Abstract
In recent years, bone fracture detection and classification has been a widely discussed topic and many researchers have proposed different methods to tackle this problem. Despite this, a universal approach able to classify all the fractures in the human body has not yet been defined. We aim to analyze and evaluate a selection of papers, chosen according to their representative approach, where the authors applied different deep learning techniques to classify bone fractures, in order to select the strengths of each of them and try to delineate a generalized strategy. Each study is summarized and evaluated using a radar graph with six values: area under the curve (AUC), test accuracy, sensitivity, specificity, dataset size and labelling reliability. Plus, we defined the key points which should be taken into account when trying to accomplish this purpose and we compared each study with our baseline. In recent years, deep learning and, in particular, the convolution neural network (CNN), has achieved results comparable to those of humans in bone fracture classification. Adopting a correct generalization, we are reasonably sure that a computer-aided diagnosis (CAD) system, correctly designed to assist doctors, would save a considerable amount of time and would limit the number of wrong diagnoses.
1. Introduction
Bone fractures are one of the most common injuries nowadays. Every year, 2.7 million fractures occur across the EU6 nations, France, Germany, Italy, Spain, Sweden, and the UK [1]; an incredible number of people suffers from this disorder and the implications of an untreated fracture may lead to permanent damage or even death. A great responsibility for this lies with the doctors, who have to evaluate tens of X-ray images a day. The technology utilized for first diagnosis is mostly X-ray, which is a modality that has been used for more than one hundred years and is still frequently used. It is challenging for doctors to evaluate X-ray images: firstly, X-ray could hide certain particularities of the bone; secondly, a lot of experience is needed to correctly classify different types of fractures; thirdly, doctors have often to act in emergency situations and may be constrained by fatigue. Actually, it has been shown that the performance of radiologists in the interpretation of musculoskeletal radiographs decrease in fracture detection by the end of the work day compared to the beginning of the work day [2]. In addition, radiographic interpretation often takes place in environments without the availability of qualified colleagues for second opinions [3]. The success of the treatment and prognosis strongly depends on an accurate classification of the fracture among standard types, such as those defined by the Arbeitsgemeinschaft für Osteosynthesefragen (AO foundation). In that context, a computer-aided diagnosis (CAD) system able to help doctors might have a direct impact in the outcome of the patients. In this paper, we reviewed some selected papers concerning this topic, starting from basic approaches to the main advanced solutions. Initial prior works on the detection and classification of fractures [4,5,6] have focused on conventional machine learning processes consisting of pre-processing, feature extraction and classification steps. Recently, impressive results have been obtained using deep learning [7] methods. A total of 232 records were identified through database searching and other sources. At total of 107 records were screened and 65 of them excluded, resulting in 42 full-text articles assessed for eligibility. Among these papers, we selected 11 records for analysis. This whole process is shown in Figure 1 with a PRISMA flow diagram [8]. The majority of them pursue the classification between fractured and not fractured bones, while just two of them tried to classify the different types of fractures. We have chosen papers which, in our personal opinion, contain the strengths given by a deep learning approach that should be used in order to develop a generic tool able to classify every type of fracture in each bone of the human body.
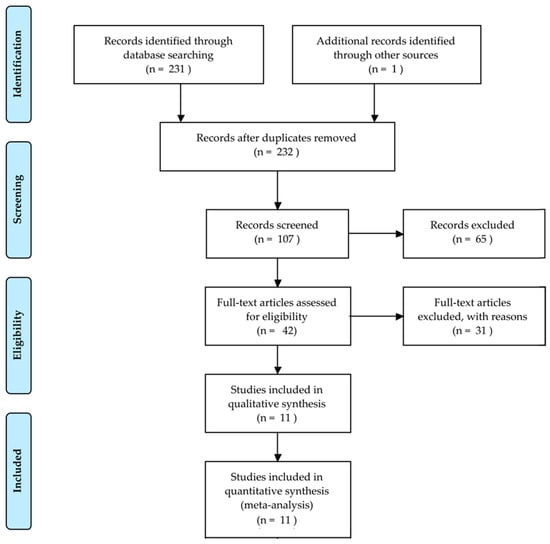
Figure 1.
PRISMA flow diagram to describe the study selection.
2. Methods
To the best of our knowledge, a work that tries to define an ideal method to classify valid fractures for every bone in the human body does not exist. In our opinion, the best way to pursue this is to evaluate different papers and select the strengths which could be mixed together to define a baseline approach. We decided to divide the evaluation of each study into different parts: aim, dataset, pre-processing, methods, results and conclusion. In the aim section, the purpose of the work is stated; in the dataset section, we focus on the number of images used and how the labelling was carried out; in the pre-processing section, we explain how images were pre-processed before feeding the network; in the methods section we detail the process the authors proposed in order to tackle the problem; and, in the results section, we outline the performance of the tool. Finally, in the conclusion section, we select the strengths of the discussed paper that should be used in a universal approach and we evaluate them using a radar graph with the following parameters:
- The area under the receiver operating characteristic (ROC) curve (AUC), where an AUC of 1.0 would indicate that the system perfectly predicts the reference standard and an AUC of 0.5 would indicate that the system is no better than chance. The ROC curve relates the specificity (or true negative rate (TNR)) and the sensitivity (or true positive rate (TPN)) of a model. AUC values range from 0 to 1 and we mapped them to the range from 1 to 5 to fit into the radar graph.
- Test accuracy—i.e., the number of correct predictions among all the predictions. The accuracy values range from 0 to 1 and we mapped them to the range from 1 to 5, as we did for the AUC value.
- Sensitivity and specificity: as accuracy does not properly reflect the performances for unbalanced dataset, we also outlined sensitivity/specificity values, where available. The first measures the proportion of actual positives that are correctly identified as such (e.g., the percentage of fractured bones which are correctly identified as having the condition). The second measures the proportion of actual negatives that are correctly identified as such (e.g., the percentage of healthy bones that are correctly identified as not having the condition). Moreover, in this case, these two values range from 0 to 1 and we mapped them to the range from 1 to 5.
- The reliability of the labelling—i.e., how carefully the dataset was labelled. As this is the only subjective parameter, we tried to be as objective as possible. The variables taken into consideration were the number of specialists who labelled the dataset, their level of expertise and how many times the labelling phase was done. These three variables were weighted accordingly to their importance and mapped to the range from 1 to 5.
- Dataset size: to scale this value, we considered the three biggest datasets as outliers and we assigned to them the values of 4 and 5 in the range, proportionated to their dimensions. We then took the number of images used in the remaining papers and mapped them in a range from 1 to 3.
As mentioned, all parameters were remapped to the range between 1 and 5, while a value of 0 means that the parameter is not assessable for the specific method.
3. Approaches
3.1. Intelligent Bone Fracture Detection System (IBFDS)
This subsection focuses on the work of Dimililer [9].
Aim: The final aim of this paper was to classify whether a bone in an X-ray image is fractured or not. The authors did not focus on a specific type of bone, instead the dataset is composed of various body parts. The system was composed of a neural network following a pre-processing phase.
Dataset: The tool was trained with 30 images and tested with 70. The images contained different fractures in size and illumination conditions for each subject.
Pre-processing: In the pre-processing phase, the images were processed using techniques such as Haar wavelet transform and scale-invariant feature transform (SIFT). Haar wavelet transform is needed to pre-process images in order to compress them and save memory space; SIFT is a powerful method to detect feature points with a high resilience to several issues, like rotation, compression, and scaling.
Methods: In the classification phase, the author implemented a three-layer neural network with 1024 input neurons. In order to achieve the required minimum error value, the learning rate was monitored and adjusted, consequently, during various experiments.
Results: The accuracy of this tool is stated to be 94.3%, 64 out of 70 images have been correctly classified as showing broken bones.
Conclusion: Even if the number of images in the dataset is scant, the results are quite impressive: this accuracy has been obtained by the well-implemented pre-processing phase. The dataset is composed of different bones and orientations, but the pre-processing phase makes the network really robust to the inputs. On the other hand, the number of images in the dataset is low for this kind of task and it would have been better to use 70 images for training and 30 for testing, not the other way around. Data augmentation would have been helpful as well. We decided to evaluate this paper because it describes the technology used before the advent of the convolutional layers. The pre-processing phase was a fundamental part of the process in order to feed the fully connected layers with the correct information. All the remaining papers use convolutional layers to extract features before feeding the fully connected layers.
3.2. Deep Neural Network Improves Fracture Detection by Clinicians
This subsection focuses on the work of Lindsey et al. [10].
Aim: The aim of this work is to implement a tool that could help doctors in diagnosis, in order to distinguish if a wrist bone is fractured or not and which part of the bone is fractured.
Dataset: The dataset was annotated by 18 senior sub-specialized orthopedic surgeons and consisted of 135,845 radiographs of a variety of body parts. Of these, 34,990 radiographs (training set) were posterior–anterior or lateral wrist views. The remaining 100,855 radiographs (pre-training set) belonged to 11 other body parts: foot, elbow, shoulder, knee, spine, femur, ankle, humerus, pelvis, hip, and tibia. Two datasets were used for clinical tests of the model. The first dataset (Test Set 1) consisted of 3500 wrist radiographs, randomly picked from the wrist dataset. The second dataset (Test Set 2) consisted of 1400 wrist radiographs, consecutively collected over a three-month period to ensure that the dataset was representative of a real-world clinical environment. Every training image was labelled with a bounding box drawn by a group of senior orthopedic surgeons who specialized in fractures. The training set has been split into 90% of the radiographs for training and 10% for validation. The data augmentation alterations included random rotations, cropping, horizontal mirroring, and lighting and contrast adjustments.
Pre-processing: The images were pre-processed by rotating, cropping, and applying an aspect ratio, preserving rescaling operation to yield a fixed resolution of 1024 × 512.
Methods: The model is a deep CNN, whose architecture is an extension of the common U-Net [11] model. The CNN has two outputs: the probability that the radiograph has a visible fracture and a heat map indicating for each location in the image the probability that the fracture is present in that location. The training of the model can be divided into two stages. In the first stage, the model was pre-trained on the pre-training set. In the second stage, the obtained model was fine-tuned using the training set, to specialize it to the task of detecting and localizing wrist fractures. The authors used a number of techniques to prevent overfitting, including early stopping, data augmentation and dropout with a probability of 0.5.
Results: The model was tested on two different datasets: on Test Set 1, the model achieved an AUC of 0.967; on Test Set 2, the model achieved an AUC of 0.975. With the same set of images used for clinicians’ evaluations, the model operated at 93.9% sensitivity and 94.5% specificity. After the training and testing phases of the CNN, the authors ran a controlled experiment with 40 emergency medicine clinicians, to evaluate each clinician’s ability to detect fractures in wrist radiographs, both with and without the help of the system. Among these, 16 were physicians’ assistants (PAs) and 24 were medical doctors (MDs). With the use of the proposed system, the MDs’ average sensitivity and specificity improved by 10% and 7%, respectively, the PAs’ average sensitivities and specificities improved by 12% and 6% respectively.
Conclusion: This study showed that specialists’ evaluations may be improved with the use of this system. This procedure should be applied in each work aimed at showing that a CAD system could help humans in evaluation. Pre-training the model before training it with the wrist bone images seem like a good procedure to adjust the parameters for the task, instead of using weights taken from a network that has been trained with a completely different dataset, e.g., ImageNet [12].
3.3. Automated Detection and Classification of the Proximal Humerus Fracture
This subsection focuses on the work of Chung et al. [13].
Aim: This work addresses the problem of the classification of different types of fractures in the proximal humerus bone. To evaluate the performance of fracture classification, the authors refer to Neer’s classification, which is the most commonly used classification for the proximal humerus fracture and distinguish 4 different types of fracture: greater tuberosity (B), surgical neck (C), three-part (D), and four-part (E). The healthy humerus group is named A.
Dataset: A total of 1891 plain shoulder radiographs (1376 proximal humerus fracture cases and 515 normal shoulders) were used as the total dataset in this study. No more than one image was used from each patient. Fracture classification was performed by two shoulder orthopedic specialists with 14 and 17 years of experience and one radiologist with expertise in musculoskeletal diseases and 15 years of medical experience. A total of 515 cases were labelled as A, 346 cases as B, 514 cases as C, 269 cases as D, and 247 cases as E. The training dataset was augmented (shifting in the upward, downward, left, and right directions, scale transformations with 15% magnification, and 90°, 180°, 270° rotation) by a factor of 24 to increase the number of training images to more than 40,000 at a time. The dataset of the 1891 images was divided into 10 partitions without overlapping images: one partition was used as a test dataset, while all other images were used as training datasets.
Pre-processing: Each plain shoulder radiograph was manually cropped into a square in which the humeral head and neck were centered and constituted approximately 50% of the square image, resized to 256 × 256 pixels, and stored as a JPEG file.
Methods: The authors used the open source pre-trained ResNet-152 [14] as a deep CNN model. As the dataset was divided into 10 partitions, 10 experiments were performed in order to obtain an averaged performance.
Results: The deep learning CNN model for distinguishing between normal and proximal humerus fractured shoulders showed more than 95% accuracy and an AUC of 0.996 and a sensitivity/specificity of 0.99/0.97. For fracture type classification, performance was measured only in the fracture images after excluding the normal shoulder images to evaluate the actual performance of fracture classification. The accuracy of the CNN model for distinguishing each fracture type from the other types was 86% with an AUC of 0.98 and sensitivity/specificity of 0.97/0.94 for B, 80% with an AUC of 0.94 and sensitivity/specificity of 0.90/0.85 for C, 65% with an AUC of 0.90 and sensitivity/specificity of 0.88/0.83 for D, and 75% with an AUC of 0.94 and sensitivity/specificity of 0.93/0.85 for E.
Conclusion: The ResNet-152 showed superior performance to that of general physicians and general orthopedists and similar performance to that of the shoulder orthopedists. To avoid overfitting, the authors decided not to use healthy images for fracture classification in order to let the CNN focus on different types of fractures, and just one image per person in order to decrease the over-performance that might be given by including very similar images. On the contrary, in the data augmentation phase, the authors used 90°, 180° and 270° rotations to increase the dataset. This might lead to overfitting because the majority of X-ray images are taken with the same orientation. It would have been (probably) better to slightly change the degree, keeping the main direction of the bone unchanged, for example using a range from -10° to 10°. The authors also used 10-fold cross validation, which is a good practice to reduce evaluation biases. Finally, as stated at the end of the paper, the lossy JPEG compression may influence the image quality. Thus, it may be better to use non-lossy compression, such as PNG or TIFF.
3.4. Artificial Intelligence for Analysing Orthopaedic Trauma Radiographs
This subsection focuses on the work of Olczak et al. [15].
Aim: The aim of this study was to assess if standard deep learning networks can be trained to identify if a bone is fractured or not in orthopedic radiographs. Secondarily, the authors also examined whether deep learning could be used to determine additional features such as body part, exam view, and laterality. We focused the discussion only on the fracture classification in this review.
Dataset: The dataset was composed of 256,458 hand, wrist and ankle radiographs, with associated radiologist reports. Labels were extracted from digital imaging and communications in medicine (DICOM) headers. In total, 56% of the images contained fractures. The data was split into 70% training images, 20% validation images and 10% test images.
Pre-processing: Each image was cropped and rescaled to 256 × 256 pixels, about 10–20% of original size. As the authors tested different network architecture, an additional rescaling was performed to match the predefined image size of each network.
Methods: The authors selected five common deep networks for this task: a BVLC Reference CaffeNet network (eight layers), a VGG CNN S network (eight layers), a VGG CNN (16 and 19 layer networks) and a network-in-network (14 layers). The networks were pre-trained on the ImageNet dataset and the last fully connected layer was replaced in order adapt the network for this specific task. The learning rate was adapted at the end of each epoch.
Results: Comparing the result of the five different networks, the best raw performance was exhibited by the VGG16 [16] network, with an accuracy of 83%. To assess a network’s performance and understand where it fails, the authors tested 400 images from the test dataset and compared the network with human performance by allowing two senior orthopedic consultants to identify fractures in the same 400 images at the same resolution as the network. As VGG16 had the best performance in the fracture class, the authors selected it for manual review. When comparing the network with the two senior orthopedic surgeons, they found that the network performed in a similar manner to the humans.
Conclusion: Testing different existing networks and choosing the one that performs best is a good practice in the field of neural networks. This approach might be useful for classifying fractures at different skeletal sites. The dataset contained a really high number of images, the highest among the datasets used in the papers we reviewed. This is obviously one of the most fundamental aspects when working with deep learning. With a huge dataset, is not easy to label the images manually, and that is why the authors decided to label them automatically from the hospital information. This procedure is subject to errors and a second review may be useful.
3.5. Artificial Intelligence in Fracture Detection: Transfer Learning from Deep Convolutional Neural Networks
This subsection focuses on the work of Kim and MacKinnon [17].
Aim: The aim of this paper is to use transfer learning in order to classify wrist fractures in two classes: broken and unbroken. According to the authors, this was the first work where transfer learning from pre-trained CNNs has been successfully applied to the problem of fracture detection in plain radiographs.
Dataset: The final dataset was composed of 1389 images, 695 wrist radiographs showing a fracture and 694 showing no fracture. This classification was obtained from the radiological report and subsequently checked and verified by a radiology registrar with three years’ radiology experience. Subsequently, data augmentation was applied: horizontal flip, rotation (between 0° and 25°), width and height shift (by a factor of 0%–15%), shearing (between 0%–10%) and zoom (between 0%–15%). This resulted in an overall amplification by a factor of eight, with 5560 images in the fracture group and 5552 images in the no-fracture group. The dataset was then split in a train-validation-test with a ratio of 80:10:10.
Pre-processing: Images were converted to JPEG by a trained radiologist, ensuring the most appropriate windowing was selected.
Methods: The network used for this purpose was InceptionV3 [18], originally trained with the ImageNet dataset and then adapted and re-trained for the broken/unbroken classification, modifying the top-layer of the network. Hyper-parameters, such as learning rate and number of epochs, were optimized iteratively.
Results: The resulting AUC value was 0.954.
Conclusion: This study demonstrates that transfer learning from deep CNN pre-trained on non-medical images can easily be applied with a modest dataset size. Data augmentation was computed offline and the same augmented images were used for the whole computation. It is a good practice to do data augmentation in real time, in order to produce different augmented images for every epoch.
3.6. Detection of Distal Radius Fractures Trained by a Small Set of X-Ray Images and Faster R-CNN (Region Based Convolutional Neural Network)
This subsection focuses on the work of Yahalomi et al. [19].
Aim: Distal radius fractures are the most common fractures of the upper extremity of the human body. The authors trained Faster R-CNN [20], a machine vision neural network for object detection, to identify and locate distal radius fractures.
Dataset: The initial dataset was composed of 55 images of distal radius fractures and 40 images of hands without fractures. In addition, 25 images not showing hand bones were used for the negative test set. The images were divided into 80% for training and 20% for testing. Another particularity was that images of the same hand in the dataset came in couples: anteroposterior position image (AP) and lateral position image. However, after some testing, the authors discovered that the detection neural network had better results if trained to exclude lateral images. The reason is that the two types of images are substantially different and that, in some lateral images, the fracture was hidden by other bones. Data augmentation was applied to increase the number of images to 4476 using mirroring, sharpness, brightness and contrast augmentation. Each image was labelled with bounding boxes around the fractured area.
Pre-processing: Images have been converted to different resolutions from 500 × 500 to 1600 × 1600 pixels in order to assess the performance of the tool.
Methods: The authors used Faster R-CNN to achieve two different tasks: classifying whether the fracture is present or not and finding the fracture’s location. Faster R-CNN is an evolution of R-CNN and Fast R-CNN where region proposals are generated by CNNs rather than using selective searches. Faster R-CNN has three different phases. At first, the input images go through a CNN that extract feature maps. Secondly, a region proposal network (RPN) is used for generating region proposals, i.e., to pre-check which location contains an object without classifying the entity of the object. The output is then passed through a region of interest (ROI) pooling to perform max pooling oF inputs of non-uniform sizes and obtain fixed-size feature maps. Finally, the pooled area goes through CNN and two fully connected branches for class softmax and bounding box regression, in order to detect the object class and return the bounding box of that object. The neural network used in this work was VGG16. New layers were randomly initialized by zero-mean Gaussian distributions while all other layers are initialized using the pre-trained model of ImageNet classification.
Results: The classification accuracy obtained was 96% and mAP (mean average precision) score of 0.866, which is the network’s precision in finding the location of the fractures.
Conclusion: This is the only work we found that implements the technology of R-CNN not only to classify fractures, but also to detect the exact region of the fracture with a high accuracy (the results were demonstrated to be significantly more accurate than the detection achieved by physicians and radiologists). The authors, smartly, did not use shear, strain or spot noise augmentation since these transformations could cause a normal hand image to be classified as a hand with a fracture. The authors tested the network with different resolution images as inputs in order to understand what the image size should be to feed into the network; this proved to be 1300 × 1300 pixels. The images have been labelled by just one specialist; however, it would have been better to have a second opinion.
3.7. Application of a Deep Learning Algorithm for Detection and Visualization of Hip Fractures on Plain Pelvic Radiographs
This subsection focuses on the work of Cheng et al. [21].
Aim: The aim of this work is to use a CNN (pre-trained with a dataset of medical images) to classify and localize hip fractures in plain frontal pelvic radiographs (PXRs). The localization phase is implemented by the use of gradient-weighted class activation mapping (Grad-CAM) [22] to confirm the validity of the model.
Dataset: We can distinguish three different datasets: the first is the pre-training dataset (named A), composed of 25,505 limb radiographs (6019 ankles, 3832 elbows, 4134 feet, and 3378 wrists). The second is the plain frontal pelvic dataset (named B), composed of 3605 images. The last one (named C) is composed of 100 independent PRXs (25 femoral neck fractures, 25 intertrochanteric fractures, 50 without hip fractures) acquired subsequently for a second test. Images were initially labelled as fracture or no-fracture according to the previous diagnosis, then a group of radiologists reviewed every images to confirm the labelling. During the training process, image augmentation was applied with a zoom of 10%, horizontal flip, vertical flip, and rotation of 10°.
Pre-processing: Poor-quality images were excluded. The input images were resized to 512 × 512 pixels to reduce the complexity and computation.
Methods: The authors used the DenseNet [23] network for the classification task. The majority of the papers from this review used ImageNet to pre-train the network; instead, in this work, the authors decided to pre-train the network with the A dataset. Hence, the network was initially trained to identify the body part in each limb radiograph. The pre-trained network was then fed with the B dataset to recognize if the bone was broken or not. Finally, to demonstrate that the CNN was actually focusing on the right area of the images, the authors implemented Grad-CAM to generate a heat map in the images that the network classified as fractured.
Results: During the pre-training phase, the model showed an accuracy of 99.5% for the test dataset in recognizing specific body parts. When the task was changed to detect hip fracture, the final test accuracy was 90% and the AUC was 0.98. After applying the hip model to the C dataset, the accuracy, sensitivity and specificity of the model were 91%, 98% and 84%, respectively. A web-based questionnaire was made to compare these results with a total of 21 specialists. The range of sensitivity of primary physicians (except radiologists and orthopedic surgeons) was 84% to 100%, and the specificity ranged from 46% to 94%. The experts, including two radiologists and four orthopedic surgeons, completed the questionnaire and achieved a mean sensitivity of 99.3% and a specificity of 87.7%. Furthermore, the heat maps computed with Grad-CAM were reviewed: after analyzing 49 heat map images, only two images identified the wrong activation site.
Conclusion: Pre-training the network with limb images instead of ImageNet increased the accuracy from 79% to 91%. This shows that is better to pre-train a network with more related images. The use of Grad-CAM confirmed that the network was actually focusing on the correct area of the images. For the sake of data augmentation, the authors also implemented vertical flip. In our opinion, this could be useless because upside-down X-rays are rare and this added extra information may lead to overfitting.
3.8. Fracture Detection with Artificial Intelligence: Improved Accuracy with Region of Interest Focusing
This subsection focuses on the work of Thurston et al. [24].
Aim: The aim of this paper is to improve the performance of the system described and already discussed from Kim and MacKinnon (Section 3.5). The original paper discussed the implementation of a deep learning network to recognize whether a bone was fractured or not. The improvement of this work is given by removing unnecessary parts of the image with a semi-automated cropping process.
Dataset: A total of 11,112 lateral wrist radiographs were used to retrain the network. Plus, a set of 50 fracture and 50 no-fracture radiographs were used in final testing of the model.
Pre-processing: In an attempt to reduce overfitting, an automated technique was employed to identify the anatomical region of interest and crop the non-necessary image portions. The region of interest was defined using the Python OpenCV [25] matchTemplate() function. The method takes a template image and slides it across every position in the subject image (the wrist radiograph), returning the position in which the closest match was calculated. In this study, the region of interest was the distal radius. The template was, therefore, an anatomical representation of the distal radius. The template was produced by using a representative lateral wrist radiograph and applying a smoothing algorithm, followed by a binary threshold to segment the bone. A scaled template matching approach was adopted to account for different wrist sizes.
Methods: The network used is the InceptionV3 network.
Results: This tool was able to crop the image appropriately in 92.4% of cases. The remainder of the cases were cropped manually and, therefore, the process was described as semi-automated. The accuracy of the model was improved when the region of interest focusing was applied, reaching an AUC of 0.978 compared with an AUC of 0.954 without region of interest. Sensitivity and specificity values were 96% and 94%, respectively.
Conclusion: This extension study has demonstrated that the accuracy of the network to predict fractures can be increased by removing surplus imaging data. The process is still semi-automated, it should be possible to make it fully automated using different techniques, for example using feature matching that tries to match features between the template and the image.
3.9. MURA: Large Dataset for Abnormality Detection in Musculoskeletal Radiographs
This subsection focuses on the work of Rajpurkar et al. [26].
Aim: The aim of this work was to train a baseline model that is able to classify whether a radiographic study is normal or abnormal: a study interpreted as normal can eliminate the need for patients to undergo further diagnostic procedures or interventions. The authors introduced the MURA dataset, one of the largest public radiographic image datasets.
Dataset: MURA dataset contains 14,863 musculoskeletal studies of the upper extremity. Each study contains one or more images taken from different views, with a total of 40,561 images, and was manually labelled by radiologists as normal or abnormal. The studies are divided in 9045 normal and 5818 abnormal for seven different extremities, including the shoulder, humerus, elbow, forearm, wrist, hand, and finger. The dataset was split into training (36,808 images), validation (3197 images), and test (556 images) sets. Data augmentation was also applied with lateral inversions and rotations (maximum 30°).
Pre-processing: Each image was normalized to have the same mean and standard deviation and scaled to 320 × 320 pixels.
Methods: The model used is a 169-layer convolutional neural network based on the architecture of DenseNet that takes as input one or more views for the same study. The output is a binary prediction, where a bone is abnormal if the output is greater than 0.5. For this reason, the final fully connected layer was replaced with a single output layer and a sigmoid non-linearity. The weights were initialized after training with the ImageNet dataset. Finally, the authors implemented CAM to visualize where the network focused during classification.
Results: The AUC of the model was 0.929. Sensitivity and specificity were 81% and 89% respectively. Plus, the authors chose three radiologists to create a gold standard, defined as the majority vote of the labels of the radiologists and then compared radiologists and model performances with the Cohen’s kappa statistic (K) [27], which expresses the agreement of each radiologist/model with the gold standard. On studies of fractures of fingers, the model’s performance was 0.389, which was comparable to the best radiologist performance of 0.410. Similarly, on the wrist, the model’s performance was 0.931, which was the same for the best radiologist. On all other skeletal sites and overall, the model’s performance was lower than the best radiologist’s performance. The AUC of the model was 0.929.
Conclusion: The dataset used by the authors is freely available, which implies that may be used by other researchers in order to improve the performance of the model. The dataset could also be used to pre-train a model for bone classification that can rely only on a small dataset.
3.10. Towards an Interactive and Interpretable (computer-aided diagnosis) CAD System to Support Proximal Femur Fracture Classification
This subsection focuses on the work of Jiménez-Sánchez et al. [28].
Aim: This work proposed a fully automatic CAD tool that is able to identify, localize and finally classify proximal femur fractures on X-rays images according to the AO classification. Following AO classification, proximal femur fractures were divided into three main groups: A, B and C, depending on the area that is involved. Each of these classes are subsequently divided into sub-groups.
Dataset: The dataset is composed of a total of 1347 X-ray images. For the two-class problem, 780 fracture images and 567 normal images were considered. The same dataset was used for the three-class problem, considering 327 images of type A fractures, 453 of type B fractures and 567 normal X-rays. Three clinical experts participated in the evaluation: one fifth-year resident trauma surgeon, one trauma surgery attendant and one senior radiologist. The dataset was split into three parts with the ratio 70:10:20 to build, respectively the training, validation and the test set. In order to overcome the problem of class imbalance, data augmentation techniques, such as translation, scaling and rotation were used.
Pre-processing: Initially, the images were parted into two, obtaining two images containing one femur each. All images were resized from 2500 × 2048 pixels (original size) to dimensions compatible to the input size of the deep learning model used. For each image, the coordinates of a squared bounding box (marked by specialists) surrounding the femur head area were stored.
Methods: At first, the task was addresses in a two-class approach between fracture and no-fracture, secondly between no-fracture, A type and B type and thirdly between no-fracture and each subclass of A and B fractures. The authors did not consider class C fractures defined in the AO classification. At the beginning, the classification task has been pursued using full X-ray half-images, down-sampled to 224 × 224 pixel, as the network used was ResNet50. CAM were also used to confirm the network’s learning. Next, they investigated the influence of localizing a relevant ROI prior to the classification. The cropped image containing only the ROI was then used as input to the same network instead of the full image. To make this process automated, the authors leveraged the bounding box annotations manually provided by our experts to formulate the problem as a regression, in which the goal was to find the ROI in the radiograph. For this purpose, an auxiliary network based on AlexNet [29] was trained to predict the extreme points of the bounding box. For this architecture, full X-ray images were down-sampled to 227 × 227 pixel.
Results: Using full images, accuracies for each type of classification were: 0.83 for binary classification, 0.89 for three-class classification, and 0.72 for the fine-grained classification. Using cropped images, accuracy for binary classification improved to 0.94 and to 0.91 for three-class classification. Improvements for fine-grained classification with cropped images are not stated in the paper. Results are slightly lower using the automated-cropping method.
Conclusion: Using AO foundation classification is a perfect approach to define a generalized method. In addition, as we already notice in previous papers, it is shown that using ROI is useful for improving classification accuracy. The main obstacle broached by the authors is the large imbalance in the frequency of appearance of the classes in the fine-grained classification. A method to deal with unbalanced data must be defined.
3.11. Classification of Atypical Femur Fracture with Deep Neural Networks
This subsection focuses on the work of Chen [30].
Aim: The objective of this project was to develop a deep neural network to assist specialists in classifying femur bones in two classes: atypical femur fracture (AFF) and normal femur fracture (NFF), where AFF is a type of stress fracture that occurs in conjunction with prolonged bisphosphonate treatment. Plus, this work aimed to visualize the learning features using class activation mapping (CAM).
Dataset: After a manual screening to exclude bad quality images, the final number of input samples in the dataset was 796 (397 with AFF, 399 with NFF). The images are then augmented with rotation (between −10° and +10°), width and height shift and zooming (both by a factor of 0%–10%).
Pre-processing: The dataset is pre-processed with standardized image processing methods, such as down-sampling and normalization. The X-ray images are of different shapes and all the input images are reshaped accordingly to the input size requested from the network used.
Methods: The author tested VGG19, ResNet50 and InceptionV3 architecture pre-trained with ImageNet for the classification task. Two pipelines were implemented: the first is a fully automated approach where the images were directly inputted into the network; the second presented a manual intervention in order to move the fracture towards the center before feeding the network. During the learning, CAM was also used to see where the network was focusing.
Results: Using five-fold cross validation, using the fully automated approach, the averaged accuracy was 82.7%, 89.4% and 90.5% for VGG19, InceptionV3 and ResNet50, respectively. On the other hand, using the interactive approach resulted in an improved accuracy of 92.2%, 93.4% and 94.4% for VGG19, Inception and ResNet50, respectively. At the beginning of this project, the author obtained a suspiciously high performance, but after visualization with CAM, she found that the network was focusing on the notations converted from digital imaging and communications in medicine (DICOM) and not on the fracture region.
Conclusion: This work affirmed, once again, that transfer learning from ImageNet could be easily used when working with X-ray images. The most interesting aspect of this paper is the result given by CAM, which confirms the importance of this tool in demonstrating that the network is actually learning.
4. Discussion and Conclusions
In order to summarize the methods, Table 1 shows the main feature values of every study. Starting from this table, we mapped each value with the criteria already outlined from 1 to 5, showed in Table 2 and Figure 2.

Table 1.
Summary of the six different features of fracture classification methods, related to the corresponding paper. Legend: not assessable (NA); pre-training (P-TR); training (TR); testing (TE); binary (B); multi-class (M); number of specialists (N); average years of expertise (Y); number of subsequent checks (C).

Table 2.
Values extracted from Table 1 and mapped in the range from 1 to 5. In case of multiple and binary results we used just the binary one in order to relate different studies. For the dataset size, we considered [10,15,26] as outliers and mapped them to the values of 5 and 4. We then mapped the other values from 1 to 3. For the labelling, we computed the values using the formula and a subsequent mapping from 1 to 5.
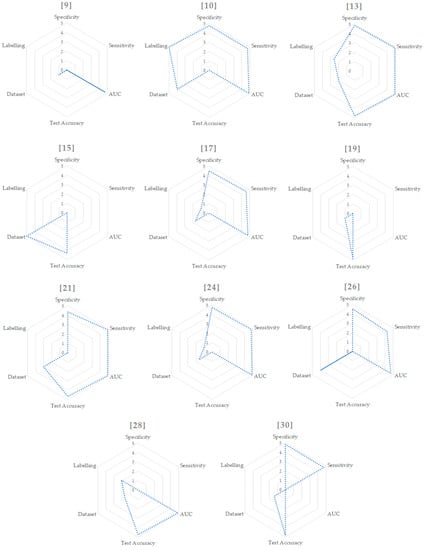
Figure 2.
Radar graphs for each paper.
Starting from these values, we were able to put together all the pros found in the different articles and define what should be the correct approach to tackle this task. Most of the work focused on classification between broken and unbroken bones, without extending the task to different types of fractures. We think that a generalized tool, able to distinguish different types of fractures, should follow the classification stated by the AO foundation. The AO classification is hierarchical and is determined by the localization and configurations of the fracture lines, where each bone is divided in subsequent sub-groups of fracture, as shown in Figure 3 for the case of proximal femur. In the common literature, the AO classification was claimed to present a better reproducibility compared to other classification systems [31] and its configuration made it optimal for a classification task. Plus, the structure is the same for different bones in the human body, so the approach could be easily extended. Once the correct classification system is defined, an adequate dataset is certainly one of the most important aspects for a deep learning-based application to operate efficiently. Even if, in some studies, good results have been obtained for the fracture/no-fracture classification without using a large dataset [9,19], a correct number of images is suggested when the network has to distinguish between different sub-groups of fractures. The dataset could be increased and balanced with data augmentation techniques, if needed, but without adding useless or misleading information. For example, using shear, strain or spot noise augmentation could cause a normal bone image to be classified as a bone with a fracture [19]. Thus, data augmentation is not always enough to balance irregular datasets. One idea to tackle this could be to assign different weights to different classes when computing the cost function—classes with few images will be associated with higher weights. In addition, technologies such as generative adversarial networks [32] might be used to generate “fake” fractured bones, but this would probably be unfeasible, as the fractures might appear unrealistic. Concerning the pre-processing phase, the dataset should be cleaned from images containing prosthetics or other evident defects, and we recommend using no more than one image per person to decrease over-performance by the inclusion of very similar images of the same patient. It is also demonstrated that selecting the fractured area and feeding the neural network with cropped images instead of the full image improves the network performance [24,28,30]. For this reason, a fully automated tool to select the fractured regions should be designed for the pre-processing phase. In addition, it is suggested that a lossless format is used, such as PNG or TIFF, and that images are resized to different sizes in order to see which one works best [19]. Unfortunately, this is not feasible when using an existing network with transfer learning, because the input images must have a fixed size. Transfer learning allows the use of a network pre-trained on a different dataset for your own dataset. The most-used network architectures were VGG, ResNet, DenseNet and Inception, pre-trained with an ImageNet dataset. As demonstrated in different papers, pre-training the network using a larger dataset of X-ray bone images may improve the performance [10,26]. For example, the MURA dataset [26] is one of the biggest bone datasets freely available. For this reason, we recommend trying different networks pre-trained with the MURA dataset and testing which one works best for the specific problem at hand. Moreover, the hyperparameters, which must be set correctly after some trials, are the learning rate (with transfer learning it is better to use a low learning rate in order not to change the pre-trained weights too much), the learning rate decay and the batch size. Cross-validation should be used to demonstrate that the network correctly generalizes the dataset features [13,30]. Another improvement could be introduced by removing surplus imaging data [13]. For example, if a network has to classify between the no-fracture class and three different types of fractures (A, B and C) it works at its best if trained to exclude no-fracture images. Following these results and the AO foundation classification, one idea could be to apply a hierarchical approach. To be clearer, an initial network that classifies between fracture and no-fracture and a subsequent one that takes the images predicted to be fracture and classifies them into three types (A, B and C). Finally, class activation mapping or similar technologies should be used to see where the network is focusing [21,26,28]. A clear example is found in [30], where the author understood, with the help of CAM, that the network was learning the wrong features. Last but not least, the specialists play a fundamental role: the dataset must be correctly labelled, and different years of experience are needed to properly classify the types of fractures following the AO classification, especially the subgroups. Both for labelling and evaluation, if possible, more than one expert, coming from different specializations, should be enrolled in order to obtain multiple opinions. The final aim of this tool would be to prove that the CAD system effectively helps doctors in their diagnoses, it would be also important to evaluate the performance of the specialists with and without the help of the tool [10]. The main characteristics of our proposed baseline are stated in Table 3 and compared with each evaluated paper.
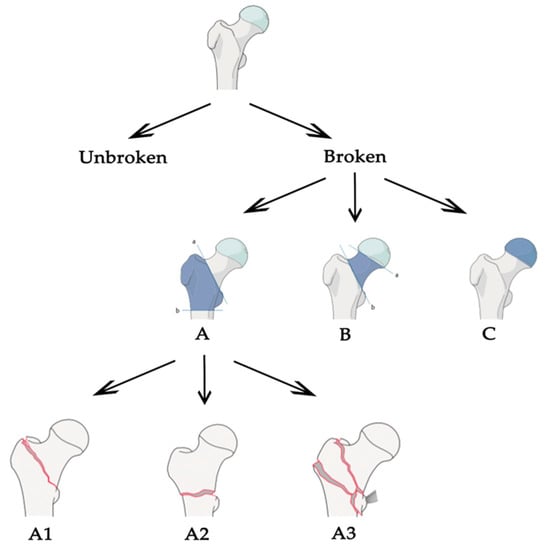
Figure 3.
Arbeitsgemeinschaft für Osteosynthesefragen (AO) partial classification for proximal femur fracture.

Table 3.
Comparison between our proposed baseline and each paper.
Author Contributions
Conceptualization, L.T. and E.V.; methodology, L.T. and S.M.; validation, L.T.; formal analysis, L.T., R.M. and S.M.; investigation, L.T.; resources, L.T., R.M. and E.V.; data curation, L.T.; writing—original draft preparation, L.T.; writing—review and editing, R.M., E.V. and S.M.; supervision, R.M. and E.V.; project administration, E.V. and S.M.; funding acquisition, E.V., R.M. and S.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- International Osteoporosis Foundation. Broken Bones, Broken Lives: A Roadmap to Solve the Fragility Fracture Crisis in Europe; International Osteoporosis Foundation, 2018. Available online: http://share.iofbonehealth.org/EU-6-Material/Reports/IOF%20Report_EU.pdf (accessed on 21 February 2020).
- Krupinski, E.A.; Berbaum, K.S.; Caldwell, R.T.; Schartz, K.M.; Kim, J. Long Radiology Workdays Reduce Detection and Accommodation Accuracy. J. Am. Coll. Radiol. 2010, 7, 698–704. [Google Scholar] [CrossRef]
- Hallas, P.; Ellingsen, T. Errors in fracture diagnoses in the emergency department—Characteristics of patients and diurnal variation. BMC Emerg. Med. 2006, 6, 4. [Google Scholar] [CrossRef]
- Al-Ayyoub, M.; Hmeidi, I.; Rababah, H. Detecting hand bone fractures in x-ray images. JMPT 2013, 4, 155–168. [Google Scholar]
- Cao, Y.; Wang, H.; Moradi, M.; Prasanna, P.; Syeda-Mahmood, T.F. Fracture detection in x-ray images through stacked random forests feature fusion. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), New York, NY, USA, 16–19 April 2015; IEEE: Brooklyn, NY, USA, 2015; pp. 801–805. [Google Scholar]
- Myint, W.W.; Tun, H.M.; Tun, K.S. Analysis on Detecting of Leg Bone Fracture from X-ray Images. Int. J. Sci. Res. Publ. IJSRP 2018, 8, 371–377. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. Int. J. Surg. 2010, 8, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Dimililer, K. IBFDS: Intelligent bone fracture detection system. Procedia Comput. Sci. 2017, 120, 260–267. [Google Scholar] [CrossRef]
- Lindsey, R.; Daluiski, A.; Chopra, S.; Lachapelle, A.; Mozer, M.; Sicular, S.; Hanel, D.; Gardner, M.; Gupta, A.; Hotchkiss, R.; et al. Deep neural network improves fracture detection by clinicians. Proc. Natl. Acad. Sci. USA 2018, 115, 11591–11596. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; Volume 9351, pp. 234–241. ISBN 978-3-319-24573-7. [Google Scholar]
- Fei-Fei, L.; Deng, J.; Li, K. ImageNet: Constructing a large-scale image database. J. Vis. 2010, 9, 1037. [Google Scholar] [CrossRef]
- Chung, S.W.; Han, S.S.; Lee, J.W.; Oh, K.-S.; Kim, N.R.; Yoon, J.P.; Kim, J.Y.; Moon, S.H.; Kwon, J.; Lee, H.-J.; et al. Automated detection and classification of the proximal humerus fracture by using deep learning algorithm. Acta Orthop. 2018, 89, 468–473. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; IEEE: Las Vegas, NV, USA, 2016; pp. 770–778. [Google Scholar]
- Olczak, J.; Fahlberg, N.; Maki, A.; Razavian, A.S.; Jilert, A.; Stark, A.; Sköldenberg, O.; Gordon, M. Artificial intelligence for analyzing orthopedic trauma radiographs: Deep learning algorithms—Are they on par with humans for diagnosing fractures? Acta Orthop. 2017, 88, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:14091556. [Google Scholar]
- Kim, D.H.; MacKinnon, T. Artificial intelligence in fracture detection: Transfer learning from deep convolutional neural networks. Clin. Radiol. 2018, 73, 439–445. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; IEEE: Las Vegas, NV, USA, 2016; pp. 2818–2826. [Google Scholar]
- Yahalomi, E.; Chernofsky, M.; Werman, M. Detection of Distal Radius Fractures Trained by a Small Set of X-Ray Images and Faster R-CNN. In Intelligent Computing; Arai, K., Bhatia, R., Kapoor, S., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 997, pp. 971–981. ISBN 978-3-030-22870-5. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-T.; Ho, T.-Y.; Lee, T.-Y.; Chang, C.-C.; Chou, C.-C.; Chen, C.-C.; Chung, I.-F.; Liao, C.-H. Application of a deep learning algorithm for detection and visualization of hip fractures on plain pelvic radiographs. Eur. Radiol. 2019, 29, 5469–5477. [Google Scholar] [CrossRef] [PubMed]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; IEEE: Venice, Italy, 2017; pp. 618–626. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; IEEE: Honolulu, HI, USA, 2017; pp. 2261–2269. [Google Scholar]
- Thurston, M.; MacKinnon, T.; Kim, D.H. Fracture detection with artificial intelligence: Improved accuracy with region of interest focusing. In Proceedings of the 2018 European Congress of Radiology, Vienna, Austria, 28 February–4 March 2018. [Google Scholar]
- Bradski, G. The OpenCV Library. Dr. Dobb J. Softw. Tools 2000, 25, 120–125. [Google Scholar]
- Rajpurkar, P.; Irvin, J.; Bagul, A.; Ding, D.; Duan, T.; Mehta, H.; Yang, B.; Zhu, K.; Laird, D.; Ball, R.L.; et al. MURA: Large Dataset for Abnormality Detection in Musculoskeletal Radiographs. arXiv 2018, arXiv:171206957. [Google Scholar]
- McHugh, M.L. Interrater reliability: The kappa statistic. Biochem. Medica 2012, 22, 276–282. [Google Scholar] [CrossRef]
- Jiménez-Sánchez, A.; Kazi, A.; Albarqouni, S.; Kirchhoff, C.; Biberthaler, P.; Navab, N.; Mateus, D.; Kirchhoff, S. Towards an Interactive and Interpretable CAD System to Support Proximal Femur Fracture Classification. arXiv 2019, arXiv:190201338. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Chen, Y. Classification of Atypical Femur Fracture with Deep Neural Networks; KTH University: Stockholm, Sweden, 2019. [Google Scholar]
- Jin, W.-J.; Dai, L.-Y.; Cui, Y.-M.; Zhou, Q.; Jiang, L.-S.; Lu, H. Reliability of classification systems for intertrochanteric fractures of the proximal femur in experienced orthopaedic surgeons. Injury 2005, 36, 858–861. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. arXiv 2014, arXiv:14062661. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).