Simple Summary
The study presents the molecular detection of two bacterial agents in a hard tick (Amblyomma pseudoconcolor) collected on a large hairy armadillo (Chaetophractus villosus) from the Argentinian Patagonia. Molecular detection of bacterial agents was performed by polymerase chain reaction (PCR). One tick, determined morphologically and genetically as A. pseudoconcolor, was collected on C. villosus. The bacterial agents detected in the hard tick were identified as Candidatus Rickettsia andeanae and Ehrlichia sp. The results of this study and previous findings suggest that A. pseudoconcolor may be a potential vector of some Rickettsia and Ehrlichia bacteria of unknown pathogenicity.
Abstract
This study presents the molecular detection of Candidatus Rickettsia andeanae and Ehrlichia sp. in Amblyomma pseudoconcolor Aragão, 1908 (Acari: Ixodidae) collected on a large hairy armadillo (Chaetophractus villosus (Desmarest, 1804)). On 12 October 2020, a specimen of C. villosus was found dead on the road in Río Negro province, Argentina. Molecular detection of Rickettsia and Ehrlichia agents was performed amplifying the gltA and 16S rRNA gene, respectively. One tick, determined morphologically and genetically as A. pseudoconcolor, was collected on C. villosus. The rickettsial agent detected in A. pseudoconcolor was identified as Candidatus Rickettsia andeanae. The Ehrlichia sp. strain showed high sequence similarity to different uncultured Ehrlichia sp. detected in horses, capybaras and Ixodes ornithorhynchi from Nicaragua, Brazil and Australia, respectively. The results of this study and previous findings suggest that A. pseudoconcolor may be a potential vector of some Rickettsia and Ehrlichia bacteria of unknown pathogenicity.
1. Introduction
The large hairy armadillo, Chaetophractus villosus (Desmarest, 1804) (Cingulata: Chlamyphoridae) is one of the largest species of the order Cingulata in South America and the most abundant species of armadillos in Argentina [1]. The geographical distribution of this mammal reaches from the southeast of Bolivia and west of Paraguay to the south of Argentina, including one population situated in the Province of Tierra del Fuego in the extreme south of the Southern Cone of South America [1]. Chaetophractus villosus prefers inhabiting mostly sandy and open soils such as steppes and hills, but also intermountain valleys, plains and grasslands. Further, it could be found in human-modified environments, including peri-urban areas [1,2]. Chaetophractus villosus is frequently parasitized by ticks from the genus Amblyomma, especially the species Amblyomma auricularium (Conil, 1878) and Amblyomma pseudoconcolor Aragão, 1908 [3,4].
Amblyomma pseudoconcolor Aragão, 1908 (Acari: Ixodidae) is an endemic tick species in Argentina that is distributed from the Neotropical Region to the Andean Region of the Southern Cone of America, including the biogeographic provinces as defined by Morrone [5]: Chaco, Pampa, Monte and Central Patagonia, from the north to the south [3,6]. Its main hosts for all life stages (larvae, nymphs, adults) are mammals of the family Dasypodidae and Chlamyphoridae [4]. Parasitism of A. pseudoconcolor on C. villosus from the Argentinean Patagonia was described by Ezquiaga et al. [7]. Amblyomma pseudoconcolor is often confused morphologically with A. auricularium that also parasites on armadillos (Cingulata) (Nava et al., 2017; Guglielmone et al., 2021) [3,4]. However, A. pseudoconcolor presents a typical scutal ornamentation, which is not seen in A. auricularium [4]. Three different rickettsial agents of unknown pathogenicity were detected in A. pseudoconcolor: Candidatus Rickettsia andeanae [8,9], Rickettsia amblyommatis [10], and Rickettsia bellii [11]. In addition, human parasitism of A. pseudoconcolor is described in one case [8].
The aim of this study was to detect and identify the possible presence of bacteria belonging to the order Rickettsiales in a female specimen of A. pseudoconcolor collected on C. villosus from Río Negro province, Patagonia, Argentina.
2. Materials and Methods
On 12 October 2020, a male adult specimen of C. villosus was found dead on the road Ruta Provinicial Nº1 between the locations of Viedma and Balneario El Cóndor (40°56′44.88″ S; 62°51′40.75″ W), Río Negro, Argentina. The animal was examined manually for the presence of ticks. The tick collected free on the animal was sent to the Instituto Nacional de Tecnología Agropecuaria Rafaela, Santa Fe, Argentina, for identification and detection of tick-borne bacteria. Firstly, the tick was identified morphologically according to Nava et al. [3]. Afterwards, complete DNA was extracted using the High Pure PCR Template Preparation Kit (Roche, Mannheim, Germany) according to the manufacturer’s instructions. To confirm the morphological identification, a PCR detecting a specific fragment of the mitochondrial 16S rRNA gene of members of the order Ixodidae was processed using the primers 16S + 1 (5′-CCG GTC TGA ACT CAG ATC AAG T-3′; [12]) and 16S-1 (5′-GCT CAA TGA TTT TTT AAA TTG CTG T-3′; [12]) following Mangold et al. [13]. To detect bacteria from the order Rickettsiales, three different conventional PCR assays were used: (I) Amplification of a 345 bp fragment of the 16S rRNA gene of the family Anaplasmataceae (used primers: GE2-F2 (5′- GTT AGT GGC AGA CGG GTG AGT-3′) and HE3 (5′-TAT AGG TAC CGT CAT TAT CTT CCC TAT-3′) [14,15,16], (II) amplifying an 830 bp fragment of the Rickettsia genus specific gene for a citrate synthase—gltA (primers: CS-239: 5′-GCT CTT CTC ATC CTA TGG CTA TTA T-3′; CS-1069: 5′-CAG GGT CTT CGT GCA TTT CTT) [17], and (III) amplification of an approximate 530 bp fragment of the gene for a 190-kDa outer membrane protein (ompA) that is specific for Rickettsia sp. of the spotted fever group Rickettsiae (primer names and sequences: Rr 190.70p; 5′-ATG GCG AAT ATT TCT CCA AAA-3′ and Rr 190.602n; 5′-AGT GCA GCA TTC GCT CCC CCT-3′) [18]. In all PCR reactions, ultra-pure water was used as negative control while DNA of Ehrlichia canis and Rickettsia massiliae acted as positive control for the detection of the genes for 16S rRNA and gltA, respectively. Positive PCR amplicons of the three assays were purified using the High Pure PCR Product Purification Kit (Roche, Mannheim, Germany) and sent to INTA Castelar (Genomics Unit, Buenos Aires, Argentina) for sequencing. Obtained partial sequences were edited using BioEdit Sequence Alignment Editor [19] with manual edition whenever it was necessary, aligned with the program Clustal W [20], and compared with sequences deposited in GenBank. Phylogenetic analyses were performed with maximum-likelihood (ML) methods by using the program Mega X [21]. Best-fitting substitution models were determined with the Akaike Information Criterion using the ML model test implemented in MEGA X. Support for the topologies was tested by bootstrapping over 1.000 replications, and gaps were excluded from the comparisons.
The present work was executed with the permission from the Secretariat of Environment and Climate Change of the Province of Río Negro (File No. 08526SAYDS 2015/218/222).
3. Results and Discussion
The female tick that was collected on C. villosus was identified morphologically as A. pseudoconcolor based on the typical scutum ornate, with small pale spots on the yellowish-brown ground with small, moderately deep punctuations, the dental formula (3/3) and the spurs on the trochanters [3] (see Figure 1). Further, the identification was confirmed by sequencing a fragment of the mitochondrial 16S rRNA gene. The obtained partial sequence (GenBank accession number: OP744428) showed sequence identities of 99.30% and 99.53% to A. pseudoconcolor from Argentina (GenBank accession numbers AY628134 and AY628135). The tick specimen was deposited in the tick collection of the Instituto Nacional de Tecnología Agropecuaria (INTA; Rafaela, Santa Fe, Argentina) with the collection number INTA 2521 (see Figure 1).
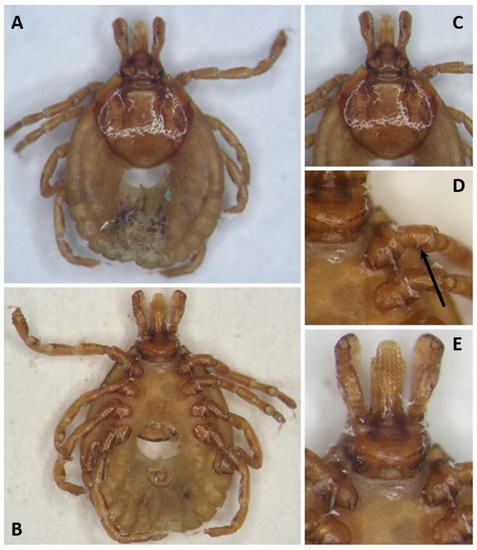
Figure 1.
Amblyomma pseudoconcolor female (Tick collection Instituto Nacional de Tecnología Agropecuaria (INTA; Rafaela, Santa Fe, Argentina); collection number INTA 2521). (A) dorsal view; (B) ventral view; (C) scutum ornamentation; (D) coxae and trochanters; (E) capitulum ventral view. The black arrow in (D) indicates the typical spur on the trochanter.
The tick sample showed positive PCR results in two of the three applied assays: 16S rRNA gene for Anaplasmataceae and gltA for Rickettsia spp. The two amplicons were purified and partial gene sequences were obtained. The partial sequence of the gltA gene (GenBank accession number: OP753007) showed a sequence identity of 100% (653/653bp) to different Ca. R. andeanae sequences from isolates made from Amblyomma maculatum Koch, 1844, Amblyomma parvum Aragão, 1908 and Amblyomma tigrinum Koch, 1844 from Argentina, Brazil, Peru and the United States (GenBank accession numbers: EF451001, GU131156, GU169050, KT153033 and KX576677). In the ML tree based on GenBank sequences of different species of Rickettsia, the gltA sequence generated in this study forms part of a clade including sequences of Ca. R. andeanae detected in different species of Amblyomma from the Southern cone of South America (see Figure 2). This clade separates clearly (bootstrap value 97) from other species of Rickettsia. Candidatus R. andeanae is a member of the spotted fever group Rickettsiae (SFGR) of unknown pathogenicity [9,22,23]. Interestingly, the PCR assay amplifying the ompA gene that is specific for SFGR resulted negative in this study. However, previous studies have shown [24,25] that the applied PCR assay in this study is not appropriate for the detection of the ompA gene of Ca. R. andeanae and therefore must be replaced by another assay described by Ermeeva et al. [26] in future studies. Jiang et al. [27] firstly described the detection of Ca. R. andeanae in A. maculatum from Peru. Further, this rickettsial agent could be detected in A. parvum from Argentina and Brazil [24,25,28] and A. tigrinum from Argentina and Chile [29,30]. DNA of Ca. R. andeanae (named as Rickettsia sp. strain Argentina by the authors) was previously detected in A. pseudoconcolor collected in Santiago del Estero province, Argentina [8]. Santiago del Estero province belongs to the Chaco biogeographic province, which is ecologically distinct from the Central Patagonia biogeographic province where A. pseudoconcolor was sampled in this study [5]. Further, Ca. R. andeanae was also detected in Ixodes boliviensis collected from a horse in Peru [27]. Nevertheless, the results of the present study together with the reports from literature suggest that this SFGR is closely associated with the hard tick genus Amblyomma and widely distributed in South America.
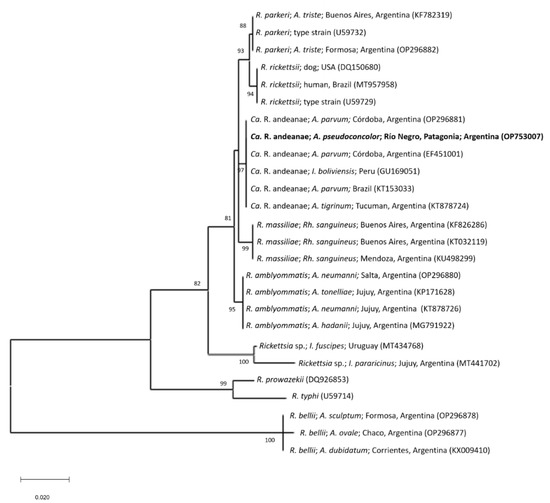
Figure 2.
Maximum-likelihood tree constructed from gltA partial sequences of different Rickettsia species (Substitution model: Tamura-Nei 93 model with Gamma distribution). The sequence generated in this study is written in bold letters. Numbers represent bootstrap support generated from 1000 replications. GenBank accession numbers are given in brackets. Abbreviations: A.: Amblyomma; An.: Anaplasma; Ca.: Candidatus; I.: Ixodes; R.: Rickettsia; Rh.: Rhipicephalus.
The partial sequence of the 16S rRNA gene from the tick simple that was generated in this study showed a sequence identity of 99.53% (305/307bp) to an uncultured Ehrlichia sp. detected in a horse from Nicaragua (GenBank accession number: KJ434178) and 98.69% to uncultured Ehrlichia spp. from Ixodes ornithorhynchi Lucas, 1846 (Australia; 301/305bp; GenBank accession number: MF069159) and Hydrochoerus hydrochaeris Linnaeus, 1766 (Brazil; 301/305bp; GenBank accession number: MW785880). Figure 3 shows the phylogenetic tree constructed of partial 16S rRNA gene sequences of different Ehrlichia spp. Based on the used fragment of the 16S rRNA gene, the phylogenetic position of the Ehrlichia sp. detected in this study (GenBank accession number: OP744461) in relation to other Ehrlichia spp. remains unresolved. In Argentina, five species of Amblyomma have been reported infected with different strains of undetermined Ehrlichia sp. so far.: Amblyomma neumanni Ribaga, 1902 [31]; Amblyomma ovale Koch, 1844 [32]; A. parvum [33,34]; A. tigrinum [33,34,35,36]; and Amblyomma triste Koch, 1844 [37]. The Ehrlichia strain detected in A. pseudoconcolor in this work is not related to the remaining Ehrlichia strains previously detected in Argentina, at least considering those from which 16S rRNA gene sequences are available. Based on the results of this studies together with previous reports, it must be assumed that the diversity of Ehrlichia spp. in Amblyomma ticks from Argentina is greater than previously suggested. However, further studies using genetic markers with a higher level of polymorphism should be performed for a more accurate phylogenetic characterization of these Ehrlichia spp. In addition, the Ehrlichia–host relationship and the possible pathogenicity of these strains must be studied more in detail.
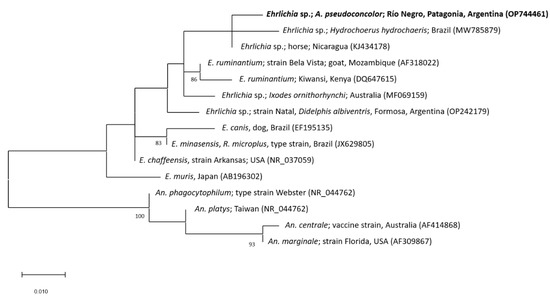
Figure 3.
Maximum-likelihood tree constructed from 16S rRNA gene partial sequences of different Ehrlichia species (Substitution model: Hasegawa–Kishino–Yano model with Gamma distribution). Sequences of Anaplasma spp. were used as outgroup. The sequence generated in this study is written in bold letters. Numbers represent bootstrap support generated from 1000 replications. Bootstrap values minor to 80 are not shown. GenBank accession numbers are given in brackets. Abbreviations: A.: Amblyomma; An.: Anaplasma; E.: Ehrlichia; R.: Rhipicephalus.
4. Conclusions
The results of this study demonstrate the presence of a spotted fever group Rickettsia—Ca. R. andeanae—and the first detection of a putative novel strain of Ehrlichia sp. in A. pseudoconcolor. Further studies must be executed to investigate if A. pseudoconcolor may act as a vector for these bacteria and which is the role of the C. villosus in this bacteria–tick–host relationship.
Author Contributions
Conceptualization, M.W. and P.S.S.; methodology, M.W. and P.S.S.; investigation, P.S.S., E.L.T., S.D.A. and M.W.; resources, S.D.A. and M.W.; data curation, P.S.S. and S.N.; writing—original draft preparation, P.S.S. and M.W.; writing—review and editing, P.S.S., E.L.T. and S.N.; visualization, E.L.T. and P.S.S.; supervision, S.N.; project administration, P.S.S.; funding acquisition, P.S.S. and S.N. All authors have read and agreed to the published version of the manuscript.
Funding
The study was financed by Universidad Nacional de Río Negro (PI-JI-40-C-869), Instituto Nacional de Tecnología Agropecuaria (PE-E5-I109), Asociación Cooperadora INTA Rafaela and Agencia Nacional de Promoción Científica y Tecnológica (PICT 2018-2579; PICT 2019-970).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank Macarena Sarli and Fernando Sebastian Flores for their help in the laboratory.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nigro, A.N.; Gasparri, B.; Steger, E.P. Xenartros Argentinos: Guía Para su Identificación, 1st ed.; Fundación de Historia Natural Félix de Azara: Ciudad de Buenos Aires, Argentina; Universidad Maimónides: Buenos Aires, Argentina, 2021; 134p. [Google Scholar]
- Abba, A.M.; Vizcaíno, S.F.; Cassini, M.H. Effects of land use on the distribution of three species of armadillos in the Argentinean pampas. J. Mammal. 2007, 88, 502–507. [Google Scholar] [CrossRef]
- Nava, S.; Venzal, J.M.; González-Acuña, D.; Martins, T.F.; Guglielmone, A.A. Ticks of the Southern Cone of America: Diagnosis, Distribution and Hosts with Taxonomy, Ecology and Sanitary Importance; Elsevier: Amsterdam, The Netherlands; Academic Press: London, UK, 2017; 352p. [Google Scholar]
- Guglielmone, A.A.; Nava, S.; Robbins, R.G. Neotropical Hard Ticks (Acari: Ixodida: Ixodidae): A Critical Analysis of Their Taxonomy, Distribution, and Host Relationships; Springer: Berlin/Heidelberg, Germany, 2021; 486p. [Google Scholar]
- Morrone, J.J. Biogeographic areas and transition zones of Latin America and the Caribbean Islands based on panbiogeographic and cladistics analyses of the entomofauna. Annu. Rev. Entomol. 2006, 51, 467–494. [Google Scholar] [CrossRef] [PubMed]
- Guglielmone, A.A.; Estrada-Peña, A.; Luciani, C.A.; Mangold, A.J.; Keirans, J.E. Hosts and distribution of Amblyomma auricularium (Conil 1878) and Amblyomma pseudoconcolor Aragão, 1908 (Acari: Ixodidae). Exp. Appl. Acarol. 2003, 29, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Ezquiaga, M.C.; Gallo, J.A.; D’Agostino, R.L.; Udrizar Sauthier, D.E.; Abba, A.M.; Sanchez, J. Fleas and ticks in armadillos from Argentinean Patagonia: Diversity, abundance and distribution. Acta Trop. 2021, 219, 105911. [Google Scholar] [CrossRef] [PubMed]
- Tomassone, L.; Nuñez, P.; Ceballos, L.A.; Gürtler, R.E.; Kitron, U.; Farber, M. Detection of “Candidatus Rickettsia sp. strain Argentina” and Rickettsia bellii in Amblyomma ticks (Acari: Ixodidae) from Northern Argentina. Exp. Appl. Acarol. 2010, 52, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Parola, P.; Paddock, C.D.; Socolovschi, C.; Labruna, M.B.; Mediannikov, O.; Kernif, T.; Abdad, M.Y.; Stenos, J.; Bitam, I.; Fournier, P.E.; et al. Update on tick-borne rickettsioses around the world: A geographic approach. Clin. Microbiol. Rev. 2013, 26, 657–702. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.B.; Cardoso, K.M.; de Oliveira, S.V.; Costa, R.M.F.; Oliveira, G.; Amorim, M.; Alves, L.C.; Monteiro, M.F.M.; Gazeta, G.S. Rickettsia amblyommatis infecting Amblyomma pseudoconcolor in area of new focus of spotted fever in northeast Brazil. Acta Trop. 2018, 182, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.B.; Barbieri, A.R.; Szabó, M.P.J.; Ramos, V.N.; Piovezan, U.; Labruna, M.B. New records of Rickettsia bellii-infected ticks in Brazil. Braz. J. Vet. R. Anim. Sci. 2017, 54, 92–95. [Google Scholar] [CrossRef]
- Norris, D.E.; Klompen, J.S.H.; Keirans, J.E.; Black, W.C. Population genetics of Ixodes scapularis (Acari: Ixodidae) based on mitochondrial 16S and 12S genes. J. Med. Entomol. 1996, 33, 78–89. [Google Scholar] [CrossRef]
- Mangold, A.J.; Bargues, M.D.; Mas-Coma, S. Mitochondrial 16S rDNA sequences and phylogenetic relationships of species of Rhipicephalus and other tick genera among Metastriata (Acari: Ixodidae). Parasitol. Res. 1998, 84, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.E.; Sumner, J.W.; Dawson, J.E.; Tzianabos, T.; Greene, C.R.; Olson, J.G.; Fishbein, D.B.; Olsen-Rasmussen, M.; Holloway, B.P.; George, E.H. Detection of the etiologic agent of human ehrlichiosis by polymerase chain reaction. J. Clin. Microbiol. 1992, 30, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Breitschwerdt, E.B.; Hegarty, B.C.; Hancock, S.I. Sequential evaluation of dogs naturally infected with Ehrlichia canis, Ehrlichia chaffeensis, Ehrlichia equi, Ehrlichia ewingii or Bartonella vinsoni. J. Clin. Microbiol. 1998, 36, 2645–2651. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, D.M.; Hagiwara, M.K.; Labruna, M.B. In vitro isolation and molecular characterization of an Ehrlichia canis strain from São Paulo, Brazil. Braz. J. Microbiol. 2008, 39, 489–493. [Google Scholar] [CrossRef] [PubMed]
- Labruna, M.B.; Whitworth, T.; Horta, M.C.; Bouyer, D.H.; McBride, J.W.; Pinter, A.; Popov, V.; Gennari, S.M.; Walker, D.H. Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of São Paulo, Brazil, where Brazilian spotted fever is endemic. J. Clin. Microbiol. 2004, 42, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Regnery, R.L.; Spruill, C.L.; Plikaytis, B.D. Genotypic identification of Rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J. Bacteriol. 1991, 173, 1576–1589. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Fournier, P.E.; Raoult, D. Current knowledge on phylogeny and taxonomy of Rickettsia spp. Ann. N. Y. Acad. Sci. 2009, 1166, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Merhej, V.; Raoult, D. Rickettsial evolution in the light of comparative genomics. Biol. Rev. Camb. Philos. Soc. 2011, 86, 379–405. [Google Scholar] [CrossRef]
- Nieri-Bastos, F.A.; Lopes, M.G.; Cançado, P.H.; Rossa, G.A.; Faccini, J.L.; Gennari, S.M.; Labruna, M.B. Candidatus Rickettsia andeanae, a spotted fever group agent infecting Amblyomma parvum ticks in two Brazilian biomes. Memórias Do Inst. Oswaldo Cruz 2014, 109, 259–261. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, P.S.; Tarragona, E.L.; Bottero, M.N.; Mangold, A.J.; Mackenstedt, U.; Nava, S. Bacteria of the genera Ehrlichia and Rickettsia in ticks of the family Ixodidae with medical importance in Argentina. Exp. Appl. Acarol 2017, 71, 87–96. [Google Scholar] [CrossRef]
- Eremeeva, M.E.; Bosserman, E.A.; Demma, L.J.; Zambrano, M.L.; Blau, D.M.; Dasch, G.A. Isolation and identification of Rickettsia massiliae from Rhipicephalus sanguineus ticks collected in Arizona. Appl. Environ. Microbiol. 2006, 72, 5569–5577. [Google Scholar] [CrossRef]
- Jiang, J.; Blair, P.J.; Felices, V.; Moron, C.; Cespedes, M.; Anaya, E.; Schoeler, G.B.; Sumner, J.W.; Olson, J.G.; Richards, A.L. Phylogenetic analysis of a novel molecular isolate of spotted fever group Rickettsiae from northern Peru: Candidatus Rickettsia andeanae. Ann. N. Y. Acad. Sci. 2005, 1063, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, R.C.; Moraes-Filho, J.; Nava, S.; Brandao, P.E.; Richtzenhain, L.J.; Labruna, M.B. Detection of a novel spotted fever group Rickettsia in Amblyomma parvum ticks (Acari: Ixodidae) from Argentina. Exp. Appl. Acarol. 2007, 43, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Abarca, K.; López, J.; Acosta-Jamett, G.; Martinez-Valdebenito, C. Identificación de Rickettsia andeanae en dos regiones de Chile [Detection of Rickettsia andeanae in two regions of Chile]. Rev. Chilena. Infectol. 2013, 30, 388–394. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Saracho Bottero, M.N.; Tarragona, E.L.; Nava, S. Spotted fever group rickettsiae in Amblyomma ticks likely to infest humans in rural areas from northwestern Argentina. Medicina 2015, 75, 391–395. [Google Scholar] [PubMed]
- Fargnoli, L.; Fernadez, C.; Monje, L.D. Novel Ehrlichia strain infecting cattle tick Amblyomma neumanni, Argentina 2018. Emerg. Infect. Dis. 2020, 26, 1027–1030. [Google Scholar] [CrossRef] [PubMed]
- Tarragona, E.L.; Flores, F.S.; Lamattina, D.; Torrents, J.; Sebastian, P.S.; Nava, S. Two novel Ehrlichia (Rickettsiales: Anaplasmataceae) strains detected in ticks (Ixodida, Ixodidae) and opossums (Didelphimorphia: Didelphidae) in Argentina. Ticks Tick Borne Dis. 2022, 13, 102043. [Google Scholar] [CrossRef] [PubMed]
- Cicuttin, G.L.; De Salvo, M.N.; Nava, S. Two novel Ehrlichia strains detected in Amblyomma tigrinum ticks associated to dogs in peri-urban areas of Argentina. Comp. Immunol. Microbiol. Infect. Dis. 2017, 53, 40–44. [Google Scholar] [CrossRef]
- Monje, L.D.; Fernandez, C.; Percara, A. Detection of Ehrlichia sp. strain San Luis and Candidatus Rickettsia andeanae in Amblyomma parvum ticks. Ticks Tick-Borne Dis. 2019, 307, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Tomassone, L.; Nuñez, P.; Gürtler, R.E.; Ceballos, L.A.; Orozco, M.M.; Kitron, U.D.; Farber, M. Molecular detection of Ehrlichia chaffeensis in Amblyomma parvum ticks, Argentina. Emerg. Infect. Dis. 2008, 14, 1953–1955. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, A.T.; Fernandez, C.; Fargnoli, L.; Beldomenico, P.M.; Monje, L.D. A putative novel strain of Ehrlichia infecting Amblyomma tigrinum associated with Pampas fox (Lycalopex gymnocercus) in Esteros del Ibera ecoregion, Argentina. Ticks Tick Borne Dis. 2020, 11, 101318. [Google Scholar] [CrossRef] [PubMed]
- Cicuttin, G.L.; De Salvo, M.N.; Pérez, P.D.; Silva, D.; Félix, M.L.; Venzal, J.M.; Nava, S. A novel Ehrlichia strain (Rickettsiales: Anaplasmataceae) detected in Amblyomma triste (Acari: Ixodidae), a tick species of public health importance in the Southern Cone of America. Pathog. Glob. Health 2020, 114, 318–322. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).