However, [Ru(bpy)2(dppz)]2+ showed negligible photocytotoxicity since, despite its high affinity for DNA, the singlet O2 production efficiency was very low [169].
In order to intervene in this direction and improve the photosensitizing properties of Ru-dppz derivatives, Gasser designed a series of derivatives containing functional subunits on the dppz ligand [170,171], by maintaining the affinity for DNA, on the one hand but improving the photocytotoxicity values as PDT agents (improving the performance of the 3MLCT state), on the other hand.
The use of the dinuclear [Ru(bpy)2tpphzRu(bpy)2]4+ and [Ru(phen)2tpphzRu(phen)2]4+ molecular complexes as multifunctional biological imaging agent for DNA was reported. In particular, the efficiency of these species to “switch -on” nuclear DNA of living cells without requiring prior membrane permeabilization was demonstrated for both luminescence and transition electron microscopy [172].
Recently, a new derivative of [Ru(bpy)2(dppz)], demonstrating a dramatic improvement in photobiological activity by introducing a pyrenyl substituent, was proposed [173]. The studies demonstrated that the population of an LC triplet state was much more performant in terms of singlet oxygen yield and potentially usable in clinical PDT applications.
In addition to DNA, Ru-dppz-type complexes are able to interact with proteins. One example is the heteroleptic ruthenium species based on the ligands 7,8-dimethyl dppz, 1,10-phenanthroline-4-carboxamide, and 5,6-dimethyl-1,10-phenanthroline [174], which is able to bind to acetylcholinesterase (AChE), exhibiting the typical light-switch behavior when binding to the hydrophobic domains of the protein. A further example is the more classic [Ru(bpy)2dppz]2+, which is not photoluminescent in aqueous solution, even in the presence of monomeric amyloid beta (Aβ) (a peptide fragment containing 36–43 amino acids.) Monomeric Aβ self-assembles to form aggregates (amyloid fibrils) that are crucially involved in Alzheimer′s disease. In the presence of aggregates of Aβ fibrils, the photoluminescence of [Ru(bpy)2dppz]2+ is recovered [175,176]. A further interesting application arises from the modification of Ru-dppz complexes by introducing an aptamer fragment into the system. Although the aptamers are single-stranded DNA oligonucleotides, they are able to fold into three-dimensional structures through base pairing. This process leads to protection of the dppz system from the solvent, which results in luminescence. More specifically, the system is applicable by exploiting the combination of the light switch effect with the selective ability of aptamers towards specific analytes. Recognition by the aptamers of a specific analyte (e.g., immunoglobulin E (IgM)) [177], abrin toxin [178], and ATP [179,180]) leads to the unfolding of its structure and the exposure of the dppz complex to the aqueous solvent, with consequent extinction of the luminescence.
4. Conclusions
Ru-dppz type complexes were extensively designed, synthesized, and studied. The combination of their photophysical properties and the peculiarities of the light-switch effect on DNA makes them powerful tools for analyzing the behavior of molecules of great biological impact. Despite the fact that the first example in this direction appeared more than 30 years ago, derivatives of Ru-dppz complexes are still of very wide interest, thanks to the possibility of crossing their characteristic and peculiar specificity towards biological material with a sharp and modulable light response. Over time, we progressed from simple interaction studies with DNA or specific sequences in vitro to direct applications, to investigate the selectivity of these molecules for different DNA structures, including B-form, Z-form, duplex, triplex, G-quadruplex, defective DNA, etc. Research in this field, together with the expansion and emergence of increasingly high-performance spectroscopic and microscopic techniques, continues to revive the applications of these apparently simple, but highly versatile molecular derivatives, in fields as diverse as cell nuclei imaging, photoactivatable, and DNA-targeted anticancer drugs, nucleic acid sequencing, protein aggregate recognition, and virus labelling. In only thirty years, there has been an ever-increasing number of applications that seem to be constrained only by the growth of experimental techniques of investigation and the imagination of researchers.
Author Contributions
Writing—original draft preparation, M.L.D.P., G.L.G., F.N. and F.P.; Writing—review and editing, M.L.D.P. and F.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and cited refs.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rodley, G.A.; Scobie, R.S.; Bates, R.H.; Lewitt, R.M. A possible conformation for double-stranded polynucleotides. Proc. Natl. Acad. Sci. USA 1976, 73, 2959–2963. [Google Scholar] [CrossRef]
- Zeglis, B.M.; Pierre, V.C.; Barton, J.K. Metallo-intercalators and metallo-insertors. Chem. Commun. 2007, 44, 4565–4579. [Google Scholar] [CrossRef]
- Liu, H.K.; Sadler, P.J. Metal Complexes as DNA Intercalators. Acc. Chem. Res. 2011, 44, 349–359. [Google Scholar] [CrossRef]
- Pages, B.J.; Ang, D.L.; Wright, E.P.; Aldrich-Wright, J.R. Metal complex interactions with DNA. Dalton Trans. 2015, 44, 3505–3526. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, B.; Vancamp, L.; Trosko, J.E.; Mansour, V.H. Platinum Compounds: A New Class of Potent Antitumour Agents. Nature 1969, 222, 385–386. [Google Scholar] [CrossRef]
- Jung, Y.; Lippard, S.J. Direct Cellular Responses to Platinum-Induced DNA Damage. Chem. Rev. 2007, 107, 1387–1407. [Google Scholar] [CrossRef] [PubMed]
- Florea, A.-M.; Büsselberg, D. Cisplatin as an Anti-Tumor Drug: Cellular Mechanisms of Activity, Drug Resistance and Induced Side Effects. Cancers 2011, 3, 1351–1371. [Google Scholar] [CrossRef] [PubMed]
- Barton, J.K.; Goldberg, J.M.C.; Kumar, V.; Turro, N.J. Binding modes and base specificity of tris(phenanthroline)ruthenium(II) enantiomers with nucleic acids: Tuning the stereoselectivity. J. Am. Chem. Soc. 1986, 108, 2081–2088. [Google Scholar] [CrossRef]
- Kelly, J.M.; Tossi, A.B.; McConnell, D.J.; OhUigin, C. A study of the interactions of some polypyridylruthenium(II) complexes with DNA using fluorescence spectroscopy, topoisomerisation and thermal denaturation. Nucleic Acids Res. 1985, 13, 6017–6034. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayana, S.; Dabrowiak, J.C.; Chaires, J.B. Tris(phenanthroline)ruthenium(II) enantiomer interactions with DNA: Mode and specificity of binding. Biochemistry 1993, 32, 2573–2584. [Google Scholar] [CrossRef]
- Hiort, C.; Nordén, B.; Rodger, A. Enantiopreferential DNA binding of [ruthenium(II)(1,10-phenanthroline)3]2+ studied with linear and circular dichroism. J. Am. Chem. Soc. 1990, 112, 1971–1982. [Google Scholar] [CrossRef]
- Lincoln, P.; Nordén, B. DNA Binding Geometries of Ruthenium(II) Complexes with 1,10-Phenanthroline and 2,2‘-Bipyridine Ligands Studied with Linear Dichroism Spectroscopy. Borderline Cases of Intercalation. J. Phys. Chem. B 1998, 102, 9583–9594. [Google Scholar] [CrossRef]
- Barton, J.K. Tris(phenanthroline)metal complexes: Probes for DNA helicity. J. Biomol. Struct. Dyn. 1983, 1, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Barton, J.K.; Dannenberg, J.J.; Raphael, A.L. Enantiomeric selectivity in binding tris(phenanthroline)zinc(II) to DNA. J. Am. Chem. Soc. 1982, 104, 4967–4969. [Google Scholar] [CrossRef]
- Nordén, B.; Tjerneld, F. Binding of inert metal complexes to deoxyribonucleic acid detected by linear dichroism. FEBS Lett. 1976, 67, 368–370. [Google Scholar] [CrossRef]
- Li, G.; Sun, L.; Ji, L.; Chao, H. Ruthenium(II) complexes with dppz: From molecular photoswitch to biological applications. Dalton Trans. 2016, 45, 13261–13276. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H.C. Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Br. J. Nat. 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walters, P. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Leslie, A.G.; Arnott, S.; Chandrasekaran, R.; Ratliff, R.L. Polymorphism of DNA double helices. J. Mol. Biol. 1980, 143, 49–72. [Google Scholar] [CrossRef]
- Carrier, A.; Le Ber, P.; Auclain, C. Spin-labeled oxazolopyridocarbazole as a probe for studying nonintercalating DNA groove binding ligands. Biochemistry 1990, 29, 6002–6009. [Google Scholar] [CrossRef]
- Matsuzawa, Y.; Minagawa, K.; Yoshikawa, K.; Matsumoto, M.; Doi, M. Conformational dynamics of DNA affected by intercalation and minor groove binding: Direct observation of large DNA. Nucleic Acids Symp. Ser. 1991, 25, 131–132. [Google Scholar]
- Hurley, L.H. DNA and associated targets for drug design. J. Med. Chem. 1989, 32, 2027–2033. [Google Scholar] [CrossRef]
- Iida, H.; Jia, G.; Lown, J.W. Rational recognition of nucleic acid sequences. Curr. Opin. Biotechnol. 1999, 10, 29–33. [Google Scholar] [CrossRef]
- Pasternack, R.F.; Bustamante, C.; Collings, P.J.; Giannetto, A.; Gibbs, E.J. Porphyrin assemblies on DNA as studied by a resonance light-scattering technique. J. Am. Chem. Soc. 1993, 115, 5393–5399. [Google Scholar] [CrossRef]
- Pasternack, R.F.; Schaefer, K.F. Resonance Light-Scattering Studies of Porphyrin Diacid Aggregates. Inorg. Chem. 1994, 33, 2062–2065. [Google Scholar] [CrossRef]
- Hudson, B.P.; Sou, J.; Berger, D.J.; McMillin, D.R. Luminescence studies of the intercalation of Cu(TMpyP4) into DNA. J. Am. Chem. Soc. 1992, 114, 8997–9002. [Google Scholar] [CrossRef]
- Santoro, A.; Holub, J.; Fik-Jaskółka, M.A.; Vantomme, G.; Lehn, J.-M. Dynamic Helicates Self-Assembly from Homo- and Heterotopic Dynamic Covalent Ligand Strands. Chem. Eur. J. 2020, 26, 15664–15671. [Google Scholar] [CrossRef] [PubMed]
- Kano, K.; Fukuda, K.; Wakami, H.; Nishiyabu, R.; Pasternack, R.F. Factors Influencing Self-Aggregation Tendencies of Cationic Porphyrins in Aqueous Solution. J. Am. Chem. Soc. 2000, 122, 7494–7502. [Google Scholar] [CrossRef]
- Arena, G.; Monsù Scolaro, L.; Pasternack, R.F.; Romeo, R. Synthesis, Characterization, and Interaction with DNA of the Novel Metallointercalator Cationic Complex (2,2′:6′,2″-terpyridine)methylplatinum(II). Inorg. Chem. 1995, 34, 2994–3002. [Google Scholar] [CrossRef]
- Casamento, M.; Arena, G.E.; Lo Passo, C.; Pernice, I.; Romeo, A.; Monsù Scolaro, L. Interaction of organometallic cationic complex ions containing terpyridine ligands with nucleic acids: An investigation on aggregative phenomena. Inorg. Chim. Acta 1998, 275, 242–249. [Google Scholar] [CrossRef]
- Cusumano, M.; Di Pietro, M.L.; Giannetto, A. DNA interaction of platinum(II) complexes with 1,10-phenanthroline and extended phenanthrolines. Inorg. Chem. 2006, 45, 230–235. [Google Scholar] [CrossRef]
- Pasternack, R.F.; Gibbs, E.J.; Villafranca, J.J. Interactions of porphyrins with nucleic acids. Biochemistry 1983, 22, 2406–2414. [Google Scholar] [CrossRef]
- Gibbs, E.J.; Mauer, M.C.; Zhang, H.F.; Reiff, W.M.; Hill, D.T.; Blaszkiewicz, M.M.; Mckinnie, R.E.; Liu, H.-Q.; Pasternack, R.F. Interactions of porphyrins with purified DNA and more highly organized structures. J. Inorg. Biochem. 1988, 32, 39–65. [Google Scholar] [CrossRef]
- Lerman, L.S. Structural considerations in the interaction of DNA and acridines. J. Mol. Biol. 1961, 3, 18–30. [Google Scholar] [CrossRef]
- Haq, I. Thermodynamics of drug-DNA interactions. Arch. Biochem. Biophys. 2002, 403, 1–15. [Google Scholar] [CrossRef]
- Bauer, W.; Vinograd, J. The interaction of closed circular DNA with intercalative dyes. I. The superhelix density of SV40 DNA in the presence and absence of dye. J. Mol. Biol. 1968, 33, 141–171. [Google Scholar] [CrossRef]
- Bauer, W.; Vinograd, J. Interaction of closed circular DNA with intercalative dyes. II. The free energy of superhelix formation in SV40 DNA. J. Mol. Biol. 1970, 47, 419–435. [Google Scholar] [CrossRef]
- Waring, M.J. Variation of the supercoils in closed circular DNA by binding of antibiotics and drugs: Evidence for molecular models involving intercalation. J. Mol. Biol. 1970, 54, 247–279. [Google Scholar] [CrossRef]
- Cairns, J. The application of autoradiography to the study of DNA viruses. Cold Spring Harb. Symp. Quant. Biol. 1962, 27, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Waring, M.J. DNA Modification and Cancer. Ann. Rev. Biochem. 1981, 50, 159–192. [Google Scholar] [CrossRef] [PubMed]
- Drug-nucleic acid interactions. In Methods in Enzymology; Chairs, J.B., Waring, M.J., Eds.; Academic Press: San Diego, CA, USA, 2001; Volume 340. [Google Scholar]
- Cantor, C.R.; Schimmel, P.R. Biophysical Chemistry; Freeman, W.H., Ed.; John Wiley & Sons: San Francisco, CA, USA, 1980. [Google Scholar]
- Sauer, K. Methods of Enzymology: Biochemical Spectroscopy; Academic Press: San Diego, CA, USA, 1995; Volume 246. [Google Scholar]
- Utsuno, K.; Tsuboi, M. Degree of DNA unwinding caused by the binding of aclacinomycin A. Chem. Pharm. Bull. 1997, 45, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Crothers, D.M. Calculation of binding isotherms for heterogeneous polymers. Biopolymers 1975, 6, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Kreft, D.; Wang, Y.; Rattay, M.; Toensing, K.; Anselmetti, D. Binding mechanism of anti-cancer chemotherapeutic drug mitoxantrone to DNA characterized by magnetic tweezers. J. Nanobiotechnol. 2018, 16, 56. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, A.; Barattucci, A.; Bonaccorsi, P.; Giannetto, A.; La Ganga, G.; Musarra-Pizzo, M.; Salerno, T.M.G.; Santoro, A.; Sciortino, M.T.; Puntoriero, F.; et al. Carbohydrates and Charges on Oligo(phenylenethynylenes): Towards the Design of Cancer Bullets. Chem. Eur. J. 2018, 24, 16972–16976. [Google Scholar] [CrossRef]
- Barton, D.; Nakanishi, K.; Meth-Cohn, O. DNA Intercalators. In Comprehensive Natural Products Chemistry; Wilson, W.D., Ed.; Pergamon: Oxford, UK, 1999; Volume 427. [Google Scholar]
- Balzani, V.; Credi, A.; Venturi, M. Photochemistry and photophysics of coordination compounds: An extended view. Coord. Chem. Rev. 1998, 171, 3–16. [Google Scholar] [CrossRef]
- Turro, N.J. From molecular chemistry to supramolecular chemistry to superdupermolecular chemistry. Controlling covalent bond formation through non-covalent and magnetic interactions. Chem. Commun. 2002, 2279–2292. [Google Scholar] [CrossRef]
- Sun, S.S.; Lees, A.J. Transition metal based supramolecular systems: Synthesis, photophysics, photochemistry and their potential applications as luminescent anion chemosensors. Coord. Chem. Rev. 2002, 230, 171–192. [Google Scholar] [CrossRef]
- Balzani, V.; Bergamini, G.; Campagna, S.; Puntoriero, F. Photochemistry and Photophysics of Coordination Compounds I; Balzani, V., Campagna, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 1–36. [Google Scholar]
- Puntoriero, F.; Arrigo, A.; Santoro, A.; La Ganga, G.; Tuyèras, F.; Campagna, S.; Dupeyre, G.; Lainé, P.P. Photoinduced Intercomponent Processes in Selectively Addressable Bichromophoric Dyads Made of Linearly Arranged Ru(II) Terpyridine and Expanded Pyridinium Components. Inorg. Chem. 2019, 58, 5807–5817. [Google Scholar] [CrossRef]
- Santoni, M.-P.; Santoro, A.; Salerno, T.M.G.; Puntoriero, F.; Nastasi, F.; Di Pietro, M.L.; Galletta, M.; Campagna, S. Photoinduced Charge Separation in a Donor–Spacer–Acceptor Dyad with N-Annulated Perylene Donor and Methylviologen Acceptor. ChemPhysChem 2015, 15, 3147–3150. [Google Scholar] [CrossRef] [PubMed]
- Jenette, K.W.; Lippard, S.J.; Vassiliades, G.A.; Bauer, W.R. Metallointercalation Reagents. 2-Hydroxyethanethiolato(2,2′,2″-terpyridine)-platinum(II) Monocation Binds Strongly to DNA By Intercalation. Proc. Natl. Acad. Sci. USA 1974, 71, 3839–3843. [Google Scholar] [CrossRef]
- Cusumano, M.; Di Pietro, M.L.; Giannetto, A.; Nicolò, F.; Rotondo, E. Noncovalent Interactions of Platinum(II) Square Planar Complexes Containg Ligands Out-of-Plane with DNA. Inorg. Chem. 1998, 37, 563–568. [Google Scholar] [CrossRef]
- Cusumano, M.; Di Pietro, M.L.; Giannetto, A. Stacking Surface Effect in the DNA Intercalation of Some Polypiridine Platinum(II) Complexes. Inorg. Chem. 1999, 38, 1754–1758. [Google Scholar] [CrossRef]
- Cusumano, M.; Di Pietro, M.L.; Giannetto, A.; Vainiglia, P.A. The intercalation to DNA of bipyridyl complexes of platinum(II) with thioureas. J. Inorg. Biochem. 2005, 99, 560–565. [Google Scholar] [CrossRef]
- Marverti, G.; Cusumano, M.; Ligabue, A.; Di Pietro, M.L.; Vainiglia, P.A.; Ferrari, A.; Bergomi, M.; Moruzzi, M.S.; Frassineti, C. Studies on the anti-proliferative effects of novel DNA-intercalating bipyridyl–thiourea–Pt(II) complexes against cisplatin-sensitive and -resistant human ovarian cancer cells. J. Inorg. Biochem. 2008, 102, 699–712. [Google Scholar] [CrossRef]
- Marverti, G.; Ligabue, A.; Montanari, M.; Guerrieri, D.; Cusumano, M.; Di Pietro, M.L.; Troiano, L.; Di Vono, E.; Iotti, S.; Farruggia, G.; et al. Characterization of the cell growth inhibitory effects of a novel DNA-intercalating bipyridyl-thiourea-Pt(II) complex in cisplatin-sensitive and—Resistant human ovarian cancer cells. Investig. New Drugs 2011, 29, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Campagna, S.; Puntoriero, F.; Nastasi, F.; Bergamini, G.; Balzani, V. Photochemistry and Photophysics of Coordination Compounds: Ruthenium. Top. Curr. Chem. 2007, 280, 117–214. [Google Scholar]
- Santoni, M.-P.; Hanan, G.S.; Hasenknopf, B.; Proust, A.; Nastasi, F.; Serroni, S.; Campagna, S. Dinuclear Ru(II) complexes of bis-(dipyrid-2-yl)triazine (bis-dpt) ligands as efficient electron reservoirs. Chem. Commun. 2011, 47, 3586–3588. [Google Scholar] [CrossRef] [PubMed]
- Kumar, C.V.; Barton, J.K.; Turro, N.J. Photophysics of ruthenium complexes bound to double helical DNA. J. Am. Chem. Soc. 1985, 107, 5518–5523. [Google Scholar] [CrossRef]
- Barton, J.K.; Danishefsky, A.T.; Goldberg, J.M. Tris(phenanthroline)ruthenium(II): Stereoselectivity in binding to DNA. J. Am. Chem. Soc. 1984, 106, 2172–2176. [Google Scholar] [CrossRef]
- Rehmann, J.P.; Barton, J.K. 1H NMR studies of tris(phenanthroline) metal complexes bound to oligonucleotides: Characterization of binding modes. Biochemistry 1990, 29, 1701–1709. [Google Scholar] [CrossRef]
- Lippard, S.J.; Bond, P.J.; Wu, K.C.; Bauer, W.R. Stereochemical requirements for intercalation of platinum complexes into double-stranded DNA’s. Science 1976, 194, 726–728. [Google Scholar] [CrossRef]
- Wang, A.H.J.; Nathans, J.; van der Marel, G.; van Boom, J.H.; Rich, A. Molecular structure of a double helical DNA fragment intercalator complex between deoxy CpG and a terpyridine platinum compound. Nature 1978, 276, 471–474. [Google Scholar] [CrossRef]
- Mei, H.-Y.; Barton, J.K. Chiral probe for A-form helixes of DNA and RNA: Tris(tetramethylphenanthroline)ruthenium(II). J. Am. Chem. Soc. 1986, 108, 7414–7416. [Google Scholar] [CrossRef]
- Jenkins, Y.; Friedman, A.E.; Turro, N.J.; Barton, J.K. Characterization of dipyridophenazine complexes of ruthenium(II): The light switch effect as a function of nucleic acid sequence and conformation. Biochemistry 1992, 31, 10809–10816. [Google Scholar] [CrossRef]
- Friedman, A.E.; Chambron, J.C.; Sauvage, J.-P.; Turro, N.J.; Barton, J.K. A molecular light switch for DNA: Ru(bpy)2(dppz)2+. J. Am. Chem. Soc. 1990, 112, 4960–4962. [Google Scholar] [CrossRef]
- Hiort, C.H.; Lincoln, P.; Nordén, B. DNA binding of DELTA- and LAMBDA-[Ru(phen)2DPPZ]2+. J. Am. Chem. Soc. 1993, 115, 3448–3454. [Google Scholar] [CrossRef]
- Moucheron, C.; Kirsch-De Mesmaeker, A. New DNA-binding ruthenium(II) complexes as photo-reagents for mononucleotides and DNA. J. Phys. Org. Chem. 1998, 11, 577–583. [Google Scholar] [CrossRef]
- Hartshorn, R.M.; Barton, J.K. Novel dipyridophenazine complexes of ruthenium(II): Exploring luminescent reporters of DNA. J. Am. Chem. Soc. 1992, 114, 5919–5925. [Google Scholar] [CrossRef]
- Önfelt, B.; Lincoln, P.; Nordén, B. A Molecular Staple for DNA: Threading Bis-intercalating [Ru(phen)2dppz]2+ Dimer. J. Am. Chem. Soc. 1999, 121, 10846–10847. [Google Scholar] [CrossRef]
- Önfelt, B.; Lincoln, P.; Nordén, B. Enantioselective DNA Threading Dynamics by Phenazine-Linked [Ru(phen)2dppz]2+ Dimers. J. Am. Chem. Soc. 2001, 123, 3630–3637. [Google Scholar] [CrossRef] [PubMed]
- Metcalfe, C.; Adams, H.; Haq, I.; Thomas, J.A. A ruthenium dipyridophenazine complex that binds preferentially to GC sequences. Chem. Commun. 2003, 1152–1153. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, Y.; Ihara, T.; Okada, K.; Tsujimura, Y.; Shirasaka, Y.; Tazaki, M.; Jyo, A. Asymmetric cooperativity in tandem hybridization of enantiomeric metal complex-tethered short fluorescent DNA probes. Chem. Commun. 2005, 4523–4525. [Google Scholar] [CrossRef]
- Monczak, K.; Peuntinger, K.; Sorsche, D.; Heinemann, F.W.; Guldi, D.M.; Rau, S. Synthesis and Characterization of a Trisheteroleptic RuII-Based Molecular Switch. Chem. Eur. J. 2014, 20, 15426–15433. [Google Scholar] [CrossRef]
- Metcalfe, C.; Webb, M.; Thomas, J.A. A facile synthetic route to bimetallic ReI complexes containing two dppz DNA intercalating ligands. Chem. Commun. 2002, 2026–2027. [Google Scholar] [CrossRef] [PubMed]
- Olmon, E.D.; Sontz, P.A.; Blanco-Rodríguez, A.M.; Towrie, M.; Clark, I.P.; Vlček, A., Jr.; Barton, J.K. Charge Photoinjection in Intercalated and Covalently Bound [Re(CO)3(dppz)(py)]+–DNA Constructs Monitored by Time-Resolved Visible and Infrared Spectroscopy. J. Am. Chem. Soc. 2011, 133, 13718–13730. [Google Scholar] [CrossRef]
- Thorp-Greenwood, F.L.; Coogan, M.P.; Mishra, L.; Kumari, N.; Rai, G.; Saripella, S. The importance of cellular localisation of probes: Synthesis, photophysical properties, DNA interactions and cellular imaging properties of rhenium dppz complexes with known cellular localisation vectors. New J. Chem. 2012, 36, 64–72. [Google Scholar] [CrossRef]
- Fumanal, M.; Vela, S.; Gattuso, H.; Monari, A.; Daniel, C. Absorption Spectroscopy and Photophysics of a ReI-dppz Probe for DNA-Mediated Charge Transport. Chem. Eur. J. 2018, 24, 14425–14435. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.L.; Reddy, Y.H.K.; Kumar, K.A.; Vidhisha, S.; Satyanarayana, S. Synthesis, characterization, DNA-binding, and DNA-photocleavage properties of [Co(bpy)2(7-NO2-dppz)]3+, [Co(dmb)2(7-NO2-dppz)]3+, and [Co(phen)2(7-NO2-dppz)]3+ complexes: (7-nitro-dppz = 7-nitrodipyrido[3,2-a:2′-3′-c]phenazine; bpy = 2,2′-bipyridine; dmb = 4,4′-dimethyl-2,2′-bipyridine; phen = 1,10-phenanthroline) and their toxicity on different microorganisms. Nucleosides Nucleotides Nucleic Acids 2009, 28, 204–219. [Google Scholar] [PubMed]
- Shahabadi, N.; Kashanian, S.; Mahdavi, M. DNA interaction studies of cobalt (II) mixed-ligand complexes containing dimethyl-1,10-phenanthroline and dipyrido[3,2-a:2′,3′-c]phenazine: The role of methyl substitutions on the mode of binding. DNA Cell Biol. 2011, 30, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Gupta, T.; Dhar, S.; Nethaji, M.; Chakravarty, A.R. Bis(dipyridophenazine)copper(II) complex as major groove directing synthetic hydrolase. Dalton Trans 2004, 12, 1896–1900. [Google Scholar] [CrossRef]
- Madureira, J.; Ramos, C.I.V.; Marques, M.; Maia, C.; de Sousa, B.; Campino, L.; Santana-Marques, M.G.; Farrell, N. Nonclassic metallointercalators with dipyridophenazine: DNA interaction studies and leishmanicidal activity. Inorg. Chem. 2013, 52, 8881–8894. [Google Scholar] [CrossRef] [PubMed]
- Angeles-Boza, A.M.; Bradley, P.M.; Fu, P.K.-L.; Wicke, S.E.; Bacsa, J.; Dunbar, K.R.; Turro, C. DNA binding and photocleavage in vitro by new dirhodium(II) dppz complexes: Correlation to cytotoxicity and photocytotoxicity. Inorg. Chem. 2004, 43, 8510–8519. [Google Scholar] [CrossRef]
- Menon, E.L.; Perera, R.; Navarro, M.; Kuhn, R.J.; Morrison, H. Phototoxicity against tumor cells and sindbis virus by an octahedral rhodium bisbipyridyl complex and evidence for the genome as a target in viral photoinactivation. Inorg. Chem. 2004, 43, 5373–5381. [Google Scholar] [CrossRef]
- Petitjean, A.; Puntoriero, F.; Campagna, S.; Juris, A.; Lehn, J.-M. Multicomponent supramolecular devices: Synthesis, optical, and electronic properties of bridged bis-dirhodium and -diruthenium complexes. Eur. J. Inorg. Chem. 2006, 19, 3878–3892. [Google Scholar] [CrossRef]
- Liang, H.; Hao, T.; Yin, C.; Yang, X.; Fu, H.; Zheng, X.; Li, R.; Xiao, D.; Chen, H. Cyclometalated Rhodium(III) Complexes Based on Substituted 2-Phenylpyridine Ligands: Synthesis, Structures, Photophysics, Electrochemistry, and DNA-Binding Properties. Eur. J. Inorg. Chem. 2017, 36, 4149–4157. [Google Scholar] [CrossRef]
- Trovato, E.; Di Pietro, M.L.; Puntoriero, F. Shining a new light on an old game—An OsII-based near-IR light switch. Eur. J. Inorg. Chem. 2012, 3984–3988. [Google Scholar] [CrossRef]
- Wragg, A.; Gill, M.R.; Hill, C.J.; Su, X.; Meijer, A.J.H.M.; Smythe, C.; Thomas, J.A. Dinuclear osmium (II) probes for high-resolution visualisation of cellular DNA structure using electron microscopy. Chem. Commun. 2014, 50, 14494–14497. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.L.; Che, C.M.; Yan, S.C. Platinum (II) complexes with dipyridophenazine ligands as human telomerase inhibitors and luminescent probes for G-quadruplex DNA. J. Am. Chem. Soc. 2009, 131, 1835–1846. [Google Scholar] [CrossRef]
- Puntoriero, F.; Campagna, S.; Di Pietro, M.L.; Giannetto, A.; Cusumano, M. Luminescence of a Pt(II) complex in the presence of DNA. Dependence of luminescence changes on the interaction binding mode. Photochem. Photobiol. Sci. 2007, 6, 357–360. [Google Scholar] [CrossRef]
- Cusumano, M.; Di Pietro, M.L.; Giannetto, A.; Nicolò, F.; Nordén, B.; Lincoln, P. Ambivalent Intercalators for DNA: L-Shaped Platinum(II) Complexes. Inorg. Chem. 2004, 43, 2416–2421. [Google Scholar] [CrossRef]
- Jacques, A.; Kirsch-De Mesmaeker, A.; Elias, B. Selective DNA Purine Base Photooxidation by Bis-terdentate Iridium(III) Polypyridyl and Cyclometalated Complexes. Inorg. Chem. 2014, 53, 1507–1512. [Google Scholar] [CrossRef]
- Sreedharan, S.; Sinopoli, A.; Jarman, P.J.; Robinson, D.; Clemmet, C.; Scattergood, P.A.; Rice, C.R.; Smythe, C.G.W.; Thomas, J.A.; Elliott, P.I.P. Mitochondria-localising DNA-binding biscyclometalated phenyltriazole iridium(III) dipyridophenazene complexes: Syntheses and cellular imaging properties. Dalton Trans. 2018, 47, 4931–4940. [Google Scholar] [CrossRef] [PubMed]
- Dreyse, P.; Santander-Nelli, M.; Zambrano, D.; Rosales, L.; Sanhueza, L. Electron-donor substituents on the dppz-based ligands to control luminescence from dark to bright emissive state in Ir(III) complexes. Int. J. Quantum. Chem. 2020, 120, e26167. [Google Scholar] [CrossRef]
- Olson, E.J.C.; Hu, D.; Hörmann, A.; Jonkman, A.M.; Arkin, M.R.; Stemp, E.D.A.; Barton, J.K.; Barbara, P.F. First Observation of the Key Intermediate in the “Light-Switch” Mechanism of [Ru(phen)2dppz]2+. J. Am. Chem. Soc. 1997, 119, 11458–11467. [Google Scholar] [CrossRef]
- Coates, C.G.; Olofsson, J.; Coletti, M.; McGarvey, J.J.; Önfelt, B.; Lincoln, P.; Nordén, B.; Tuite, E.; Matousek, P.; Parker, A.W. Picosecond Time-Resolved Resonance Raman Probing of the Light-Switch States of [Ru(Phen)2dppz]2+. J. Phys. Chem. B 2001, 105, 12653–12664. [Google Scholar] [CrossRef]
- Olofsson, J.; Onfelt, B.; Lincoln, P. Three-State Light Switch of [Ru(phen)2dppz]2+: Distinct Excited-State Species with Two, One, or No Hydrogen Bonds from Solvent. J. Phys. Chem. A 2004, 108, 4391–4398. [Google Scholar] [CrossRef]
- Onfelt, B.; Olofsson, J.; Lincoln, P.; Norden, B. Picosecond and Steady-State Emission of [Ru(phen)2dppz]2+ in Glycerol: Anomalous Temperature Dependence. J. Phys. Chem. A 2003, 107, 1000–1009. [Google Scholar] [CrossRef]
- Olofsson, J.; Wilhelmsson, L.M.; Lincoln, P. Effects of Methyl Substitution on Radiative and Solvent Quenching Rate Constants of [Ru(phen)2dppz]2+ in Polyol Solvents and Bound to DNA. J. Am. Chem. Soc. 2004, 126, 15458–15465. [Google Scholar] [CrossRef]
- Pourtois, G.; Beljonne, D.; Moucheron, C.; Schumm, S.; Kirsch-De Mesmaeker, A.; Lazzaroni, R.; Bredas, J.L. Photophysical properties of ruthenium(II) polyazaaromatic compounds: A theoretical insight. J. Am. Chem. Soc. 2004, 126, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Brennaman, M.K.; Meyer, T.J.; Papanikolas, J.M. [Ru(bpy)2dppz]2+ Light-Switch Mechanism in Protic Solvents as Studied through Temperature-Dependent Lifetime Measurements. J. Phys. Chem. A 2004, 108, 9938–9944. [Google Scholar] [CrossRef]
- Nair, R.B.; Cullum, B.M.; Murphy, C.J. Optical Properties of [Ru(phen)2dppz]2+ as a Function of Nonaqueous Environment. Inorg. Chem. 1997, 36, 962–965. [Google Scholar] [CrossRef] [PubMed]
- Reichardt, C. Solvents and Solvent Effects in Organic Chemistry, 3rd ed.; VCH Publishers: Weinheim, Germany, 1988. [Google Scholar]
- Stimpson, S.; Jenkinson, D.R.; Sadler, A.; Latham, M.; Wragg, A.; Meijer, A.J.H.M.; Thomas, J.A. Tuning the Excited State of Water-Soluble IrIII-Based DNA Intercalators that are Isostructural with [RuII(NN)2(dppz)] Light-Switch Complexes. Angew. Chem. Int. Ed. 2015, 54, 3000–3003. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.B.; Yeung, L.K.; Murphy, C.J. Synthesis and Solvent-Dependent Properties of Ru(acac)2dppz. Inorg. Chem. 1999, 38, 2536–2538. [Google Scholar] [CrossRef]
- Keller, C.E.; Pollard, C.; Yeung, L.K.; Plessinger, W.D.; Murphy, C.J. Optical sensing properties of [Ru (CN)4dppz]2− (dppz = dipyrido [3,2-a:2,3-c] phenazine). Inorg. Chim. Acta 2000, 298, 209–215. [Google Scholar] [CrossRef]
- Liu, J.G.; Zhang, Q.L.; Shi, X.F.; Ji, L.N. Interaction of [Ru(dmp)2(dppz)]2+ and [Ru(dmb)2(dppz)]2+ with DNA: Effects of the Ancillary Ligands on the DNA-Binding Behaviors. Inorg. Chem. 2001, 40, 5045–5050. [Google Scholar] [CrossRef] [PubMed]
- Greguric, A.; Greguric, I.D.; Hambley, T.W.; Aldrich-Wright, J.R.; Collins, J.G. Minor groove intercalation of Δ-[Ru(Me2phen)2dppz]2+ to the hexanucleotide d(GTCGAC)2. J. Chem. Soc. Dalton Trans. 2002, 849–855. [Google Scholar] [CrossRef]
- Moucheron, C; Kirsch-De Mesmaeker, A; Choua, S; Photophysics of Ru(phen)2(PHEHAT)2+: A Novel “Light Switch” for DNA and Photo-oxidant for Mononucleotides. Inorg. Chem. 1997, 36, 584–592. [CrossRef]
- Boisdenghien, A.; Moucheron, C.; Kirsch-De Mesmaeker, A. Ru(phen)2(PHEHAT)]2+ and [Ru(phen)2(HATPHE)]2+: Two Ruthenium(II) Complexes with the Same Ligands but Different Photophysics and Spectroelectrochemistry. Inorg. Chem. 2005, 44, 7678–7685. [Google Scholar] [CrossRef] [PubMed]
- Jacquet, L.; Kirsch-De Mesmaeker, A. Spectroelectrochemical Characteristics and Photophysics of a Series of RuII Complexes with 1,4,5,8,9,12-hexaatatriphenylene: Effects of Polycomplexation. J. Chem. Soc. Faraday Trans. 1992, 88, 2471–2480. [Google Scholar] [CrossRef]
- Masschelein, A.; Jacquet, L.; Kirsch-DeMesmaeker, A.; Nasielski, J. Ruthenium Complexes with 1,4,5,8-Tetraazaphenanthrene. Unusual Photophysical Behavior of the Tris-Homoleptic Compound. Inorg. Chem. 1990, 29, 855–860. [Google Scholar] [CrossRef]
- Ortmans, I.; Elias, B.; Kelly, J.M.; Moucheron, C.; Kirsch-DeMesmaeker, A. [Ru(TAP)2(dppz)]2+: A DNA intercalating complex, which luminesces strongly in water and undergoes photo-induced proton-coupled electron transfer with guanosine-5′-monophosphate. Dalton Trans. 2004, 668–676. [Google Scholar] [CrossRef]
- Nair, R.B.; Teng, E.S.; Kirkland, S.L.; Murphy, C.J. Synthesis and DNA-Binding Properties of [Ru(NH3)4dppz]2+. Inorg. Chem. 1998, 37, 139–141. [Google Scholar] [CrossRef]
- Liu, J.-G.; Ye, B.-H.; Li, H.; Zhen, Q.-X.; Ji, L.-N.; Fu., Y.-H. Polypyridyl ruthenium(II) complexes containing intramolecular hydrogen-bond ligand: Syntheses, characterization, and DNA-binding properties. J. Inorg. Biochem. 1999, 76, 265–271. [Google Scholar] [CrossRef]
- Schwalbe, M.; Karnahl, M.; Tschierlei, S.; Uhlemann, U.; Schmitt, M.; Dietzek, B.; Popp, J.; Groake, R.; Vos, J.G.; Rau, S. The switch that wouldn’t switch–unexpected luminescence from a ruthenium(II)-dppz-complex in water. Dalton Trans. 2010, 39, 2768–2771. [Google Scholar] [CrossRef] [PubMed]
- Whittemore, T.J.; White, T.A.; Turro, C. New Ligand Design Provides Delocalization and Promotes Strong Absorption throughout the Visible Region in a Ru(II) Complex. J. Am. Chem. Soc. 2018, 140, 229–234. [Google Scholar] [CrossRef]
- Toupin, N.P.; Nadella, S.; Steinke, S.J.; Turro, C.; Kodanko, J. Dual-Action Ru(II) Complexes with Bulky π-Expansive Ligands: Phototoxicity without DNA Intercalation. J. Inorg. Chem. 2020, 59, 3919–3933. [Google Scholar] [CrossRef]
- Foxon, S.P.; Green, C.; Walker, M.G.; Wragg, A.; Adams, H.; Weinstein, J.A.; Parker, S.C.; Meijer, A.J.H.M.; Thomas, J.A. Synthesis, Characterization, and DNA Binding Properties of Ruthenium(II) Complexes Containing the Redox Active Ligand Benzo[i]dipyrido[3,2-a:2′,3′-c]phenazine-11,16-quinone. Inorg. Chem. 2012, 51, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Puckett, C.A.; Barton, J.K. Fluorescein Redirects a Ruthenium-Octaarginine Conjugate to the Nucleus. J. Am. Chem. Soc. 2009, 131, 8738–8739. [Google Scholar] [CrossRef] [PubMed]
- Puckett, C.A.; Barton, J.K. Targeting a ruthenium complex to the nucleus with short peptides. Bioorg. Med. Chem. 2010, 18, 3564–3569. [Google Scholar] [CrossRef]
- Liu, Y.; Chouai, A.; Degtyareva, N.N.; Lutterman, D.A.; Dunbar, K.R.; Turro, C. Chemical Control of the DNA Light Switch: Cycling the Switch ON and OFF. J. Am. Chem. Soc. 2005, 127, 10796–10797. [Google Scholar] [CrossRef]
- Bolger, J.; Gourdon, A.; Ishow, E.; Launay, J.-P. Electronic structure of the “molecular light switch” bis(bipyridine)dipyrido[3,2-a:2′,3′-c]phenazineruthenium(2+). Cyclic voltammetric, UV/visible and EPR/ENDOR study of multiply reduced complexes and ligands. Inorg. Chem. 1996, 35, 2937–2944. [Google Scholar] [CrossRef]
- Shade, C.M.; Kennedy, R.D.; Rouge, J.L.; Rosen, M.S.; Wang, M.X.; Seo, S.E.; Clingerman, D.J.; Mirkin, C.A. Duplex-Selective Ruthenium-Based DNA Intercalators. Chem. Eur. J. 2015, 21, 10983–10987. [Google Scholar] [CrossRef]
- Boynton, A.N.; Marcélis, L.; Barton, J.K. [Ru(Me4phen)2dppz]2+, a Light Switch for DNA Mismatches. J. Am. Chem. Soc. 2016, 138, 5020–5023. [Google Scholar] [CrossRef]
- Campagna, S.; Cavazzini, M.; Cusumano, M.; Di Pietro, M.L.; Giannetto, A.; Puntoriero, F.; Quici, S. Luminescent Ir(III) Complex Exclusively Made of Polypyridine Ligands Capable of Intercalating into Calf-Thymus DNA. Inorg. Chem. 2011, 50, 10667–10672. [Google Scholar] [CrossRef]
- Di Pietro, M.L.; Puntoriero, F.; Tuyéras, F.; Ochsenbein, P.; Lainé, P.P.; Campagna, S. Photochemically driven intercalation of small molecules into DNA by in situ irradiation. Chem. Commun. 2010, 46, 5169–5171. [Google Scholar] [CrossRef] [PubMed]
- Trovato, E.; Di Pietro, M.L.; Giannetto, A.; Dupeyre, G.; Lainé, P.P.; Nastasi, F.; Puntoriero, F.; Campagna, S. Designing expanded bipyridinium as redox and optical probes for DNA. Photochem. Photobiol. Sci. 2020, 19, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Vos, J.G.; Kelly, J.M. Ruthenium polypyridyl chemistry; from basic research to applications and back again. Dalton Trans. 2006, 4869–4883. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.M.; McConnell, D.J.; OhUigin, C.; Tossi, B.; Kirsch-De Maesmaeker, A.; Masschelein, A.; Nasielski, J. Ruthenium polypyridyl complexes; their interaction with DNA and their role as sensitisers for its photocleavage. J. Chem. Soc. Chem. Commun. 1987, 1821–1823. [Google Scholar] [CrossRef]
- Blasius, R.; Moucheron, C.; Kirsch-De Mesmaeker, A. Photoadducts of Metallic Compounds with Nucleic Acids−Role Played by the Photoelectron Transfer Process and by the TAP and HAT Ligands in the RuII Complexes. Eur. J. Inorg. Chem. 2004, 3971–3979. [Google Scholar] [CrossRef]
- Le Gac, S.; Foucart, M.; Gerbaux, P.; Defrancq, E.; Moucheron, C.; Kirsch-De Mesmaeker, A. Photo-reactive RuII-oligonucleotide conjugates: Influence of an intercalating ligand on the inter- and intra-strand photo-ligation processes. Dalton Trans. 2010, 9672–9683. [Google Scholar] [CrossRef]
- Keane, P.M.; Poynton, F.E.; Hall, J.P.; Sazanovich, I.V.; Towrie, M.; Gunnlaugsson, T.; Quinn, S.J.; Cardin, C.J.; Kelly, J.M. Reversal of a Single Base-Pair Step Controls Guanine Photo-Oxidation by an Intercalating Ruthenium(II) Dipyridophenazine Complex. Angew. Chem. Int. Ed. 2015, 127, 8484–8488. [Google Scholar] [CrossRef]
- Vanderlinden, W.; Blunt, M.; David, C.C.; Moucheron, C.; Kirsch-De Mesmaeker, A.; De Feyter, S. Mesoscale DNA Structural Changes on Binding and Photoreaction with Ru[(TAP)2PHEHAT]2+. J. Am. Chem. Soc. 2012, 134, 10214–10221. [Google Scholar] [CrossRef]
- Blasius, R.; Nierengarten, H.; Luhmer, M.; Constant, J.F.; Defrancq, E.; Dumy, P.; van Dorsselaer, A.; Moucheron, C.; Kirsch-De Mesmaeker, A. Photoreaction of [Ru(HAT)2phen]2+ with Guanosine-5′-Monophosphate and DNA: Formation of New Types of Photoadducts. Chem. Eur. J. 2005, 11, 1507–1517. [Google Scholar] [CrossRef]
- Shi, S.; Geng, X.; Zhao, J.; Yao, T.; Wang, C.; Yang, D.; Zheng, L.; Ji, L. Interaction of [Ru(bpy)2(dppz)]2+ with human telomeric DNA: Preferential binding to G-quadruplexes over i-motif. Biochimie 2010, 92, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Zhao, J.; Geng, X.; Yao, T.; Huang, H.; Liu, T.; Zheng, L.; Li, Z.; Yang, D.; Ji, L. Molecular “light switch” for G-quadruplexes and i-motif of human telomeric DNA: [Ru(phen)2(dppz)]2+. Dalton Trans. 2010, 39, 2490–2493. [Google Scholar] [CrossRef] [PubMed]
- Wachter, E.; Moyá, D.; Parkin, S.; Glazer, E.C. Ruthenium Complex “Light Switches” that are Selective for Different G-Quadruplex Structures. Chem. Eur. J. 2016, 22, 550–559. [Google Scholar] [CrossRef]
- Zeraati, M.; Langley, D.B.; Schofield, P.; Moye, A.L.; Rouet, R.; Hughes, W.E.; Bryan, T.M.; Dinger, M.E.; Christ, D. I-motif DNA structures are formed in the nuclei of human cells. Nat. Chem. 2018, 10, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Gueron, M.; Leroy, J.L. The i-motif in nucleic acids. Curr. Opin. Struct. Biol. 2000, 10, 326–331. [Google Scholar] [CrossRef]
- Baptista, R.; Devereux, S.J.; Gurung, S.P.; Hall, J.P.; Sazanovich, I.V.; Towrie, M.; Cardin, C.J.; Brazier, J.A.; Kelly, J.M.; Quinn, S.J. The influence of loops on the binding of the [Ru(phen)2dppz]2+ light-switch compound to i-motif DNA structures revealed by time-resolved spectroscopy. Chem. Commun. 2020, 56, 9703–9706. [Google Scholar] [CrossRef]
- Devereux, S.J.; Poynton, F.E.; Baptista, F.R.; Gunnlaugsson, T.; Cardin, C.J.; Sazanovich, I.V.; Towrie, M.; Kelly, J.M.; Quinn, S.J. Caught in the Loop: Binding of the [Ru(phen)2(dppz)]2+ Light-Switch Compound to Quadruplex DNA in Solution Informed by Time-Resolved Infrared Spectroscopy. Chem. Eur. J. 2020, 26, 17103–17109. [Google Scholar] [CrossRef]
- Fang, Y.-Q.; Taylor, N.J.; Hanan, G.S.; Loiseau, F.; Passalacqua, R.; Campagna, S.; Nierengarten, H.; Van Dorsselaer, A. A Strategy for Improving the Room-Temperature Luminescence Properties of Ru(II) Complexes with Tridentate Ligands. J. Am. Chem. Soc. 2002, 124, 7912–7913. [Google Scholar] [CrossRef]
- Fang, Y.-Q.; Taylor N., J.; Laverdière, F.; Hanan, G.S.; Loiseau, F.; Nastasi, F.; Campagna, S.; Nierengarten, H.; Leize, E.; Van Dorsselaer, A. Ruthenium(II) Complexes with Improved Photophysical Properties Based on Planar 4′-(2-Pyrimidinyl)-2,2′:6′,2″-terpyridine Ligands. Inorg. Chem. 2007, 46, 2854–2863. [Google Scholar] [CrossRef]
- Liu, Y.; Hammitt, R.; Lutterman, D.A.; Thummel, R.P.; Turro, C. Marked Differences in Light-Switch Behavior of Ru(II) Complexes Possessing a Tridentate DNA Intercalating Ligand. Inorg. Chem. 2007, 46, 6011–6021. [Google Scholar] [CrossRef] [PubMed]
- Kober, E.M.; Caspar, J.V.; Sullivan, B.P.; Meyer, T. Synthetic routes to new polypyridyl complexes of osmium(II). Inorg. Chem. 1988, 27, 4587–4598. [Google Scholar] [CrossRef]
- Holmlin, R.E.; Stemp, E.D.A.; Barton, J.K. Os(phen)2dppz2+ in Photoinduced DNA-Mediated Electron Transfer Reactions. J. Am. Chem. Soc. 1996, 118, 5236–5244. [Google Scholar] [CrossRef]
- Holmlin, R.E.; Dandliker, P.J.; Barton, J.K. Charge Transfer through the DNA Base Stack. Angew. Chem. 1997, 109, 2830–2848, Angew. Chem. Int. Ed. Engl.1997, 36, 2714–2730. [Google Scholar] [CrossRef]
- Petralia, S.; Castagna, M.E.; Cappello, E.; Puntoriero, F.; Trovato, E.; Gagliano, A.; Conoci, S. A miniaturized silicon based device for nucleic acids electrochemical detection. Sens. Bio-Sens. Res. 2015, 6, 90–94. [Google Scholar] [CrossRef][Green Version]
- Petralia, S.; Sciuto, E.L.; Di Pietro, M.L.; Zimbone, M.; Grimaldi, M.G.; Conoci, S. An innovative chemical strategy for PCR-free genetic detection of pathogens by an integrated electrochemical biosensor. Analyst 2017, 142, 2090–2093. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, B.C.; Estalayo-Adrián, S.; Blasco, S.; Bright, S.A.; Kelly, J.M.; Williams, D.C.; Gunnlaugsson, T. Luminescent ruthenium polypyridyl complexes with extended ‘dppz’ like ligands as DNA targeting binders and cellular agents. Dalton Trans. 2016, 45, 18208–18220. [Google Scholar] [CrossRef]
- Neugebauer, U.; Pellegrin, Y.; Devocelle, M.; Forster, R.J.; Signac, W.; Moran, N.; Keyes, T.E. Ruthenium polypyridyl peptide conjugates: Membrane permeable probes for cellular imaging. Chem. Commun. 2008, 2, 5307–5309. [Google Scholar] [CrossRef]
- Brennaman, M.K.; Alstrum-Acevedo, J.H.; Fleming, C.N.; Jang, P.; Meyer, T.J.; Papanikolas, J.M. Turning the [Ru(bpy)2(dppz)]2+ Light-Switch On and Off with Temperature. J. Am. Chem. Soc. 2002, 124, 15094–15098. [Google Scholar] [CrossRef]
- Gill, M.R.; Thomas, J.A. Ruthenium(II) polypyridyl complexes and DNA–from structural probes to cellular imaging and therapeutics. Chem. Soc. Rev. 2012, 41, 3179–3192. [Google Scholar] [CrossRef]
- Chambron, J.-C.; Sauvage, J.-P. Ru(bipy)2dppz2+: A highly sensitive luminescent probe for micellar sodium dodecyl sulfate solutions. Chem. Phys. Lett. 1991, 182, 603–607. [Google Scholar] [CrossRef]
- De la Cadena, A.; Pascher, T.; Davydova, D.; Akimov, D.; Herrmann, F.; Presselt, M.; Wächtler, M.; Dietzek, B. Intermolecular exciton-exciton annihilation in phospholipid vesicles doped with [Ru(bpy)2dppz]2+. Chem. Phys. Lett. 2016, 644, 56–61. [Google Scholar] [CrossRef]
- Schatzschneider, U.; Niesel, J.; Ott, I.; Gust, R.; Alborzinia, H.; Wölfl, S. Cellular uptake, cytotoxicity, and metabolic profiling of human cancer cells treated with ruthenium(II) polypyridyl complexes [Ru(bpy)2(N-N)]Cl2 with N-N = bpy, phen, dpq, dppz, and dppn. ChemMedChem 2008, 3, 1104–1109. [Google Scholar] [CrossRef]
- Byrne, A.; Burke, C.S.; Keye, T.E. Precision targeted ruthenium(II) luminophores; highly effective probes for cell imaging by stimulated emission depletion (STED) microscopy. Chem. Sci. 2016, 7, 6551–6562. [Google Scholar] [CrossRef]
- Puckett, C.A.; Barton, J.K. Methods to Explore Cellular Uptake of Ruthenium Complexes. J. Am. Chem. Soc. 2007, 129, 46–47. [Google Scholar] [CrossRef]
- Cloonan, S.M.; Elmes, R.B.P.; Erby, M.L.; Bright, S.A.; Poynton, F.E.; Nolan, D.E.; Quinn, S.J.; Gunnlaugsson, T.; Williams, D.C. Detailed Biological Profiling of a Photoactivated and Apoptosis Inducing pdppz Ruthenium(II) Polypyridyl Complex in Cancer Cells. J. Med. Chem. 2015, 58, 4494–4505. [Google Scholar] [CrossRef]
- Jia, F.; Wang, S.; Man, Y.; Kumar, P.; Liu, B. Recent Developments in the Interactions of Classic Intercalated Ruthenium Compounds: [Ru(bpy)2dppz]2+ and [Ru(phen)2dppz]2+ with a DNA Molecule. Molecules 2019, 24, 769. [Google Scholar] [CrossRef]
- Matson, M.; Svensson, F.R.; Nordén, B.; Lincoln, P. Correlation Between Cellular Localization and Binding Preference to RNA, DNA, and Phospholipid Membrane for Luminescent Ruthenium(II) Complexes. J. Phys. Chem. B 2011, 115, 1706–1711. [Google Scholar] [CrossRef]
- Svensson, F.R.; Abrahamsson, M.; Strömberg, N.; Ewing, A.G.; Lincoln, P.J. Ruthenium(II) Complex Enantiomers as Cellular Probes for Diastereomeric Interactions in Confocal and Fluorescence Lifetime Imaging Microscopy. Phys. Chem. Lett. 2011, 2, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Pierroz, V.; Joshi, T.; Leonidova, A.; Mari, C.; Schur, J.; Ott, I.; Spiccia, L.; Ferrari, S.; Gasser, G. Molecular and Cellular Characterization of the Biological Effects of Ruthenium(II) Complexes Incorporating 2-Pyridyl-2-pyrimidine-4-carboxylic Acid. J. Am. Chem. Soc. 2012, 134, 20376–20387. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Joyce, L.E.; Dickson, N.M.; Turro, C. Efficient DNA photocleavage by [Ru(bpy)2(dppn)]2+ with visible light. Chem. Commun. 2010, 46, 2426–2428. [Google Scholar] [CrossRef] [PubMed]
- Pierroz, V.; Rubbiani, R.; Gentili, C.; Patra, M.; Mari, C.; Gasser, G.; Ferrari, S. Dual mode of cell death upon the photo-irradiation of a RuII-polypyridyl complex in interphase or mitosis. Chem. Sci. 2016, 7, 6115–6124. [Google Scholar] [CrossRef]
- Hess, J.; Huang, H.; Kaiser, A.; Pierroz, V.; Blacque, O.; Chao, H.; Gasser, G. Evaluation of the Medicinal Potential of Two Ruthenium(II) Polypyridine Complexes as One-and Two-Photon Photodynamic Therapy Photosensitizers. Chem. Eur. J. 2017, 23, 9888–9896. [Google Scholar] [CrossRef]
- Gill, M.; Garcia-Lara, J.; Foster, S.; Smythe, C.; Battaglia, G.; Thomas, J.A. A ruthenium(II) polypyridyl complex for direct imaging of DNA structure in living cells. Nat. Chem. 2009, 1, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Hua, W.; Xu, G.; Zhao, J.; Wang, Z.; Lu, J.; Sun, W.; Gou, S. DNA-Targeting RuII-Polypyridyl Complex with a Long-Lived Intraligand Excited State as a Potential Photodynamic Therapy Agent. Chem. Eur. J. 2020, 26, 17495–17503. [Google Scholar] [CrossRef]
- Mulcany, S.P.; Li, S.; Korn, R.; Xie, X.; Meggers, E. Solid-phase synthesis of tris-heteroleptic ruthenium(II) complexes and application to acetylcholinesterase inhibition. Inorg. Chem. 2008, 47, 5030–5032. [Google Scholar] [CrossRef] [PubMed]
- Shoji, M.; Golde, T.E.; Ghiso, J.; Cheung, T.T.; Estus, S.; Shaffer, L.M.; Cai, X.D.; McKay, D.M.; Tintner, R.; Frangione, B.; et al. Production of the Alzheimer amyloid beta protein by normal proteolytic processing. Science 1992, 258, 126–129. [Google Scholar] [CrossRef]
- Cook, N.P.; Torres, V.; Jain, D.; Martí, A.A. Sensing Amyloid-β Aggregation Using Luminescent Dipyridophenazine Ruthenium(II) Complexes. J. Am. Chem. Soc. 2011, 133, 11121. [Google Scholar] [CrossRef]
- Jiang, Y.; Fang, X.; Bai, C. Signaling Aptamer/Protein Binding by a Molecular Light Switch Complex. Anal. Chem. 2004, 76, 5230–5235. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yu, T.; Guo, L.; Xie, J.; Shao, N.; He, Z. In vitro selection of DNA aptamer against abrin toxin and aptamer-based abrin direct detection. Biosens. Bioelectron. 2007, 22, 2456–2463. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, Y.; Zhou, C.; Fang, X. Aptamer-Based ATP Assay Using a Luminescent Light Switching Complex. Anal. Chem. 2005, 77, 3542–3546. [Google Scholar] [CrossRef]
- Hu, L.; Bian, Z.; Li, H.; Han, S.; Yuan, Y.; Gao, L.; Xu, G. [Ru(bpy)2dppz]2+ electrochemiluminescence switch and its applications for DNA interaction study and label-free ATP aptasensor. Anal. Chem. 2009, 81, 9807–9811. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).