Screening of circRNAs Associated with Secondary Wool Follicle Development in Fine-Wool Sheep and Construction of Their ceRNA Network
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Moral Declaration
2.2. Sample Collection and Animal Processing
2.3. circRNA Sequencing
2.3.1. Extraction of Total RNA from the Skin and RNA Inversion
2.3.2. Library Preparation and Sequencing
2.3.3. Identification and Characteristic Analysis of circRNA
2.4. DE circRNA Screening and Functional Enrichment Analysis of Source Genes
2.5. Validation of DE circRNAs
2.6. Construction of circRNA-miRNA-mRNA Interaction Network
2.7. Statistical Analysis—Quantification of Gene Expression and Analysis
3. Results
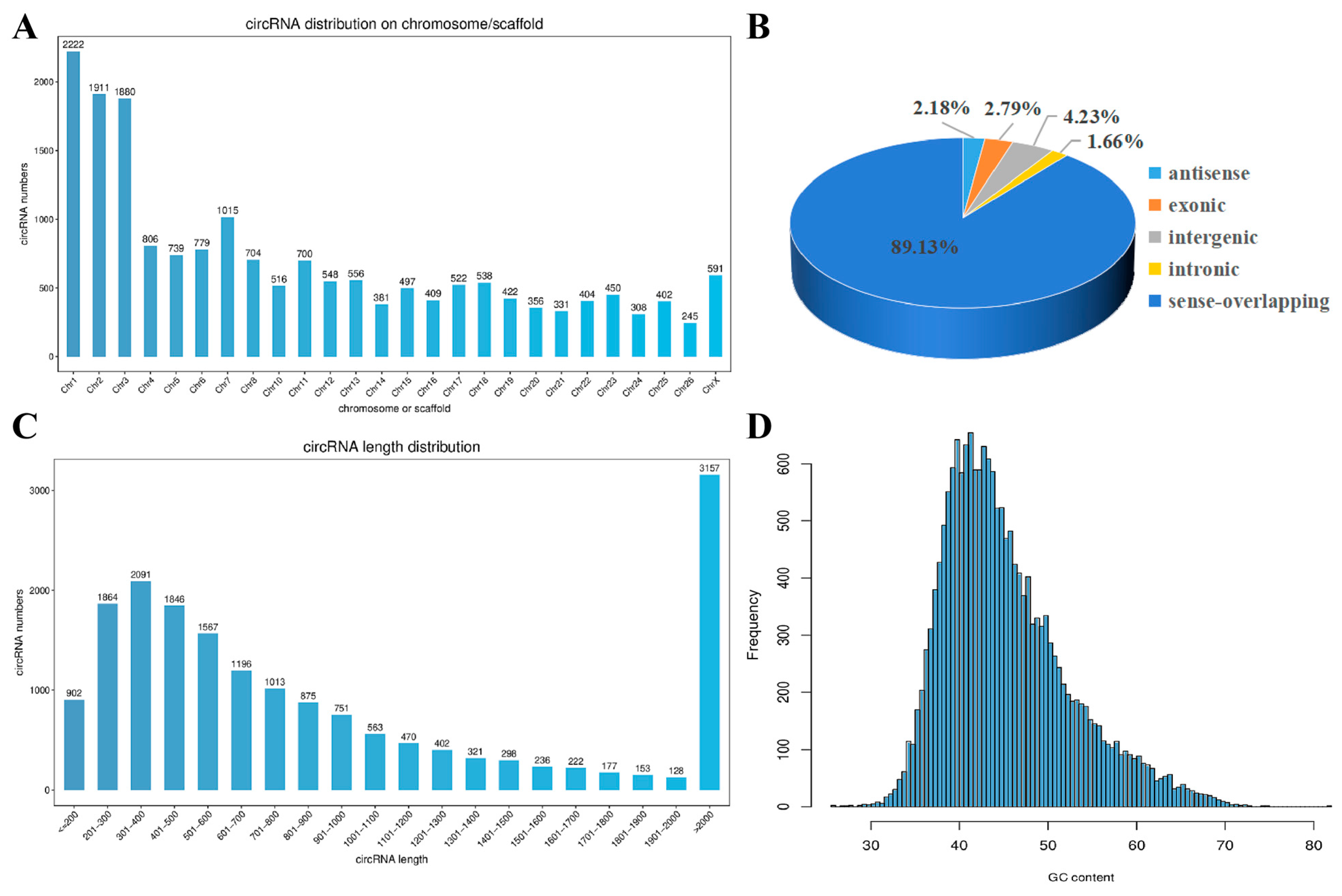
3.1. High-Throughput Sequencing Quality Assessment
3.2. circRNA Identification and Expression Analysis
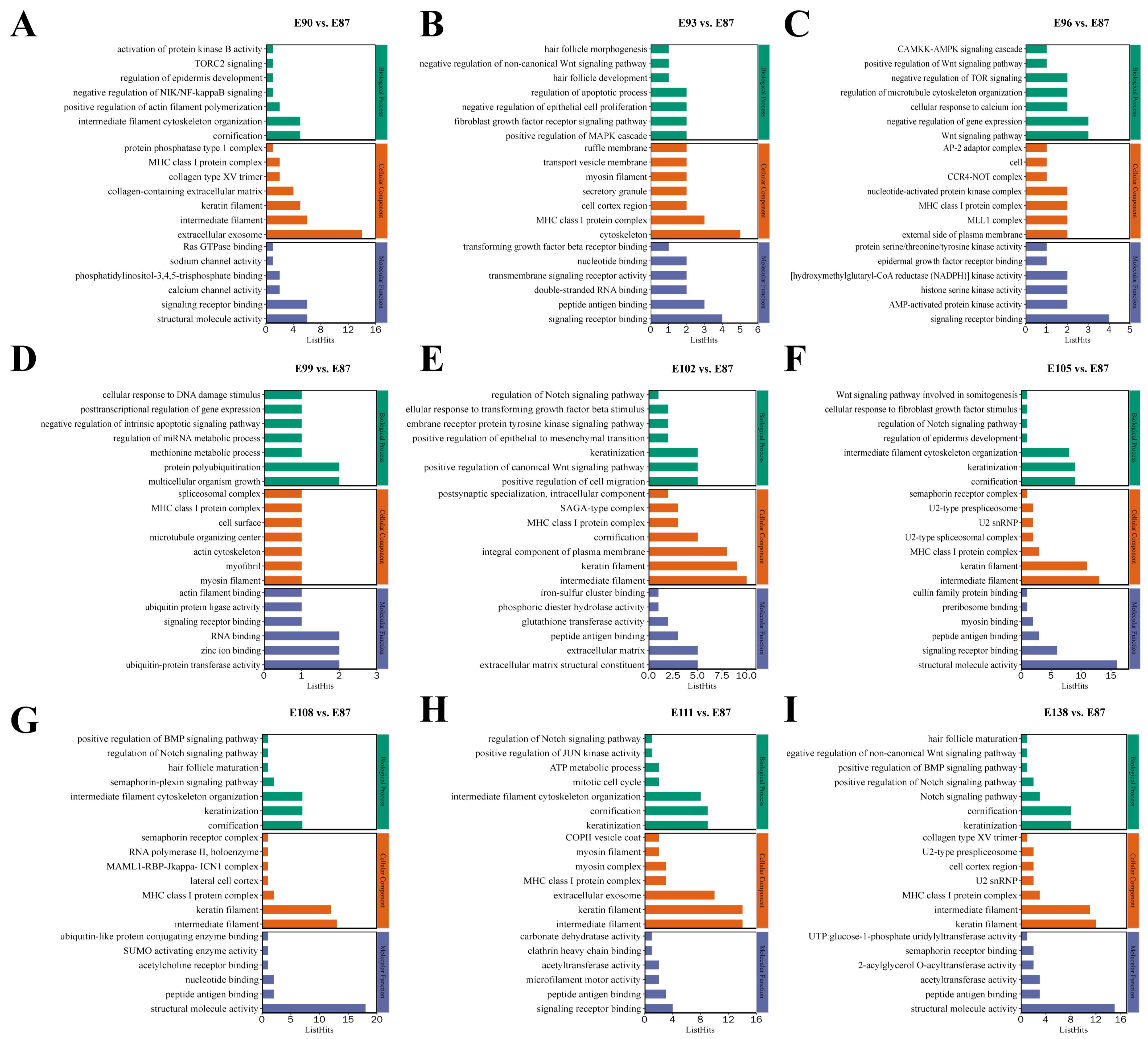
3.3. Functional Enrichment Analysis of Target Genes
3.4. Validation of the RNA-Seq Data
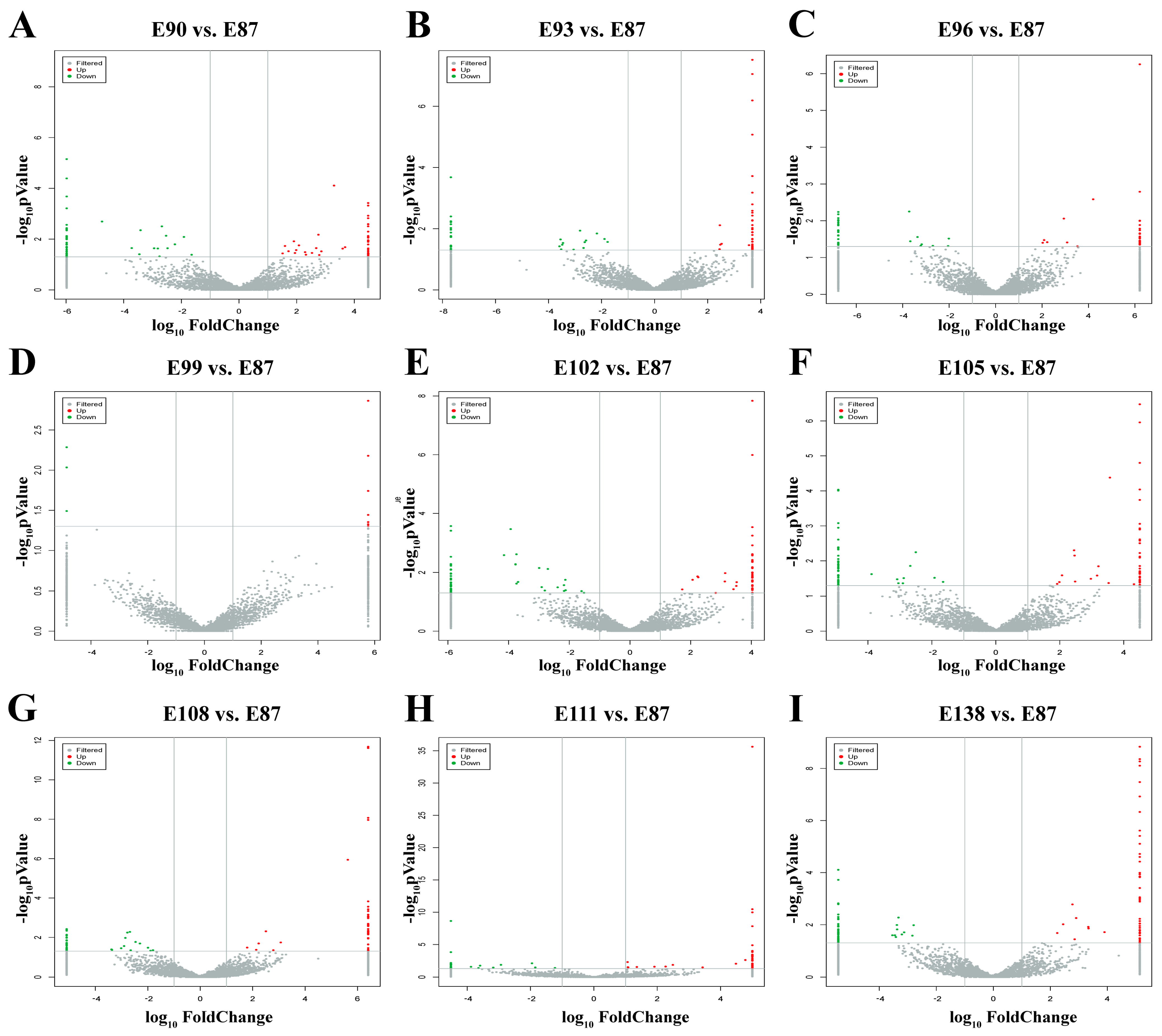
3.5. Screening of DE circRNAs Related to Secondary Hair Follicle Development
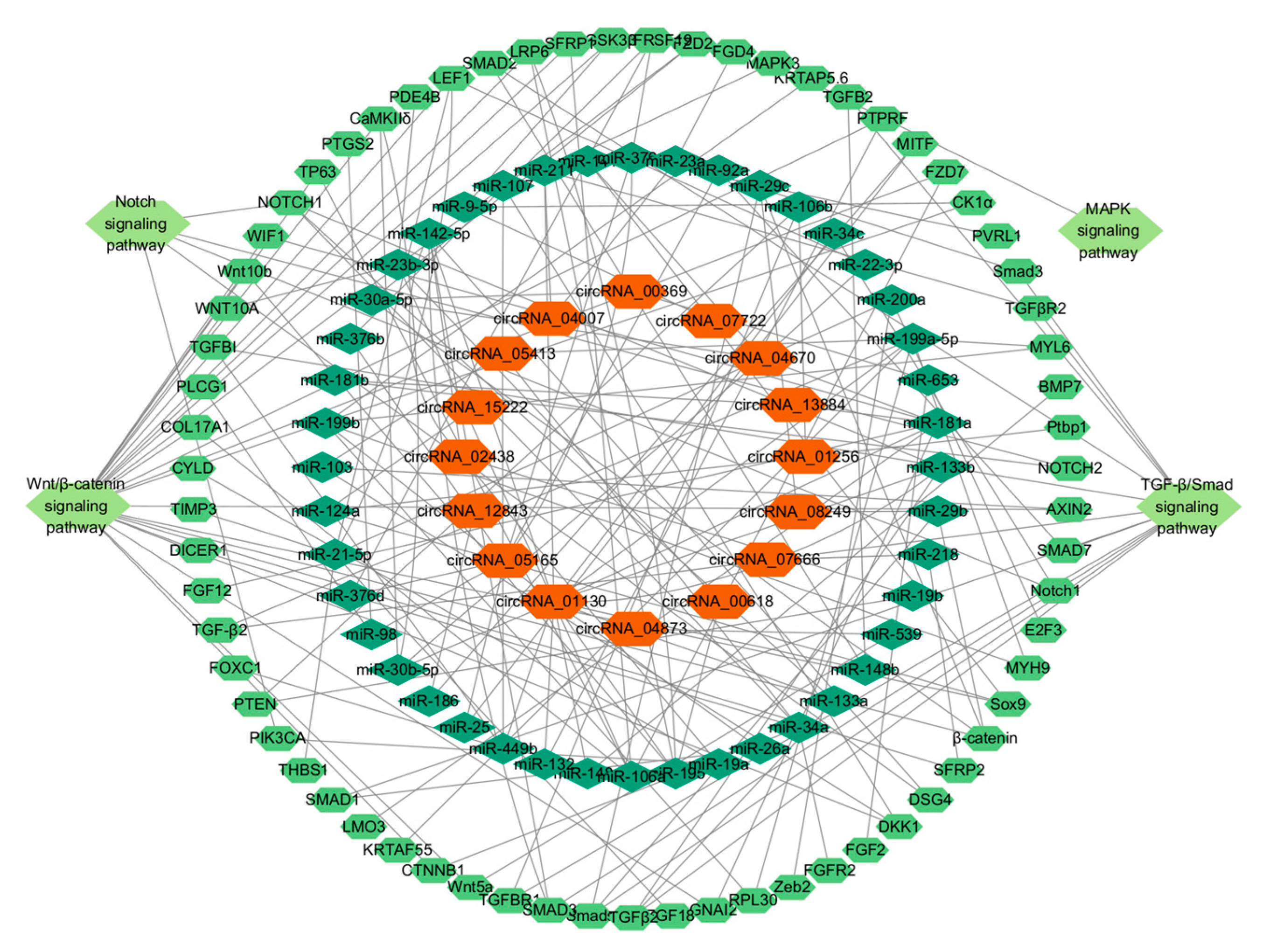
3.6. Analysis of circRNA-miRNA-mRNA Interaction Network
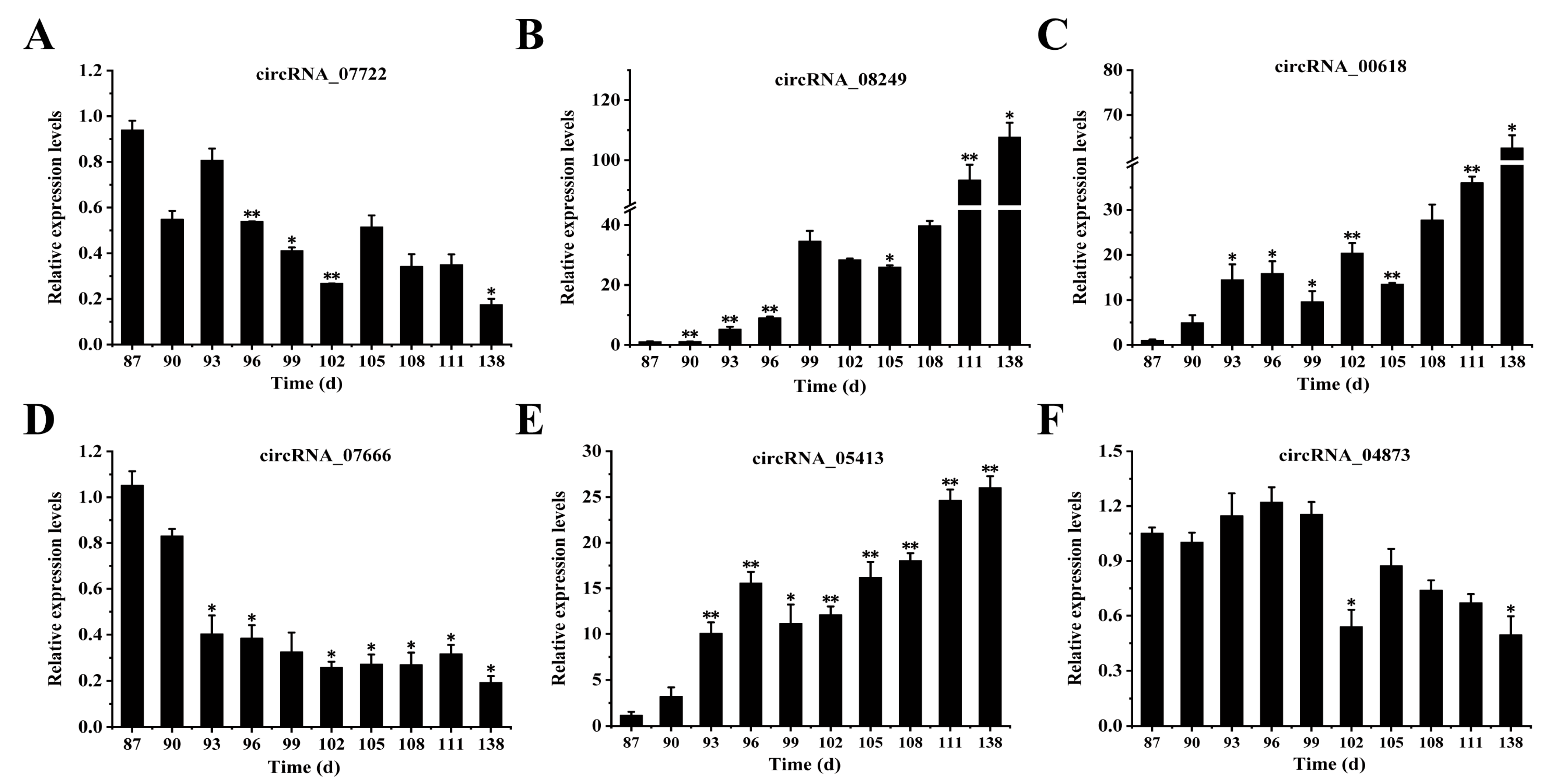
3.7. Analysis of the Expression Levels of circRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, H.Y.; Yang, H.; Shi, G.Q.; Shen, M.; Yang, J.Q.; Yang, Y.L.; Liu, X.J. Expression profile analysis of microRNAs during hair follicle development in the sheep foetus. Biosci. Biotechnol. Biochem. 2019, 83, 1045–1061. [Google Scholar] [CrossRef] [PubMed]
- Sperling, L.C. Hair anatomy for the clinician. J. Am. Acad. Dermatol. 1991, 25, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Lyne, A. The development of the epidermis and hair canals in the merino sheep foetus. Aust. J. Biol. Sci. 1957, 10, 390. [Google Scholar] [CrossRef]
- Parry, A.L.; Nixon, A.J.; Craven, A.J.; Pearson, A.J. The microanatomy, cell replication, and keratin gene expression of hair follicles during a photoperiod-induced growth cycle in sheep. Cells Tissues Organs. 1995, 154, 283–299. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, X.; Yan, H.; Zeng, J.; Ma, S.; Niu, Y.; Zhou, G.; Jiang, Y.; Chen, Y. Comparative transcriptome analysis of fetal skin reveals key genes related to hair follicle morphogenesis in cashmere goats. PLoS ONE 2016, 11, e0151118. [Google Scholar] [CrossRef]
- Li, G.; Ji, Y.; Li, Y. Molecular mechanism of hair follicle morphogenesis. Int. J. Dermatol. Venereol. 2004, 30, 38. [Google Scholar]
- McMahon, A.P.; Ingham, P.W.; Tabin, C.J. Developmental roles and clinical significance of Hedgehog signaling. Curr. Top. Dev. Biol. 2003, 53, 1–114. [Google Scholar] [CrossRef]
- Crowe, R.; Henrique, D.; Ish-Horowicz, D.; Niswander, L. A new role for Notch and Delta in cell fate decisions: Patterning the feather array. Development 1998, 125, 767–775. [Google Scholar] [CrossRef]
- Schmidt-Ullrich, R.; Paus, R. Molecular principles of hair follicle induction and morphogenesis. BioEssays 2005, 27, 247–261. [Google Scholar] [CrossRef]
- Thomadakis, G.; Ramoshebi, L.N.; Crooks, J.; Rueger, D.C.; Ripamonti, U. Immunolocalization of bone morphogenetic protein-2 and-3 and osteogenic protein-1 during murine tooth root morphogenesis and in other craniofacial structures. Eur. J. Oral. Sci. 1999, 107, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Millar, S.E. Molecular mechanisms regulating hair follicle development. J. Invest. Dermatol. 2002, 118, 216–225. [Google Scholar] [CrossRef]
- Chen, L.-L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.-O.; Chen, T.; Xiang, J.-F.; Yin, Q.-F.; Xing, Y.-H.; Zhu, S.; Yang, L.; Chen, L.-L. Circular intronic long noncoding RNAs. Mol. Cell. 2013, 51, 792–806. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Vlatkovic, I.; Babic, A.; Will, T.; Epstein, I.; Tushev, G.; Akbalik, G.; Wang, M.; Glock, C.; Quedenau, C.; et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015, 18, 603–610. [Google Scholar] [CrossRef]
- Xu, Z.; Li, P.; Fan, L.; Wu, M. The potential role of circRNA in tumor immunity regulation and immunotherapy. Front. Immunol. 2018, 9, 9. [Google Scholar] [CrossRef]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Westholm, J.O.; Miura, P.; Olson, S.; Shenker, S.; Joseph, B.; Sanfilippo, P.; Celniker, S.E.; Graveley, B.R.; Lai, E.C. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014, 9, 1966–1980. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Wang, J.; Xu, Y.; Zhou, H.; Li, Y.; Sun, W. CircCSPP1 Competitively Binds miR-10a to Regulate BMP7 Expression and Affects the Proliferation of Dermal Papilla Cells. Int. J. Mol. Sci. 2024, 25, 11547. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, Y.; Zhao, J.; Shen, J.; Wang, Z.; Bai, M.; Fan, Y.; Yin, R.; Mao, Y.; Bai, W. CircRNA-1967 participates in the differentiation of goat SHF-SCs into hair follicle lineage by sponging miR-93-3p to enhance LEF1 expression. Anim. Biotechnol. 2023, 34, 482–494. [Google Scholar] [CrossRef]
- Wu, Y.Y.; Yue, Y.J.; Guo, T.T.; Wang, T.X.; Guo, J.; Li, G.Y.; Han, J.L.; Yang, M.; Liu, J.B.; Sun, X.P. Fetal skin hair follicle development and morphological structure of Chinese super-fine wool sheep (Gansu type). Sci. Agric. Sin. 2013, 46, 1923. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef]
- Frazee, A.C.; Sabunciyan, S.; Hansen, K.D.; Irizarry, R.A.; Leek, J.T. Differential expression analysis of RNA-seq data at single-base resolution. Biostatistics 2014, 15, 413–426. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Mao, Y.; Wang, J.; Chen, R.; Libing, Z.; Su, Y.-Q.; Chen, J.; Zheng, W.-Q. Analysis of liver and gill miRNAs of Larimichthys crocea against Cryptocryon irritans challenge. Fish. Shellfish Immunol. 2016, 59, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.-D.; Lin, F.-M.; Wu, W.-Y.; Liang, C.; Huang, W.-C.; Chan, W.-L.; Tsai, W.-T.; Chen, G.-Z.; Lee, C.-J.; Chiu, C.-M.; et al. miRTarBase: A database curates experimentally validated microRNA–target interactions. Nucleic Acids Res. 2011, 39 (Suppl. S1), D163–D169. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- McDonald, B.; Hoey, W.; Hopkins, P.S. Cyclical fleece growth in cashmere goats. Aust. J. Agric. Res. 1987, 38, 597. [Google Scholar] [CrossRef]
- Zhao, B.; Chen, Y.; Hu, S.; Yang, N.; Wang, M.; Liu, M.; Li, J.; Xiao, Y.; Wu, X. Systematic analysis of non-coding RNAs involved in the Angora rabbit (Oryctolagus cuniculus) hair follicle cycle by RNA sequencing. Front. Genet. 2019, 10, 407. [Google Scholar] [CrossRef]
- Zhu, Z.; Li, Y.; Liu, W.; He, J.; Zhang, L.; Li, H.; Li, P.; Lv, L. Comprehensive circRNA expression profile and construction of circRNA-associated ceRNA network in fur skin. Exp. Dermatol. 2018, 27, 251–257. [Google Scholar] [CrossRef]
- Tan, W.L.W.; Lim, B.T.S.; Anene-Nzelu, C.G.O.; Ackers-Johnson, M.; Dashi, A.; See, K.; Tiang, Z.; Lee, D.P.; Chua, W.W.; Luu, T.D.A.; et al. A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 2017, 113, 298–309. [Google Scholar] [CrossRef]
- Zhu, L.; Jiang, Q.; Meng, J.; Zhao, H.; Lin, J. Pan-cancer analysis of COL15A1: An immunological and prognostic biomarker. Discov. Oncol. 2024, 15, 325. [Google Scholar] [CrossRef]
- Nakamuta, S.; Yang, Y.-T.; Wang, C.-L.; Gallo, N.B.; Yu, J.-R.; Tai, Y.; Van Aelst, L. Dual role for DOCK7 in tangential migration of interneuron precursors in the postnatal forebrain. J. Cell Biol. 2017, 216, 4313–4330. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Dalal, K. ADAMTS-4 and ADAMTS-5: Key enzymes in osteoarthritis. J. Cell. Biochem. 2011, 112, 3507–3514. [Google Scholar] [CrossRef] [PubMed]
- Foomani, F.H.; Jarzembowski, J.A.; Mostaghimi, S.; Mehrvar, S.; Kumar, S.N.; Ranji, M. Optical metabolic imaging of mitochondrial dysfunction on HADH mutant newborn rat hearts. IEEE J. Transl. Eng. Health Med. 2021, 9, 1800407. [Google Scholar] [CrossRef]
- Kuivaniemi, H.; Tromp, G. Type III collagen (COL3A1): Gene and protein structure, tissue distribution, and associated diseases. Gene 2019, 707, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Yeung, W.; Qi, X.; Pang, L.; Liu, H.; Ng, K.; Liu, J.; Lo, M.; Man, K. Type III TGF-β Receptor Down-Regulation Promoted Tumor Progression via Complement Component C5a Induction in Hepatocellular Carcinoma. Cancers 2021, 13, 1503. [Google Scholar] [CrossRef]
- Gao, G.; Li, X.; Jiang, Z.; Osorio, L.; Tang, Y.L.; Yu, X.; Jin, G.; Zhou, Z. Isthmin-1 (Ism1) modulates renal branching morphogenesis and mesenchyme condensation during early kidney development. Nat. Commun. 2023, 14, 2378. [Google Scholar] [CrossRef]
- Higginbotham, H.; Guo, J.; Yokota, Y.; Umberger, N.L.; Su, C.-Y.; Li, J.; Verma, N.; Hirt, J.; Ghukasyan, V.; Caspary, T.; et al. Arl13b-regulated cilia activities are essential for polarized radial glial scaffold formation. Nat. Neurosci. 2013, 16, 1000–1007. [Google Scholar] [CrossRef]
- Ge, W.; Zhang, W.; Zhang, Y.; Zheng, Y.; Li, F.; Wang, S.; Liu, J.; Tan, S.; Yan, Z.; Wang, L.; et al. A Single-cell Transcriptome Atlas of Cashmere Goat Hair Follicle Morphogenesis. Genom. Proteom. Bioinform. 2021, 19, 437–451. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, P.; Chen, T.; Yan, W.; Guan, X.; Shen, G.; Luo, X.; Wan, X.; Ning, Q. Kctd9 deficiency impairs natural killer cell development and effector function. Front. Immunol. 2019, 10, 744. [Google Scholar] [CrossRef]
- Gao, W.Z.; Xue, H.L.; Yang, J.C. Proteomics analysis of the secondary hair follicle cycle in Liaoning cashmere goat. Small Rumin. Res. 2021, 201, 106408. [Google Scholar] [CrossRef]
- Mumblat, Y.; Kessler, O.; Ilan, N.; Neufeld, G. Full-length semaphorin-3C is an inhibitor of tumor lymphangiogenesis and metastasis. Cancer Res. 2015, 75, 2177–2186. [Google Scholar] [CrossRef]
- McCrea, P.D.; Park, J.-I. Developmental functions of the P120-catenin sub-family. Biochim. Biophys. Acta 2007, 1773, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Sennett, R.; Wang, Z.; Rezza, A.; Grisanti, L.; Roitershtein, N.; Sicchio, C.; Mok, K.W.; Heitman, N.J.; Clavel, C.; Ma’ayan, A.; et al. An integrated transcriptome atlas of embryonic hair follicle progenitors, their niche, and the developing skin. Dev. Cell 2015, 34, 577–591. [Google Scholar] [CrossRef]
- Merkestein, M.; Laber, S.; McMurray, F.; Andrew, D.; Sachse, G.; Sanderson, J.; Li, M.; Usher, S.; Sellayah, D.; Ashcroft, F.M.; et al. FTO influences adipogenesis by regulating mitotic clonal expansion. Nat. Commun. 2015, 6, 6792. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Romero-Carvajal, A.; Navajas Acedo, J.N.; Jiang, L.; Kozlovskaja-Gumbrienė, A.; Alexander, R.; Li, H.; Piotrowski, T. Regeneration of sensory hair cells requires localized interactions between the Notch and Wnt pathways. Dev. Cell 2015, 34, 267–282. [Google Scholar] [CrossRef]
- Diike, P.; Heiden, C.H. Smad Signal Transduction: Smads in Proliferation, Differentiation and Disease; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, H.T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002, 12, 9–18. [Google Scholar] [CrossRef]
- Akilli Öztürk, Ö.A.; Pakula, H.; Chmielowiec, J.; Qi, J.; Stein, S.; Lan, L.; Sasaki, Y.; Rajewsky, K.; Birchmeier, W. Gab1 and Mapk signaling are essential in the hair cycle and hair follicle stem cell quiescence. Cell Rep. 2015, 13, 561–572. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zang, K.; Tang, Z.; Yang, T.; Ye, X.; Dang, Y. Hordenine activated dermal papilla cells and promoted hair regrowth by activating Wnt signaling pathway. Nutrients 2023, 15, 694. [Google Scholar] [CrossRef]
- Foitzik, K.; Lindner, G.; Mueller-Roever, S.; Maurer, M.; Botchkareva, N.; Botchkarev, V.; Handjiski, B.; Metz, M.; Hibino, T.; Soma, T.; et al. Control of murine hair follicle regression (catagen) by TGF-β1 in vivo. FASEB J. 2000, 14, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Foitzik, K.; Paus, R.; Doetschman, T.; Dotto, G.P. The TGF-β2 isoform is both a required and sufficient inducer of murine hair follicle morphogenesis. Dev. Biol. 1999, 212, 278–289. [Google Scholar] [CrossRef]
- Dai, B.; Sha, R.N.; Yuan, J.L.; Liu, D.J. Multiple potential roles of thymosin β4 in the growth and development of hair follicles. J. Cell Mol. Med. 2021, 25, 1350–1358. [Google Scholar] [CrossRef] [PubMed]
- RunQing, C.; HaiYin, H.; Peng, W.; KaiYang, L.; MingXing, C.; YuFang, L. Estrogen mediates CircZNF423 as a sponge for oar-miR-541-3p to target CALM3 for regulating myoblast proliferation in sheep. Sci. Agric. Sin. 2024, 57, 597. [Google Scholar] [CrossRef]
- Yin, R.H.; Zhao, S.J.; Jiao, Q.; Wang, Z.Y.; Bai, M.; Fan, Y.X.; Zhu, Y.B.; Bai, W.L. CircRNA-1926 promotes the differentiation of goat SHF stem cells into hair follicle lineage by miR-148a/b-3p/CDK19 axis. Animals 2020, 10, 1552. [Google Scholar] [CrossRef]
- Tian, D.; Pei, Q.; Jiang, H.; Guo, J.; Ma, X.; Han, B.; Li, X.; Zhao, K. Comprehensive analysis of the expression profiles of mRNA, lncRNA, circRNA, and miRNA in primary hair follicles of coarse sheep fetal skin. BMC Genom. 2024, 25, 574. [Google Scholar] [CrossRef]
- Chen, F.; Feng, Z.; Zhu, J.; Liu, P.; Yang, C.; Huang, R.; Deng, Z. Emerging roles of circRNA_NEK6 targeting miR-370-3p in the proliferation and invasion of thyroid cancer via Wnt signaling pathway. Cancer Biol. Ther. 2018, 19, 1139–1152. [Google Scholar] [CrossRef] [PubMed]


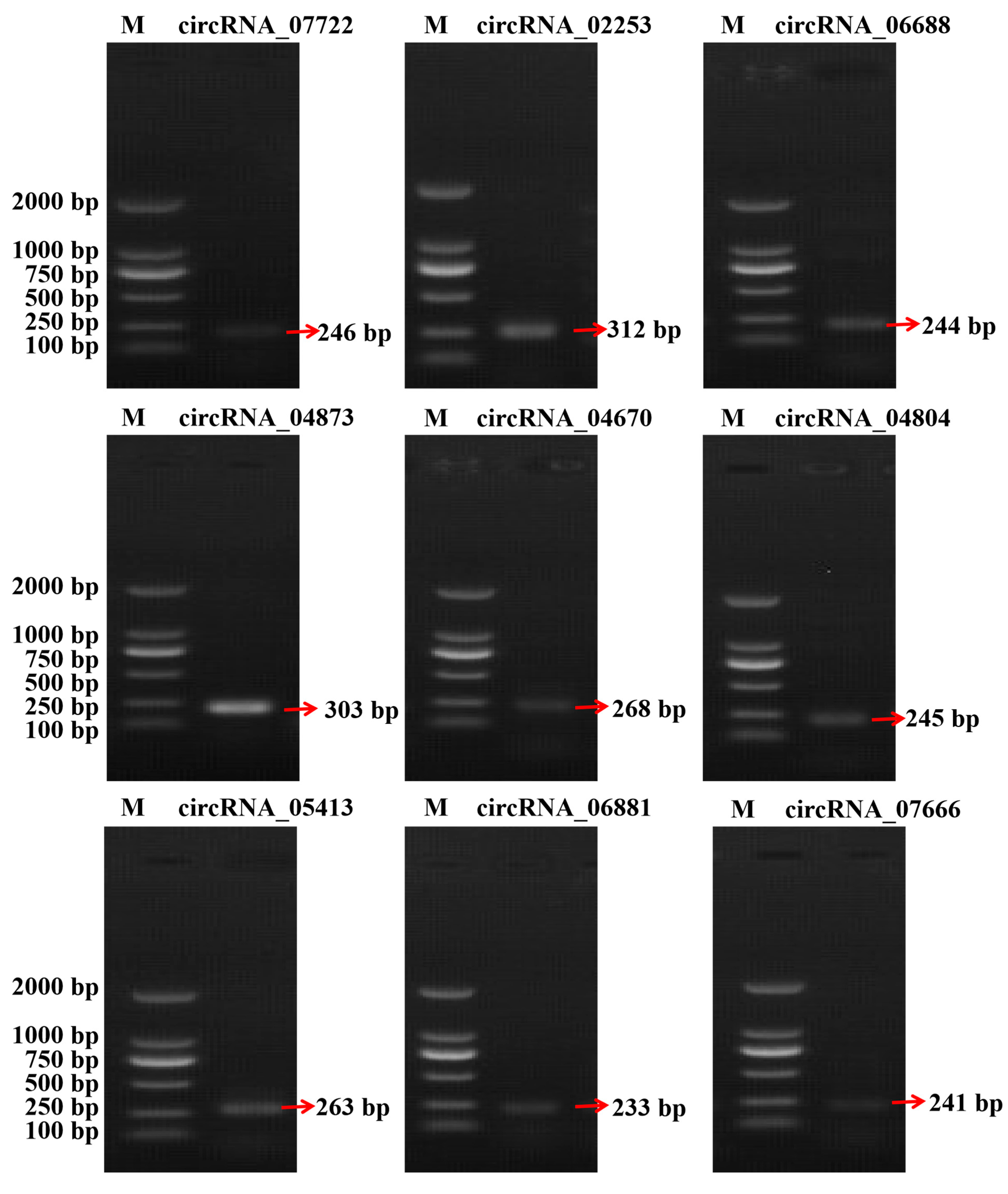
| circRNAs | Forward Primer Sequence | Reverse Primer Sequence |
|---|---|---|
| circRNA_07722 | AGCCTTCCAATAGTGAACCTCA | GCAACCAAGTGTAGTGCAGG |
| circRNA_02253 | TGAAACACAGTTGGCAGAGTCT | ACGGCAAGGCACCAAACT |
| circRNA_06688 | TGGTGAAGACATTGCAGACGA | AGCTCATTCACTTGTTCTCTCCA |
| circRNA_04873 | GCGGATGAAAGGAATGGGGA | TGCTCTGTTGACTCCCTGAAA |
| circRNA_06881 | GAGAACCAGTGGCTGCGG | GCTTCTCCAACTTGTCCTCCT |
| circRNA_04670 | TCGAGTTTGAATGGCTGAGACA | AGATGTATTCCAAGGTCCCCG |
| circRNA_04804 | GTTGGAGCAGGAAGAGGAGC | TGAGATAGCAGGAGTTTGGAAGAC |
| circRNA_07666 | TTTCTCGATTGGACCTGCGA | GGGTGAATGATCCTCTGGTGG |
| circRNA_05413 | GGAGATCCCGCAAGCAGAG | AGCTTGCTATCCGAGTCTTTCT |
| circRNA_03494 | ATTGTTGCCTACGCCCACTT | GCCATTCGTGAACAGCATCG |
| circRNA_08249 | AAGGGTGATTCTGGTGCTCC | CAGACCAGGTGTACCAGCAG |
| β-actin | CAGTCGGTTGGATGGAGCAT | AGGCAGGGACTTCCTGTAAC |
| circRNA ID | Source Gene | p-Value | Up/ Down | circRNA Size | Isoform Name |
|---|---|---|---|---|---|
| circRNA_07722 | COL15A1 | 2.29 × 10−2 | Down | 362 | XM_027964395.1 |
| circRNA_00369 | DOCK7 | 7.36 × 10−3 | Down | 429 | XM_015092012.2 |
| circRNA_01130 | ADAMTS5 | 2.03 × 10−3 | Down | 301 | XM_015092381.2 |
| circRNA_15222 | HADH | 4.82 × 10−2 | Down | 504 | XM_004009637.4 |
| circRNA_08249 | COL3A1 | 7.73 × 10−3 | Up | 315 | XM_004004514.4 |
| circRNA_00618 | TGFBR3 | 4.99 × 10−2 | Up | 871 | XM_012176015.3 |
| circRNA_04007 | ISM1 | 2.15 × 10−2 | Up | 739 | XM_027976478.1 |
| circRNA_01256 | ARL13B | 1.43 × 10−2 | Up | 1077 | XM_004002849.4 |
| circRNA_02438 | POSTN | 4.29 × 10−2 | Down | 570 | XM_004012110.4 |
| circRNA_07666 | KCTD9 | 3.92 × 10−2 | Down | 314 | XM_027964318.1 |
| circRNA_12843 | KRT77 | 8.74 × 10−4 | Up | 12,347 | XM_027967277.1 |
| circRNA_13884 | SEMA3C | 1.06 × 10−2 | Down | 1027 | XM_004007807.4 |
| circRNA_05413 | CTNND1 | 2.03 × 10−2 | Up | 272 | XM_027979807.1 |
| circRNA_05165 | DKK3 | 2.54 × 10−2 | Down | 222 | XM_027979387.1 |
| circRNA_04873 | ZNF114 | 4.36 × 10−2 | Down | 1359 | XM_027978554.1 |
| circRNA_04670 | FTO | 4.35 × 10−2 | Down | 344 | XM_027977360.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Y.; Xi, B.; Song, Y.; Xiao, T.; Lu, Z.; Liu, J.; Yuan, C.; Guo, T. Screening of circRNAs Associated with Secondary Wool Follicle Development in Fine-Wool Sheep and Construction of Their ceRNA Network. Animals 2025, 15, 2629. https://doi.org/10.3390/ani15172629
Luo Y, Xi B, Song Y, Xiao T, Lu Z, Liu J, Yuan C, Guo T. Screening of circRNAs Associated with Secondary Wool Follicle Development in Fine-Wool Sheep and Construction of Their ceRNA Network. Animals. 2025; 15(17):2629. https://doi.org/10.3390/ani15172629
Chicago/Turabian StyleLuo, Yu, Binpeng Xi, Yufang Song, Tong Xiao, Zengkui Lu, Jianbin Liu, Chao Yuan, and Tingting Guo. 2025. "Screening of circRNAs Associated with Secondary Wool Follicle Development in Fine-Wool Sheep and Construction of Their ceRNA Network" Animals 15, no. 17: 2629. https://doi.org/10.3390/ani15172629
APA StyleLuo, Y., Xi, B., Song, Y., Xiao, T., Lu, Z., Liu, J., Yuan, C., & Guo, T. (2025). Screening of circRNAs Associated with Secondary Wool Follicle Development in Fine-Wool Sheep and Construction of Their ceRNA Network. Animals, 15(17), 2629. https://doi.org/10.3390/ani15172629





