Effects of a Bacillus licheniformis Fermentation Extract and Monensin on the Rumen and Hindgut Microbiota Composition of Lactating Dairy Cows
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Experimental Design, and Treatments
2.2. Rumen-Fluid and Feces Collection
Rumen Fluid and Feces Volatile Fatty Acid (VFA) Analyses
2.3. DNA Extraction, 16S rRNA Gene Amplicon Sequencing, and Bioinformatics Analysis
2.4. Calculations and Statistical Analyses
3. Results
3.1. Production Performance
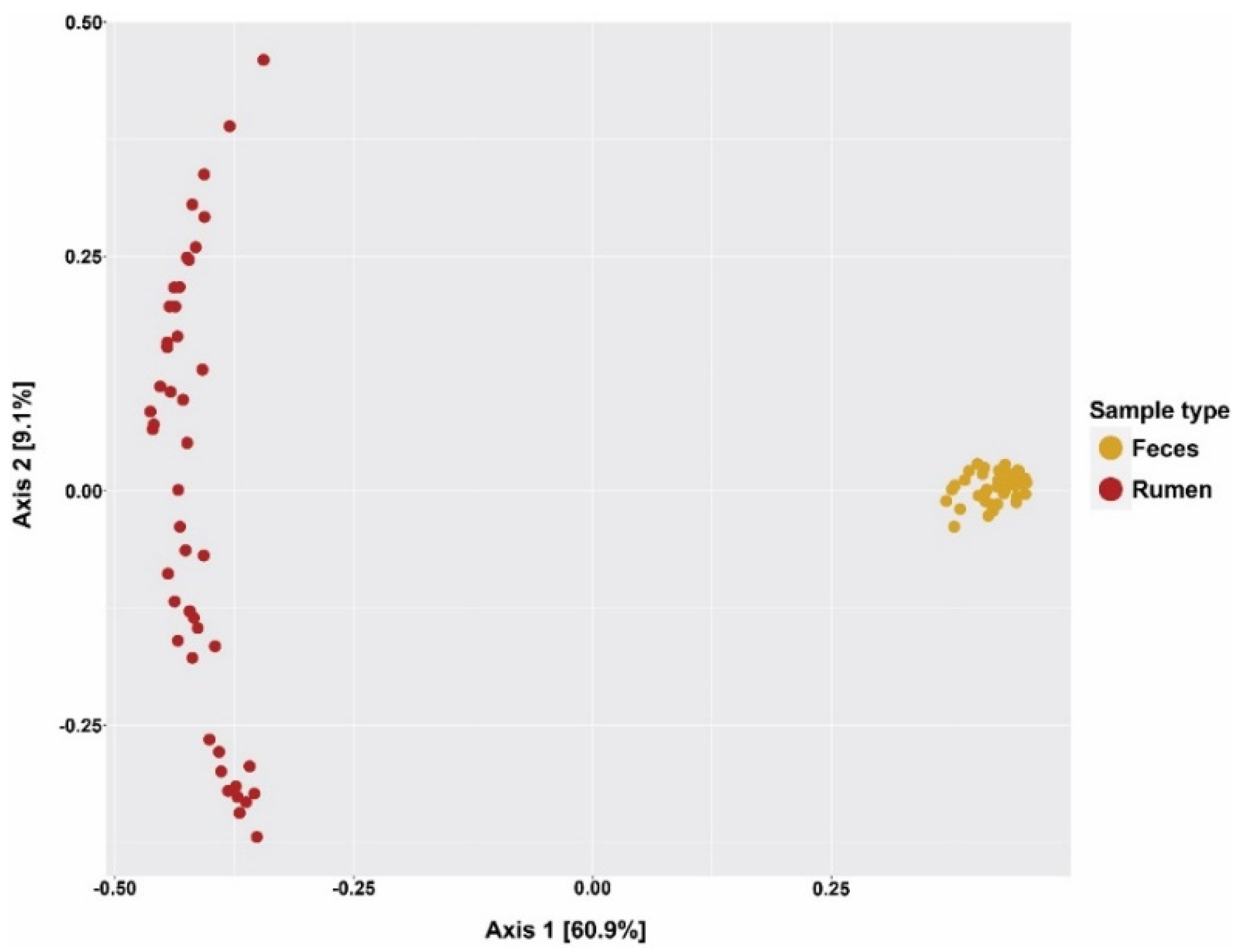
3.2. Alpha and Beta Diversity of Rumen and Fecal Microbial Communities
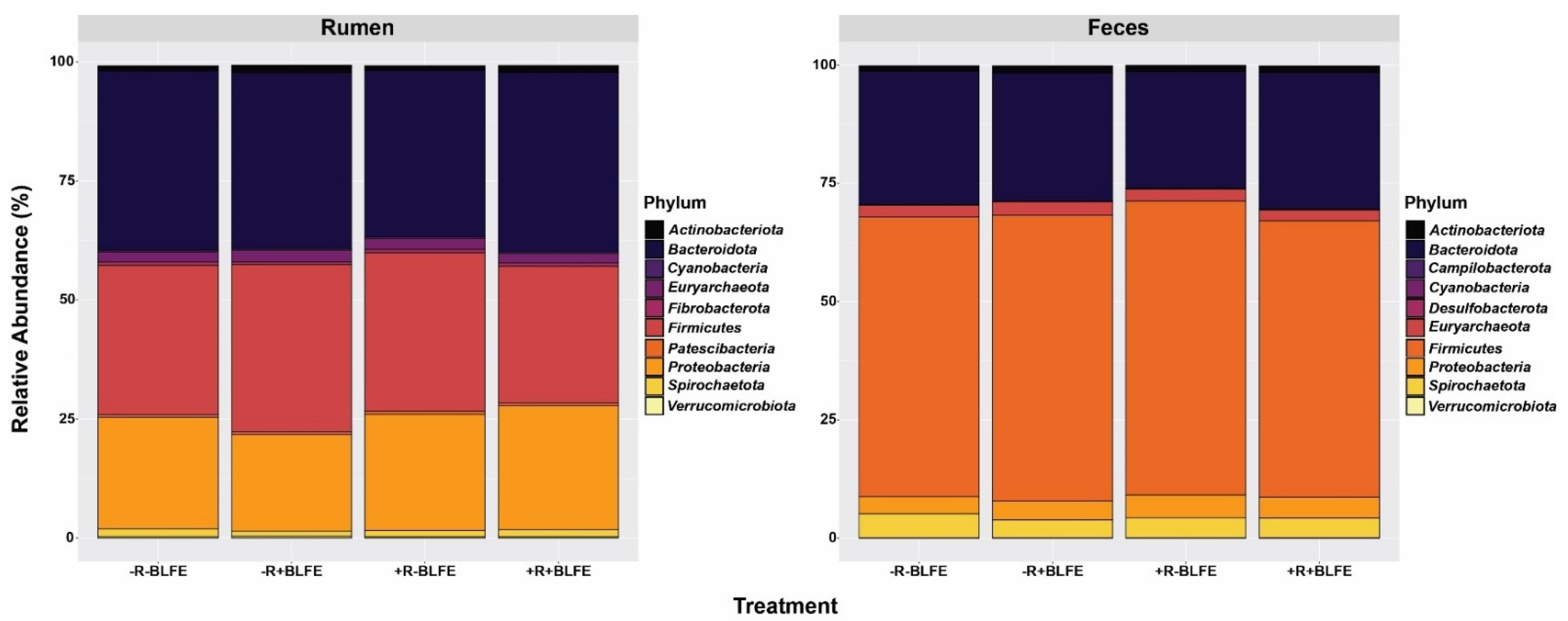
3.3. Kingdom and Phylum Abundance
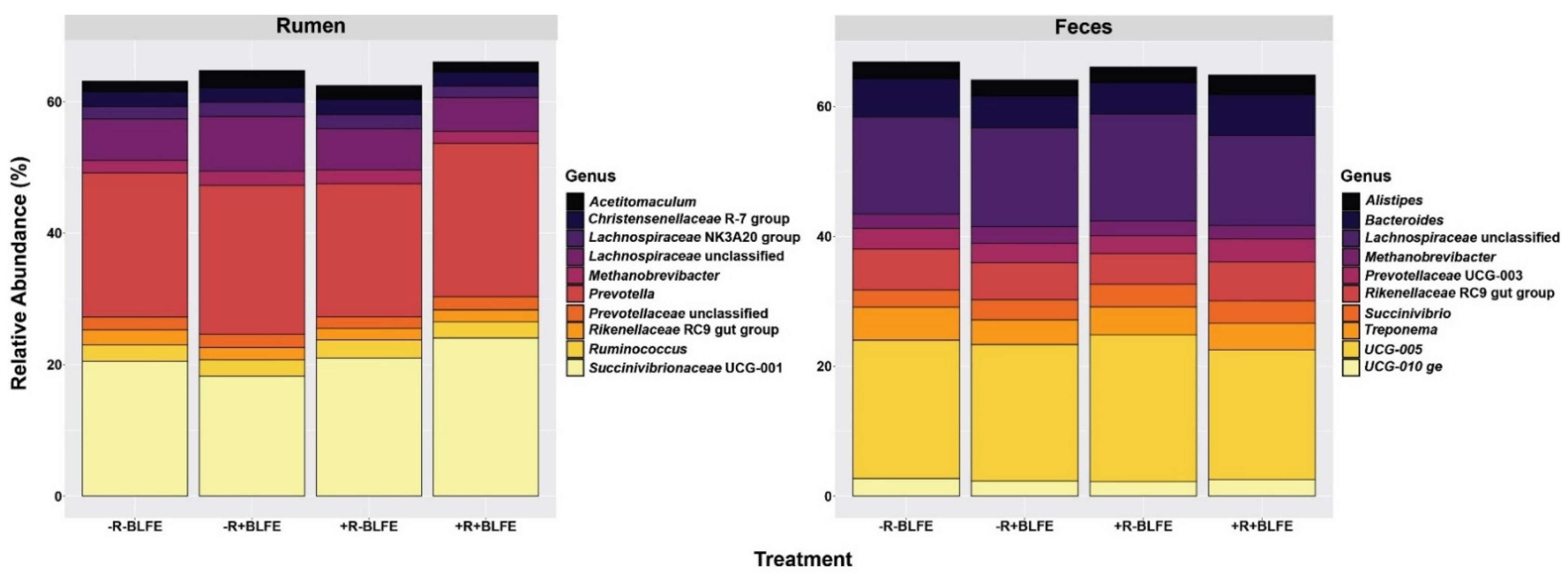
3.4. Relative Abundances of Dominant Genera in Rumen Fluid and Feces
3.5. Treatments’ Effects on Ruminal and Fecal Genera Abundances
3.6. Correlations Between Ruminal and Fecal Genera Abundances and Feed Efficiency
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mizrahi, I.; Jami, E. Review: The compositional variation of the rumen microbiome and its effect on host performance and methane emission. Animal 2018, 12, s220–s232. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, H.F.; Zhou, Z.; Gomes, M.S.; Peixoto, P.M.G.; Bonsaglia, E.C.R.; Canisso, I.F.; Weimer, B.C.; Lima, F.S. Rumen and lower gut microbiomes relationship with feed efficiency and production traits throughout the lactation of Holstein dairy cows. Sci. Rep. 2022, 12, 4904. [Google Scholar] [CrossRef]
- Shabat, S.K.; Sasson, G.; Doron-Faigenboim, A.; Durman, T.; Yaacoby, S.; Berg Miller, M.E.; White, B.A.; Shterzer, N.; Mizrahi, I. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 2016, 10, 2958–2972. [Google Scholar] [CrossRef]
- Li, D.; Wang, P.; Wang, P.P.; Hu, X.S.; Chen, F. Gut microbiota promotes production of aromatic metabolites through degradation of barley leaf fiber. J. Nutr. Biochem. 2018, 58, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Paz, H.A.; Hales, K.E.; Wells, J.E.; Kuehn, L.A.; Freetly, H.C.; Berry, E.D.; Flythe, M.D.; Spangler, M.L.; Fernando, S.C. Rumen bacterial community structure impacts feed efficiency in beef cattle. J. Anim. Sci. 2018, 96, 1045–1058. [Google Scholar] [CrossRef]
- McLoughlin, S.; Spillane, C.; Claffey, N.; Smith, P.E.; O’Rourke, T.; Diskin, M.G.; Waters, S.M. Rumen Microbiome Composition Is Altered in Sheep Divergent in Feed Efficiency. Front. Microbiol. 2020, 11, 01981. [Google Scholar] [CrossRef]
- Zhang, J.; Jin, W.; Jiang, Y.; Xie, F.; Mao, S. Response of Milk Performance, Rumen and Hindgut Microbiome to Dietary Supplementation with Aspergillus oryzae Fermentation Extracts in Dairy Cows. Curr. Microbiol. 2022, 79, 113. [Google Scholar] [CrossRef]
- Carberry, C.A.; Kenny, D.A.; Han, S.; McCabe, M.S.; Waters, S.M. Effect of Phenotypic Residual Feed Intake and Dietary Forage Content on the Rumen Microbial Community of Beef Cattle. Appl. Environ. Microbiol. 2012, 78, 4949–4958. [Google Scholar] [CrossRef]
- Ellison, M.J.; Conant, G.C.; Lamberson, W.R.; Cockrum, R.R.; Austin, K.J.; Rule, D.C.; Cammack, K.M. Diet and feed efficiency status affect rumen microbial profiles of sheep. Small Rumin. Res. 2017, 156, 12–19. [Google Scholar] [CrossRef]
- Costa-Roura, S.; Villalba, D.; Blanco, M.; Casasus, I.; Balcells, J.; Seradj, A.R. Ruminal microbiota is associated with feed-efficiency phenotype of fattening bulls fed high-concentrate diets. Anim. Prod. Sci. 2022, 62, 1344–1352. [Google Scholar] [CrossRef]
- Yang, H.; Huang, X.C.; Fang, S.M.; He, M.Z.; Zhao, Y.Z.; Wu, Z.F.; Yang, M.; Zhang, Z.Y.; Chen, C.Y.; Huang, L.S. Unraveling the Fecal Microbiota and Metagenomic Functional Capacity Associated with Feed Efficiency in Pigs. Front. Microbiol. 2017, 8, 1555. [Google Scholar] [CrossRef]
- Quan, J.P.; Cai, G.Y.; Yang, M.; Zeng, Z.H.; Ding, R.R.; Wang, X.W.; Zhuang, Z.W.; Zhou, S.P.; Li, S.Y.; Yang, H.Q.; et al. Exploring the Fecal Microbial Composition and Metagenomic Functional Capacities Associated with Feed Efficiency in Commercial DLY Pigs. Front. Microbiol. 2019, 10, 52. [Google Scholar] [CrossRef]
- McGuffey, R.K.; Richardson, L.F.; Wilkinson, J. Ionophores for Dairy Cattle: Current Status and Future Outlook. J. Dairy Sci. 2001, 84, E194–E203. [Google Scholar] [CrossRef]
- Horst, E.A.; Kvidera, S.K.; Hagerty, S.; French, P.D.; Carlson, D.B.; Dhuyvetter, K.; Holloway, A.W. Effect of Monensin on Milk Production Efficiency and Milk Composition in Lactating Dairy Cows fed Modern Diets. J. Dairy Sci. 2023, 107, 1441–1449. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.B.; Strobel, H.J. Effect of Ionophores on Ruminal Fermentation. Appl. Environ. Microbiol. 1989, 55, 1–6. [Google Scholar] [CrossRef]
- Mammi, L.M.E.; Guadagnini, M.; Mechor, G.; Cainzos, J.M.; Fusaro, I.; Palmonari, A.; Formigoni, A. The Use of Monensin for Ketosis Prevention in Dairy Cows during the Transition Period: A Systematic Review. Animals 2021, 11, 1988. [Google Scholar] [CrossRef]
- Marques, R.D.; Cooke, R.F. Effects of Ionophores on Ruminal Function of Beef Cattle. Animals 2021, 11, 2871. [Google Scholar] [CrossRef]
- Scharen, M.; Drong, C.; Kiri, K.; Riede, S.; Gardener, M.; Meyer, U.; Hummel, J.; Urich, T.; Breves, G.; Danicke, S. Differential effects of monensin and a blend of essential oils on rumen microbiota composition of transition dairy cows. J. Dairy Sci. 2017, 100, 2765–2783. [Google Scholar] [CrossRef]
- Linde, D.A.; Schokker, D.; du Toit, C.J.L.; Ramkilawon, G.D.; van Marle-Koester, E. The Effect of a Bacillus Probiotic and Essential Oils Compared to an Ionophore on the Rumen Microbiome Composition of Feedlot Cattle. Animals 2023, 13, 2927. [Google Scholar] [CrossRef] [PubMed]
- Khochamit, N.; Siripornadulsil, S.; Sukon, S.; Siripornadulsil, W. Antibacterial activity and genotypic-phenotypic characteristics of bacteriocin-producing Bacillus subtilis KKU213: Potential as a probiotic strain. Microbiol. Res. 2015, 170, 36–50. [Google Scholar] [CrossRef]
- Chang, M.N.; Ma, F.T.; Wei, J.Y.; Liu, J.H.; Nan, X.M.; Sun, P. Live Bacillus subtilis natto Promotes Rumen Fermentation by Modulating Rumen Microbiota In Vitro. Animals 2021, 11, 1519. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Cui, K.; Ma, T.; Wan, F.; Wang, W.; Yang, D.; Wang, Y.; Guo, B.; Zhao, L.; Diao, Q. Influence of dietary supplementation with Bacillus licheniformis and Saccharomyces cerevisiae as alternatives to monensin on growth performance, antioxidant, immunity, ruminal fermentation and microbial diversity of fattening lambs. Sci. Rep. 2018, 8, 16712. [Google Scholar] [CrossRef]
- Qiao, G.; Shan, H.A.S.; Ma, N.; Ma, Q.Q.; Sun, Z.W. Effect of supplemental Bacillus cultures on rumen fermentation and milk yield in Chinese Holstein cows. J. Anim. Physiol. Anim. Nutr. 2010, 94, 429–436. [Google Scholar] [CrossRef]
- Andreazzi, A.S.R.; Pereira, M.N.; Reis, R.B.; Pereira, R.A.N.; Junior, N.N.M.; Acedo, T.S.; Hermes, R.G.; Cortinhas, C.S. Effect of exogenous amylase on lactation performance of dairy cows fed a high-starch diet. J. Dairy Sci. 2018, 101, 7199–7207. [Google Scholar] [CrossRef]
- Fernando, S.C.; Purvis II, H.T.; Horn, G.W.; Fieser, B.G.; DeSilva, U. Alterations in Ruminal Microflora of Steers Consuming Wheat Pasture Dosed with an Ionophore. 2005. Available online: https://extension.okstate.edu/programs/beef-extension/research-reports/site-files/documents/2005/fernando-2.pdf (accessed on 10 September 2022).
- McGarvey, J.A.; Place, S.; Palumbo, J.; Hnasko, R.; Mitloehner, F. Dosage-dependent effects of monensin on the rumen microbiota of lactating dairy cattle. Microbiologyopen 2019, 8, e00783. [Google Scholar] [CrossRef]
- Fuerniss, L.K.; Kreikemeier, K.K.; Reed, L.D.; Cravey, M.D.; Johnson, B.J. Cecal microbiota of feedlot cattle fed a four-species Bacillus supplement. J. Anim. Sci. 2022, 100, skac258. [Google Scholar] [CrossRef]
- Rigert, S.L.; Hartoonian, P.; Omale, S.E.; Rodriguez-Jimenez, S.; Bradley, C.M.K.; Horst, E.A.; Holloway, A.W.; Baumgard, L.H.; Appuhamy, J.A.D.R.N. Effects of feeding a Bacillus licheniformis fermentation extract with or without monensin on milk production and feed efficiency of lactating dairy cows. J. Dairy Sci. 2024, 107 (Suppl. S1), 85. [Google Scholar]
- Dhuyvetter, K.C.; Prentice, D.L.; Kvidera, S.K. Effect of removing Rumensin from diet on milk production efficiency. J. Dairy Sci. 2023, 106 (Suppl. S1), 452. [Google Scholar]
- Wickramasinghe, J.; Anderson, C.J.; Kaya, C.A.; Gorden, P.J.; Ribeiro, F.R.B.; Dohms, J.; Rigert, S.; Schmitz-Esser, S.; Appuhamy, R. Evaluating Ruminal and Small Intestinal Morphology and Microbiota Composition of Calves Fed a Macleaya cordata Extract Preparation. Animals 2023, 13, 54. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef]
- Kers, J.G.; Saccenti, E. The Power of Microbiome Studies: Some Considerations on Which Alpha and Beta Metrics to Use and How to Report Results. Front. Microbiol. 2022, 12, 796025. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer Publishing Company Incorporated: New York, NY, USA, 2009. [Google Scholar]
- National Research Council. Nutrient Requirements of Dairy Cattle, 7th ed.; The National Academies Press: Washington, DC, USA, 2001. [Google Scholar]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzale, A.; Kosciolek, T.; McCall, L.I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef]
- Guo, X.; Liu, Y.Q.; Jiang, Y.; Yao, J.H.; Li, Z.J. Ruminal Bacterial Community Successions in Response to Monensin Supplementation in Goats. Animals 2022, 12, 2291. [Google Scholar] [CrossRef]
- Kim, M.; Eastridge, M.L.; Yu, Z. Investigation of ruminal bacterial diversity in dairy cattle fed supplementary monensin alone and in combination with fat, using pyrosequencing analysis. Can. J. Microbiol. 2014, 60, 65–71. [Google Scholar] [CrossRef]
- Weiss, C.P.; Beck, P.A.; Richeson, J.T.; Tomczak, D.J.; Chai, J.M.; Hess, T.; Hubbell, D.; Zhao, J.C. Effect of monensin intake during a stocker phase and subsequent finishing phase on rumen bacterial diversity of beef steers. J. Anim. Sci. 2019, 97, 163–164. [Google Scholar] [CrossRef]
- Chai, J.; Weiss, C.P.; Beck, P.A.; Zhao, W.; Li, Y.; Zhao, J. Diet and monensin influence the temporal dynamics of the rumen microbiome in stocker and finishing cattle. J. Anim. Sci. Biotechnol. 2024, 15, 12. [Google Scholar] [CrossRef]
- Williamson, J.R.; Callaway, T.R.; Lourenco, J.M.; Ryman, V.E. Characterization of rumen, fecal, and milk microbiota in lactating dairy cows. Front. Microbiol. 2022, 13, 984119. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Zhang, M.L.; Zhang, R.Y.; Zhu, W.Y.; Mao, S.Y. Comparative studies of the composition of bacterial microbiota associated with the ruminal content, ruminal epithelium and in the faeces of lactating dairy cows. Microb. Biotechnol. 2016, 9, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Hagey, J.V.; Bhatnagar, S.; Heguy, J.M.; Karle, B.M.; Price, P.L.; Meyer, D.; Maga, E.A. Fecal Microbial Communities in a Large Representative Cohort of California Dairy Cows. Front. Microbiol. 2019, 10, 1093. [Google Scholar] [CrossRef]
- Clemmons, B.A.; Voy, B.H.; Myer, P.R. Altering the Gut Microbiome of Cattle: Considerations of Host-Microbiome Interactions for Persistent Microbiome Manipulation. Microb. Ecol. 2019, 77, 523–536. [Google Scholar] [CrossRef]
- Hanigan, M.D.; Appuhamy, J.A.D.R.N.; Gregorini, P. Revised digestive parameter estimates for the Molly cow model. J. Dairy Sci. 2013, 96, 3867–3885. [Google Scholar] [CrossRef]
- Sze, M.A.; Schloss, P.D. Looking for a Signal in the Noise: Revisiting Obesity and the Microbiome. mBio 2016, 7, e01018-16. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Stojanov, S.; Berlec, A.; Strukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Zhang, M.; Liu, J.; Zhu, W. Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: Membership and potential function. Sci. Rep. 2015, 5, 16116. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, M.N.V.; Jewell, K.A.; Freitas, F.S.; Benjamin, L.A.; Tótola, M.R.; Borges, A.C.; Moraes, C.A.; Suen, G. Characterizing the microbiota across the gastrointestinal tract of a Brazilian Nelore steer. Vet. Microbiol. 2013, 164, 307–314. [Google Scholar] [CrossRef]
- Pitta, D.; Indugu, N.; Vecchiarelli, B.; Rico, D.; Harvatine, K. Alterations in ruminal bacterial populations at induction and recovery from diet-induced milk fat depression in dairy cows. J. Dairy Sci. 2018, 101, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Lopes, D.R.G.; de Souza Duarte, M.; La Reau, A.J.; Chaves, I.Z.; de Oliveira Mendes, T.A.; Detmann, E.; Bento, C.B.P.; Mercadante, M.E.Z.; Bonilha, S.F.M.; Suen, G.; et al. Assessing the relationship between the rumen microbiota and feed efficiency in Nellore steers. J. Anim. Sci. Biotechnol. 2021, 12, 79. [Google Scholar] [CrossRef]
- Li, F.; Guan, L.L. Metatranscriptomic Profiling Reveals Linkages between the Active Rumen Microbiome and Feed Efficiency in Beef Cattle. Appl. Environ. Microbiol. 2017, 83, e00061-17. [Google Scholar] [CrossRef]
- Xue, M.Y.; Xie, Y.Y.; Zhong, Y.; Ma, X.J.; Sun, H.Z.; Liu, J.X. Integrated meta-omics reveals new ruminal microbial features associated with feed efficiency in dairy cattle. Microbiome 2022, 10, 32. [Google Scholar] [CrossRef]
- Appuhamy, J.A.; France, J.; Kebreab, E. Models for predicting enteric methane emissions from dairy cows in North America, Europe, and Australia and New Zealand. Glob. Chang. Biol. 2016, 22, 3039–3056. [Google Scholar] [CrossRef]
- Nalla, K.; Manda, N.K.; Dhillon, H.S.; Kanade, S.R.; Rokana, N.; Hess, M.; Puniya, A.K. Impact of Probiotics on Dairy Production Efficiency. Front. Microbiol. 2022, 13, 805963. [Google Scholar] [CrossRef] [PubMed]
- Jinno, C.; Li, X.D.; Liu, Y.H. Dietary supplementation of Bacillus subtilis or antibiotics modified intestinal microbiome of weaned pigs under enterotoxigenic infection. Front. Microbiol. 2022, 13, 1064328. [Google Scholar] [CrossRef] [PubMed]
- Trovatelli, L.D.; Matteuzzi, D. Presence of bifidobacteria in the rumen of calves fed different rations. Appl. Environ. Microbiol. 1976, 32, 470–473. [Google Scholar] [CrossRef]
- van den Bossche, T.; Goossens, K.; Ampe, B.; Tamassia, L.F.M.; de Boever, J.L.; Vandaele, L. Effect of supplementing an α-amylase enzyme or a blend of essential oil components on the performance, nutrient digestibility, and nitrogen balance of dairy cows. J. Dairy Sci. 2024, 107, 4509–4523. [Google Scholar] [CrossRef]
- Wu, Y.; Jiao, C.; Diao, Q.; Tu, Y. Effect of Dietary and Age Changes on Ruminal Microbial Diversity in Holstein Calves. Microorganisms 2023, 12, 12. [Google Scholar] [CrossRef]
- Cristobal-Carballo, O.; McCoard, S.A.; Cookson, A.L.; Laven, R.A.; Ganesh, S.; Lewis, S.J.; Muetzel, S. Effect of Divergent Feeding Regimes During Early Life on the Rumen Microbiota in Calves. Front. Microbiol. 2021, 12, 711040. [Google Scholar] [CrossRef]
- Haney, M.E.; Knox, N.G.; Hoehn, M.M. Monensin, a new biologically active compound. I. Discovery and isolation. Antimicrob Agents Chemother 1967, 7, 353–358. [Google Scholar]
- Stepanchenko, N.; Stefenoni, H.; Hennessy, M.; Nagaraju, I.; Wasson, D.E.; Cueva, S.F.; Räisänen, S.; Dechow, C.D.; Pitta, D.W.; Hristov, A.N. Microbial composition, rumen fermentation parameters, enteric methane emissions, and lactational performance of phenotypically high and low methane-emitting dairy cows. J. Dairy Sci. 2023, 106, 6146–6170. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Su, X.; Bai, H.; Yang, Y.; Wang, H.; Dan, Z.; Lu, J.; Wu, S.; Cai, C.; Cao, Y.; et al. Specific enrichment of microbes and increased ruminal propionate production: The potential mechanism underlying the high energy efficiency of Holstein heifers fed steam-flaked corn. AMB Express 2019, 9, 209. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Fernandez, M.V.; Daniel, J.-B.; Seymour, D.J.; Kvidera, S.K.; Bester, Z.; Doelman, J.; Martín-Tereso, J. Targeting the Hindgut to Improve Health and Performance in Cattle. Animals 2020, 10, 1817. [Google Scholar] [CrossRef] [PubMed]

| Variable | −R 1 | +R | SEM | p-Value | ||||
|---|---|---|---|---|---|---|---|---|
| −BLFE | +BLFE | −BLFE | +BLFE | R | BLFE | R × BLFE | ||
| DMI, kg/d | 30.3 | 31.1 | 30.8 | 31.0 | 0.62 | 0.74 | 0.38 | 0.62 |
| Milk yield, kg/d | 35.4 b | 38.7 a | 38.0 ab | 37.3 ab | 0.63 | 0.51 | 0.05 | <0.01 |
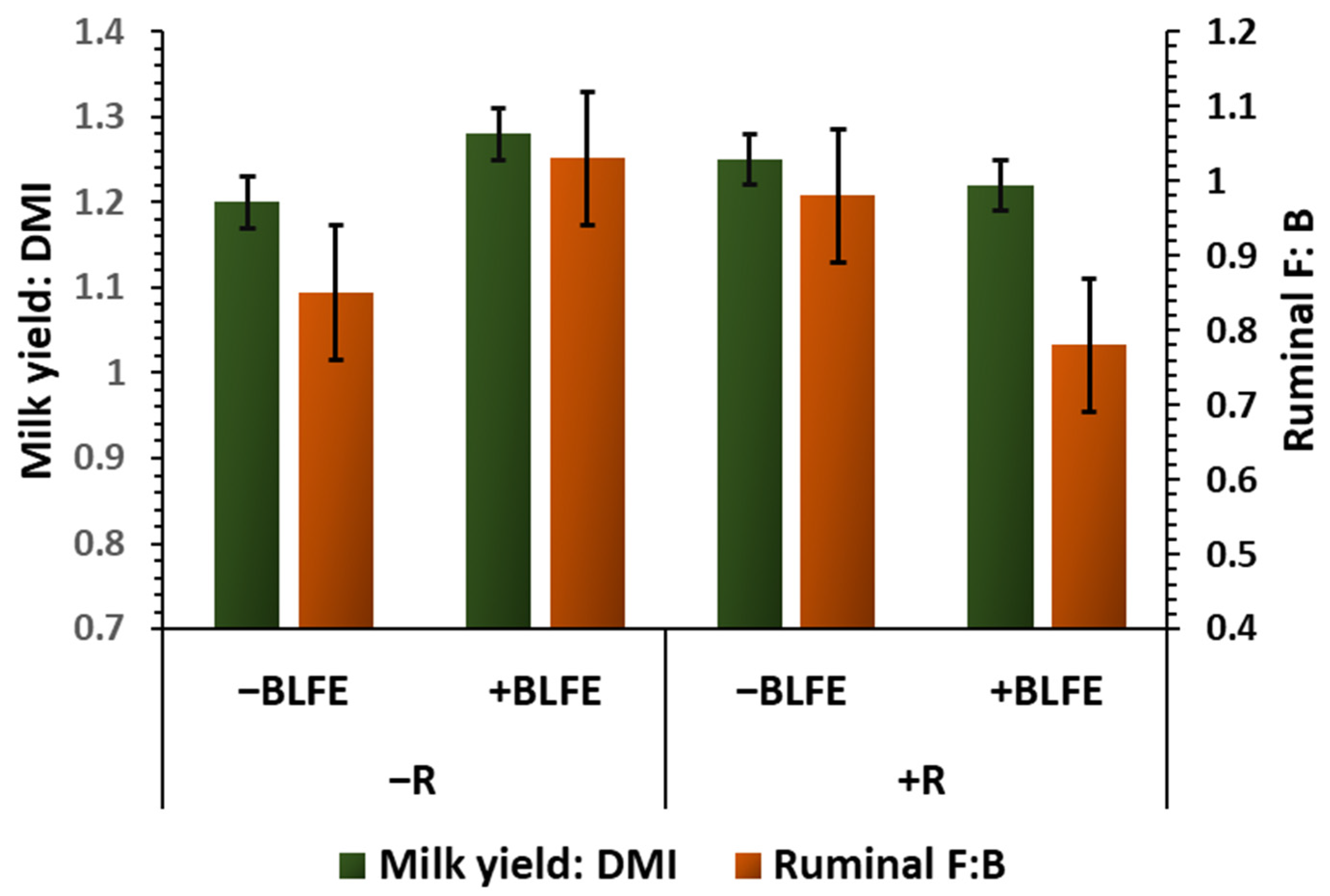
| Milk yield/DMI 2 | 1.20 b | 1.28 a | 1.25 ab | 1.22 ab | 0.03 | 0.65 | 0.38 | 0.03 |
| Milk protein, % | 3.40 | 3.48 | 3.46 | 3.47 | 0.03 | 0.27 | 0.09 | 0.14 |
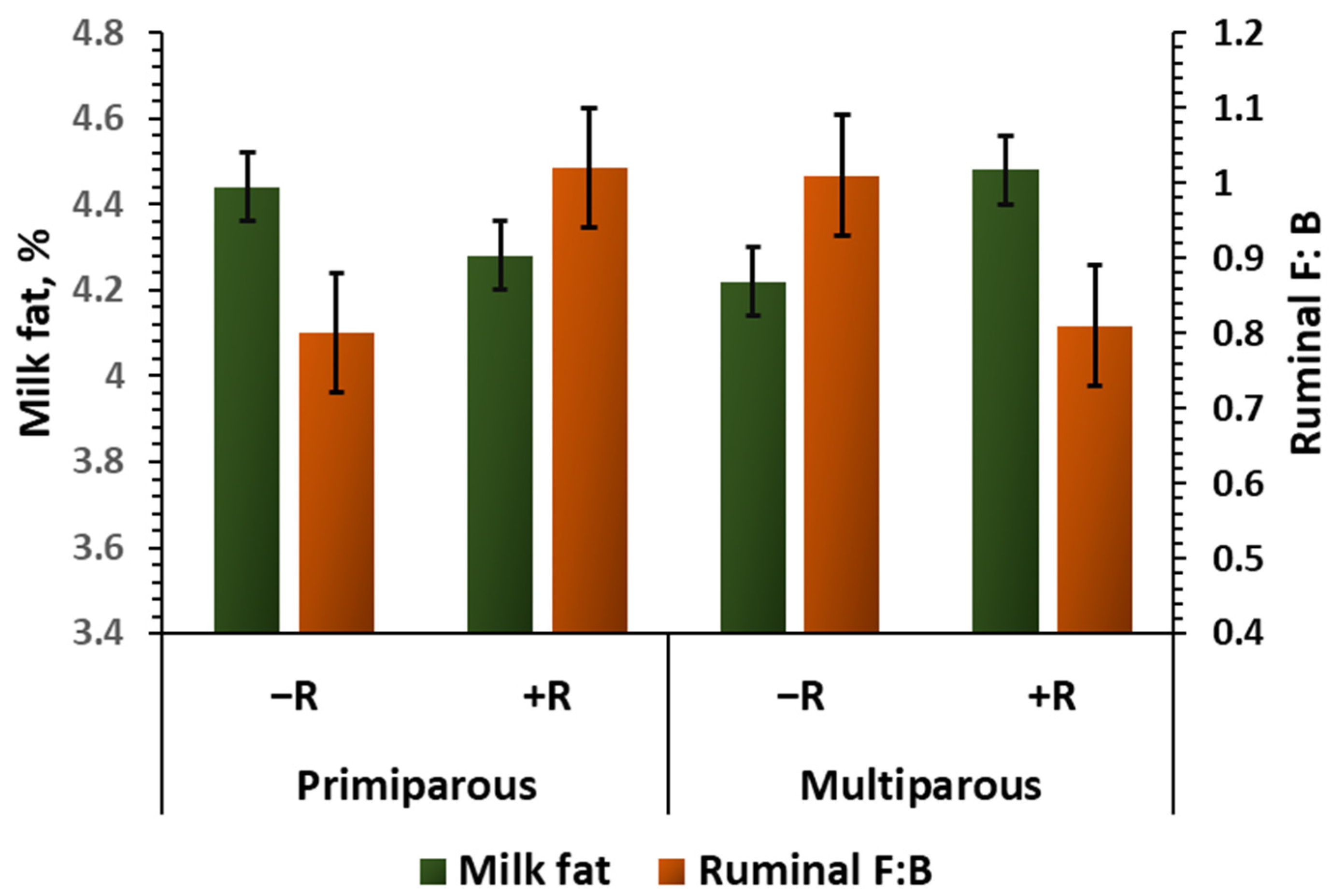
| Milk fat, % | 4.33 | 4.26 | 4.46 | 4.37 | 0.07 | 0.09 | 0.27 | 0.95 |
| ECM, kg/d | 40.4 | 41.9 | 44.1 | 43.1 | 1.02 | 0.02 | 0.75 | 0.23 |
| Parameter | −R 1 | +R | SEM | p-Value | ||||
|---|---|---|---|---|---|---|---|---|
| −BLFE | +BLFE | −BLFE | +BLFE | R | BLFE | R × BLFE | ||
| Rumen fluid | ||||||||
| Obs. OTU 2 | 2269 | 2174 | 2235 | 2123 | 105 | 0.68 | 0.32 | 0.94 |
| Chao1 | 2937 | 2783 | 2907 | 2731 | 119 | 0.73 | 0.17 | 0.92 |
| Shannon | 5.36 | 5.34 | 5.33 | 5.20 | 0.21 | 0.68 | 0.72 | 0.80 |
| Simpson | 0.93 | 0.94 | 0.94 | 0.93 | 0.02 | 0.75 | 0.93 | 0.62 |
| Feces | ||||||||
| Obs. OUT 2 | 1596 | 1542 | 1542 | 1596 | 37.4 | 0.31 | 0.74 | 0.28 |
| Chao1 | 1972 | 1952 | 2006 | 2064 | 52.1 | 0.17 | 0.72 | 0.46 |
| Shannon | 5.35 | 5.28 | 5.35 | 5.40 | 0.05 | 0.28 | 0.80 | 0.23 |
| Simpson | 0.98 | 0.98 | 0.98 | 0.98 | < 0.01 | 0.28 | 0.70 | 0.10 |
| Variable | Microbial Community | SEM | p-Value | |
|---|---|---|---|---|
| Rumen Fluid | Feces | |||
| Alpha diversity | ||||
| Observed OTUs | 2195 | 1599 | 39.4 | <0.01 |
| Chao 1 | 2837 | 2013 | 45.2 | <0.01 |
| Shannon | 5.29 | 5.34 | 0.08 | 0.65 |
| Simpson | 0.93 | 0.98 | 0.01 | <0.01 |
| Relative abundance, % | ||||
| Kingdom | ||||
| Bacteria | 97.7 | 97.7 | 0.14 | 0.15 |
| Archaea | 2.29 | 2.60 | 0.14 | 0.10 |
| Phylum | ||||
| Firmicutes (F) | 32.2 | 59.9 | 1.15 | <0.01 |
| Bacteroidetes (B) | 36.7 | 27.0 | 0.90 | <0.01 |
| F: B | 0.89 | 2.34 | 0.08 | <0.01 |
| VFA, molar % | ||||
| Acetate | 61.8 | 67.9 | 0.63 | <0.01 |
| Propionate | 23.9 | 18.1 | 0.59 | <0.01 |
| Propionate/acetate | 0.39 | 0.27 | 0.01 | <0.01 |
| –R 1 | +R | SEM | p-Value | ||||||
|---|---|---|---|---|---|---|---|---|---|
| –BLFE | +BLFE | –BLFE | +BLFE | R | BLFE | R × BLFE | |||
| Rumen Fluid | |||||||||
| Kingdom abundance | |||||||||
| Bacteria | 97.7 | 97.5 | 97.5 | 97.9 | 0.27 | 0.76 | 0.89 | 0.30 | |
| Archaea | 2.20 | 2.46 | 2.41 | 2.09 | 0.27 | 078 | 0.91 | 0.18 | |
| Phylum abundance | |||||||||
| Firmicutes | 31.1 | 36.2 | 33.1 | 28.5 | 2.67 | 0.28 | 0.93 | 0.07 | |
| Bacteroidetes | 37.4 | 36.3 | 34.6 | 37.4 | 1.89 | 0.65 | 0.64 | 0.30 | |
| F: B 2 | 0.85 ab | 1.03 a | 0.98 ab | 0.78 b | 0.09 | 0.54 | 0.93 | 0.04 | |
| Feces | |||||||||
| Kingdom abundance | |||||||||
| Bacteria | 97.4 | 97.3 | 97.4 | 97.6 | 0.29 | 0.54 | 0.78 | 0.64 | |
| Archaea | 2.63 | 2.73 | 2.63 | 2.43 | 0.29 | 060 | 0.88 | 0.60 | |
| Phylum abundance | |||||||||
| Firmicutes | 59.6 | 60.2 | 61.9 | 58.0 | 1.74 | 0.98 | 0.34 | 0.20 | |
| Bacteroidetes | 27.8 | 27.3 | 24.8 | 29.0 | 1.69 | 0.71 | 0.29 | 0.16 | |
| F: B2 | 2.20 | 2.33 | 2.58 | 2.19 | 0.21 | 0.54 | 0.53 | 0.20 | |
| Taxonomy (SILVA v138) | −R 1 | +R | SEM | p-Value | ||||
|---|---|---|---|---|---|---|---|---|
| −BLFE | +BLFE | −BLFE | +BLFE | R | BLFE | R × BLFE | ||
| Rumen Fluid | ||||||||
| Bifidobacterium | 0.43 | 0.91 | 0.36 | 0.65 | 0.16 | 0.30 | 0.03 | 0.56 |
| Erysipelotrichaceae_UCG-002 | 0.08 | 0.21 | 0.08 | 0.18 | 0.04 | 0.85 | 0.01 | 0.71 |
| Oscillospirales_ge | 0.32 | 0.36 | 0.42 | 0.38 | 0.03 | 0.02 | 0.99 | 0.19 |
| unclassified_Clostridia | 0.36 | 0.43 | 0.30 | 0.20 | 0.06 | 0.03 | 0.86 | 0.19 |
| Feces | ||||||||
| Bacteroides | 6.00 ab | 4.87 b | 4.82 b | 6.23 a | 0.48 | 0.86 | 0.77 | 0.01 |
| Prevotella | 1.19 b | 2.67 a | 2.43 ab | 1.17 b | 0.52 | 0.80 | 0.83 | 0.01 |
| Rikenellaceae_RC9_gut_group | 6.32 a | 5.31 ab | 4.72 b | 6.03 ab | 0.49 | 0.37 | 0.76 | 0.02 |
| Christensenellaceae_R-7_group | 1.66 ab | 1.35 ab | 1.28 b | 1.74 a | 0.14 | 0.99 | 0.57 | 0.01 |
| Unclassified_Oscillospiraceae | 0.94 a | 0.71 ab | 0.55 b | 0.94 a | 0.10 | 0.36 | 0.41 | <0.01 |
| Frisingicoccus | 0.29 b | 0.46 a | 0.41 ab | 0.30 ab | 0.06 | 0.69 | 0.65 | 0.03 |
| Parasutterella | 0.30 a | 0.23 ab | 0.18 b | 0.30 a | 0.03 | 0.36 | 0.36 | <0.01 |
| Anaerosporobacter | 0.29 ab | 0.42 a | 0.35 ab | 0.19 b | 0.07 | 0.19 | 0.84 | 0.03 |
| Lachnospiraceae_UCG-001 | 0.14 b | 0.28 a | 0.24 ab | 0.19 ab | 0.04 | 0.98 | 0.27 | 0.02 |
| Monoglobus | 0.64 ab | 0.51 b | 0.48 b | 0.75 a | 0.10 | 0.68 | 0.47 | 0.04 |
| Turicibacter | 0.13 c | 0.31 a | 0.24 abc | 0.18 bc | 0.04 | 0.84 | 0.22 | 0.01 |
| Oscillibacter | 0.20 | 0.29 | 0.24 | 0.27 | 0.02 | 0.56 | <0.01 | 0.10 |
| Taxonomy (SILVA v138) | Milk Yield/DMI | Rumen Fluid P: A |
|---|---|---|
| Rumen fluid | ||
| Among the 20 most abundant | ||
| Succinivibrionaceae_UCG-001 | 0.55 (<0.01) | 0.84 (<0.01) |
| Prevotellaceae_UCG-004 | −0.43 (<0.01) | −0.67 (<0.01) |
| Lachnospiraceae_NK3A20 | −0.31 (0.04) | −0.40 (0.01) |
| Succiniclasticum | −0.34 (0.02) | −0.59 (<0.01) |
| Acetitomaculum | −0.43 (<0.01) | −0.37 (0.01) |
| Bacteroidales p-251-o5_ge | −0.47 (<0.01) | −0.60 (<0.01) |
| Unclassified_Ruminococcaceae | −0.45 (<0.01) | −0.62 (<0.01) |
| Rikenellaceae_RC9_gut_group | −0.44 (<0.01) | −0.79 (<0.01) |
| Bacteroidales_RF16_group_ge | −0.33 (0.02) | −0.54 (<0.01) |
| Christensenellaceae_R-7_group | −0.44 (<0.01) | −0.78 (<0.01) |
| Among those affected by treatments (Table 5) | ||
| Unclassified_Clostridia | −0.30 (0.04) | −0.66 (<0.01) |
| Feces | ||
| Among the 20 most abundant | ||
| Succinivibrio | −0.35 (0.02) | −0.16 (0.32) |
| Unclassified_Lachnospiraceae | −0.31 (0.04) | −0.16 (0.30) |
| Prevotella | −0.37 (0.01) | −0.13 (0.39) |
| Bacteroidales_RF16_group_ge | 0.36 (0.02) | 0.29 (0.06) |
| Christensenellaceae_R-7_group | 0.30 (0.04) | −0.12 (0.42) |
| Among those affected by treatments (Table 5) | ||
| Prevotella | −0.37 (0.01) | −0.13 (0.30) |
| Christensenellaceae_R-7_group | 0.30 (0.04) | −0.12 (0.42) |
| Frisingicoccus | −0.35 (0.02) | −0.16 (0.30) |
| Monoglobus | 0.38 (0.01) | 0.08 (0.65) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hartoonian, P.; Jonas, L.C.; Omale, S.; Rigert, S.; Bradley, C.; Horst, E.; Beitz, D.; Schmitz-Esser, S.; Appuhamy, R. Effects of a Bacillus licheniformis Fermentation Extract and Monensin on the Rumen and Hindgut Microbiota Composition of Lactating Dairy Cows. Animals 2025, 15, 2980. https://doi.org/10.3390/ani15202980
Hartoonian P, Jonas LC, Omale S, Rigert S, Bradley C, Horst E, Beitz D, Schmitz-Esser S, Appuhamy R. Effects of a Bacillus licheniformis Fermentation Extract and Monensin on the Rumen and Hindgut Microbiota Composition of Lactating Dairy Cows. Animals. 2025; 15(20):2980. https://doi.org/10.3390/ani15202980
Chicago/Turabian StyleHartoonian, Phoebe, Lucille C. Jonas, Shedrack Omale, Sydney Rigert, Catherine Bradley, Erin Horst, Donald Beitz, Stephan Schmitz-Esser, and Ranga Appuhamy. 2025. "Effects of a Bacillus licheniformis Fermentation Extract and Monensin on the Rumen and Hindgut Microbiota Composition of Lactating Dairy Cows" Animals 15, no. 20: 2980. https://doi.org/10.3390/ani15202980
APA StyleHartoonian, P., Jonas, L. C., Omale, S., Rigert, S., Bradley, C., Horst, E., Beitz, D., Schmitz-Esser, S., & Appuhamy, R. (2025). Effects of a Bacillus licheniformis Fermentation Extract and Monensin on the Rumen and Hindgut Microbiota Composition of Lactating Dairy Cows. Animals, 15(20), 2980. https://doi.org/10.3390/ani15202980







