Machine Learning-Based Co-Expression Network Analysis Unravels Potential Fertility-Related Genes in Beef Cows
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
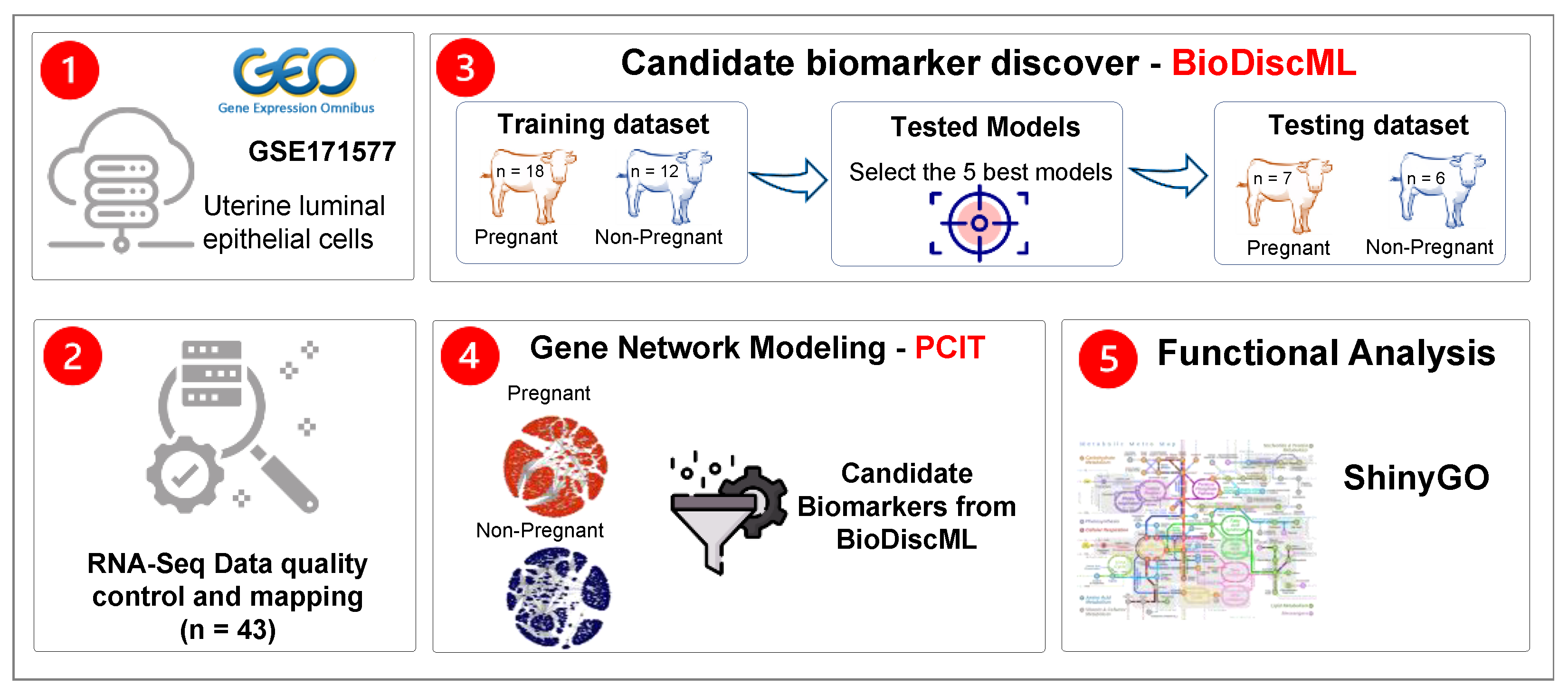
2.1. Data Retrieval and Quality Control
2.2. Gene Expression Normalization and Supervised Machine Learning
2.3. Gene Co-Expression Network Analysis
2.4. Functional Over-Representation Analysis
3. Results
3.1. Identification of Potential Biomarker Genes through ML
3.2. Gene Network Analysis
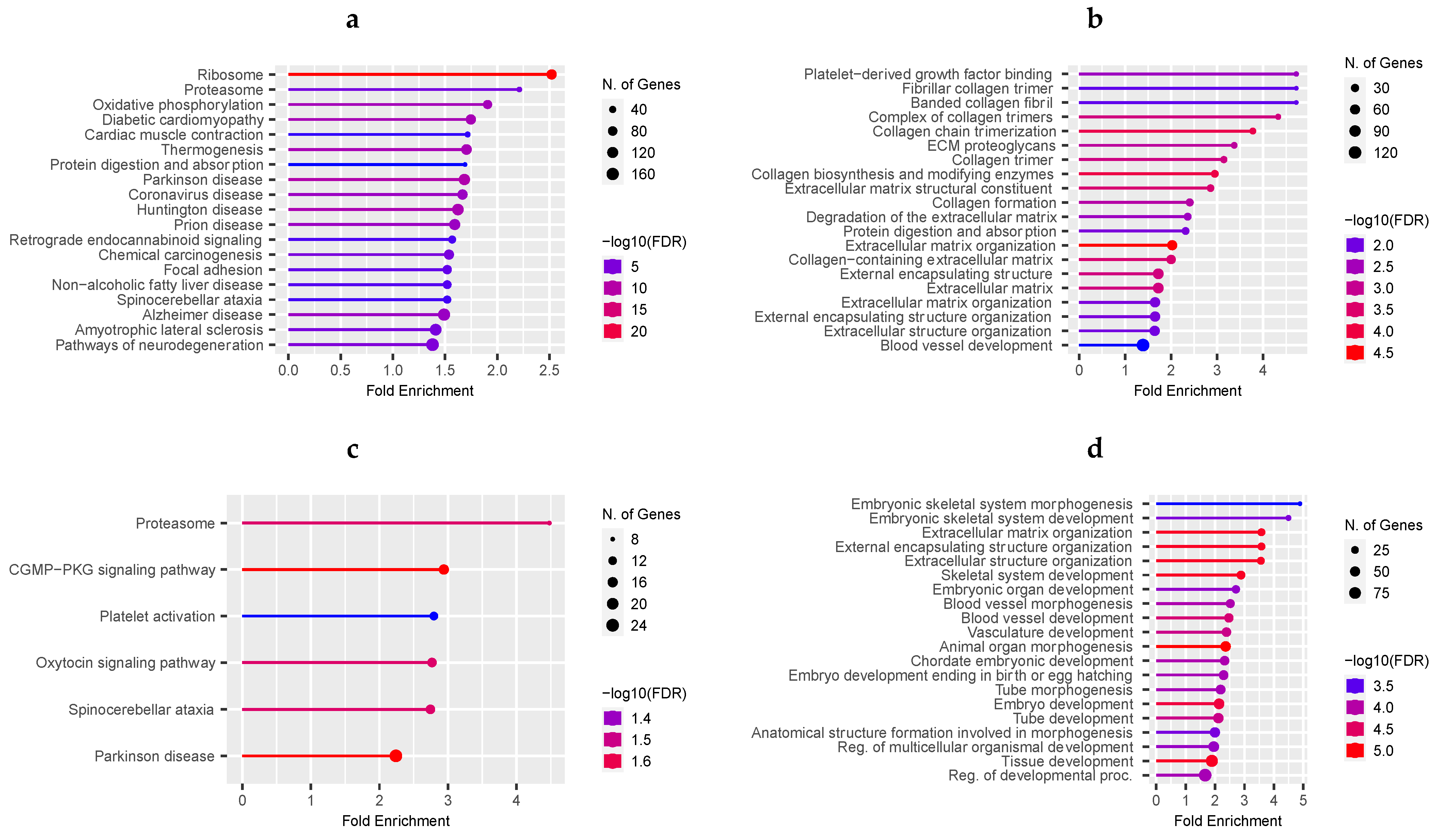
3.3. Functional Over-Representation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Han, Y.; Peñagaricano, F. Unravelling the genomic architecture of bull fertility in Holstein cattle. BMC Genet. 2016, 17, 143. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.F.; Schnabel, R.D.; Sutovsky, P. Review: Genomics of bull fertility. Animal 2018, 12, s172–s183. [Google Scholar] [CrossRef] [PubMed]
- Mercadante, V.R.G.; Dias, N.W.; Timlin, C.L.; Pancini, S. 375 Economic consequences of pregnancy loss in beef cattle. J. Anim. Sci. 2020, 98, 124. [Google Scholar] [CrossRef]
- Bach, À.; Bach, À. Effects of nutrition and genetics on fertility in dairy cows. Reprod. Fertil. Dev. 2019, 31, 40–54. [Google Scholar] [CrossRef]
- Berry, D.P.; Wall, E.; Pryce, J.E. Genetics and genomics of reproductive performance in dairy and beef cattle. Animal 2014, 8, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Moorey, S.E.; Biase, F.H. Beef heifer fertility: Importance of management practices and technological advancements. J. Anim. Sci. Biotechnol. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ortega, M.S. Identification of genes associated with reproductive function in dairy cattle. Anim. Reprod. 2018, 15, 923–932. [Google Scholar] [CrossRef]
- Olasege, B.S.; Tahir, M.S.; Gouveia, G.C.; Kour, J.; Porto-Neto, L.R.; Hayes, B.J.; Fortes, M.R.S. Genetic parameter estimates for male and female fertility traits using genomic data to improve fertility in Australian beef cattle. Anim. Prod. Sci. 2021, 61, 1863. [Google Scholar] [CrossRef]
- Ponsart, C.; Le Bourhis, D.; Knijn, H.; Fritz, S.; Guyader-Joly, C.; Otter, T.; Lacaze, S.; Charreaux, F.; Schibler, L.; Dupassieux, D.; et al. Reproductive technologies and genomic selection in dairy cattle. Reprod. Fertil. Dev. 2013, 26, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Moorey, S.E.; Walker, B.N.; Elmore, M.F.; Elmore, J.B.; Rodning, S.P.; Biase, F.H. Rewiring of gene expression in circulating white blood cells is associated with pregnancy outcome in heifers (Bos taurus). Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Phillips, K.M.; Read, C.C.; Kriese-Anderson, L.A.; Rodning, S.P.; Brandebourg, T.D.; Biase, F.H.; Marks, M.L.; Elmore, J.B.; Stanford, M.K.; Dyce, P.W. Plasma metabolomic profiles differ at the time of artificial insemination based on pregnancy outcome, in Bos taurus beef heifers. Sci. Rep. 2018, 8, 13196. [Google Scholar] [CrossRef] [PubMed]
- Cánovas, A.; Reverter, A.; DeAtley, K.L.; Ashley, R.L.; Colgrave, M.L.; Fortes, M.R.S.; Islas-Trejo, A.; Lehnert, S.; Porto-Neto, L.; Rincón, G.; et al. Multi-tissue omics analyses reveal molecular regulatory networks for puberty in composite beef cattle. PLoS ONE 2014, 9, e102551. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Reverter, A.; Cánovas, A.; Venus, B.; Islas-Trejo, A.; Porto-Neto, L.R.; Lehnert, S.A.; Medrano, J.F.; Moore, S.S.; Fortes, M.R.S. Global differential gene expression in the pituitary gland and the ovaries of pre- and postpubertal Brahman heifers. J. Anim. Sci. 2017, 95, 599–615. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Geary, T.W.; Burns, G.W.; Moraes, J.G.N.; Moss, J.I.; Denicol, A.C.; Dobbs, K.B.; Ortega, M.S.; Hansen, P.J.; Wehrman, M.E.; Neibergs, H.; et al. Identification of beef heifers with superior uterine capacity for pregnancy. Biol. Reprod. 2016, 95, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gómez, E.; Salvetti, P.; Gatien, J.; Carrocera, S.; Martín-González, D.; Muñoz, M. Blood plasma metabolomics predicts pregnancy in Holstein cattle transferred with fresh and vitrified/warmed embryos produced in vitro. J. Proteome Res. 2020, 19, 1169–1182. [Google Scholar] [CrossRef]
- Gaiteri, C.; Ding, Y.; French, B.; Tseng, G.C.; Sibille, E. Beyond modules and hubs: The potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav. 2014, 13, 13–24. [Google Scholar] [CrossRef]
- Hudson, N.J.; Dalrymple, B.P.; Reverter, A. Beyond differential expression: The quest for causal mutations and effector molecules. BMC Genom. 2012, 13, 356. [Google Scholar] [CrossRef]
- Xu, C.; Jackson, S.A. Machine learning and complex biological data. Genome Biol. 2019, 20, 76. [Google Scholar] [CrossRef]
- Rabaglino, M.B.; Kadarmideen, H.N. Machine learning approach to integrated endometrial transcriptomic datasets reveals biomarkers predicting uterine receptivity in cattle at seven days after estrous. Sci. Rep. 2020, 10, 16981. [Google Scholar] [CrossRef]
- Fonseca, P.A.S.; Suárez-Vega, A.; Cánovas, A. Weighted gene correlation network meta-analysis reveals functional candidate genes associated with high- and sub-fertile reproductive performance in beef cattle. Genes 2020, 11, 543. [Google Scholar] [CrossRef]
- Martins, T.; Sponchiado, M.; Silva, F.A.C.C.; Estrada-Cortés, E.; Hansen, P.J.; Peñagaricano, F.; Binelli, M. Progesterone-dependent and progesterone-independent modulation of luminal epithelial transcription to support pregnancy in cattle. Physiol. Genom. 2022, 54, 71–85. [Google Scholar] [CrossRef]
- Ewels, P. SRA-Explorer. Available online: https://sra-explorer.info/ (accessed on 13 May 2022).
- Andrews, S. FASTQC. A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 6 January 2022).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Rosen, B.D.; Bickhart, D.M.; Schnabel, R.D.; Koren, S.; Elsik, C.G.; Tseng, E.; Rowan, T.N.; Low, W.Y.; Zimin, A.; Couldrey, C.; et al. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience 2020, 9, giaa021. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. 2018. Available online: https://www.R-project.org (accessed on 6 January 2022).
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Di Pietro, A.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, M.; Vittrant, B.; Martin-Magniette, M.L.; Scott Boyer, M.P.; Perin, O.; Bergeron, A.; Fradet, Y.; Droit, A. Large-scale automatic feature selection for biomarker discovery in high-dimensional OMICS data. Front. Genet. 2019, 10, 452. [Google Scholar] [CrossRef]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98140-6. [Google Scholar]
- Reverter, A.; Chan, E.K.F. Combining partial correlation and an information theory approach to the reversed engineering of gene co-expression networks. Bioinformatics 2008, 24, 2491–2497. [Google Scholar] [CrossRef] [PubMed]
- Diniz, W.J.S.; Crouse, M.S.; Cushman, R.A.; McLean, K.J.; Caton, J.S.; Dahlen, C.R.; Reynolds, L.P.; Ward, A.K. Cerebrum, liver, and muscle regulatory networks uncover maternal nutrition effects in developmental programming of beef cattle during early pregnancy. Sci. Rep. 2021, 11, 2771. [Google Scholar] [CrossRef]
- Assenov, Y.; Ramírez, F.; Schelhorn, S.-E.; Lengauer, T.; Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef]
- Shannon, P. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Goenawan, I.H.; Bryan, K.; Lynn, D.J. DyNet: Visualization and analysis of dynamic molecular interaction networks. Bioinformatics 2016, 32, 2713–2715. [Google Scholar] [CrossRef]
- Fuller, T.F.; Ghazalpour, A.; Aten, J.E.; Drake, T.A.; Lusis, A.J.; Horvath, S. Weighted gene coexpression network analysis strategies applied to mouse weight. Mamm. Genome 2007, 18, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Pryce, J.E.; Veerkamp, R.F. The incorporation of fertility indices in genetic improvement programmes. BSAP Occas. Publ. 2001, 26, 237–249. [Google Scholar] [CrossRef]
- Spencer, T.E. Early pregnancy: Concepts, challenges, and potential solutions. Anim. Front. 2013, 3, 48–55. [Google Scholar] [CrossRef]
- Binelli, M.; Scolari, S.C.; Pugliesi, G.; Van Hoeck, V.; Gonella-Diaza, A.M.; Andrade, S.C.S.; Gasparin, G.R.; Coutinho, L.L. The transcriptome signature of the receptive bovine uterus determined at early gestation. PLoS ONE 2015, 10, e0122874. [Google Scholar] [CrossRef]
- Mazzoni, G.; Pedersen, H.S.; Rabaglino, M.B.; Hyttel, P.; Callesen, H.; Kadarmideen, H.N. Characterization of the endometrial transcriptome in early diestrus influencing pregnancy status in dairy cattle after transfer of in vitro-produced embryos. Physiol. Genom. 2020, 52, 269–279. [Google Scholar] [CrossRef]
- Estrada-Cortés, E.; Ortiz, W.G.; Chebel, R.C.; Jannaman, E.A.; Moss, J.I.; De Castro, F.C.; Zolini, A.M.; Staples, C.R.; Hansen, P.J. Embryo and cow factors affecting pregnancy per embryo transfer for multiple-service, lactating Holstein recipients. Transl. Anim. Sci. 2019, 3, 60–65. [Google Scholar] [CrossRef] [PubMed]
- França, M.R.; da Silva, M.I.S.; Pugliesi, G.; Van Hoeck, V.; Binelli, M. Evidence of endometrial amino acid metabolism and transport modulation by peri-ovulatory endocrine profiles driving uterine receptivity. J. Anim. Sci. Biotechnol. 2017, 8, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, B.; Oliveira, M.L.; Pugliesi, G.; Batista, E.O.S.; Binelli, M. Cytobrush: A tool for sequential evaluation of gene expression in bovine endometrium. Reprod. Domest. Anim. 2017, 52, 1153–1157. [Google Scholar] [CrossRef] [PubMed]
- Law, R.H.P.; Zhang, Q.; McGowan, S.; Buckle, A.M.; Silverman, G.A.; Wong, W.; Rosado, C.J.; Langendorf, C.G.; Pike, R.N.; Bird, P.I.; et al. An overview of the serpin superfamily. Genome Biol. 2006, 7, 216. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sanrattana, W.; Maas, C.; de Maat, S. SERPINs—From trap to treatment. Front. Med. 2019, 6, 25. [Google Scholar] [CrossRef]
- Bédard, J.; Brûlé, S.; Price, C.A.; Silversides, D.W.; Lussier, J.G. Serine protease inhibitor-E2 (SERPINE2) is differentially expressed in granulosa cells of dominant follicle in cattle. Mol. Reprod. Dev. 2003, 64, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Li, H.; Cao, D.; Chen, Y. The development of endometrial hyperplasia in aged PD-1-deficient female mice. Diagn. Pathol. 2014, 9, 97. [Google Scholar] [CrossRef]
- Taglauer, E.S.; Trikhacheva, A.S.; Slusser, J.G.; Petroff, M.G. Expression and function of PDCD1 at the human maternal-fetal interface. Biol. Reprod. 2008, 79, 562–569. [Google Scholar] [CrossRef]
- Dickinson, S.E.; Griffin, B.A.; Elmore, M.F.; Kriese-Anderson, L.; Elmore, J.B.; Dyce, P.W.; Rodning, S.P.; Biase, F.H. Transcriptome profiles in peripheral white blood cells at the time of artificial insemination discriminate beef heifers with different fertility potential. BMC Genom. 2018, 19, 129. [Google Scholar] [CrossRef]
- Kishi, T.; Mayanagi, T.; Iwabuchi, S.; Akasaka, T.; Sobue, K. Myocardin-related transcription factor A (MRTF-A) activity-dependent cell adhesion is correlated to focal adhesion kinase (FAK) activity. Oncotarget 2016, 7, 72113–72130. [Google Scholar] [CrossRef]
- Di-Luoffo, M.; Daems, C.; Bergeron, F.; Tremblay, J.J. Novel targets for the transcription factors MEF2 in MA-10 Leydig cells. Biol. Reprod. 2015, 93, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Rubin, L.P.; Gong, X. Translational Physiology: MEF2 transcription factors in human placenta and involvement in cytotrophoblast invasion and differentiation. Physiol. Genom. 2018, 50, 10. [Google Scholar] [CrossRef] [PubMed]
- Velasquez, L.S.; Sutherland, L.B.; Liu, Z.; Grinnell, F.; Kamm, K.E.; Schneider, J.W.; Olson, E.N.; Small, E.M. Activation of MRTF-A-dependent gene expression with a small molecule promotes myofibroblast differentiation and wound healing. Proc. Natl. Acad. Sci. USA 2013, 42, 16850–16855. [Google Scholar] [CrossRef] [PubMed]
- Holtz, M.L.; Misra, R.P. Serum response factor is required for cell contact maintenance but dispensable for proliferation in visceral yolk sac endothelium. BMC Dev. Biol. 2011, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Scolari, S.C.; Pugliesi, G.; Strefezzi, R.D.F.; Andrade, S.C.D.S.; Coutinho, L.L.; Binelli, M. Dynamic remodeling of endometrial extracellular matrix regulates embryo receptivity in cattle. Reproduction 2017, 153, 49–61. [Google Scholar] [CrossRef]
- Banerjee, P.; Rodning, S.P.; Diniz, W.J.S.; Dyce, P.W. Co-expression network and integrative analysis of metabolome and transcriptome uncovers biological pathways for fertility in beef heifers. Metabolites 2022, 12, 708. [Google Scholar] [CrossRef]
- Calamita, P.; Gatti, G.; Miluzio, A.; Scagliola, A.; Biffo, S. Translating the game: Ribosomes as active players. Front. Genet. 2018, 9, 533. [Google Scholar] [CrossRef]
- Plaks, V.; Gershon, E.; Zeisel, A.; Jacob-Hirsch, J.; Neeman, M.; Winterhager, E.; Rechavi, G.; Domany, E.; Dekel, N. Blastocyst implantation failure relates to impaired translational machinery gene expression. Reproduction 2014, 148, 87–98. [Google Scholar] [CrossRef]
- Xin, L.; Xu, B.; Ma, L.; Hou, Q.; Ye, M.; Meng, S.; Ding, X.; Ge, W. Proteomics study reveals that the dysregulation of focal adhesion and ribosome contribute to early pregnancy loss. PROTEOMICS—Clin. Appl. 2016, 10, 554–563. [Google Scholar] [CrossRef]
- Zhao, X.; Guan, J.L. Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv. Drug Deliv. Rev. 2011, 63, 610–615. [Google Scholar] [CrossRef]
- Klohonatz, K.M.; Nulton, L.C.; Hess, A.M.; Bouma, G.J.; Bruemmer, J.E. The role of embryo contact and focal adhesions during maternal recognition of pregnancy. PLoS ONE 2019, 14, e0213322. [Google Scholar] [CrossRef] [PubMed]
- Burghardt, R.C.; Burghardt, J.R.; Taylor, J.D.; Reeder, A.T.; Nguen, B.T.; Spencer, T.E.; Bayless, K.J.; Johnson, G.A. Enhanced focal adhesion assembly reflects increased mechanosensation and mechanotransduction at maternal–conceptus interface and uterine wall during ovine pregnancy. Reproduction 2009, 137, 567–582. [Google Scholar] [CrossRef] [PubMed]
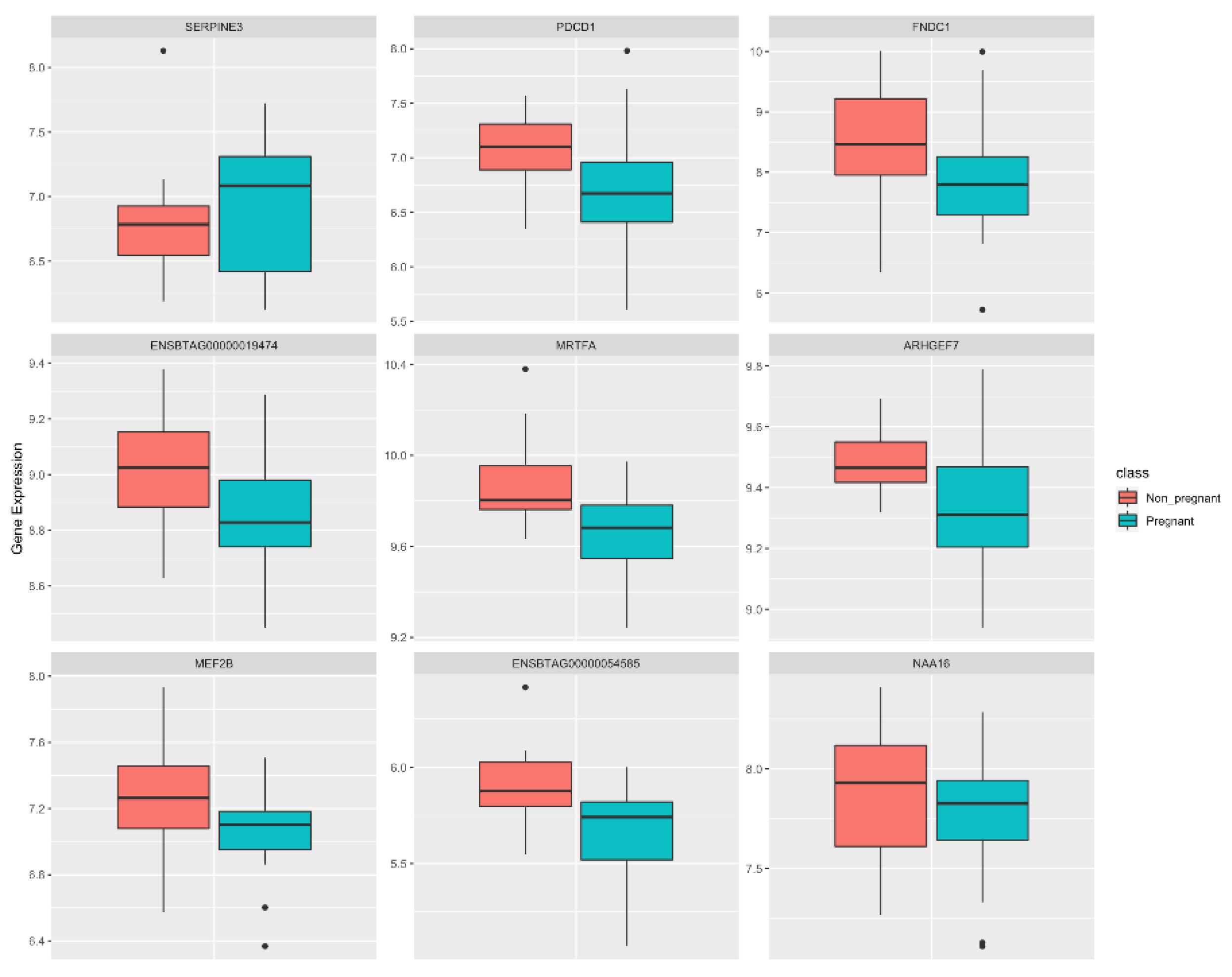
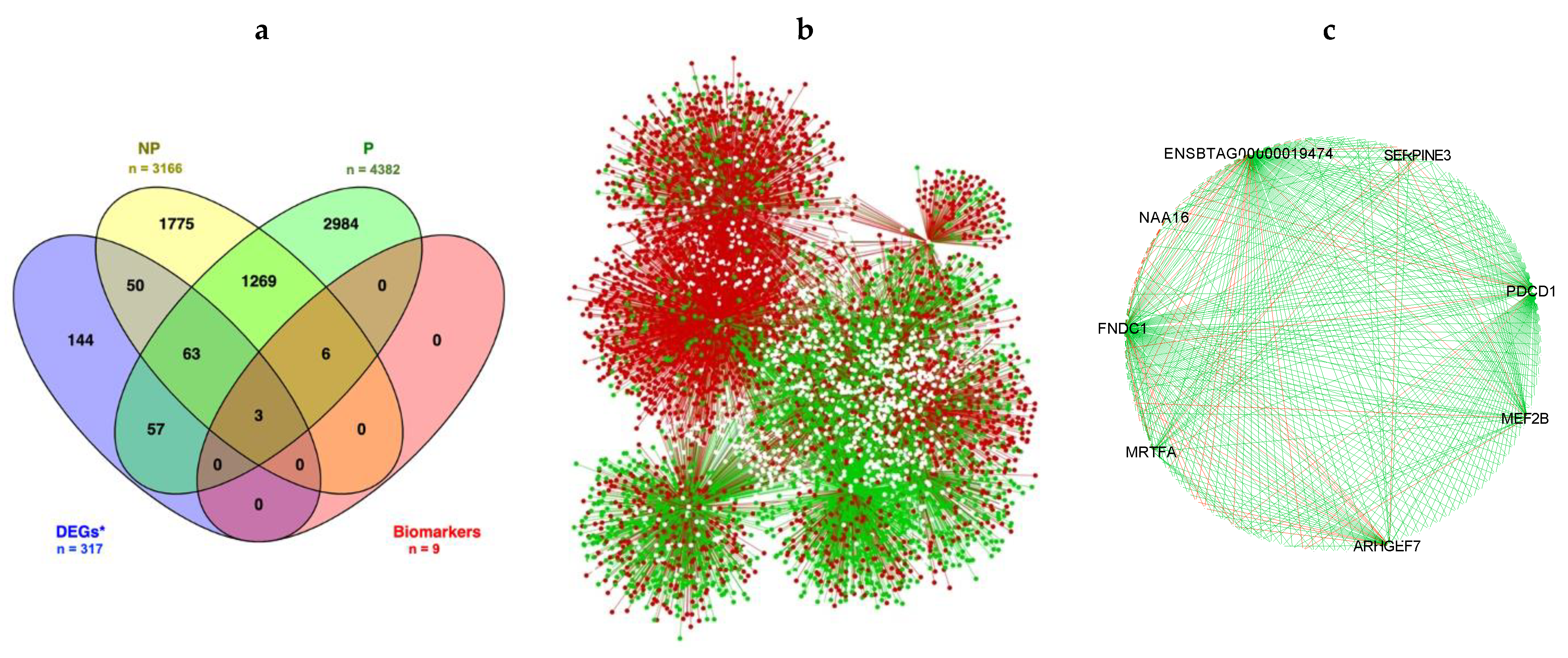
| Ensembl Gene ID | Gene Symbol | Nodes in NP | Nodes in P | DIFFK | z-Score * |
|---|---|---|---|---|---|
| ENSBTAG00000001818 | MEF2B | 794 | 169 | 0.88983 | 45.428 |
| ENSBTAG00000003938 | FNDC1 | 670 | 342 | 0.62088 | 31.6887 |
| ENSBTAG00000005284 | SERPINE3 | 646 | 401 | 0.55219 | 28.1798 |
| ENSBTAG00000019474 | ENSBTAG00000019474 | 577 | 507 | 0.39619 | 20.2104 |
| ENSBTAG00000002630 | MRTFA | 373 | 127 | 0.38698 | 19.74 |
| ENSBTAG00000038251 | NAA16 | 384 | 1488 | −0.4864 | −24.876 |
| ENSBTAG00000020726 | ARHGEF7 | 331 | 1534 | −0.5831 | −29.818 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diniz, W.J.S.; Banerjee, P.; Rodning, S.P.; Dyce, P.W. Machine Learning-Based Co-Expression Network Analysis Unravels Potential Fertility-Related Genes in Beef Cows. Animals 2022, 12, 2715. https://doi.org/10.3390/ani12192715
Diniz WJS, Banerjee P, Rodning SP, Dyce PW. Machine Learning-Based Co-Expression Network Analysis Unravels Potential Fertility-Related Genes in Beef Cows. Animals. 2022; 12(19):2715. https://doi.org/10.3390/ani12192715
Chicago/Turabian StyleDiniz, Wellison J. S., Priyanka Banerjee, Soren P. Rodning, and Paul W. Dyce. 2022. "Machine Learning-Based Co-Expression Network Analysis Unravels Potential Fertility-Related Genes in Beef Cows" Animals 12, no. 19: 2715. https://doi.org/10.3390/ani12192715
APA StyleDiniz, W. J. S., Banerjee, P., Rodning, S. P., & Dyce, P. W. (2022). Machine Learning-Based Co-Expression Network Analysis Unravels Potential Fertility-Related Genes in Beef Cows. Animals, 12(19), 2715. https://doi.org/10.3390/ani12192715






