Simple Summary
Sexual dimorphism is a phenomenon commonly existing in animals. Chinese tongue sole Cynoglossus semilaevis is an economical marine fish with obvious female-biased size dimorphism. So, it is important to explore the molecular mechanism beyond gonadal development for sex control in aquaculture industry. RNA-binding protein Ewing Sarcoma protein-like (ewsr1) gene is important for mouse gonadal development and reproduction, however there are limited studies on this gene in teleost. In this study, two ewsr1 genes were cloned and characterized from C. semilaevis. The ewsr1-w gene, located in W chromosomes, showed female-biased expression during C. semilaevis gonadal development. In addition, knock-down effect and transcriptional regulation of Cs-ewsr1-w further suggested its essential role in ovarian development. This study broadened our understanding on ewsr1 function in teleost, and provided genetic resources for the further development of sex control breeding techniques in C. semilaevis aquaculture.
Abstract
Ewsr1 encodes a protein that acts as a multifunctional molecule in a variety of cellular processes. The full-length of Cs-ewsr1-w and Cs-ewsr1-z were cloned in Chinese tongue sole (Cynoglossus semilaevis). The open reading frame (ORF) of Cs-ewsr1-w was 1,767 bp that encoded 589 amino acids, while Cs-ewsr1-z was 1,794 bp that encoded 598 amino acids. Real-time PCR assays showed that Cs-ewsr1-w exhibited significant female-biased expression and could be hardly detected in male. It has the most abundant expression in ovaries among eight healthy tissues. Its expression in ovary increased gradually from 90 d to 3 y with C. semilaevis ovarian development and reached the peak at 3 y. After Cs-ewsr1-w knockdown with siRNA interference, several genes related to gonadal development including foxl2, sox9b and pou5f1 were down-regulated in ovarian cell line, suggesting the possible participation of Cs-ewsr1-w in C. semilaevis ovarian development. The dual-luciferase reporter assay revealed that the -733/-154 bp Cs-ewsr1-w promoter fragment exhibited strong transcription activity human embryonic kidney (HEK) 293T cell line. The mutation of a MAF BZIP Transcription Factor K (Mafk) binding site located in this fragment suggested that transcription factor Mafk might play an important role in Cs-ewsr1-w basal transcription. Our results will provide clues on the gene expression level, transcriptional regulation and knock-down effect of ewsr1 gene during ovarian development in teleost.
1. Introduction
Sexual dimorphism is widespread in mammals, fish, birds and reptiles that characterized by body size, physiological and color differences between females and males [1,2,3,4,5,6]. This phenomenon has been found in a lot of fish species, of which Chinese tongue sole Cynoglossus semilaevis shows typically female-biased size dimorphism [2]. Female C. semilaevis can reach over twice in size and weight of males at the same age [7].
Sexual dimorphism is mainly resulted by genetic selection during the evolutionary process and is the consequence of differential expression of sex-biased genes in development stages [8,9,10]. The previous transcriptome analysis revealed thousands of sex-biased genes in somatotropic and reproductive tissues of C. semilaevis [11]. Cyp19a gene was expressed higher in ovary than testis and rose along the gonadal development, implying its participation in sex determination [12]. Dhcr24 (24-dehydrocholesterol reductase) gene, involved in steroid hormones and PI3K/Akt pathway and IGF-1 system, had the highest expression in liver and gonad of females [13]. The female-biased gonadal gene igfbp7 (insulin-like growth factor binding protein 7) might be involved in growth regulation of C. semilaevis by influencing insulin-like growth factor 1 receptor (igf1r), serine/threonine kinase 1 (akt) and NFκB (the nuclear factor kappa B) signal [14]. To better understand the molecular mechanism refining sex determination and differentiation in C. semilaevis, comparative transcriptome analysis was performed to reveal 156 genes correlated with ovary differentiation, including RNA-binding protein Ewing Sarcoma protein-like (ewsr1) gene [15].
The ewsr1 gene encodes a multifunctional RNA binding protein that regulates transcription and RNA splicing by interacting with other proteins and other cellular processes [16,17,18]. It is one of Translocated in liposarcoma, Ewing’s sarcoma and TATA-binding protein-associated factor 15 (TET, also named as FET) protein family members that also contains Fused in Sarcoma (FUS) and TATA-box binding protein Associated Factor 15 (TAF15) [17]. These proteins share high homology amino acid sequences in vertebrates [19]. EWSR1 regulates gene transcription by interacting with CREB binding proteins, basic transcription factors (TFs) TFIID and RNA polymerase II [17,20]. In zebrafish, ewsr1a and ewsr1b were required for mitotic stability and cellular survival in central nervous system (CNS) during early embryonic development [21]. Moreover, ewsr1 regulated the transcription of HNF4, oct4 and BRN3A, which involved with development and hormone regulation [16,22,23,24,25,26,27]. The offspring of ewsr1-deficient mice caused the abnormal gonadal development and subsequently sterile [22,28]. However, there has been rare focus on its function in gonadal development in teleost.
In C. semilaevis genome, it was found that two allele genes of ewsr1 were located on chromosomes W and Z, named as Cs-ewsr1-w and Cs-ewsr1-z, respectively. Based on the transcriptome dataset, we cloned and characterized two genes. Cs-ewsr1-w gene was chosen for further analysis on its transcriptional regulation and knock-down effect. These results could improve our understanding on the role of ewsr1 genes in C. semilaevis.
2. Materials and Methods
2.1. Ethics Approval
All the animal experiments were performed under the inspection of Yellow Sea Fisheries Research Institute’s animal care and use committee (Approval number, YSFRI-2022023). MS222 (Sigma-Aldrich, Oakville, ON, Canada) was used for anesthesia to minimize fish suffering (solubilized in seawater, final concentration 20 mg/L, fish was treated for 5 min) during experimental procedure [29]. The 293T cell line was purchased from ATCC (CRL-3216TM) (American Type Culture Collection, Manassas, VA, USA). The C. semilaevis ovarian cell line was previously established and cultured in our laboratory [30].
2.2. Samples Collection
All fish samples used in this experiment have been approved by the Care and Use of Laboratory Animals of the Chinese Academy of Fishery Sciences. Before sampling, genomic DNA was extracted from cut fins by TIANamp Marine Animals DNA Kit (TIANGEN, Beijing, China) for genetic sex identification by using PCR amplification with primers sex-F and sex-R (Table 1) [31]. After anesthesia with MS-222 (20 mg/L) [29], gonads were dissected from different developmental stages of C. semilaevis, including 90-day post hatching (90 d), 6-month post hatching (6 m), and 1.5-year post hatching (1.5 y). Six females and males were sampled at each stage. Brain, gonad, liver, spleen, heart, kidney, intestine and muscle were collected from three 3 y females and males. Tissues were put into RNAwait RNAlater solution (Solarbio, Beijing, China) quickly and stored in a refrigerator at −80 °C for subsequent RNA extraction.

Table 1.
All primers used in this study.
2.3. Gene Cloning of Cs-ewsr1-w and Cs-ewsr1-z
RNA was extracted from each sample by using Trizol Reagent (Invitrogen, Carlsbad, CA, USA). The quality and quantity of RNA was checked with agarose gel electrophoresis and P100 Series Spectrophotometers (Pultton, San Jose, CA, USA). The first strand cDNA was synthesized with 800 ng RNA as the template by using PrimeScript RT Kit with gDNA eraser (TaKaRa, Tokyo, Japan). Gene specific primers for gene cloning were designed by Primer Premier 5.0 (Table 1) based on partial sequences of ewsr1-w and ewsr1-z from C. semilaevis genome [32]. The mixed cDNA of females and males was used as the template for PCR amplification. The 25 μL PCR mixture contained 12.5 μL Ex Taq Mix (TaKaRa, Tokyo, Japan), 0.5 μL forward/ reverse primers, and 1 μL cDNA template. The PCR program was set as follows: 95 °C for 5 min, 40 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, and 72 °C for 10 min. PCR products were purified by FastPure Gel DNA Extraction Mini Kit (Vazyme, Nanjing, China), connected to pEASY-T1 vector, transformed, and sequenced in Beijing Ribioco Biotechnology Co., Ltd. (Ribioco, Beijing, China). The 3′ and 5′ untranslated regions (UTR) were amplified by using SMARTer RACE 5′/3′ Kit (TaKaRa, Tokyo, Japan) with the primers listed in Table 1 and the PCR program was followed as mentioned above.
2.4. Characterization of Cs-ewsr1-w and Cs-ewsr1-z
The characters including open reading frame (ORF), amino acid sequence, molecular weight, protein domains, and phosphorylation sites were predicted and analyzed by DNAstar (V7.1.0) (Bioinformatics Software, Madison, WI, USA), SMART (V9.0, Letunic et al. [33], Heidelberg, Germany) (http://smart.embl.de/, accessed on 26 March 2022), and NetPhos-3.1 (https://services.healthtech.dtu.dk/service.php?NetPhos-3.1) (Department of Health Technology, Technical University of Denmark, Kongens Lyngby, Denmark). The BLASTP Program (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) (National Center for Biotechnology Information, National Institutes of Health, Bethesda, MD, USA) was used for multiple sequence alignment. The MEGA X (Kumar et al. [34], Philadelphia, PA, USA) was used to construct phylogenetic tree by neighbour-joining algorithm (NJ). The NCBI accession numbers of amino acid sequences used in this study were listed in Table 2.

Table 2.
Accession numbers of Ewsr1 proteins used in this study.
2.5. Gene Expression Patterns of Cs-ewsr1-w and Cs-ewsr1-z in Different Tissues and Stages
The expressions of Cs-ewsr1-w and Cs-ewsr1-z in different development stages and tissues were analyzed with gene specific primers (Table 1) via qPCR assays on a 7500 Fast Real Time PCR platform (Applied Biosystems, Foster City, CA, USA). β-actin was set as the internal control. The 20 μL reactions contained 10 μL SYBR Premix Ex TaqTM (TaKaRa, Tokyo, Japan), 2 μL cDNA, 0.4 μL of each sense and anti-sense primers, and 0.4 μL ROX Dye II. The qPCR program was set as default settings followed by the dissociation curve, that is, 95 °C for 30 s, 40 cycles of 95 °C for 5 s, 60 °C for 30 s. The relative mRNA expression of Cs-ewsr1-w and Cs-ewsr1-z were processed by using 2−ΔΔCt method [35]. The data were analyzed by one-way ANOVA followed by Duncan’s multiple comparison in SPSS 25.0 (IBM Corp., Armonk, NY, USA), and the differences were considered significant when p < 0.05.
2.6. Promoter Activities Analysis of Cs-ewsr1-w
Based on the TFs prediction, six promoter plasmids with luciferase report were constructed by serial-deletion to detect the promoting activity of regulatory elements of Cs-ewsr1-w. The primers were listed in Table 1. The fragments were inserted into pGL3-basic vector (Promega, Madison, WI, USA) for the recombinant plasmid construction of pGL3-Cs-ewsr1-w-F1~F6 by using TSV-S1 Trelief® SoSoo Cloning Kit (Tsingke, Beijing, China).
Human embryonal kidney (HEK) 293T cells were maintained in DME/F-12 containing 10% fetal bovine serum (FBS, Gibco, New York, NY, USA) and 1% bFGF (Invitrogen, Carlsbad, CA, USA) in 5% CO2 at 37 °C. The pGL3-Cs-ewsr1-w~F1~F6 were transfected into HEK293T cells by using Lipo8000TM Transfection Reagent (Beyotime, Shanghai, China). Meanwhile, pGL3-basic and PGL3-control plasmids were used as the negative control and the positive control, respectively. The pRL-TK plasmid was transfected at the mean time as the internal reference. Dual Luciferase Reporter Gene Assay Kit (Beyotime, Shanghai, China) was employed to measure the promoter activities. Each experiment was performed in triplicates following the standard protocol provided by the manufacturer. Data obtained from Varioskan Flash spectral scanning multimode reader (Thermo Fisher Scientific, Vantaa, Finland) were analyzed by LSD (Least-significant difference) in SPSS 25.0, and the significance was regarded at p < 0.05.
The TF binding sites were predicted by PROMO (http://alggen.lsi.upc.es/, accessed on 2 April 2022) (Messeguer et al. [36], Barcelona, Spain) and JASPAR 2022 (https://jaspar.genereg.net/, accessed on 2 April 2022) (Castro-Mondragon et al. [37], Oslo, Norway). Nucleotides were mutated within TF binding sites (Mafk, c-MYC, MAC1, and POU1F1a-binding sites) following the protocols of Fast Site-Directed Mutagenesis Kit (TIANGEN, Beijing, China). After successful mutations were confirmed by sequencing, transfection and dual luciferase assays detection were performed as mentioned above.
2.7. The Knockdown Effect of Cs-ewsr1-w siRNA in C. semilaevis Ovarian Cells
The specific siRNA of Cs-ewsr1-w gene, the negative control siRNA, and siR transfect control (5cy3) were synthesized in Sangon Biotech (Sangon, Shanghai, China). C. semilaevis ovarian cells were cultured in L-15 medium supplemented with 1% bFGF and 15% FBS at 24 °C. Cs-ewsr1-w siRNA was transfected into the cells by using riboFECTTM CP Transfection Kit (Ribobio, Beijing, China) following the protocol described in the previous study [13]. Three replicates were set for both Cs-ewsr1-w-siRNA and negative control (NC) groups. At 48 h post transfection, the cellular status and the florescence of 5cy3-transfected cells would be checked. When it reached 90–95% of cell confluency and the percentage of transfection reached ~80%, it would be a good timing to harvest ovarian cells for the following experiments. After cell collection and RNA extraction, reverse transcription and qPCR assays were performed following the methods mentioned above. The relative expression levels of sex-related genes, such as Forkhead Box L2 (foxl2), SRY-box transcription factor 9b (sox9b), POU Class 5 Homeobox 1 (pou5f1) were measured with the primers listed in Table 1.
3. Results
3.1. Gene Cloning and Characterization of Cs-ewsr1s
Cs-ewsr1-w (GenBank accession no. 103397238) located in W chromosomes, with the full length of 2297 bp containing the ORF region of 1767 bp encoding 588 amino acids (Figure 1A). Functional domain prediction showed that Cs-Ewsr1-W contained RNA recognition motif in 313–393 residues and a Ran binding protein zinc finger domain in 454–480 residues. Cs-ewsr1-z (GenBank accession no. 103398620), located in Z chromosome, was 2230 bp in full length. It contains 1794 bp ORF region encoding 598 amino acids (Figure 1B). The same functional domains were located at 313–399 and 460–486 in Cs-Ewsr1-Z, respectively.

Figure 1.
The ORF and predicted amino acid sequences of Cs-ewsr1-w gene (A) and Cs-ewsr1-z gene (B). The UTR region sequence is represented in lowercase letters. The ORF sequence is represented in uppercase letters. The start codon and stop codon are bold in red. An asterisk (*) represents the stop codon at the end of the ORF. The RNA recognition motif is represented by an underscore and the Ran binding protein zinc finger domain is represented by a dot-dash underline.
The phylogenetic tree was constructed by using Ewsr1 proteins from 17 different species. The results showed that two Cs-Ewsr1 proteins were clustered together and then embedded with the teleost clade with Paralichthys olivaceus, Scophthalmus maximus and Poecilia formosa. The mammalian and the other species were clustered together (Figure 2). RNA recognition motif and Ran binding protein zinc finger domain were conserved in all the aligned species, including teleost, amphibians, and mammalians.
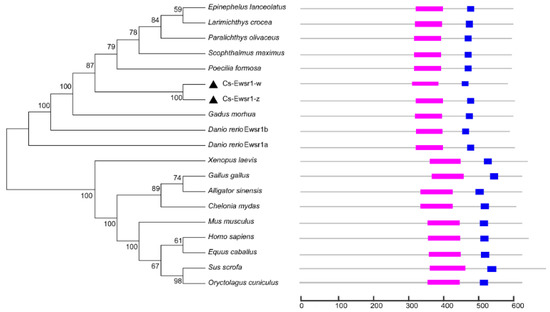
Figure 2.
Phylogenetic analysis of Ewsr1 proteins in multiple species. “ ” represents RNA recognition motif, “
” represents RNA recognition motif, “ ” respresents Ran binding protein zinc finger domain, “▲” represents Ewsr1 proteins in C. semilaevis.
” respresents Ran binding protein zinc finger domain, “▲” represents Ewsr1 proteins in C. semilaevis.
 ” represents RNA recognition motif, “
” represents RNA recognition motif, “ ” respresents Ran binding protein zinc finger domain, “▲” represents Ewsr1 proteins in C. semilaevis.
” respresents Ran binding protein zinc finger domain, “▲” represents Ewsr1 proteins in C. semilaevis.
3.2. The Expression Patterns of Cs-ewsr1s in Different Tissues and Developmental Stages
qPCR assays revealed that the sex-biased expression patterns of two Cs-ewsr1s in Chinese tongue sole. Cs-ewsr1-w was expressed in all tissues of female tongue sole with the highest expression in gonad, but was hardly detected in any tissue of male tongue sole (Figure 3A). In comparison, Cs-ewsr1-z was prevalently expressed in all tissues of female and male tongue sole (Figure 3B). It exhibited the highest expression in male gonad, followed by female gonad, brains of male and female, livers of male and female, and female heart and intestine (Figure 3).
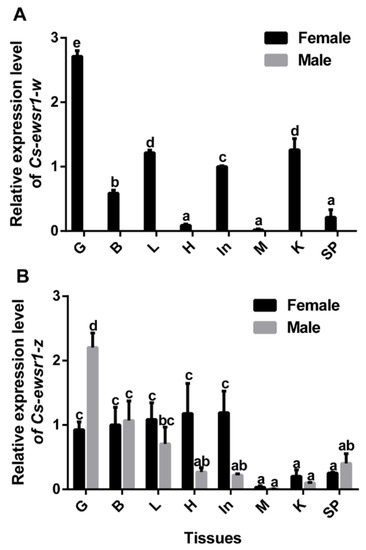
Figure 3.
The gene expression patterns of Cs-ewsr1-w (A) and Cs-ewsr1-z (B) in various tissues of healthy C. semilaevis, including gonad (G), brain (B), liver (L), heart (H), intestines (In), muscle (M), kidney (K), and spleen (SP). Different letters represent significant differences among species (p < 0.05).
Cs-ewsr1-w gradually increased with ovary development, and reached the peak at 3 y (Figure 4A). However, its expression was hardly detected during testis development because of no expression in testis. Cs-ewsr1-z gene was expressed in all tested developmental stages of ovaries and testes. Its expression was relatively low in 6 m female and male, and was significantly higher in testes of 1.5 y male (Figure 4B).
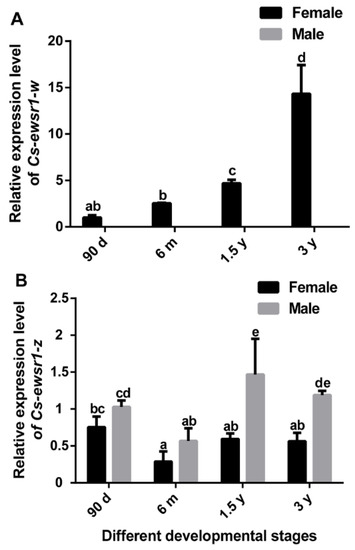
Figure 4.
The relative expression patterns of Cs-ewsr1-w (A) and Cs-ewsr1-z (B) genes in female and male gonads from different developmental stages. Different letters represent significant differences among species (p < 0.05).
3.3. Promoter Activity of Cs-ewsr1-w Detection and Analysis
The Cs-ewsr1-w promoter sequence of 2690 bp (−2590/+99) was cloned by genomic DNA with specific primers Cs-ewsr1-w-P-F/R (Table 1). Cs-ewsr1-w promoter region had only one CpG island, which was located from −1215 to −1101 bp.
A series of promoter fragments with different length deletion were generated to explore the promoter activity of Cs-ewsr1-w gene. The promoter activities of all Cs-ewsr1-w fragments were significantly higher than that of pGL3-basic (p < 0.05, Figure 5), among which the activity of Cs-ewsr1-w-P-F2/R fragment was the highest. The relative activity significantly decreased by 2.7-fold from Cs-ewsr1-w-P-F5/R fragment to Cs-ewsr1-w-P-F6/R fragment (p < 0.05, Figure 5), indicated region −733 to −154 positively affected the promoter activity of Cs-ewsr1-w gene. Similarly, the positive effect was detected from other two region as well, which were −2190 to −1692 bp and −154 to +99 bp (Figure 5).
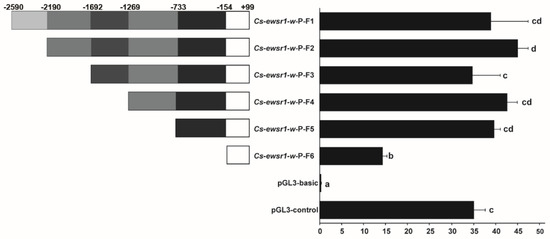
Figure 5.
Fluorescence activity of different fragment lengths in the promoter region of Cs-ewsr1-w gene. The left panel exhibited the schematic map of promoter fragments with different deletion regions. The right panel showed the luciferase activities of the promoter fragments with different deletions. Different letters represent significant differences among species (p < 0.05).
Prediction of TF binding sites in the -733/-154 bp interval of Cs-ewsr1-w promoter revealed numerous TFs binding sites, including STAT4, HOXA3, c-Myc, GAGA factor, MAC1, POU1F1a, Mafk, and PRA (Figure 6). The interval containing TF Mafk, c-MYC, MAC1 and POU1F1a were mutated and transfected into 293T cells for detection after 48 h. The results indicated mutation of Mafk binding site led to the significant decrease by 46% in the activity of Cs-ewsr1-w-P-F5/R fragment (p < 0.05, Figure 7), which showed no significant difference compared with the activity of Cs-ewsr1-w-P-F6/R fragment. Mutations on other TF binding sites showed no significant effect (Figure 7).

Figure 6.
Nucleotide sequence of Cs-ewsr1-w promoter region (−911/+50) and the predicted transcription factor binding sites. The highly active region (-733/-154) was highlighted in yellow. Star codon “ATG” was labelled in red. Underlined boldface letters indicated the predicted transcription factors.
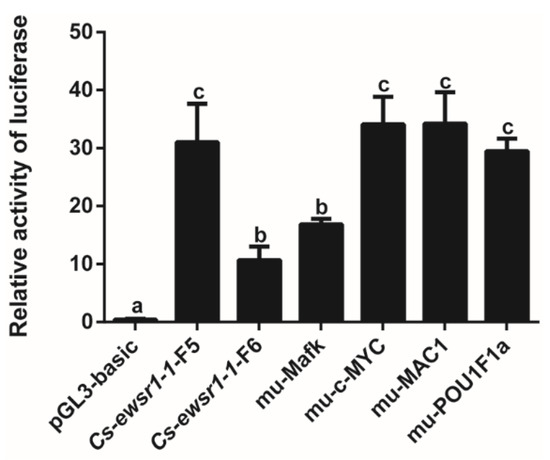
Figure 7.
Fluorescence activity of the mutated transcription factor binding sites in Cs-ewsr1-w promoter compared with that of Cs-ewsr1-w promoter region with different deletion. Different letters represent significant differences among species (p < 0.05).
3.4. Expression Patterns of Sex-Related Genes in Cs-ewsr1-w Knockdown Ovarian Cells
Cs-ewsr1-w expression was significantly reduced by 80% (p < 0.05) after in vitro Cs-ewsr1-w siRNA interference (RNAi), while no significant variation of Cs-ewsr1-z gene was detected. The down-regulation of sex-related genes was detected after Cs-ewsr1-w RNAi (p < 0.05), including foxl2, sox9b and pou5f1. Among that, foxl2 expression significantly dropped by18.8-fold compared with the control group. The expression levels of sox9b and pou5f1 were significantly reduced by half (p < 0.05, Figure 8).
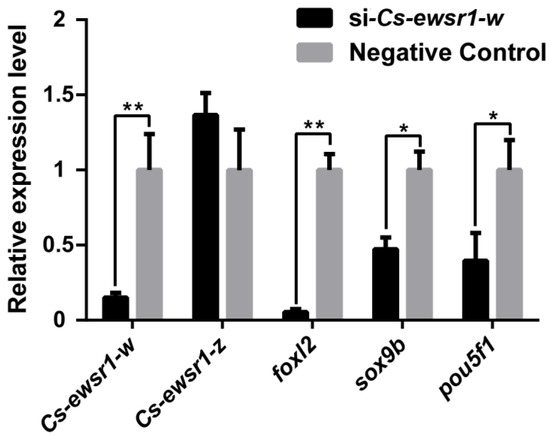
Figure 8.
The knockdown effect of Cs-ewsr1-w siRNA in C. semilaevis ovarian cells. The relative expression variations of Cs-ewsr1-z, Forkhead Box L2 (foxl2), SRY-box transcription factor 9b (sox9b), POU Class 5 Homeobox 1 (pou5f1) were measured. An asterisk (*) indicates a significant difference between negative control group and siRNA-treated group (p < 0.05). Two asterisks (**) indicates an extremely significant difference (p < 0.01).
4. Discussion
Ewsr protein, a key player in cancer, is involved in RNA metabolism and DNA repair [38]. However, studies on teleost EWS protein are rare. Based on the comparative transcriptome analysis on early developmental stages of gonad in C. semilaevis, ewsr1-w was specifically expressed in ovary and continuously up-regulated with ovarian differentiation [15]. The amino acid sequences of Cs-Ewsr1-w and Cs-Ewsr1-z proteins have high similarity of 91.41%, both of which contained the conserved RNA recognition motif and Ran binding protein-zinc finger domain, suggesting that Cs-Ewsr1s might have similar function with Ewsr1s in other vertebrates. RNA recognition motif is associated with the interaction of protein with RNA. Meanwhile, this protein has a variable number of RGG (arginine-glycine-glycine) repeats that are regarded as a RNA-binding region as well [39]. Based on the phylogenetic analysis, the sequences we obtained from C. semilaevis fell in a well-supported clade, suggesting that we obtained the ewsr1 gene orthologs.
Based on our qPCR results, Cs-ewsr1-w gene was uniquely expressed in females with the highest transcriptional level in ovary. Its expression increased gradually with ovarian development from 90 d to 3 y. These results indicated the possible involvement of Cs-ewsr1-w gene in ovarian development. After Cs-ewsr1-w gene expression was interfered in the ovarian cells of C. semilaevis, several gonadal development-related genes were down-regulated, including foxl2, sox9b and pou5f1. Foxl2 gene, belonging to winged helix transcription factor, is one of the crucial players in ovarian development [40]. Many studies have shown that this gene functions in sex differentiation and gonadal development in teleost [41,42,43,44]. C. semilaevis foxl2 was significantly expressed in 12-month old ovary of phase II fish ovary development stages, suggesting its possible involvement in oocyte development [45]. Sox9b, another important gene related to gonadal development, were significantly expressed in ovaries of fugu (Takifugu rubripes) and zebrafish (Danio rerio) [46,47]. It facilitated sex differentiation and gonadal development in medaka and Japanese flounder [48,49,50]. Besides, prominent expression of sox9b was detected in gonads of early-stage C. semilaevis, indicating its potential involvement in gonadal differentiation [51]. Pou5f1 (also known as oct4) is a key TF regulating embryonic stem cell pluripotency, primordial germ cell formation, early embryonic and gonadal germ cell development [27]. Pou5f1 analogue in teleost plays a post-embryonic role in adult gonad and gametes development [52,53,54]. Based on the suppressive effect of Cs-ewsr1-w knockdown on several sex-related genes, we proposed that it might be a positive regulator in ovarian development of C. semilaevis. In the future, analysis including in vivo trials would be conducted for further investigation on its mechanism.
Promoters contain sequence-specific binding sites for many TFs, and the TFs could recognize and bind target sequences to guide underlying transcription and regulate transcriptional activity [55,56,57]. In Cs-ewsr1-w promoter -733 to -154 bp was the core region that had a great effect on transcription regulation. After site-direct mutagenesis on the TF-binding sites, the activity of TF Mafk-binding site decreased significantly, suggesting potential involvement of Mafk in Cs-ewsr1-w transcription. Mafk, a member of small MAFs family, was essential for mice embryonic development [58]. It regulates genes involved in several cellular processes, including ubiquitination/proteasome [59]. Numerous ubiquitin-conjugating enzyme genes showed female-biased gene expressions in early developmental stages of C. semilaevis, revealing the indispensable involvement of ubiquitination pathway in female differentiation [15]. Based on the above analysis, we deduced that TF Mafk might be a positive regulator in Cs-ewsr1-w transcription during C. semilaevis ovarian development. The regulation mechanism between them are worthy for further functional studies. In the future, further studies will be performed to get a clearer picture on the regulation of Cs-ewsr1-w by TF Mafk during female differentiation and ovarian development.
5. Conclusions
In this study, two ewsr1 genes were cloned and characterized from Chinese tongue sole (Cs-ewsr1-w and Cs-ewsr1-z). The female-biased gonad expression of Cs-ewsr1-w was observed from 90 d to 3 y, suggesting its potential roles in ovarian development. Its knockdown significantly down-regulated the expressions of foxl2, sox9b and pou5f1. The activity analysis, and the prediction and verification of transcription factors for Cs-ewsr1-w promoter shed some lights on the transcription regulation of this gene. Our findings suggested the potential roles of Cs-ewsr1-w in C. semilaevis ovarian development, providing fundamental information for further exploration on its biological functions in teleost.
Author Contributions
Conceptualization, Z.C. (Zhangfan Chen) and N.W.; methodology, P.C. and Z.C. (Zhangfan Chen).; validation, R.S. and X.L.; formal analysis, W.X.; investigation, P.C., Z.C. (Zhangfan Chen), N.W. and W.X.; resources, Q.Y., Z.C. (Zhongkai Cui) and J.C.; writing—original draft preparation, P.C.; writing—review and editing, Z.C. (Zhangfan Chen); project administration, S.C.; funding acquisition, N.W., W.X., Z.C. (Zhongkai Cui) and S.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31730099, 31873037), National Key R&D Program of China (2018YFD0900202), Key Research and Development Project of Shandong Province (2021LZGC028, Special Grants for Academicians), the Central Public-interest Scientific Institute Basal Research Fund, CAFS (No.2020TD20), Science and Technology Research Program, Caofeidian District, Tangshan, China (202111).
Institutional Review Board Statement
The animal study protocol was approved by Yellow Sea Fisheries Research Institute’s animal care and use committee YSFRI-2022023.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Naqvi, S.; Godfrey, A.K.; Hughes, J.F.; Goodheart, M.L.; Mitchell, R.N.; Page, D.C. Conservation, acquisition, and functional impact of sex-biased gene expression in mammals. Science 2019, 365, eaaw7317. [Google Scholar] [CrossRef]
- Mei, J.; Gui, J. Genetic basis and biotechnological manipulation of sexual dimorphism and sex determination in fish. Sci. China Life Sci. 2015, 58, 124–136. [Google Scholar] [CrossRef]
- Parker, G.A. The evolution of sexual size dimorphism in fish*. J. Fish Biol. 1992, 41, 1–20. [Google Scholar] [CrossRef]
- Foellmer, M.W.; Moya-Laraño, J. Chater 7. Sexual size dimorphism in spiders: patterns and processes. In Sex, Size and Gender Roles: Evolutionary Studies of Sexual Size Dimorphism; Daphne, J.F., Wolf, U.B., Tamás, S., Eds.; Oxford University Press: New York, NY, USA, 2007; pp. 71–81. ISBN 9780191709036. [Google Scholar]
- Lindenfors, P.; Gittleman, J.; Jones, K. Chapter 2. Sexual size dimorphism in mammals. In Sex, Size and Gender Roles: Evolutionary Studies of Sexual Size Dimorphism; Daphne, J.F., Wolf, U.B., Tamás, S., Eds.; Oxford University Press: New York, NY, USA, 2007; pp. 16–26. ISBN 9780191709036. [Google Scholar]
- Székely, T.; Lislevand, T.; Figuerola, J. Chapter 3. Sexual size dimorphism in birds. In Sex, Size and Gender Roles: Evolutionary Studies of Sexual Size Dimorphism; Daphne, J.F., Wolf, U.B., Tamás, S., Eds.; Oxford University Press: New York, NY, USA, 2007; pp. 27–37. ISBN 9780191709036. [Google Scholar]
- Sun, Y.; Yu, H.; Zhang, Q.; Qi, J.; Zhong, Q.; Chen, Y.; Li, C. Molecular characterization and expression pattern of two zona pellucida genes in half-smooth tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2010, 155, 316–321. [Google Scholar] [CrossRef]
- Bonduriansky, R.; Chenoweth, S. Intralocus sexual conflict. Trends Ecol. Evol. 2009, 24, 280–288. [Google Scholar] [CrossRef]
- Parsch, J.; Ellegren, H. The evolutionary causes and consequences of sex-biased gene expression. Nat. Rev. Genet. 2013, 14, 83–87. [Google Scholar] [CrossRef]
- Williams, T.M.; Carroll, S.B. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat. Rev. Genet. 2009, 10, 797–804. [Google Scholar] [CrossRef]
- Wang, N.; Wang, R.K.; Wang, R.Q.; Chen, S.L. Transcriptomics analysis revealing candidate networks and genes for the body size sexual dimorphism of Chinese tongue sole (Cynoglossus semilaevis). Funct. Integr. Genom. 2018, 18, 327–339. [Google Scholar] [CrossRef]
- Wang, K.L.; Zhang, H.; Hu, Q.M.; Shao, C.W.; Chen, S.L. Expression and purification of half-smooth tongue sole (Cynoglossus semilaevis) CSDAZL protein. Protein Expr. Purif. 2014, 102, 8–12. [Google Scholar] [CrossRef]
- Wang, N.; Gong, Z.H.; Wang, J.; Xu, W.T.; Yang, Q.; Chen, S.L. Characterization of Chinese tongue sole (Cynoglossus semilaevis) 24-dehydrocholesterol reductase: Expression profile, epigenetic modification, and its knock-down effect. Gen. Comp. Endocrinol. 2021, 312, 113870. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Li, X.H.; Shi, R.; Cheng, P.; Wang, N.; Chen, S.L. The female-biased expression, transcriptional regulation and knock-down effect of insulin-like growth factor binding protein 7 in Chinese tongue sole, Cynoglossus semilaevis. Aquaculture 2022, 551, 737956. [Google Scholar] [CrossRef]
- Xu, W.T.; Cui, Z.K.; Wang, N.; Zhang, M.Q.; Wang, J.; Xu, X.T.; Liu, Y.; Chen, S.L. Transcriptomic analysis revealed gene expression profiles during the sex differentiation of Chinese tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100919. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Nguyen, P.T.; Shim, H.S.; Hyeon, S.J.; Im, H.; Choi, M.H.; Chung, S.; Kowall, N.W.; Lee, S.B.; Ryu, H. EWSR1, a multifunctional protein, regulates cellular function and aging via genetic and epigenetic pathways. Biochim. Biophys. Acta-(BBA) Mol. Basis Dis. 2019, 1865, 1938–1945. [Google Scholar] [CrossRef] [PubMed]
- Rossow, K.L.; Janknecht, R. The Ewing’s sarcoma gene product functions as a transcriptional activator. Cancer Res. 2001, 61, 2690–2695. [Google Scholar]
- Erkizan, H.; Uversky, V.; Toretsky, J. Oncogenic Partnerships: EWS-FLI1 Protein Interactions Initiate Key Pathways of Ewing’s Sarcoma. Clin. Cancer Res. 2010, 16, 4077–4083. [Google Scholar] [CrossRef] [PubMed]
- Morohoshi, F.; Ootsuka, Y.; Arai, K.; Ichikawa, H.; Mitani, S.; Munakata, N.; Ohki, M. Genomic structure of the human RBP56/hTAF(II)68 and FUS/TLS genes. Gene 1998, 221, 191–198. [Google Scholar] [CrossRef]
- Bertolotti, A.; Melot, T.; Acker, J.; Vigneron, M.; Delattre, O.; Tora, L. EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: Interactions between two members of the tet family, EWS and HTAF(II)68, and subunits of TFIID and RNA polymerase II complexes. Mol. Cell. Biol. 1998, 18, 1489–1497. [Google Scholar] [CrossRef]
- Azuma, M.; Embree, L.J.; Sabaawy, H.; Hickstein, D.D. Ewing sarcoma protein ewsr1 maintains mitotic integrity and proneural cell survival in the zebrafish embryo. PLoS ONE 2007, 2, e979. [Google Scholar] [CrossRef]
- Li, H.; Watford, W.; Li, C.; Parmelee, A.; Bryant, M.A.; Deng, C.; O’Shea, J.; Lee, S.B. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J. Clin. Investig. 2007, 117, 1314–1323. [Google Scholar] [CrossRef]
- Araya, N.; Hirota, K.; Shimamoto, Y.; Miyagishi, M.; Yoshida, E.; Ishida, J.; Kaneko, S.; Kaneko, M.; Nakajima, T.; Fukamizu, A. Cooperative interaction of EWS with CREB-binding protein selectively activates hepatocyte nuclear factor 4-mediated transcription. J. Biol. Chem. 2003, 278, 5427–5432. [Google Scholar] [CrossRef]
- Lee, J.; Rhee, B.K.; Bae, G.Y.; Han, Y.M.; Kim, J. Stimulation of Oct-4 activity by Ewing’s sarcoma protein. Stem Cells 2005, 23, 738–751. [Google Scholar] [CrossRef] [PubMed]
- Kovar, H. Dr. Jekyll and Mr. Hyde: The Two Faces of the FUS/EWS/TAF15 Protein Family. Sarcoma 2011, 2011, 837474. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Han, K.; Chen, S.; Cai, M.; Wang, Y.; Zhang, Z. Molecular cloning and expression of Octamer-binding transcription factor (Oct4) in the large yellow croaker, Larimichthys crocea. Gene Expr. Patterns 2018, 27, 16–30. [Google Scholar] [CrossRef]
- Lanier, J.; Quina, L.A.; Eng, S.R.; Cox, E.; Turner, E.E. Brn3a target gene recognition in embryonic sensory neurons. Dev. Biol. 2007, 302, 703–716. [Google Scholar] [CrossRef]
- Tian, H.; Petkov, P.M. Mouse EWSR1 is crucial for spermatid post-meiotic transcription and spermiogenesis. Development 2021, 148, dev199414. [Google Scholar] [CrossRef]
- Sun, Y.X.; Zhu, Y.; Cheng, P.; Zhang, M.Q.; Wang, N.; Cui, Z.K.; Wei, M.; Xu, W.T. A Z-Linked E3 Ubiquitin Ligase Cs-rchy1 Is Involved in Gametogenesis in Chinese Tongue Sole, Cynoglossus semilaevis. Animals 2021, 11, 3265. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Wang, T.Z.; Wang, N.; Liu, X.F.; Sha, Z.X.; Chen, S.L. Establishment and characterization of an ovarian cell line from half-smooth tongue sole Cynoglossus semilaevis. J. Fish Biol. 2015, 86, 46–59. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, S.L.; Gao, F.T.; Meng, L.; Hu, Q.M.; Song, W.; Shao, C.W.; Lv, W.Q. SCAR-transformation of sex-specific SSR marker and its application in half-smooth tongue sole (Cynoglossus semiliaevis). Agric. Biotechnol. 2014, 6, 787–792. [Google Scholar] [CrossRef]
- Chen, S.L.; Zhang, G.J.; Shao, C.W.; Huang, Q.F.; Liu, G.; Zhang, P.; Song, W.T.; An, N.; Chalopin, D.; Volff, J.; et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014, 46, 253–260. [Google Scholar] [CrossRef]
- Leturnic, I.; Khedkar, S.; Bork, P. SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis acorss computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Messeguer, X.; Escudero, R.; Farré, D.; Nuñez, O.; Martínez, J.; Albà, M.M. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002, 18, 333–334. [Google Scholar] [CrossRef] [PubMed]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Lemma, R.B.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Pérez, N.M.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef]
- Paronetto, M.P. Ewing sarcoma protein: A key player in human cancer. Int. J. Cell Biol. 2013, 2013, 642853. [Google Scholar] [CrossRef]
- Bertolotti, A.; Lutz, Y.; Heard, D.J.; Chambon, P.; Tora, L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996, 15, 5022–5031. [Google Scholar] [CrossRef]
- Uhlenhaut, N.H.; Treier, M. Foxl2 function in ovarian development. Mol. Genet. Metab. 2006, 88, 225–234. [Google Scholar] [CrossRef]
- Okada, H.; Hagihara, S.; Yamashita, K.; Ijiri, S.; Adachi, S. Expression pattern of foxl2 and dmrt1 in gonad of Amur sturgeon Acipenser schrenckii in relation to sex differentiation. Aquaculture 2017, 479, 712–720. [Google Scholar] [CrossRef]
- Alam, M.A.; Kobayashi, Y.; Horiguchi, R.; Hirai, T.; Nakamura, M. Molecular cloning and quantitative expression of sexually dimorphic markers Dmrt1 and Foxl2 during female-to-male sex change in Epinephelus merra. Gen. Comp. Endocrinol. 2008, 157, 75–85. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Yamaguchi, S.; Hirai, T.; Kitano, T. Follicle-stimulating hormone signaling and Foxl2 are involved in transcriptional regulation of aromatase gene during gonadal sex differentiation in Japanese flounder, Paralichthys olivaceus. Biochem. Biophys. Res. Commun. 2007, 359, 935–940. [Google Scholar] [CrossRef]
- Ijiri, S.; Kaneko, H.; Kobayashi, T.; Wang, D.S.; Sakai, F.; Paul-Prasanth, B.; Nakamura, M.; Nagahama, Y. Sexual dimorphic expression of genes in gonads during early differentiation of a teleost fish, the Nile tilapia Oreochromis niloticus. Biol. Reprod. 2008, 78, 333–341. [Google Scholar] [CrossRef]
- Dong, X.L.; Chen, S.L.; Ji, X.S.; Shao, C.W. Molecular cloning, characterization and expression analysis of Sox9a and Foxl2 genes in half-smooth tongue sole (Cynoglossus semilaevis). Acta Oceanol. Sin. 2011, 30, 68–77. [Google Scholar] [CrossRef]
- Chiang, E.F.; Pai, C.I.; Wyatt, M.; Yan, Y.L.; Postlethwait, J.; Chung, B. Two sox9 genes on duplicated zebrafish chromosomes: Expression of similar transcription activators in distinct sites. Dev. Biol. 2001, 231, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Cui, J.; Yang, G.; Gong, Q.; Gu, Q. Expression detection of DMRTs and two sox9 genes in Takifugu rubripes (Tetraodontidae, Vertebrata). J. Ocean Univ. China 2007, 6, 182–186. [Google Scholar] [CrossRef]
- Nakamura, S.; Aoki, Y.; Saito, D.; Kuroki, Y.; Fujiyama, A.; Naruse, K.; Tanaka, M. Sox9b/sox9a2-EGFP transgenic medaka reveals the morphological reorganization of the gonads and a common precursor of both the female and male supporting cells. Mol. Reprod. Dev. 2008, 75, 472–476. [Google Scholar] [CrossRef]
- Nakamura, S.; Watakabe, I.; Nishimura, T.; Toyoda, A.; Taniguchi, Y.; Tanaka, M. Analysis of Medaka sox9 Orthologue Reveals a Conserved Role in Germ Cell Maintenance. PLoS ONE 2012, 7, e29982. [Google Scholar] [CrossRef]
- Li, X.; Yu, H.; Wang, Y.; Liu, X.; Liu, Y.; Qu, J.; Wang, X. Roles of Two Sox9 Genes during Gonadal Development in Japanese Flounder: Sex Differentiation, Spermatogenesis and Gonadal Function Maintenance. Int. J. Mol. Sci. 2018, 19, 512. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, M.Q.; Cheng, P.; Gong, Z.H.; Li, X.H.; Wang, N.; Wei, M.; Xu, X.W.; Xu, W.T. pitpbeta_w Encoding Phosphatidylinositol Transfer Protein Is Involved in Female Differentiation of Chinese Tongue Sole, Cynoglossus semilaevis. Front. Genet. 2022, 13, 861763. [Google Scholar] [CrossRef]
- Xiaohuan, H.; Yang, Z.; Linyan, L.; Zhenhua, F.; Linyan, Z.; Zhijian, W.; Ling, W.; Deshou, W.; Jing, W. Characterization of the POU5F1 Homologue in Nile Tilapia: From Expression Pattern to Biological Activity. Stem Cells Dev. 2016, 25, 1386–1395. [Google Scholar] [CrossRef]
- Zhong, C.; Liu, M.; Tao, Y.; Wu, X.; Yang, Y.; Wang, T.; Meng, Z.; Xu, H.; Liu, X. Pou5f1 and Nanog Are Reliable Germ Cell-Specific Genes in Gonad of a Protogynous Hermaphroditic Fish, Orange-Spotted Grouper (Epinephelus coioides). Genes 2021, 13, 79. [Google Scholar] [CrossRef]
- Gao, J.; Wang, X.; Zhang, Q. Evolutionary Conservation of pou5f3 Genomic Organization and Its Dynamic Distribution during Embryogenesis and in Adult Gonads in Japanese Flounder Paralichthys olivaceus. Int. J. Mol. Sci. 2017, 18, 231. [Google Scholar] [CrossRef] [PubMed]
- Shijun, L.; Khan, R.; Raza, S.H.A.; Jieyun, H.; Chugang, M.; Kaster, N.; Gong, C.; Chunping, Z.; Schreurs, N.M.; Linsen, Z. Function and characterization of the promoter region of perilipin 1 (PLIN1): Roles of E2F1, PLAG1, C/EBPβ, and SMAD3 in bovine adipocytes. Genomics 2020, 112, 2400–2409. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Richmond, T. Eukaryotic transcription factors. Curr. Opin. Struct. Biol. 1998, 8, 41–48. [Google Scholar] [CrossRef]
- de Vooght, K.M.; van Solinge, W.W. Gene promoter analysis in molecular diagnostics: Do or don’t? Expert Rev. Mol. Diagn. 2009, 9, 403–405. [Google Scholar] [CrossRef]
- Yamazaki, H.; Katsuoka, F.; Motohashi, H.; Engel, J.D.; Yamamoto, M. Embryonic lethality and fetal liver apoptosis in mice lacking all three small Maf proteins. Mol. Cell. Biol. 2012, 32, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Kannan, M.B.; Solovieva, V.; Blank, V. The small MAF transcription factors MAFF, MAFG and MAFK: Current knowledge and perspectives. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2012, 1823, 1841–1846. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).