Simple Summary
This article identifies candidate genes associated with economically useful carcass traits in a population of Aberdeen Angus cattle using a GWAS. The study revealed a number of significant SNPs associated with economically useful traits (slaughter weight, meat yield, meat mar-bling). Functional annotation detected 33 genes associated with biological processes (GO BP) and 5 genes registered in the Cattle QTL database. For the OSBPL3 and RBFOX3 genes, within which a number of economically useful trait SNPs Hapmap39351-BTA-71190 and ARS-BFGL-NGS-11938 are localized, related with slaughter weight and meat marbling, we recommend developing tests for their inclusion in the genomic selection of beef cattle. Of interest is the OSBPL3 gene, within which the SNPs are located, so that tests that are unique and breed-specific can be developed. Our results provide useful information for understanding the genetic architecture and its further use in breeding.
Abstract
The use of GWASs in agriculture allows associations between SNPs and quantitative or qualitative traits in cattle to be estimated. This study investigated the relationships among a number of economically useful carcass traits (slaughter weight, meat yield, and meat marbling) with a group of SNPs that can act as DNA markers. Blood samples from Aberdeen Angus bulls (n = 260) were used as material for SNP genotyping. Genetic architecture for the kill yield trait showed the presence of 31 SNPs. Three SNPs were found to be the most reliable: BTB-00197584 (p = 7.20 × 10−5), Hapmap46735-BTA-86653 (p = 5.05 × 10−5), and BTB-00676077 (p = 7.02 × 10−5). There were 10 SNPs associated with meat yield. For meat yield, 7 SNPs were identified, four of them exceeding the threshold of established validity: ARS-BFGL-NGS-30557 (p = 1.28 × 10−12), ARS-BFGL-NGS-68920, ARS-BFGL-NGS-30466 on chromosome 8 (p = 1.20 × 10−6 and 3.03 × 10−6, respectively) and ARS-BFGL-NGS-40640 (p = 7.10 × 10−6). For marbling, 11 SNPs were identified on 6 of 29 chromosomes. These SNPs can act as potential markers for productivity assessment in cattle.
1. Introduction
Sequencing technologies have made it possible to study the genomes of cattle of different breeds and to analyze and compare their genetic structure [1]. The discovery of quantitative trait loci (QTL) has led to advances in the identification and comprehension of genetic modifications associated with economically significant phenotypic traits. Genome-wide association studies (GWASs), which use powerful techniques for the genetic analysis of single nucleotide polymorphism (SNP), are a relatively new approach to cattle genetic studies, which have identified different groups of candidate genes responsible for economically useful traits. This is especially important for research on cattle breeds whose genetic potential has not yet been fully realized. This is particularly true of the Aberdeen Angus breed.
The Aberdeen Angus breed of cattle is one of the most popular in the production of marbled meat. Animals of this breed are characterized by calm and malleable temperament, which makes them easy to handle and convenient for keeping on farms [2]. They also adapt easily to different housing and climatic conditions, making them suitable for different regions and farming systems. In terms of economic efficiency, the Aberdeen Angus breed has a good feed-to-meat conversion, which enables farmers to obtain higher yields at lower costs, while good fertility and high calf survival make them profitable for breeding. Earlier marker-assisted selection on Angus bulls in Hungary revealed the influence of a number of SNPs (DGAT1, TG, and Lep) on meat marbling [3]. There are relatively high frequencies of desirable alleles of Lep and CAPN1 genes in Russian beef cattle populations and a positive effect of allele T of the Arg4Cys LEP polymorphism on the fatness of animals from birth to weaning [4,5]. CAPN1_316 polymorphisms were found to affect the average weight gain in Aberdeen Angus cattle [4].
Although there are studies in Europe and the US, there is very little data on the genetic structure of Aberdeen Angus cattle and their meat productivity in Russia, which makes it impossible to provide breeders with reliable information on the breed for the conservation and protection of genetic diversity and use in commercial livestock breeding.
This study identifies candidate genes associated with economically useful traits in a population of Aberdeen Angus cattle using GWASs.
2. Materials and Methods
2.1. Ethical Statement
All activities related to animal care, the collection of biological material, and phenotype characterization were carried out in exact accordance with the regulations of the Federal State Budgetary Scientific Institution “Federal Research Center for Biological Systems and Agrotechnologies of the Russian Academy of Sciences”. Copies of the minutes of the 30 April 2024, Laboratory Animal Control Commission Meeting No. 2/1, were received on 6 May 2024. Data and samples were collected, and no animals were disturbed during data collection.
2.2. Animals and Phenotypes
The research work was carried out on a specialized territory for fattening animals located in the Kostroma region, Russia. Young Aberdeen Angus cattle (n = 260) were selected as the object of study. The area required to accommodate one bull in an outdoor feedlot was estimated at about 10 square meters. All individuals had identical sex and age characteristics and were under the same conditions of nutrition and care. At the time of the experiment, the age of the studied individuals was 532.0 ± 1 days. After 43 days, their weight just before slaughter was 617.6 ± 1.68 kg.
Their diet consisted of a mixture of corn silage and high-moisture corn feed. The animals also had free access to wheat straw located in the center of the feedlot. The feeders were cleaned before morning feeding. The feeding and maintenance conditions were the same for the whole group. The feed contained 53.5% dry matter, 11.75% protein, 10.3% fiber, 2.35% fat, and 5.4% ash. The animals had free access to water.
At the end of the fattening period, the cattle were sent to slaughter at a commercial meat processing facility. Animals were identified with an RFID chip. Carcasses were identified with tags and placed in a refrigerator 24–48 h after slaughtering. After cooling, the carcass was cut into marketable pieces.
2.3. DNA Samples Collection, Genotyping and Quality Control
For the genome-wide association search, 260 blood samples were obtained for genotypic analysis. Blood samples were used to extract genomic DNA. DNA extraction and genotype determination were performed using DNA-Extran (Syntol LLC, Moscow, Russia). Genomic DNA concentration was determined using Qubit (Thermofisher, Waltham, MA, USA), while DNA abundance was measured using Nanodrop ND-1000 (Thermofisher, Waltham, MA, USA).
2.4. Genome-Wide Association Studies
The genotyping of animals was performed using a high-density microarray BovineSNP50 (Illumina Inc., San Diego, CA 92122, USA) containing 53 218 SNPs according to the manufacturer’s standard methodology.
Unmapped SNPs and SNPs located on X and Y chromosomes and in mitochondrial DNA were excluded. SNPs located on autosomes were investigated. Plink 1.90 [6] was used to check the quality of the genotyped breeds based on the following criteria: the call-rate for the SNPs for each individual sample is at least 90% (—mind); the call-rate for each SNP across all genotyped samples is at least 90% (—geno); the minor allele frequency (MAF) is more than 0.05 (—maf); the deviation of SNP genotypes from the Hardy-Weinberg equilibrium in the samples had a significance level of p < 10−6 (—hwe): --assoc and --linear, to realize the methodology of multiple linear regression of the trait depending on the minor allele coding (0—homozygote for the most frequent allele, 1—heterozygote, 2—homozygote for the minor allele). Thus, the final equation had the general form of the following:
where Y is the vector of phenotypes; μ is the overall mean; b is the vector of fixed effects, including piggyback and year effects; w is the vector of the live weight of individuals when measurement is complete, treated as a covariance; c is the vector of SNP effects; a is the vector of random additive genetic effects with e ~ N (Gσα2), where G is the matrix of genomic ratios calculated from full genomic genotyping data using medium- or high-density DNA chips, and σα2 is the variance of polygenetic additives; K is the regression coefficient of the live weight of individuals when the measurement is completed; e is the vector of residual errors with e ~ N (Iσe2), where I is the unit matrix and σe2 is the residual variance; and X, S, and Z are the incident matrices for b, c, and a, respectively.
Y = μ + Xb + Kw + Sc + Za + e,
A heterozygosity test was performed to identify those samples that differed from the mean by more than three standard deviations in order to exclude them from the study. A total of 34 289 SNPs and 258 samples underwent quality control procedures and were retained for further study.
The p value (p < 1.45 × 10−5) of significance for single nucleotide polymorphisms was determined based on the Bonferroni correction method (0.05/N) [7], where N is the total number of SNPs remaining after quality control.
Data visualization was performed in the GWAS rMVP package using the R programming language [8].
2.5. Gene Identification, Function and Pathway Enrichment and Network Analysis
The region boundaries indicated according to the ARS-UCD2.0 genome assembly were applied to recognize cattle genes using the Ensembl 103 databases through the online resource [9]. The Database for Annotation, Visualization, and Integrated Discovery (DAVID), which provides researchers with a comprehensive set of functional annotation tools to understand the biological meaning of large gene lists [10], and the Cattle QTL database (https://www.animalgenome.org/cgi-bin/QTLdb/BT/index accessed on 27 May 2024), was used for functional gene annotation. Bioinformational data processing and work with graphs were carried out using R [8].
3. Results and Discussion
3.1. Information of SNPs
Quality control of SNPs was performed using PLINK 1.9. After filtering, the number of SNPs was 34 289. The filtered SNPs were distributed unevenly across all 29 chromosomes (see Figure 1). The GWAS rMVP package was used to visualize SNP density across chromosomes in the R program.

Figure 1.
SNP density plot for the studied traits. The higher the SNP density, the redder the color.
The data revealed that some of the SNPs are associated with a number of phenotypic parameters that may be important economically. The data are presented in Table 1, showing the number and chromosome distribution of SNPs associated with slaughter weight, meat yield, and meat marbling.

Table 1.
Number and chromosome distribution of SNPs associated with the indicators.
3.2. Association Analysis and the Identification of Candidate Genes Related to Carcass Yield
The genetic architecture showed the presence of 31 SNPs located on 16 chromosomes but significantly associated with the trait. No SNPs exceeding the established threshold of the full genomic study were identified (Figure 2, Table 1).
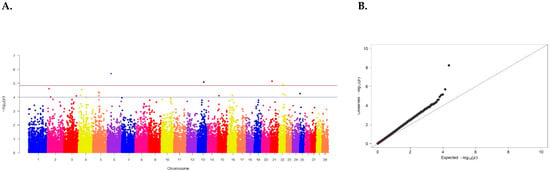
Figure 2.
(A) Distribution of SNPs across cattle chromosomes in relation to confidence level (−log10 (p)) by probability summation (blue line, p < 0.00001) and Bonferroni criterion (red line, p < 1.20 × 10−6) for slaughter weight. (B) Quartile of the probability distribution of expected and observed deviations from normal distribution for confidence values.
Three SNPs were found to be the most reliable: BTB-00197584 on chromosome 4 (p = 7.20 × 10−5), Hapmap46735-BTA-86653 on chromosome 14 (p = 5.05 × 10−5), and BTB-00676077 on chromosome 17 (p = 7.02 × 10−5) (Table 2).

Table 2.
List of SNPs significantly associated with slaughter weight.
3.3. Association Analysis and Identification of Candidate Genes Related to Meat Yield in Cattle
Ten SNPs on chromosomes 2, 5, 8, 9, 16, and 21 are associated with the trait of meat yield (Figure 3, Table 1).

Figure 3.
(A) Distribution of single nucleotide mutations on cattle chromosomes in relation to the level of confidence (−log10 (p)) by probability summation (blue line, p < 0.00001) and Bonferroni criterion (red line, p < 1.20 × 10−6) for meat yield. (B) Quartile of probability distribution of the expected and observed deviations from the normal distribution for confidence values.
For the meat yield, 7 SNPs were identified, four of them exceeding the threshold of established reliability: ARS-BFGL-NGS-30557 on chromosome 5 (p = 1.28 × 10−12), ARS-BFGL-NGS-68920, ARS-BFGL-NGS-30466 on chromosome 8 (p = 1.20 × 10−6 and 3.03 × 10−6, respectively) and ARS-BFGL-NGS-40640 on chromosome 16 (p = 7.10 × 10−6) (Table 3).

Table 3.
List of SNPs significantly associated with meat yield per bone.
3.4. Association Analysis and the Identification of Candidate Genes Related to Meat Marbling
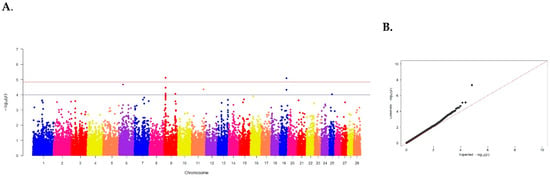
Figure 4.
(A) Distribution of single nucleotide mutations on cattle chromosomes in relation to the level of confidence (−log10 (p)) by probability summation (blue line, p < 0.00001) and Bonferroni criterion (red line, p < 1.20 × 10−6) for meat marbling. (B) Quartile of probability distribution of the expected and observed deviations from normal distribution for confidence values.
Meat marbling—the presence of intramuscular fat—is of great economic importance for beef cattle. The Aberdeen Angus breed is considered the best, and the breed purity and origin of animals are important, as only specially bred cattle have the genetic predisposition for accumulating intramuscular fat. The genetic architecture of meat marbling and the identification of genes responsible for the formation of the best grading category by full genome association study on Russian populations of the Aberdeen Angus breed has not been previously conducted.
In our studies, 6 SNPs were identified, of which 3 exceeded the threshold of established validity: ARS-BFGL-NGS-30557 located on chromosome 5 (p = 4.83 × 10−8), BovineHD0900002742 on chromosome 9 (p = 7.61 × 10−6) and ARS-BFGL-NGS-36573 on chromosome 19 (p = 8.08 × 10−6) (see Table 4).

Table 4.
List of SNPs significantly associated with meat marbling.
3.5. Structural Annotation of Genes Localized Within and/or in Close Proximity to Selected Regions of the Animal Genome
The structural annotation describes the exact location of various elements in the genome, such as open reading frames (ORFs), coding sequences (CDSs), exons, introns, repeats, splicing sites, regulatory motifs, start and stop codons, and promoters.
Table 5 presents the structural annotation of the genes identified for the studied traits. Structural annotation revealed the presence of 175 genes on 23 of 29 chromosomes.

Table 5.
Structural annotation of genes within which and/or in close proximity to SNPs.
3.6. Functional Annotation of Genes Localized Within and/or in Close Proximity to Selected Regions of the Animal Genome
Functional annotation analysis involved annotating genes with GO terms and pathway information (see Table 6).

Table 6.
Functional annotation of identified genes using the web-based DAVID program.
The HS6ST1 gene is associated with muscularity in Marchigiana and Chianina breeds and has significant additive associations with carcass weight in crossbred cattle slaughtered between 12 and 36 months of age with reported carcass phenotypes [11]. In our studies, this gene is correlated with meat yield by biological processes associated with alveolar development in the lungs (GO:0048286), neuronal development (GO:0048666), and labyrinthine layer blood vessel development (GO:0060716). The SNP ARS-BFGL-NGS-92825 was found within the HS6ST1 gene, and therefore, it can be recommended for further test development.
The UGGT1 gene is associated with characteristics including carcass weight and carcass fatness in Italian cattle breeds [12]. The OSBPL3 gene has previously been associated with the regulation of feed intake and energy expenditure in Nellore cattle, and it acts as a lipid transporter or sensor at membrane contact sites, affecting lipid metabolism [11]. In the analysis of our bull population, this gene is correlated with slaughter weight, and the SNP Hapmap39351-BTA-71190 is located where the single nucleotide substitution occurs. The GSDME gene is associated with feed conversion and feed intake, and the CFAP69 gene is associated with mammary immune system mechanisms [11].
The DDX60 gene was previously identified as predicting buffalo semen quality and fertility [13]. The ELP3 gene is significantly associated with predicted residual feed intake in commercial beef cattle littermates [14]. In our studies, the SNP ARS-BFGL-NGS-68920 was found within this gene and is correlated (p = 1.20 × 10−6) with meat yield. The SCARA5 gene is involved in iron delivery and is located on the centromere end of chromosome 8 of the bovine genome, which is essential for the genetic predisposition of muscle mineral composition in beef cattle [15].
The BTA9:43.3–43.6 Mb region, associated with meat yield, includes the ATG5 (autophagy 5) gene, which has an autophagy function mainly in preimplantation ovaries [16]. This autophagy function was previously found in significant amounts in ovaries under heat stress in pigs [17]. This gene has also been associated with embryo development after four- and eight-cell stages in mice [18]. An ATP-binding cassette, subfamily G (WHITE), member 5 (ABCG5) was identified on chromosome 11 in the 27 Mb region. ABCG family members are associated with cellular lipid transport into macrophages and hepatocytes in Nelore cattle [19].
The TMEM68 gene has been linked to feed intake and growth phenotypes in cattle [20]. In our study, it was significantly associated with the slaughter weight of animals. The TTC36 gene has been identified in cattle of the Braford breed as resistant to skin mite infestation [21].
The SPATA17 gene was previously identified in beef cattle as influencing reproduction and muscle formation [22]. In this study on Aberdeen Angus cattle, it is correlated with slaughter weight, and SNP Hapmap58079-rs29010441 was detected within it.
The RBFOX3 gene is a candidate regulator of mineral content in the muscle of Nelore cattle [23]. In our study, this gene is associated with meat marbling, and within it, SNP ARS-BFGL-NGS-11938 is localized on chromosome 19. The expression of the C1QTNF1 gene was detected in the adipose tissue of lactating Holstein cows [24]. The CLEC3B gene is involved in calcium ion binding and encodes a protein that is important for bone mineralization, cellular response to transforming growth factor beta stimulation, the positive regulation of plasminogen activation, and the development of the body’s skeletal system [25].
The ABCA14 gene is associated with the marbling category in our analysis, it was detected on chromosome 25, and within it, the SNP Hapmap43799-BTA-17809 was identified. Previously, this gene was found in a population of dairy cattle and correlated with male fertility [26]. The ZP2 gene, discovered by DNA extraction from semen straws of Holstein bulls, is responsible for oocyst maturation at different stages [27]. The analysis of commercial frozen semen from six Holstein Friesian bulls aged 2 and 4 years shows that two genes, SPADH1 and SPADH2, play a role in single fertilization [28].
By analyzing the functional annotation of the genes for the three traits studied, we can expand the understanding of the genetic architecture and select genes for further molecular validation.
Gene set enrichment analysis and gene set correlation were performed in the WebGestalt web-based program. To avoid false positives, it is necessary to designate enrichment categories using multiple tools (five enrichment tools for GO biological process, six for GO molecular function, and three for KEGG pathways) that are considered consistently enriched. Using these selection criteria, the general categories of biological, molecular, and cellular GO processes can be identified (see Figure 5).

Figure 5.
Ratio of 44 functionally identified genes across the following library groups: biological, cellular, and molecular.
The results showed the highest reliability (p = 3.91 × 10−4) for the GO:0007338 libraries responsible for the biological process, namely, predisposition to single fertilization (high fertility). It included the genes SPADH1 (spermadhesin 1), SPADH2 (spermadhesin 2), and ZP2 (zona pellucida glycoprotein 2), and the enrichment ratio was 20,57. The following annotation of the identified genes was performed in the Cattle QTL database, and the results are presented in Table 7.

Table 7.
Functional annotation of genes registered in the Cattle QTL database.
The highest number of QTLs was identified for the RBFOX3 gene (15 QTLs). This gene and the OSBPL3 gene, the results for which are presented in Table 6 and registered 1 QTL, are significant for the further development of tests for them and their introduction into genomic selection programs of beef cattle.
4. Conclusions
The results provide useful information for understanding genetic architecture and provide some foundations for molecular breeding. The overall reliability of animal genotyping was 99.5%. Functional annotation performed using the web-based program DAVID detected 33 genes associated with biological processes of the organism (GO BP) and 5 genes registered in the Cattle QTL database. For the OSBPL3 and RBFOX3 genes, within which the SNPs Hapmap39351-BTA-71190 and ARS-BFGL-NGS-11938 are localized, interrelated with slaughter weight and meat marbling, we recommend developing tests for their inclusion in the genomic selection of beef cattle. Of special interest is the OSBPL3 gene, within which the identified SNPs are located, due to which it is possible to develop unique and breed-specific tests in the future.
The study revealed a number of significant SNPs associated with economically useful traits (slaughter weight, meat yield, meat marbling) in Aberdeen Angus cattle. Genes containing SNPs (OSBPL3, ABCG5, CRYM, ZP2, and ACADSB) were identified mainly for their involvement in biological quality and metabolism.
Author Contributions
Conceptualization, V.K. and A.R.; methodology, D.K. and E.B.; software, A.R. and D.K.; validation, V.K., and E.B.; resources, V.K., A.R., and A.P; data curation, A.R., V.K., and D.K.; writing—original draft preparation, V.K., D.K., and E.B.; writing—review and editing, E.B. and V.K.; visualization, A.R. and V.K.; supervision, A.R.; project administration, A.R. Conceptualization, V.K. and E.B.; methodology, D.K. and E.B.; software, A.R. and D.K.; validation, V.R., and E.B.; resources, V.K., A.R., and V.R.; data curation, A.R., V.K., and D.K.; writing—original draft preparation, V.K., D.K., and E.B.; writing—review and editing, E.B., and D.K.; visualization, A.R. and V.R.; supervision, V.K.; project administration, V.K. All authors have read and agreed to the published version of the manuscript.
Funding
Research supported by Scientific Project No. FNWZ-2024-0003 for FSSI FRC BST RAS, Russia (Founder: Ministry of Science and Higher Education of the Russian Federation).
Institutional Review Board Statement
All animal protocols used in this study were approved by the institutional Animal Care and Use Committee of the Federal Research Centre of Biological Systems and Agrotechnologies of the Russian Academy of Sciences (Russian).
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Acknowledgments
The authors would like to express their gratitude to V.R. Kharzinova, the head of the DNA technologies laboratory in animal husbandry, and A.A. Belous, the head of the Genetic Technologies Laboratory in agro- and aquatic farming of the Ernst Federal Research Center of Animal Husbandry, for their comprehensive assistance in conducting our research.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Bhowmik, N.; Seaborn, T.; Ringwall, K.A.; Dahlen, C.R.; Swanson, K.C.; Hulsman Hanna, L.L. Genetic Distinctness and Diversity of American Aberdeen Cattle Compared to Common Beef Breeds in the United States. Genes 2023, 14, 1842. [Google Scholar] [CrossRef] [PubMed]
- de Mello Klocker Vasconcellos, L.P.; Tambasco-Talhari, D.; Pereira, A.P.; Coutinho, L.L.; Regitano, L.C. Genetic characterization of Aberdeen Angus cattle using molecular markers. Genet. Mol. Biol. 2003, 26, 133–137. [Google Scholar] [CrossRef]
- Márton, J.; Szabó, F.; Zsolnai, A.; Anton, I. Genetic diversity and phylogenetic relationship of Angus herds in Hungary and analyses of their production traits. Anim. Biosci. 2024, 37, 184–192. [Google Scholar] [CrossRef]
- Konovalova, E.N.; Selionova, M.I.; Gladyr, E.A.; Romanenkova, O.S.; Evstafeva, L.V. The analysis of the Russian beef cattle population on polymorphism of CAPN1 gene. Agric. Biol. 2023, 58, 622–637. [Google Scholar] [CrossRef]
- Konovalova, E.N.; Romanenkova, O.S.; Gladyr, E.A. Aberdeen Angus beef quality and profitability in dependence of the genotypes. Anim. Husb. Food Produc. 2024, 107, 42–50. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, R.A. When to use the Bonferroni correction. Ophthalmic Physiol. Opt. 2014, 34, 502–508. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: http://www.R-project.org/ (accessed on 14 August 2022).
- Kinsella, R.J.; Kähäri, A.; Haider, S.; Zamora, J.; Proctor, G.; Spudich, G.; Almeida-King, J.; Staines, D.; Derwent, P.; Kerhornou, A.; et al. Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database 2011, 2011, bar030. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Sherman, B.; Lempicki, R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Mota, L.F.M.; Santos, S.W.B.; Júnior, G.A.F.; Bresolin, T.; Mercadante, M.E.Z.; Silva, J.A.V.; Cyrillo, J.N.S.G.; Monteiro, F.M.; Carvalheiro, R.; Albuquerque, L.G. Meta-analysis across Nellore cattle populations identifies common metabolic mechanisms that regulate feed efficiency-related traits. BMC Genom. 2022, 23, 424. [Google Scholar] [CrossRef]
- Colombi, D.; Rovelli, G.; Luigi-Sierra, M.G.; Ceccobelli, S.; Guan, D.; Perini, F.; Sbarra, F.; Quaglia, A.; Sarti, F.M.; Pasquini, M.; et al. Population structure and identification of genomic regions associated with productive traits in five Italian beef cattle breeds. Sci. Rep. 2024, 14, 8529. [Google Scholar] [CrossRef] [PubMed]
- Swathi, D.; Ramya, L.; Archana, S.S.; Krishnappa, B.; Binsila, B.K.; Selvaraju, S. Identification of hub genes and their expression profiling for predicting buffalo (Bubalus bubalis) semen quality and fertility. Sci. Rep. 2023, 13, 22126. [Google Scholar] [CrossRef] [PubMed]
- Abo-Ismail, M.K.; Vander Voort, G.; Squires, J.J.; Swanson, K.C.; Mandell, I.B.; Liao, X.; Stothard, P.; Moore, S.; Plastow, G.; Mille, S.P. Single nucleotide polymorphisms for feed efficiency and performance in crossbred beef cattle. BMC Genet. 2014, 15, 14. [Google Scholar] [CrossRef]
- Tizioto, P.C.; Taylor, J.F.; Decker, J.E.; Gromboni, C.F.; Mudadu, M.A.; Schnabel, R.D.; Coutinho, L.L.; Mourão, G.B.; Oliveira, P.S.; Souza, M.M.; et al. Detection of quantitative trait loci for mineral content of Nelore longissimus dorsi muscle. Genet. Sel. Evol. 2015, 47, 15. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.O.; Fernandes Júnior, G.A.; Fonseca, L.F.S.; Mota, L.F.M.; Bresolin, T.; Carvalheiro, R.; de Albuquerque, L.G. Genome-wide association study for stayability at different calvings in Nellore beef cattle. BMC Genom. 2024, 25, 93. [Google Scholar] [CrossRef]
- Hale, B.J.; Hager, C.L.; Seibert, J.T.; Selsby, J.T.; Baumgard, L.H.; Keating, A.F.; Ross, J.W. Heat stress induces autophagy in pig ovaries during follicular development. Biol. Reprod. 2017, 97, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, S.; Kuma, A.; Murakami, M.; Kishi, C.; Yamamoto, A.; Mizushima, N. Autophagy is essential for preimplantation development of mouse. Science 2008, 321, 117–120. [Google Scholar] [CrossRef]
- Cesar, A.S.; Regitano, L.C.; Mourão, G.B.; Tullio, R.R.; Lanna, D.P.; Nassu, R.T.; Mudado, M.A.; Oliveira, P.S.; do Nascimento, M.L.; Chaves, A.S.; et al. Genome-wide association study for intramuscular fat deposition and composition in Nellore cattle. BMC Genet. 2014, 15, 39. [Google Scholar] [CrossRef]
- Rowan, T.N.; Schnabel, R.D.; Decker, J.E. Uncovering the architecture of selection in two Bos taurus cattle breeds. Evol. Appl. 2024, 17, e13666. [Google Scholar] [CrossRef]
- Moré, D.D.; Cardoso, F.F.; Mudadu, M.A.; Malagó, W., Jr.; Gulias-Gomes, C.C.; Sollero, B.P.; Ibelli, A.M.G.; Coutinho, L.L.; Regitano, L.C.A. Network analysis uncovers putative genes affecting resistance to tick infestation in Braford cattle skin. BMC Genom. 2019, 20, 998. [Google Scholar] [CrossRef]
- Qanbari, S.; Gianola, D.; Hayes, B.; Schenkel, F.; Miller, S.; Moore, S.; Thaller, G.; Simianer, H. Application of site and haplotype-frequency based approaches for detecting selection signatures in cattle. BMC Genom. 2011, 12, 318. [Google Scholar] [CrossRef]
- Afonso, J.; Shim, W.J.; Boden, M.; Salinas Fortes, M.R.; da Silva Diniz, W.J.; de Lima, A.O.; Rocha, M.I.P.; Cardoso, T.F.; Bruscadin, J.J.; Gromboni, C.F.; et al. Repressive epigenetic mechanisms, such as the H3K27me3 histone modification, were predicted to affect muscle gene expression and its mineral content in Nelore cattle. Biochem. Biophys. Rep. 2023, 33, 101420. [Google Scholar] [CrossRef]
- Mellouk, N.; Rame, C.; Naquin, D.; Jaszczyszyn, Y.; Touzé, J.L.; Briant, E.; Guillaume, D.; Ntallaris, T.; Humblot, P.; Dupont, J. Impact of the severity of negative energy balance on gene expression in the subcutaneous adipose tissue of periparturient primiparous Holstein dairy cows: Identification of potential novel metabolic signals for the reproductive system. PLoS ONE. 2019, 14, e0222954. [Google Scholar] [CrossRef] [PubMed]
- Buzanskas, M.E.; Grossi, D.A.; Ventura, R.V.; Schenkel, F.S.; Sargolzaei, M.; Meirelles, S.L.; Mokry, F.B.; Higa, R.H.; Mudadu, M.A.; da Silva, M.V.; et al. Genome-wide association for growth traits in Canchim beef cattle. PLoS ONE 2014, 9, e94802. [Google Scholar] [CrossRef] [PubMed]
- Hiltpold, M.; Kadri, N.K.; Janett, F.; Witschi, U.; Schmitz-Hsu, F.; Pausch, H. Autosomal recessive loci contribute significantly to quantitative variation of male fertility in a dairy cattle population. BMC Genom. 2021, 22, 225. [Google Scholar] [CrossRef] [PubMed]
- Cochran, S.D.; Cole, J.B.; Null, D.J.; Hansen, P.J. Discovery of single nucleotide polymorphisms in candidate genes associated with fertility and production traits in Holstein cattle. BMC Genet. 2013, 14, 49. [Google Scholar] [CrossRef] [PubMed]
- Westfalewicz, B.; Słowińska, M.; Judycka, S.; Ciereszko, A.; Dietrich, M.A. Comparative Proteomic Analysis of Young and Adult Bull (Bos taurus) Cryopreserved Semen. Animals 2021, 11, 2013. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).