Abstract
The application of anaerobic digestors to decentralized wastewater treatment systems (DWTS) has gained momentum worldwide due to their ease of operation, high efficiency, and ability to recycle wastewater. However, the microbial mechanisms responsible for the high efficiency and ability of DWTS to recycle wastewater are still unclear. In this study, the microbial community structure and function of two different anaerobic bioreactors (a primary sludge digestor, PSD, and anaerobic membrane bioreactor, AnMBR) of a DWTS located in Germany was investigated using 16S rRNA gene amplicon and metagenomic sequencing, respectively. The results showed that the microbial community structure was remarkably different in PSD and AnMBR. Methanobacteriaceae and Syntrophaceae were identified as the families that significantly differed in abundance between these two bioreactors. We also used genome-centered metagenomics to predict the microbial interactions and methane-generating pathway, which yielded 21 near-complete assembled genomes (MAGs) (average completeness of 93.0% and contamination of 2.9%). These MAGs together represented the majority of the microbial community. MAGs affiliated with methanogenic archaea, including Methanobacterium sp., Methanomicrobiales archaea, Methanomassiliicoccales archaea, and Methanosaeta concilii, were recruited, along with other syntrophic bacterial MAGs associated with anaerobic digestion. Key genes encoding enzymes involved in specific carbohydrate-active and methanogenic pathways in MAGs were identified to illustrate the microbial functions and interactions that occur during anaerobic digestion in the wastewater treatment. From the MAG information, it was predicted that bacteria affiliated with Bacteroidetes, Prolixibacteraceae, and Synergistaceae were the key bacteria involved in anaerobic digestion. In the methane production step, Methanobacterium sp. performed hydrogenotrophic methanogenesis, which reduced carbon dioxide to methane with hydrogen as the primary electron donor. Taken together, our findings provide a clear understanding of the methane-generating pathways and highlight the syntrophic interactions that occur during anaerobic digestion in DWTS.
1. Introduction
Anaerobic digestion (AD) is a widely applied and mature technology that is used to treat the organic fraction of municipal wastewater and produces methane for biofuel, which is considered as an effective solution to the energy crisis and global pollution threat [1]. Among the different types of AD, primary sludge digestors (PSDs) and anaerobic membrane bioreactors (AnMBRs) receive the most attention for the biological treatment of wastewater. PSDs are mostly utilized in AD for wastewater treatment and are regarded as cheap and low energy input processes, with an overall positive energy output [2]. PSDs degrade the majority of the organic content in wastewater sludge, greatly reducing the total amount of solids produced. PSDs stabilize sludge to control odors while also reducing pathogens [3]. In addition to PSDs, AnMBRs have been widely incorporated into the anaerobic biological wastewater treatment process because of their solid-liquid separation and high effluent quality [4]. Due to their high operation stability, AnMBRs are suitable for treating wastewater with a high content of COD and suspended solids, high salinity, and large variations of flow and composition, even in the presence of fat, oil, and grease or inhibitory compounds [5,6].
The complex microbial communities in AD play a significant role in maintaining stable operation and efficient biogas production [7]. To enhance biogas production in AD of organic wastes, it is vital to comprehensively understand the microbial communities involved in terms of their taxonomic composition; the interaction networks; and the relationships among operational conditions, biodiversity, and system functions [8]. AD degrades organic compounds and generates biogas and involves four sequential stages: hydrolysis, fermentation (acidogenesis), acetogenesis, and methanogenesis [9,10]. AD is reliant on the tightly coupled synergistic activities of complex microbial consortia, which are delicately balanced between their two major domains: Bacteria and Archaea. In general, bacteria are responsible for hydrolysis, acidogenesis, and acetogenesis, whereas methanogens in AD bioreactors are affiliated with Methanobacteriales, Methanomicrobiales, and Methanosarcinales [11,12]. The composition and diversity of fermenting bacteria, syntrophic bacteria, and methanogens are very related to temperature, fed substrates, and process conditions [13,14,15].
The microbiome in PSD and AnMBR in AD remains only partially elucidated because these communities are rich in uncharacterized taxa, and they are difficult to explore with traditional cultivation-based methods [16]. Metagenomics was first applied to AD in 2008 with the analysis of a German full-scale biogas plant treating farm waste [17]. Since then, several studies have examined AD metagenomes, mainly focusing on taxonomy and gene-centric functional analyses [12,18,19,20]. Recently, metagenomic analysis has shifted toward reconstructing the important metabolic pathways and genomes present in AD systems [21]. Of the AD metagenomes analyzed to date, samples have been obtained from full-scale biogas plants treating farm waste [17], industrial [19] and municipal [12,20] sludge digesters, and lab-scale reactors [18,21]. DWTS combined the advantage of PSD and AnMBR in wastewater treatment, particularly for low strength (COD < 1000 mg kg−1) municipal wastewater at relatively low temperatures (<18 °C). However, to date, no effort has been dedicated to analyzing the microbial community structure in DWTS, as well as the differences of microbial and functional divergence between PSD and AnMBR. Furthermore, the microbial interactions of the syntrophic consortium is pivotal in understanding the kinetics of bio-mediated element-cycling and methane generation in DWTS.
Therefore, based on the above considerations, we conducted 16S rRNA gene amplicon and metagenomic sequencing to characterize the community composition and reveal functional traits in PSD and AnMBR of a DWTS treating domestic wastewater. Furthermore, microbial networks, including hydrolyzing and fermenting bacteria, syntrophic bacteria, and methanogens, were identified by metagenomic binning of the dominant species of PSD and AnMBR. This study not only offers insight into the microbial traits of DWTS but also provides in-depth knowledge of the microbial networks in PSD and AnMBR during AD, thereby facilitating new process designs and operational strategies that maximize biomethane recovery from domestic wastewater.
2. Materials and Methods
2.1. Description of a Decentralized Wastewater Treatment System
The DWTS analyzed in this study was built in 2009 in Germany. The main objective of this project was to optimize the processes used for decentralized urban wastewater management. The system was designed to treat wastewater from approximately 175 inhabitants. The detailed process flow of the DWTS is presented in Figure 1. During wastewater treatment, sewage was collected from the vacuum station and pumped into an equalization tank, followed by a sedimentation tank, where the solids were settled for separating treatment. The solids, which were up to 1% to 2% of the influent quantity, were treated in a high-load PSD at approximately 37 °C. To make the treatment more efficient, water was removed during the digestion process by a rotating disk filter. Then, water without settleable solids flowed from the sedimentation tank into an AnMBR. Next, the mixed sludge circulated through an external rotating disk filter, which continuously withdrew the effluent from the bioreactor. Fouling control was achieved by rotating the shaft on which the filtration disks were fixed to create a centrifugal force, which caused the solids to flow off. Finally, the treatment divided the wastewater into a solids-free filtrate, biogas from the two anaerobic bioreactors, and stabilized solids after anaerobic digestion (Figure 1).
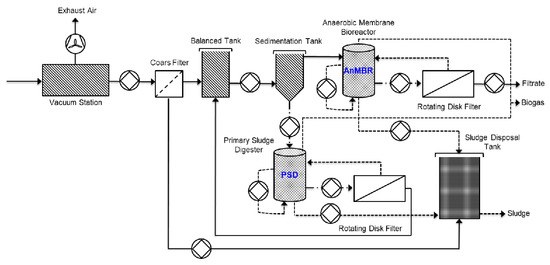
Figure 1.
Schematic diagram of the decentralized wastewater treatment system (DWTS) with a primary sludge digestor (PSD) and anaerobic membrane bioreactor (AnMBR).
2.2. Sampling and Physicochemical Analysis
Water samples were taken from the influent and effluent of the PSD and AnMBR in the DWTS. The physicochemical characteristics were measured as described previously [22]. In brief, influent and effluent chemical analyses (COD, nitrogen species, phosphorus species and alkalinity) were conducted in accordance with the Standard Methods for the Examination of Water and Wastewater, APHA [23]. The pH of the reactors was determined using a DELTA 320 (Mettler Toledo, Columbus, OH, USA). Gas samples were taken from the reactors via a small sampling port located at the top of the reactor. The biogas production was measured with a wet gas meter (COMBIMASS@ GA-s hybrid premium, Binder Group, Germany), and the CH4 production was determined using a gas chromatograph (Clarus 580 Arnel, PerkinElmer, Waltham, MA, USA) equipped with a thermal conductivity detector. Standard methods were used to measure TS, VS, TSS, and VSS [23]. Sludge samples were collected from the AnMBR and PSD and centrifuged at 10,000× g for 10 min at 4 °C. The remaining pellet of the sludge was transported on ice to the lab, where it was stored at −70 °C before DNA extraction.
2.3. DNA Extraction and 16S rRNA Gene Amplicon Analysis
We collected three samples from PSD and AnMBR, respectively. For each sample, three replicates of DNA extraction were conducted individually, and combined as a composition sample, before the PCR reaction. A PowersoilTM DNA Isolation Kit (Qiagen, Hilden, Germany) was used to extract microbial DNA from PSD and AnMBR according to the manufacturer’s protocol. The yield and quality of DNA were analyzed electrophoretically on 1% agarose gel. Prokaryotic universal primers 515F and 806R [24], where the barcode was an 12-base sequence unique to each sample, were used in PCR to amplify the V4 region of the bacterial 16S ribosomal RNA gene. The PCRs were carried out in 50 µL volume with 10 ng of template DNA, 25 µL of 2 × ExTaq PCR Master Mix (Takara Biotechnology, Dalian, China) and 1 µL of each 10 mM primer. The PCR amplification was run under the following conditions: (1) pre-denature at 95 °C for 2 min; (2) 28 cycles of denaturing at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s; and (3) a final extension at 72 °C for 5 min. The PCR products were detected by 2% denaturing agarose gels, after which the 16S rRNA gene amplicons were extracted from the gels and purified with a QIAquick PCR purification kit (Qiagen, Düsseldorf, Germany) and quantified with Fluorometer (Qubit® 3.0, ThermoFisher Scientific, Waltham, MA, USA). The purified 16S rRNA gene amplicons were paired-end sequenced on an Illumina Hiseq platform at Novogene Bioinformatics Technology, Beijing, China.
Analyses of the sequence of 16S rRNA genes were performed using QIIME2 software [25]. The following parameters were used for quality filtering: minimum/maximum length = 250/600; maximum 2 ambiguous bases; maximum 2 primer mismatches; average quality score > 25; and homopolymer < 6 nucleotides. OTUs with 97% sequence identity were clustered using the UCLUST software and the Greengenes, used as a reference 16S rRNA gene sequence database [26]. A representative sequence was chosen for each phylotype and aligned against the Greengenes using PyNAST [27]. The UCHIME algorithm using USEARCH (v7.0) was used to remove potentially chimeric sequences from the aligned representative sequences [28]. The taxonomic assignment of representative sequences from each OTUs was performed using the UCLUST consensus taxonomy assigner [29]. Finally, to estimate alpha diversity, a random sub-sampling method for each sequence library was used for microbial community diversity index calculations to control for the effects of library size. Alpha diversity indices were calculated for all samples with 1000 repetitions using a size of 16,126 sequences per sample. For principal coordinate analysis (PCoA), all samples were also subsampled to 16,126 sequences per sample to remove sample-size effects.
2.4. Metagenomic Sequencing, Assembly, and Annotation
The community genomic DNA (from triple DNA extractions from each replicate of the bioreactor) of the samples was sheared into 500- and 800-bp length fragments and multiplex-sequenced on the Illumina HiSeq 2500 platform (Illumina, Inc., San Diego, CA, USA), which generated 2 × 250-bp pair-end sequencing reads. Sequencing was carried out at Novogene Bioinformatics Technology (Beijing, China). The adapter sequences were removed from the generated reads, which were trimmed using Trimmomatic v0.30 with a quality cutoff of 30, a sliding window of 6 bp, and a minimum length cutoff of 45 bp [30]. The functional composition of the microbial communities was obtained from the results of the high-quality reads compared with the sequences in the Kyoto Encyclopedia of Genes and Genomes (KEGG, a database resource for understanding high-level functions and utilities of the biological system) [31] and Carbohydrate-Active EnZymes (CAZy, a database describes the families of structurally-related catalytic and carbohydrate-binding modules of enzymes) databases [32] using thresholds of e < e−10 and an aligned length > 150 nt. High-quality reads from the PSD and AnMBR were pooled and assembled into one metagenome using IDBA-UD 1.1.1 [33] with the following parameters: minimum k value of 60 and maximum k value of 120; increment of the k-mer of each iteration of 10 and minimum multiplicity for filtering k-mers when building the graph of 5; seed k-mer size for alignment of 5; and minimum length of contigs of 1000. The other parameters were set as default. The contigs and unmapped reads from the IDBA-UD assembly were re-assembled using the parameters as described. The contig coverage was determined by mapping the initial reads to contigs larger than 1 kbp with Bowtie2 software [34]. Subsequently, Samtools [35] was used to convert, sort, and merge the Sam files. Then, coverage (describes the number of reads that align to the contigs) was determined with the Genomecov software of the Bedtools package [36]. As low coverage and short contigs are known to be error-prone, contigs with a length < 1 kb and average coverage < 3 were discarded from the subsequent binning step.
2.5. Methanogenic Pathway of Predominant Species Determined by Genomic Binning
CONCOCT software was used for contig binning, which is based on sequence composition and coverage [37]. Subsequently, mapping of the high-quality reads was carried out using Bowtie2 [34] to determine the coverage of these contigs before conducting the CONCOCT step. CheckM [38] was used to assess the completeness and contamination of the recovered metagenome-assembled genomes (MAGs). MAGs with completeness > 90% and contamination < 10% were used for detailed downstream analyses. The abundance of each MAG of the samples was estimated by mapping the high-quality reads of individual datasets to the contigs from the MAGs.
Taxonomic analysis of the MAGs was performed by different methods, and the results were compared to obtain a consensus assignment. First, all the protein sequences translated in silico from these MAGs were input to PhyloPhlAn [39] to facilitate the phylogenetics of the MAGs, which were based on 400 universal proteins. Second, the ORFs predicted from each MAG were checked by sequence similarity to the NCBI-nr database using BlastX with a maximum allowed e-value of 1 × 10−10. The BlastX results were parsed by MEGAN [40] to access the species and genus phylum-level taxonomic annotation of each cluster of bins. The minimum bit score used for the analysis was 60, and a minimum support of 5% for each taxonomic category was used for the LCA algorithm. A taxon of a MAG was assigned when at least 85% of the identified ORFs resulted in a concordant taxonomy. We also constructed a phylogenetic tree of the recovered MAGs and their closest species’ genome using the FastTree algorithm with default settings by PhyloPhlAn.
Genes were predicted across each MAG using FragGeneScan [41], and functional prediction of the identified genes was conducted by similarity searching against the KEGG and CAZy databases using an e-value of 10−5 and an aligned length larger than 200 nt. The proteins of interest in carbohydrate and methanogenic metabolism were retrieved from the gene information in comparison with the KEGG database, and the abundance of methanogenesis-associated genes in each MAG was calculated using the mapping information of each gene.
2.6. Statistics
Statistical analyses were conducted using SPSS 17.0 Software (SPSS, Inc., Chicago, IL, USA). Statistical significance was estimated with two-way analysis of variance (ANOVA) followed by the LSD or Dunnett’s T3 test to determine the significance of differences between groups. Linear discriminant analysis (LDA) of effect size (LEfSe) was applied to determine the most discriminant taxa among the PSD and AnMBR samples [42], with a value for the statistical test equal to 0.05 and a logarithmic LDA score threshold of 4.0.
2.7. Sequence Accession
The sequence data of the 16S rRNA genes and metagenome from this study are publicly available in NCBI BioProject PRJNA578561 (with accession no. SRR10566884-SRR10566893).
3. Results and Discussion
3.1. Operational Performance of the DWTS
The detailed performance of the DWTS is summarized in Table S1. The DWTS demonstrated good performance in terms of COD (52% on the average) and total N destruction (44%). The SO42− in the effluent of the system was significantly lower (2 mg/L) than that in the effluent (34.1 mg/L). A summary of the chemical characteristics of the PSD and AnMBR in the DWTS is provided in Table 1. There was a large difference in the organic loading rate between the AnMBR and PSD, in which 0.71 g of COD and 0.16 g of VSS per L wastewater were loaded into these two bioreactors every day. The DWTS functioned efficiently in regard to biogas production. Approximately 122 L per kg COD in the AnMBR and 374 L of biogas per kg volatile suspended solids (VSS) in the PSD were observed, and the average methane composition accounted for approximately 80%~85% and 60~65% of the biogas in the AnMBR and PSD, respectively. These results implied that a considerable amount of organic matter was converted into methane in both bioreactors. Similar pH, TSS, and VSS values were monitored in both reactors (Table 1). The temperature was kept at approximately 20 °C and 35 °C for the AnMBR and PSD, respectively. In terms of methane and biogas recovery, the PSD offers potential advantages over the AnMBR.

Table 1.
Anaerobic membrane bioreactor (AnMBR) and sludge anaerobic reactor (PSD) system operating parameters.
3.2. Characteristic Microorganisms in the PSD and AnMBR
To identify the microorganisms present in the PSD and AnMBR, we conducted 16S rRNA gene amplicon sequencing on three samples from each bioreactor. After quality control, denoising and chimer removal, a total of 148,306 high-quality reads were generated from the 16S rRNA V4 sequences from 6 samples, ranging from 16,126 to 41,310 reads, with an average of 24,718 (Table S2). The sequence numbers were normalized to 16,126 reads in each sample to describe the alpha diversity and composition characteristics. All the sequences were clustered into OTUs at 97% sequence similarity. The number of OTUs per sample ranged between 386 and 608 (Table S2). Good’s coverage ranged from 0.85595 to 0.93668 in all samples (Table S2), suggesting that major microbial diversity had been captured. The average Chao1 index values were 2217 ± 295 and 1459 ± 748 for the PSD and AnMBR samples, respectively. The PD indices were 57.4 ± 1.82 for the PSD samples and 49.5 ± 7.36 for the AnMBR samples. The Shannon and Simpson diversity indices were significantly higher in the AnMBR (Shannon: 7.012 ± 0.330; Simpson 0.978 ± 0.009) than in the PSD samples (Shannon: 5.973 ± 0.040; Simpson 0.942 ± 0.05) (p < 0.05) However, the number of observed OTUs, Chao1, ACE, and Faith PD index did not differ significantly between the PSD and AnMBR (Table S3).
The 16S rRNA gene sequences were classified at both the phylum and family levels. At the phylum level, Proteobacteria (27.11 ± 0.46%), Euryarchaeota (13.72 ± 0.41%), Firmicutes (11.44 ± 0.92%), Synergistetes (11.22 ± 1.02%), and Bacteroidetes (10.89 ± 2.51%) were the predominant phyla in the PSD samples, accounting for 74.38% of the total microbiota. However, a large proportion of Proteobacteria (75.53 ± 0.46%), Firmicutes (6.37 ± 0.92%), and Bacteroidetes (5.76 ± 2.51%) were observed in the AnMBR samples (Figure 2). Samples of the PSD and AnMBR also harbored different family-level profiles, with Methanobacteriaceae (12.99 ± 0.35% in the PSD and 2.08 ± 0.35% in the AnMBR) and Syntrophaceae (4.24 ± 0.10% in the PSD and 3.61 ± 0.01% in the AnMBR) representing the most abundant genera in the PSD and AnMBR samples, respectively (Table S4). At the phylum level, the relative abundances of most predominant phyla were significantly different between the two types of samples (all p < 0.05) (Figure 2). Moreover, Methanobacteriaceae (12.99 ± 0.35% in the PSD and 2.08 ± 0.35% in the AnMBR) and Syntrophaceae (4.24 ± 0.10% in the PSD and 3.61 ± 0.01% in the AnMBR) were identified as the families that most significantly differed in abundance between the PSD and AnMBR samples (all p < 0.05) (Table S4). This result suggested a distinct difference in the microbial community structure between the PSD and AnMBR samples, which was also supported by the weighted PCoA results. The weighted PCoA showed that the PSD samples grouped together in one cluster, while the AnMBR samples grouped in another cluster (Figure S1). Methanobacteriaceae and Syntrophaceae were identified as the genera that significantly differed in abundance between the PSD and AnMBR samples. Methanobacteriaceae is recognized as a family comprising hydrogenotrophic methanogenic archaeon in anaerobic digestion and is prominent in methanogenic communities [43]. The hydrogenotrophic Methanobacteriales have been shown to correlate with biogas production in 29 full-scale digester studies, confirming their role in the maintenance of digester function [44]. Methanobacteria use the methanogenesis pathway, in which hydrogen reacts with carbon dioxide to produce methane [45]. The maintenance of the temperature (PSD at 37 °C and AnMBR at 20 °C) should be the primary reason for the significantly different abundance of Methanobacteriaceae in these two bioreactors and should also explain why biogas production was higher in the PSD. Compared with the PSD, the relative abundance of Syntrophaceae was significantly higher in the AnMBR. Bacteria affiliated with Syntrophaceae play important roles in acetate oxidation during anaerobic methane production and are seen as stabilizers that maintain AD stability, especially when the system undergoes environmental fluctuations [46]. Furthermore, the significance of Syntrophaceae in the methanogenic degradation of waste has been acknowledged [47,48]. The remarkably different abundances of Syntrophaceae and Methanobacteriaceae may reflect the functional tendencies in the AnMBR and PSD, where acetate oxidation and methane generation are the dominant microbial metabolisms, respectively, in these two types of bioreactors during wastewater treatment. The relative abundances of Desulfomonile, Smithella, and Syntrophus (all within the family Syntrophaceae) were significantly different between samples of PSD and AnMBR (p < 0.01). However, the above three genera were less abundant (<1%) in these two kinds of bioreactors. In the family Methanobacteraceae, Methanobacterium was the only identified genus with an averagely relative abundance larger than 1% in PSD and AnMBR, and also, the identified genus exhibited significantly different abundance in these two kinds of bioreactors.
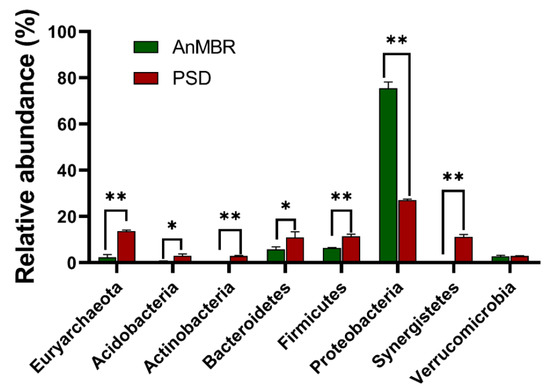
Figure 2.
Taxonomic composition of the microbial communities showing the differences between the PSD and AnMBR samples. The data reveal the phylum- and genus-level classification of the 16S rRNA gene sequences. 16S rRNA gene sequences longer than 250 bp were classified using the RDP naïve Bayesian rRNA Classifier at an 80% confidence threshold. Only the relative abundances of the identified phyla higher than 1% are listed. The average ± SD values of the samples in each group are expressed in each column. The significance of the cutoff for the corrected p-value using the Benjamini–Hochberg FDR procedure was set at 0.05. The significant differences within the sampling group are labeled **, where p < 0.05.
3.3. Variation of Gene Functional Profiles of the PSD and AnMBR
Based on the above results regarding the geochemical properties and microbial communities of the PSD and AnMBR, we hypothesize that the microbial interactions and metabolic pathways of the dominant species are very different in the two bioreactors. In addition, most of the 16S rRNA gene sequences could not be classified at the family level, indicating most of the species in the PSD and AnMBR are less well-understood. Thus, comparative metagenomics was carried out to explore the divergence of the predicted microbial metabolism during methane generation in these two bioreactors. Additionally, MAGs were reconstituted to reveal the interactions of the syntrophic consortium during methane generation. Large data sets of metagenomic DNA were produced for the PSD and AnMBR samples. We generated a total of 35.9 million 2 × 250 bp paired-end reads, with 10.34 G and 7.27 G nucleotide bases retained after quality filtering for the PSD and AnMBR samples, respectively.
The metabolic potential of the microbial communities of the PSD and AnMBR was predicted by comparison to the KEGG Ortholog (KO) database. In the PSD and AnMBR metagenomes, genes encoding metabolic functions were dominant, representing 20.22% and 22.79% within each sample, respectively, followed by genetic information processing (4.56% in the PSD and 5.07% in the AnMBR) and environmental information processing (3.30% in the PSD and 4.25% in the AnMBR) (Figure S2). In the category of metabolism, the most abundant metabolic type was carbohydrate metabolism (4.28% in the PSD and 4.82% in the AnMBR), followed by amino acid metabolism (4.19% in the PSD and 4.73% in the AnMBR) as well as energy metabolism (3.15% in the PSD and 3.35% in the AnMBR) (Figure S2). A high proportion of genes related to these functions has been previously reported in metagenomes [49,50,51], metatranscriptomes [52] and metaproteomes [53] from AD systems, indicating these metabolic activities are linked to the conversion of excess carbohydrates into methane during anaerobic digestion. When classified as KEGG level 3, the abundance of genes in methane metabolism was one of the categories with the highest abundance in the PSD and AnMBR, consisting of 1.05% and 0.88% of the total sequences (Table S5), respectively, suggesting methanogenesis was the major microbial metabolism in these two bioreactors.
To better understand the difference in carbohydrate decomposition in the PSD and AnMBR, we screened our metagenome for reads annotated as CAZyme. The PSD metagenome contained 864,945 reads (consisted of 4.17% of the total reads) associated with CAZyme that were unequally distributed among glycosyl transferases (GTs), glycoside hydrolases (GHs), carbohydrate binding modules (CBMs), carbohydrate esterases (CEs), auxiliary activities (AAs), and polysaccharide lyases (PLs) (Table S6), and their read abundances were 41.3%, 37.2%, 12.8%, 5.3%, 2.6%, and 0.8%, respectively (Figure S3). In the AnMBR metagenome, the read abundances of GTs, GHs, CBMs, CEs, AAs, and PLs were 43.4%, 37.8%, 12.0%, 4.4%, 2.6%, and 0.8%, respectively (Figure S3). A higher gene abundance of CAZyme suggests a greater role of carbohydrate metabolism in the anaerobic digestion of the PSD and AnMBR. GTs and GHs had higher percentages in terms of total reads, indicating glycosyl transferases and glycoside hydrolases play key roles in the cleavage of polymeric substrates in DWTS. The PSD metagenomes recorded a diverse profile of carbohydrate pathways for a total of 252 GHs, 85 GTs, 67 PLs, 16 CEs, 64 CBMs, and 20 AAs families, comparable to 254, 88, 66, 16, 69, and 21 in the AnMBR metagenome, respectively (Table S6). Approximately 357,389 and 321,643 reads were assigned to different GT and GH families in the PSD compared with 304,909 GTs and 258,444 GHs in the AnMBR metagenome (Table S6), implying their common abilities to form and hydrolyze glycosidic bonds when breaking down polysaccharides in the DWTS [54].
3.4. Phylogenetics of MAGs and Their Abundance in the PSD and AnMBR
A large fraction of the reads were assembled in contigs ≥1000 bp (114,558 contigs including 396,835,232 bp), yielding an N50 (defined as the minimum contig length needed to cover 50% of the total lengths of cotnigs) of 4676 with a max contig length of 323,550 and a mean size of contigs of 2139 bp. To further examine the metabolic properties of the dominant species in the PSD and AnMBR, metagenomic binning based on composition and differential coverage of contigs was performed to reconstruct the MAGs of the species that dominated these processes. Thereby, 21 MAGs (16 and 7 affiliated with Bacteria and Archaea, respectively) were observed and assigned to 15 categories, with an average completeness of 93.0% and contamination of 2.9% (Table 2). The genome sizes of the MAGs ranged from 1,551,831 bp to 5,015,715 bp, with an average of 2,650,305 bp, and the number of contigs in each MAG ranged from 142 to 629, with an average N50 size of 12,914 and mean contig length of 8695 bp (Table 2).

Table 2.
Taxonomic assignment and basic genome characteristics of the curated MAGs reconstructed from the PSD and AnMBR metagenomes a.
A detailed overview of the taxonomic classification of the MAGs is provided in Figure 3 and Table 2, in which a taxonomic classification with high confidence was possible to the species level for all the MAGs. In summary, 7 MAGs were attributed to the phylum Euryarchaeota, including 4 for Methanobacterium sp. (MAG13, MAG14, MAG16, and MAG19), 1 for Methanomassiliicoccales archaea (MAG215), 1 for Methanosaeta concilii (MAG220), and 1 for Methanomicrobiales archaea (MAG11). The second most abundant taxonomic MAGs classification was Bacteroidales bacterium, with 4 observed MAGs (MAG158, MAG217, MAG228, and MAG42). Furthermore, Synergistaceae bacterium with only 1 observed MAG (MAG200) belonging to the family Firmicutes was recruited from the metagenomic dataset. The remaining MAGs were mainly affiliated with the Aminiphilus circumscriptus (MAG74), Desulfovibrio desulfuricans (MAG68), Fusobacteriaceae bacterium (MAG142), Nitrospirae bacterium (MAG161), Prolixibacteraceae bacterium (MAG59), Succinispira mobilis (MAG159), and Sulfurovum sp. (MAG55) (Figure 3). The relationships between the MAGs and their closest species with complete genome sequences are shown in Figure 3. Most of the recruited MAGs belonged to Methanogenic archaea. MAG13, MAG14, MAG16, and MAG19 were most familiar with the genome of Methanobacterium sp. SMA-27, for which no information regarding its origin is publicly available. The sequences of MAG215 had high similarity with Methanomassiliicoccales archaeon RumEn M2, which has been reported to be a trimethylamine-utilizing archaea [55]. The closest genome affiliated with MAG11 was Methanospirillum hungatei JF-1, whereas the genome of MAG220 was affiliated with Methanosaeta harundinacea 6Ac. During the degradation of complex organic matter to carbon dioxide and methane in anaerobic environments, M. hungatei uses hydrogen or formate to produce methane, which are produced by other syntrophic microbes [56]. M. harundinacea is an acetate-scavenging methanogen isolated from a upflow anaerobic sludge blanket reactor [57]. A Bacteroidetes endosymbiont of Geopemphigus sp. exhibited the highest relationship with MAG158, MAG217, MAG228, MAG42, and MAG83. Prolixibacteraceae bacterium XSD2, Fusobacteriaceae bacterium, Nitrospirae bacterium HCH-1, and Desulfovibrio cuneatus DSM 11,391 were most similar with the genomes of MAG59, MAG142, MAG161, and MAG68, respectively. D. cuneatus is a psychrotolerant sulfate-reducing bacterium isolated from an oxic freshwater sediment [58]. In addition, MAG55, MAG100, MAG74, MAG200, and MAG159 showed the highest similarity with Sulfurovum sp. NBC37-1, Succinatimonas hippei YIT 12066, Aminiphilus circumscriptus DSM 16,581, Synergistaceae bacterium, and Succinispira mobilis DSM 6222, respectively. Sulfurovum sp. NBC37-1 is a chemolithoautotroph deep-sea Epsilonproteobacteria that obtains energy through mesophilic hydrogen- and sulfur-oxidation [59]. S. hippei YIT 12066T was isolated from human feces, and the growth of S. hippei YIT 12,066 T depends on CO2 or bicarbonate [60]. A. circumscriptus is an anaerobic amino-acid-degrading bacterium from an upflow anaerobic sludge reactor [61], and S. mobilis was proven to be a succinate-decarboxylating anaerobic bacterium in a mixed culture growing with glycolate [62].
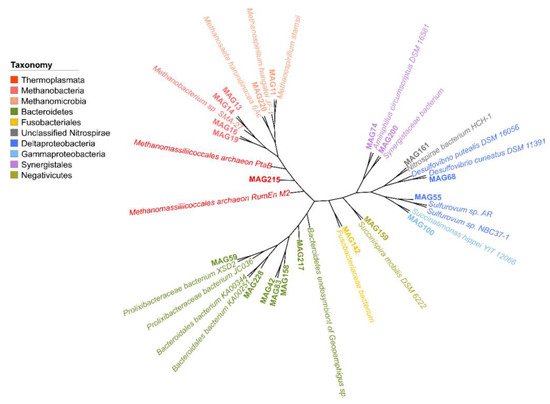
Figure 3.
Taxonomic assignment of the curated MAGs reconstructed from the PSD and AnMBR metagenomes. The lineages of the MAGs and selected related organisms are shown in different colors.
To estimate the relative abundance of the MAGs represented in the PSD and AnMBR, all the quality-filtered reads from each sample were aligned to the contigs from each MAG with Bowtie2 software. As shown in Figure 3, the relative abundance of the MAGs exhibited a large divergence between the PSD and AnMBR. For example, the highest coverage was observed in MAG16 (affiliated with Methanobacterium sp., with 5.27% reads mapped on the corresponding contigs in PSD), followed by MAG200 (affiliated with Synergistaceae bacterium, 2.79%), MAG13 (affiliated with Methanobacterium sp.), MAG74 (affiliated with A. circumscriptus) and MAG220 (affiliated with M. concilii), which were the other predominant species in the PSD, with abundances of 1.42%, 1.18%, and 0.96%, respectively. In the AnMBR metagenome, MAG83 (classified as Bacteroidetes bacterium), MAG200 (Synergistaceae bacterium), MAG217 (Bacteroidetes bacterium), MAG42 (Bacteroidales bacterium), and MAG59 (Prolixibacteraceae bacterium) were the most abundant species, with relative abundances of 4.74%, 2.93%, 2.85%, 1.59%, and 1.36%, respectively (Figure 3). Methanobacteriaceae is capable of directly converting gaseous hydrogen and carbon dioxide into methane [63], whereas microorganisms belonging to Synergistaceae family (Aminobacterium thunnarium, Aminobacterium colombiense, and Aminivibrio pyruvatiphilus) are certified as syntrophic acetate oxidizers [64,65]. This result may also derive from the functional difference between the PSD and AnMBR in the DWTS, in which wastewater was solid-liquid separated, and organic matter was degraded in the AnMBR, followed by methanogenesis in the sludge of PSD bioreactors.
3.5. Microbial Interaction and Methane-Producing Pathways of the Dominant Species in the PSD and AnMBR
To better understand the predicted function carried out by each MAG of the draft genome, genes were annotated through BlastX analyses against the KEGG and CAZy databases. The number of predicted ORFs in MAGs ranged from 1658 to 4635 with an average of 2648 (Table 2). On average, 50.8% (ranged from 36.4% to 63.7%) and 2.44% (ranged from 0.74% to 8.66%) of the ORFs were annotated in the KEGG and CAZy databases (Table S7). Genes associated with carbohydrate-active and methanogenic metabolism were recruited from each MAG, benefiting the prediction of the functions carried out by the predominant species during methane generation. The abundance of CAZyme-encoding genes varied across the MAGs. MAG59 (classified as Prolixibacteraceae bacterium), MAG158 (Bacteroidetes bacterium) and MAG127 (Bacteroidetes bacterium) had the most abundant CAZy genes, with total numbers of 269, 232, and 103, respectively. In carbohydrate-active modules, MAG59 had the highest abundance of genes encoding GHs, GTs, and PLs, with gene numbers of 191, 46, and 10 (Figure 4), respectively, providing genetic evidence for the high cellulolytic and transglycosylatic abilities of Prolixibacteraceae bacterium. The most abundant genes of CEs and CBMs were identified in MAG158. The above results suggest that polysaccharides within the DWTS habitat are hydrolyzed by the predominant Bacteroidetes bacterium and Prolixibacteraceae bacterium.
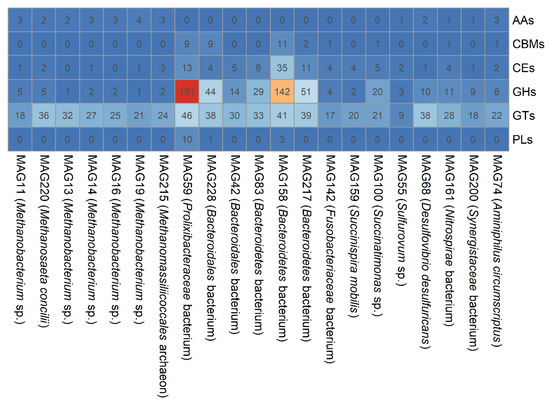
Figure 4.
Numbers of genes associated with CAZyme in bacterial and archaeal MAGs.
Methanogenic archaea can be classified into seven well-established orders: Methanobacteriales, Methanococcales, Methanomicrobiales, Methanosarcinales, Methanocellales, Methanopyrales, and Methanomassiliicoccales [66,67]. In this study, MAGs associated with Methanobacterium, Methanomicrobiales, Methanomassiliicoccales, and Methanosaeta were recruited, indicating the metabolic versatility of the methanogenesis pathway in the PSD and AnMBR of the DWTS. The annotated results of the methanogenesis genes in archaeal MGAs indicated three types of methanogenic pathways: (1) hydrogenotrophic (Figure 5), (2) acetotrophic (Figure S4), and (3) methylotrophic (Figure S5). For hydrogenotrophic methanogenesis, carbon dioxide is successively reduced to methane through a series of intermediates with a methyl group. The methyl group is then transferred to Coenzyme M, forming methyl-CoM. Thus, methyl-CoM is reduced to methane through methyl coenzyme M reductase (Mcr) at the final step. Genes encoding enzymes that participate in hydrogenotrophic methanogenesis had the highest abundance in the archaeal MAGs, followed by methylotrophic and acetotrophic methanogenesis (Figure 5, Figures S4 and S5). Generally, the genes involved in the hydrogenotrophic methanogenesis pathway had higher abundance in archaea MAGs (Figure 5), especially for formylmethanofuran dehydrogenase (K00200-K00203) and tetrahydromethanopterin S-methyltransferase (K00577-K00584). The genes of MAG14 and MAG16 (both classified as Methanobacterium sp.) included almost all the enzymes in the completely hydrogenotrophic pathway, indicating Methanobacterium-associated archaea tended to convert carbon dioxide to methane using hydrogen, which has been proven by genomic and physiological data [68,69]. It was interesting that the genes for complete typical hydrogenotrophic pathway were not detected in MAG215 (classified as Methanomassiliicoccales archaeon), while MAG215 harbored a high abundance of genes involved in methylotrophic methanogenesis; both of these results suggest that Methanomassiliicoccales archaea produced methane from methanol, in agreement with genomic insights into the methylotrophic lifestyle of Methanomassiliicoccales [70]. The acetotrophic pathway is known to be the major pathway for more than 70% of methane production in most engineering anaerobic digestion processes [71]. In the acetotrophic methanogenesis pathway (Figure S4), genes encoding acetyl-CoA synthetase (K01895) and acetyl-CoA decarbonylase/synthase (K00193, K00194, K00197) are present in all archaeal MAGs and are involved in acetate oxidation [72]. However, genes encoding phosphate acetyltransferase (K00625 and K13788) and acetate kinase (K00925), which transform CoA into acetyl-CoA and acetate into acetyl phosphate, respectively, were absent. Furthermore, no archaeal MAGs harbored all the genes encoding enzymes in the complete pathway of acetotrophic methanogenesis, suggesting the predominant pathway for methane generation was not through acetate conversion in the PSD and AnMBR bioreactors. In the methylotrophic pathway (Figure S5), almost all the archaeal MAGs (except MAG11, classified as Methanomicrobiales archaeon) had genes encoding the three subunits of methyl-coenzyme M reductase, which is the key enzyme of biological methane formation [73]. However, all the subunits of heterodisulfide reductase have not been identified in all of these archaea MAGs, posing a question regarding the ability of the affiliated archaea to convert methanol to methane. However, it should be noted that the methanogenesis pathway was predicted based on the abundance of genes in MAGs, rather than metatranscriptomics or metaproteomics, which are required to further explore the active functions involved in the methanogenesis pathway in future studies.
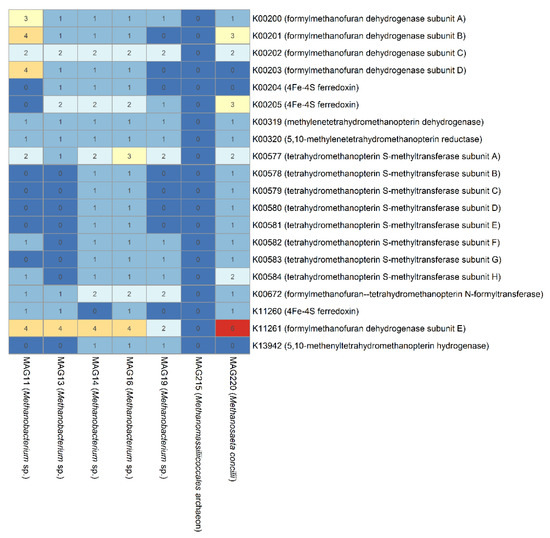
Figure 5.
Numbers of genes involved in the relevant hydrogenotrophic methanogenesis pathway in archaeal MAGs.
Various microorganisms are involved in and cooperate with each other to achieve methane formation in anaerobic digestion. According to the genome binning presented in this work, members of Bacteroidetes and Prolixibacteraceae were predicted to hydrolyze polysaccharide to acetate. Subsequently, Synergistaceae may oxidize acetate to hydrogen and carbon dioxide. Finally, Methanobacterium sp. was possible to reduced carbon dioxide to methane with hydrogen as the primary electron donor. Furthermore, sulfate was likely to be removed through desulfurization with the help of Desulfovibrio desulfuricans (Figure 6).
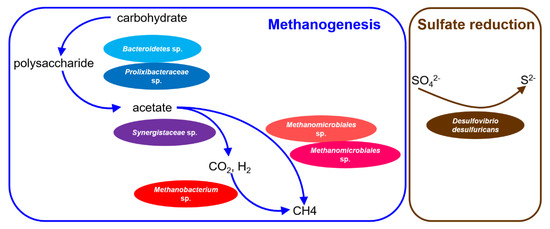
Figure 6.
Conceptual graph of the microbial network and dominant metabolic pathways of the microbial communities in the DWTS.
4. Conclusions
The in-depth 16S rRNA gene amplicon and metagenome information obtained in this study contribute to the understanding of the taxonomies and pathways involved in methanogenesis in the two types of bioreactors in the DWTS. The results obtained in this study showed that microbial composition was significantly different between the PSD and AnMBR. High-quality MAGs of predominated species were recruited, and the results of an in-depth dissection of the genes in the MAGs suggested a strong interaction of bacteria and archaea during methanogenesis that mainly occurs through the interspecific exchange of substances. The valuable insights obtained in this study are expected to provide guidance for the currently planned start-up of decentralized wastewater treatment systems that contain a primary sludge digestor and membrane bioreactor.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3417/10/1/135/s1. Table S1, physicochemical characteristics of the influent and effluent water; Table S2, sequence numbers and alpha diversity indices of 16S rRNA genes in the PSD and AnMBR samples; Table S3, differences of the alpha diversity indices of the 16S rRNA genes between the PSD and AnMBR samples; Table S4, taxonomic profiling of the 16S rRNA genes at the family level in the PSD and AnMBR; Table S5, KEGG categories of the PSD and AnMBR metagenomes; Table S6, number of reads annotated as genes affiliated with CAZyme in the PSD and AnMBR metagenomes; Table S7, rate of ORFs in MAGs annotated by the KEGG and CAZy databases; Figure S1, weighted PCoA clustering of the microbial communities in the PSD (blue triangles) and AnMBR (red triangles) samples based on the UniFrac distance; Figure S2, relative abundances of the KEGG categories of functional genes in the PSD and AnMBR metagenomes; Figure S3, read abundance of CAZyme in the PSD and AnMBR metagenomes; Figure S4, numbers of genes involved in the relevant acetotrophic methanogenesis pathway in archaeal MAGs; Figure S5, numbers of genes involved in the relevant methylotrophic methanogenesis pathway in archaeal MAGs; Figure S6, the relative abundances of Syntrophaceae and Methanobacteriaceae in PSD and AnMBR.
Author Contributions
Developed and framed research questions, K.Z., Y.-L.Z., J.-T.L., J.-L.L., and J.-J.L.; Conducted the experiments, K.Z., Y.-L.Z., X.O., J.-P.L., A.Y., and G.-J.X.; Performed the data analyses, J.-L.L. and X.Y.; Wrote the first draft of the manuscript, K.Z., Y.-L.Z., and J.-L.L.; all authors revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Provincial Natural Science Foundation of Guangdong, China (No. 2018A0303130014) and the project of Guangdong Provincial Education Department (No.2018GKTSCX091, No.2017GKQNCX004), the Excellent Youth Foundation of Guangdong Industry Polytechnic (QN2018-003), the Project of Guangzhou Science and Technology Association (K2019070101009).
Acknowledgments
The authors are grateful to ECOGENE BIOTECH CO., LTD. (Shenzhen, China) for the support on bioinformatics of 16S rRNA amplicon and metagenome data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Appels, L.; Baeyens, J.; Degrève, J.; Dewil, R. Principles and potential of the anaerobic digestion of waste-activated sludge. Prog. Energy. Combust. Sci. 2008, 34, 755–781. [Google Scholar] [CrossRef]
- Miron, Y.; Zeeman, G.; van Lier, J.B.; Lettinga, G. The role of sludge retention time in the hydrolysis and acidification of lipids, carbohydrates and proteins during digestion of primary sludge in CSTR systems. Water Res. 2000, 34, 1705–1713. [Google Scholar] [CrossRef]
- Smith, S.R.; Lang, N.L.; Cheung, K.H.M.; Spanoudaki, K. Factors controlling pathogen destruction during anaerobic digestion of biowastes. Waste Manag. 2005, 25, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Dvořák, L.; Gómez, M.; Dolina, J.; Černín, A. Anaerobic membrane bioreactors—A mini review with emphasis on industrial wastewater treatment: Applications, limitations and perspectives. Desalin. Water Treat. 2016, 57, 19062–19076. [Google Scholar] [CrossRef]
- Dereli, R.K.; Ersahin, M.E.; Ozgun, H.; Ozturk, I.; Jeison, D.; van der Zee, F.; van Lier, J.B. Potentials of anaerobic membrane bioreactors to overcome treatment limitations induced by industrial wastewaters. Bioresour. Technol. 2012, 122, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.J.; Peng, W.; Zhang, M.J.; Chen, J.R.; Hong, H.C.; Zhang, Y. A review on anaerobic membrane bioreactors: Applications, membrane fouling and future perspectives. Desalination 2013, 314, 169–188. [Google Scholar] [CrossRef]
- Kim, E.; Lee, J.; Han, G.; Hwang, S. Comprehensive analysis of microbial communities in full-scale mesophilic and thermophilic anaerobic digesters treating food waste-recycling wastewater. Bioresour. Technol. 2018, 259, 442–450. [Google Scholar] [CrossRef]
- Carabeo-Pérez, A.; Guerra-Rivera, G.; Ramos-Leal, M.; Klocke, M.; Jiménez-Hernández, J. Metagenomic approaches: Effective tools for monitoring the structure and functionality of microbiomes in anaerobic digestion systems. Appl. Microbiol. Biotechnol. 2019. [Google Scholar] [CrossRef]
- Vanwonterghem, I.; Jensen, P.D.; Ho, D.P.; Batstone, D.J.; Tyson, G.W. Linking microbial community structure, interactions and function in anaerobic digesters using new molecular techniques. Curr. Opin. Biotechnol. 2014, 27, 55–64. [Google Scholar] [CrossRef]
- Vanwonterghem, I.; Jensen, P.D.; Rabaey, K.; Tyson, G.W. Genome-centric resolution of microbial diversity, metabolism and interactions in anaerobic digestion. Environ. Microbiol. 2016, 18, 3144–3158. [Google Scholar] [CrossRef]
- Ali, N.; Gong, H.; Liu, X.; Giwa, A.S.; Wang, K. Evaluation of bacterial association in methane generation pathways of an anaerobic digesting sludge via metagenomic sequencing. Arch. Microbiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yu, K.; Xia, Y.; Lau, F.T.K.; Tang, D.T.W.; Fung, W.C.; Fang, H.H.P.; Zhang, T. Metagenomic analysis of sludge from full-scale anaerobic digesters operated in municipal wastewater treatment plants. Appl. Microbiol. Biotechnol. 2014, 98, 5709–5718. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, C.; Al-Soud, W.A.; Larsson, M.; Alm, E.; Yekta, S.S.; Svensson, B.H.; Sørensen, S.J.; Karlsson, A. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol. Ecol. 2013, 85, 612–626. [Google Scholar] [CrossRef] [PubMed]
- Rui, J.P.; Li, J.B.; Zhang, S.H.; Yan, X.F.; Wang, Y.P.; Li, X.Z. The core populations and co-occurrence patterns of prokaryotic communities in household biogas digesters. Biotechnol. Biofuels 2015, 8, 158. [Google Scholar] [CrossRef] [PubMed]
- Bialek, K.; Kim, J.; Lee, C.; Collins, G.; Mahony, T.; O’Flaherty, V. Quantitative and qualitative analyses of methanogenic community development in high-rate anaerobic bioreactors. Water Res. 2011, 45, 1298–1308. [Google Scholar] [CrossRef] [PubMed]
- Campanaro, S.; Treu, L.; Kougias, P.G.; De Francisci, D.; Valle, G.; Angelidaki, I. Metagenomic analysis and functional characterization of the biogas microbiome using high throughput shotgun sequencing and a novel binning strategy. Biotechnol. Biofuels 2016, 9, 26. [Google Scholar] [CrossRef]
- Schlüter, A.; Bekel, T.; Diaz, N.N.; Dondrup, M.; Eichenlaub, R.; Gartemann, K.-H.; Krahn, I.; Krause, L.; Krömeke, H.; Kruse, O.; et al. The metagenome of a biogas-producing microbial community of a production-scale biogas plant fermenter analysed by the 454-pyrosequencing technology. J. Biotechnol. 2008, 136, 77–90. [Google Scholar] [CrossRef]
- Wirth, R.; Kovács, E.; Maróti, G.; Bagi, Z.; Rákhely, G.; Kovács, K.L. Characterization of a biogas-producing microbial community by short-read next generation DNA sequencing. Biotechnol. Biofuels 2012, 5, 41. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, X.X.; Huang, K.L.; Miao, Y.; Shi, P.; Liu, B.; Long, C.; Li, A.M. Metagenomic profiling of antibiotic resistance genes and mobile genetic elements in a tannery wastewater treatment plant. PLoS ONE 2013, 8, e76079. [Google Scholar] [CrossRef]
- Wong, M.T.; Zhang, D.; Li, J.; Hui, R.K.H.; Tun, H.M.; Brar, M.S.; Park, T.J.; Chen, Y.; Leung, F.C. Towards a metagenomic understanding on enhanced biomethane production from waste activated sludge after pH 10 pretreatment. Biotechnol. Biofuels 2013, 6, 38. [Google Scholar] [CrossRef]
- Li, A.; Chu, Y.N.; Wang, X.M.; Ren, L.F.; Yu, J.; Liu, X.L.; Yan, J.B.; Zhang, L.; Wu, S.X.; Li, S.Z. A pyrosequencing-based metagenomic study of methane-producing microbial community in solid-state biogas reactor. Biotechnol. Biofuels 2013, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- McLeod, J.D.; Othman, M.Z.; Beale, D.J.; Joshi, D. The use of laboratory scale reactors to predict sensitivity to changes in operating conditions for full-scale anaerobic digestion treating municipal sewage sludge. Bioresour. Technol. 2015, 189, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Federation, W.E.; Association, A.P.H. Standard Methods for the Examination of Water and Wastewater; American Public Health Association (APHA): Washington, DC, USA, 2005. [Google Scholar]
- Bates, S.T.; Berg-Lyons, D.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011, 5, 908. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010, 7, 335. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2009, 26, 266–267. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2008, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Leung, H.C.; Yiu, S.M.; Chin, F.Y. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012, 9, 357. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Alneberg, J.; Bjarnason, B.S.; de Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Loman, N.J.; Andersson, A.F.; Quince, C. CONCOCT: Clustering contigs on coverage and composition. arXiv 2013, arXiv:1312.4038. [Google Scholar]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Segata, N.; Börnigen, D.; Morgan, X.C.; Huttenhower, C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 2013, 4, 2304. [Google Scholar] [CrossRef]
- Huson, D.H.; Auch, A.F.; Qi, J.; Schuster, S.C. MEGAN analysis of metagenomic data. Genome Res. 2007, 17, 377–386. [Google Scholar] [CrossRef]
- Rho, M.; Tang, H.X.; Ye, Y.Z. FragGeneScan: Predicting genes in short and error-prone reads. Nucleic Acids Res. 2010, 38, e191. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Shaw, G.T.W.; Liu, A.C.; Weng, C.Y.; Chou, C.Y.; Wang, D. Inferring microbial interactions in thermophilic and mesophilic anaerobic digestion of hog waste. PLoS ONE 2017, 12, e0181395. [Google Scholar] [CrossRef] [PubMed]
- De Vrieze, J.; Saunders, A.M.; He, Y.; Fang, J.; Nielsen, P.H.; Verstraete, W.; Boon, N. Ammonia and temperature determine potential clustering in the anaerobic digestion microbiome. Water Res. 2015, 75, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Kotsyurbenko, O.R.; Friedrich, M.W.; Simankova, M.V.; Nozhevnikova, A.N.; Golyshin, P.N.; Timmis, K.N.; Conrad, R. Shift from acetoclastic to H2-dependent methanogenesis in a west siberian peat bog at low pH values and isolation of an acidophilic Methanobacterium strain. Appl. Environ. Microbiol. 2007, 73, 2344–2348. [Google Scholar] [CrossRef] [PubMed]
- Westerholm, M.; Roos, S.; Schnürer, A. Syntrophaceticus schinkii gen. nov., sp. nov., an anaerobic, syntrophic acetate-oxidizing bacterium isolated from a mesophilic anaerobic filter. FEMS Microbiol. Lett. 2010, 309, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Gray, N.D.; Sherry, A.; Grant, R.J.; Rowan, A.K.; Hubert, C.R.J.; Callbeck, C.M.; Aitken, C.M.; Jones, D.M.; Adams, J.J.; Larter, S.R.; et al. The quantitative significance of Syntrophaceae and syntrophic partnerships in methanogenic degradation of crude oil alkanes. Environ. Microbiol. 2011, 13, 2957–2975. [Google Scholar] [CrossRef]
- Cheng, L.; Ding, C.; Li, Q.; He, Q.; Dai, L.R.; Zhang, H. DNA-SIP reveals that Syntrophaceae play an important role in methanogenic hexadecane degradation. PLoS ONE 2013, 8, e66784. [Google Scholar] [CrossRef]
- Lei, Y.Q.; Sun, D.Z.; Dang, Y.; Feng, X.L.; Huo, D.; Liu, C.Q.; Zheng, K.; Holmes, D.E. Metagenomic analysis reveals that activated carbon aids anaerobic digestion of raw incineration leachate by promoting direct interspecies electron transfer. Water Res. 2019, 161, 570–580. [Google Scholar] [CrossRef]
- Ziels, R.M.; Sousa, D.Z.; Stensel, H.D.; Beck, D.A.C. DNA-SIP based genome-centric metagenomics identifies key long-chain fatty acid-degrading populations in anaerobic digesters with different feeding frequencies. ISME J. 2018, 12, 112–123. [Google Scholar] [CrossRef]
- Delforno, T.P.; Lacerda, G.V.; Sierra-Garcia, I.N.; Okada, D.Y.; Macedo, T.Z.; Varesche, M.B.A.; Oliveira, V.M. Metagenomic analysis of the microbiome in three different bioreactor configurations applied to commercial laundry wastewater treatment. Sci. Total Environ. 2017, 587–588, 389–398. [Google Scholar] [CrossRef]
- Delforno, T.P.; Macedo, T.Z.; Midoux, C.; Lacerda, G.V.; Rué, O.; Mariadassou, M.; Loux, V.; Varesche, M.B.A.; Bouchez, T.; Bize, A.; et al. Comparative metatranscriptomic analysis of anaerobic digesters treating anionic surfactant contaminated wastewater. Sci. Total Environ. 2019, 649, 482–494. [Google Scholar] [CrossRef] [PubMed]
- Hassa, J.; Maus, I.; Off, S.; Pühler, A.; Scherer, P.; Klocke, M.; Schlüter, A. Metagenome, metatranscriptome, and metaproteome approaches unraveled compositions and functional relationships of microbial communities residing in biogas plants. Appl. Microbiol. Biotechnol. 2018, 102, 504–5063. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B.; Bairoch, A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1993, 293, 781–788. [Google Scholar] [CrossRef]
- Söllinger, A.; Schwab, C.; Weinmaier, T.; Loy, A.; Tveit, A.T.; Schleper, C.; Urich, T. Phylogenetic and genomic analysis of Methanomassiliicoccales in wetlands and animal intestinal tracts reveals clade-specific habitat preferences. FEMS Microbiol. Ecol. 2015, 92, fiv149. [Google Scholar] [CrossRef]
- Worm, P.; Stams, A.J.M.; Cheng, X.; Plugge, C.M. Growth- and substrate-dependent transcription of formate dehydrogenase and hydrogenase coding genes in Syntrophobacter fumaroxidans and Methanospirillum hungatei. Microbiology 2011, 157, 280–289. [Google Scholar] [CrossRef]
- Ma, K.; Liu, X.L.; Dong, X.Z. Methanosaeta harundinacea sp. nov., a novel acetate-scavenging methanogen isolated from a UASB reactor. Int. J. Syst. Evol. Microbiol. 2006, 56, 127–131. [Google Scholar] [CrossRef]
- Sass, H.; Berchtold, M.; Branke, J.; König, H.; Cypionka, H.; Babenzien, H.-D. Psychrotolerant sulfate-reducing bacteria from an oxic freshwater sediment description of Desulfovibrio cuneatus sp. nov. and Desulfovibrio litoralis sp. nov. Syst. Appl. Microbiol. 1998, 21, 212–219. [Google Scholar] [CrossRef]
- Yamamoto, M.; Nakagawa, S.; Shimamura, S.; Takai, K.; Horikoshi, K. Molecular characterization of inorganic sulfur-compound metabolism in the deep-sea epsilonproteobacterium Sulfurovum sp. NBC37-1. Environ. Microbiol. 2010, 12, 1144–1153. [Google Scholar] [CrossRef]
- Morotomi, M.; Nagai, F.; Watanabe, Y. CO2-dependent growth of Succinatimonas hippei YIT 12066T isolated from human feces. Microbiol. Immunol. 2012, 56, 195–197. [Google Scholar] [CrossRef]
- Díaz, C.; Baena, S.; Fardeau, M.L.; Patel, B.K.C. Aminiphilus circumscriptus gen. nov., sp. nov., an anaerobic amino-acid-degrading bacterium from an upflow anaerobic sludge reactor. Int. J. Syst. Evol. Microbiol. 2007, 57, 1914–1918. [Google Scholar] [CrossRef]
- Janssen, P.H.; Farrell, K.A. Succinispira mobilis gen. nov., sp. nov., a succinate-decarboxylating anaerobic bacterium. Int. J. Syst. Evol. Microbiol. 1999, 49, 1009–1013. [Google Scholar] [CrossRef] [PubMed]
- Jee, H.S.; Yano, T.; Nishio, N.; Nagai, S. Biomethanation of H2 and CO2 by Methanobacterium thermoautotrophicum in membrane and ceramic bioreactors. J. Ferment. Technol. 1987, 65, 413–418. [Google Scholar] [CrossRef]
- Militon, C.; Hamdi, O.; Michotey, V.; Fardeau, M.-L.; Ollivier, B.; Bouallagui, H.; Hamdi, M.; Bonin, P. Ecological significance of Synergistetes in the biological treatment of tuna cooking wastewater by an anaerobic sequencing batch reactor. Environ. Sci. Pollut. Res. 2015, 22, 18230–18238. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Wang, H.Z.; Chen, Y.; Gou, M.; Xia, Z.Y.; Hu, B.; Nie, Y.; Tang, Y.Q. Identification of novel butyrate- and acetate-oxidizing bacteria in butyrate-fed mesophilic anaerobic chemostats by DNA-based stable isotope probing. Microb. Ecol. 2019. [Google Scholar] [CrossRef]
- Thauer, R.K.; Kaster, A.-K.; Seedorf, H.; Buckel, W.; Hedderich, R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 2008, 6, 579–591. [Google Scholar] [CrossRef]
- Vanwonterghem, I.; Evans, P.N.; Parks, D.H.; Jensen, P.D.; Woodcroft, B.J.; Hugenholtz, P.; Tyson, G.W. Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat. Microbiol. 2016, 1, 16170. [Google Scholar] [CrossRef]
- Maus, I.; Wibberg, D.; Stantscheff, R.; Cibis, K.; Eikmeyer, F.-G.; König, H.; Pühler, A.; Schlüter, A. Complete genome sequence of the hydrogenotrophic Archaeon Methanobacterium sp. Mb1 isolated from a production-scale biogas plant. J. Biotechnol. 2013, 168, 734–736. [Google Scholar] [CrossRef]
- Kitamura, K.; Fujita, T.; Akada, S.; Tonouchi, A. Methanobacterium kanagiense sp. nov., a hydrogenotrophic methanogen, isolated from rice-field soil. Int. J. Syst. Evol. Microbiol. 2011, 61, 1246–1252. [Google Scholar] [CrossRef]
- Li, Y.; Leahy, S.C.; Jeyanathan, J.; Henderson, G.; Cox, F.; Altermann, E.; Kelly, W.J.; Lambie, S.C.; Janssen, P.H.; Rakonjac, J.; et al. The complete genome sequence of the methanogenic archaeon ISO4-H5 provides insights into the methylotrophic lifestyle of a ruminal representative of the Methanomassiliicoccales. Stand. Genom. Sci. 2016, 11, 59. [Google Scholar] [CrossRef]
- Zhang, L.; Loh, K.-C.; Lim, J.W.; Zhang, J. Bioinformatics analysis of metagenomics data of biogas-producing microbial communities in anaerobic digesters: A review. Renew. Sustain. Energ. Rev. 2019, 100, 110–126. [Google Scholar] [CrossRef]
- Kumari, S.; Tishel, R.; Eisenbach, M.; Wolfe, A.J. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 1995, 177, 2878–2886. [Google Scholar] [CrossRef] [PubMed]
- Ermler, U.; Grabarse, W.; Shima, S.; Goubeaud, M.; Thauer, R.K. Crystal structure of methyl-coenzyme M reductase: The key enzyme of biological methane formation. Science 1997, 278, 1457–1462. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).